Abstract
Many cancers express mutant p53 proteins that have lost wild-type tumor suppressor activity and, in many cases, have acquired oncogenic functions that can contribute to tumor progression. These activities of mutant p53 reflect interactions with several other proteins, including the p53 family members p63 and p73. Mutations in p53 that affect protein conformation (such as R175H) show strong binding to p63 and p73, whereas p53 mutants that only mildly affect the conformation (such as R273H) bind less well. A previously described aggregation domain of mutant p53 is not required for p63 or p73 binding; indeed, mutations within this region lead to the acquisition of a mutant p53 phenotype—including a conformational shift, p63/p73 binding and the ability to promote invasion. The activity of wild-type p53 is regulated by an interaction with MDM2 and we have investigated the potential role of MDM2 in the mutant p53/p63/p73 interactions. Both mutant p53 and p73 bind MDM2 well, whereas p63 binds much more weakly. We found that MDM2 can inhibit p63 binding to p53R175H but enhances the weaker p53R273H/p73 interaction. These effects on the interactions are reflected in an ability of MDM2 to relieve the inhibition of p63 by p53R175H, but enhance the inhibition of p73 activity by p53R175H and R273H. We propose a model in which MDM2 competes with p63 for binding to p53R175H to restore p63 activity, but forms a trimeric complex with p73 and p53R273H to more strongly inhibit p73 function.
Introduction
p53 is an important tumor suppressor protein that functions as a transcription factor, binding DNA sequences in the promoters of a large number of target genes that mediate responses such as cell cycle arrest, senescence and apoptosis.1 p53 is altered in the majority of human cancers, frequently resulting in the expression of mutant p53 proteins with single amino-acid substitutions in the DNA-binding domain (DBD).2 In general, these mutant p53s have lost the ability to bind the p53 binding sites in DNA and so fail to exhibit the wild-type p53 tumor suppressor activity.3 Furthermore, many of the mutant p53 proteins have also been shown to acquire a gain of function that can contribute to all stages of tumorigenesis, including the ability to promote invasion and metastasis.4, 5
p53 belongs to a family of related proteins, which also includes p63 and p73.6, 7 Each of the family members is expressed as a number of isoforms, including N-terminal variants that encode either a full-length (TA) or truncated (ΔN) p53, p63 and p73.6, 8 Alternative splicing in the C terminus of each protein can further give rise to a multitude of C-terminal isoforms such as α, β or γ.9 Regulation of the p53 family members depends on various homo- and heteromeric interactions. Each of the p53 family members contains a C-terminal oligomerization domain, allowing the formation of homotetramers to allow sequence-specific DNA binding required to function as a transcription factor.10, 11 The extreme C terminus of p63, present in TAp63α, contains a transactivation inhibitory domain that can interact with the N-terminal domain of the protein, resulting in the adoption of a closed inactive dimer.12 The TAp63α can therefore switch between an inactive dimer and an active tetramer,13 a level of control that is not seen for TAp73α.14 Although p63 and p73 can also form heterotetramers through the oligomerization domain, this region of p53 is unable to interact with p63 or p73.15, 16 However, cancer-associated point mutations within the DBD of p53 can both change the conformation of the p53 protein and allow the interaction of mutant p53 with p63 and p73.17, 18, 19, 20, 21 This interaction of mutant p53 with p63/p73 can inhibit the transcriptional activity of p63/p73 and promote invasion.22, 23, 24 Interestingly, the interaction between mutant p53 and p63 or p73 can be influenced by many proteins such as TOPBP1, Pin1, ANKRD11 and SMAD2 with functional consequences for p63 and p73 transcriptional function.5
Another important regulator of p53 is MDM2, one of the principal ubiquitin ligases responsible for targeting p53 for degradation.25, 26 The interaction between N-terminal domains in both MDM2 and p53 allows for the ubiquitination and degradation of p53, although E3 ligase-defective MDM2 mutants can retain the ability to inhibit p53 function through this interaction, which obscures the N-terminal transcriptional activation domain of p53.27, 28 MDM2 can also form an interaction with p73,29, 30, 31 although this does not lead to the ubiquitination and degradation of p73. By contrast, an MDM2/p63 interaction is seen in some32, 33 but not all studies.34, 35
In this study, we examined the interactions between the p53 family members and MDM2. We confirmed that while mutant p53 can bind to both p63 and p73, MDM2 preferentially binds to p73. This differential binding had a clear impact on the binding of mutant p53 to p63 or p73, with opposite functional consequences on the transcription activity of p63 or p73.
Results
Binding of p63 and p73 to mutant p53
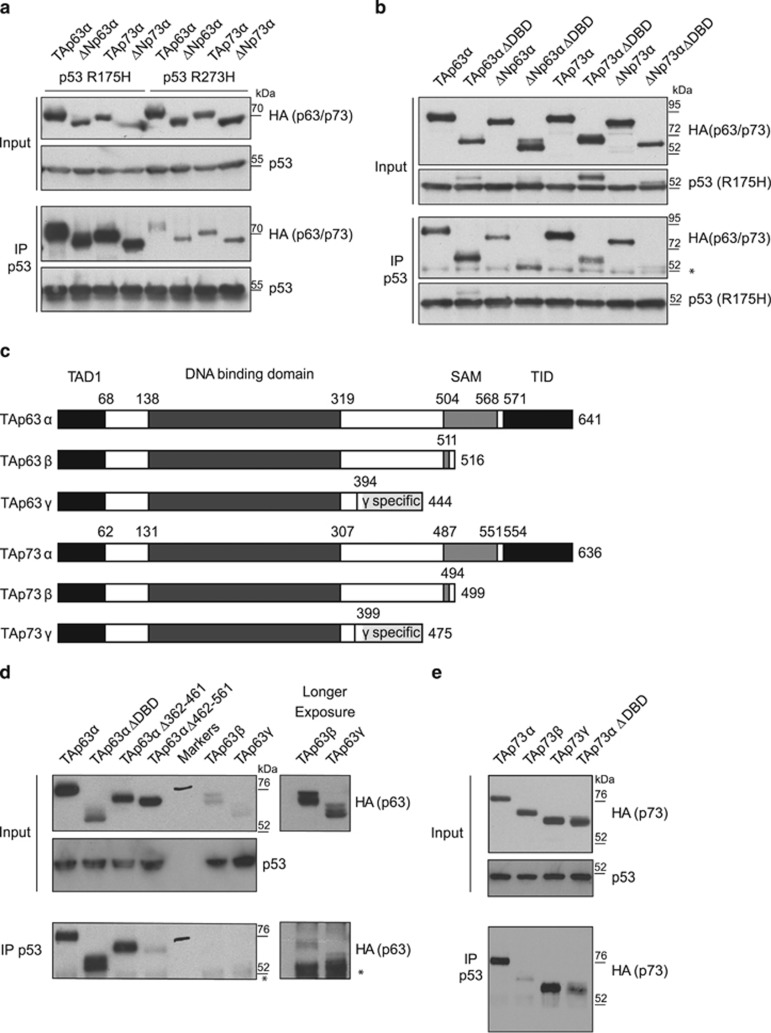
Both p63 and p73 have been shown to interact with mutant p53 to varying extents, depending on the nature of the p53 mutation.17, 18, 19, 20 We confirmed previous observations that p53R175H, a conformational mutant that results in misfolding of the p53 protein,36 binds much more strongly to both N-terminal isoforms of p63α (TA and ΔN) and p73α (TA and ΔN) than p53R273H, which directly affects a DNA-contacting residue in p53, but only slightly perturbs the wild-type conformation of the protein (Figure 1a).17, 18, 19, 20 Consistent with these reports, we observed similar binding of the TA and ΔN versions of p63 and p73 to p53R175H under conditions used for all the subsequent experiments in this study.
Figure 1.
Mutant p53 interaction with p63 and p73. (a) Immunoprecipitation (IP) of p53 with DO-1 from HCT116 p53−/− cells transfected with 3μg HA-tagged TAp63α, ΔNp63α, TAp73α or ΔNp73α in combination with 2 μg mutant p53 (R175H or R273H). Input and immunoprecipitated p53, p63 and p73 levels were determined by western blot using DO-1 and HA antibodies. (b) IP of p53 with DO-1 from HCT116 p53−/− cells transfected with 3 μg HA-tagged TAp63α, TAp63α ΔDBD, ΔNp63α, ΔNp63α ΔDBD, TAp73α, TAp73α ΔDBD, ΔNp73α or ΔNp73α ΔDBD in combination with 2 μg p53R175H. Input and immunoprecipitated p53, p63 and p73 levels were determined by western blot using DO-1 and HA antibodies. *A nonspecific band seen in all IPs. (c) Schematic representation of TAp63α, TAp63β, TAp63γ, TAp73α, TAp73β and TAp73γ, showing the major functional domains: transactivation domain 1 (TAD1), DBD, sterile α motif (SAM) domain and transactivation inhibitory domain (TID). Also indicated is the γ-specific region. Not to scale. (d) IP of p53 with DO-1 from HCT116 p53−/− cells transfected with 2 μg HA-tagged TAp63α, TAp63α ΔDBD, TAp63α Δ362–461, TAp63α Δ462–561 (LHS), TAp63β or TAp63γ (RHS) with 1.5 μg mutant p53 (R175H). Input for p53 and input and co-precipitated p63 was detected by western blotting with DO-1 and HA antibodies. A longer exposure of the last two lanes is shown to the RHS. *A nonspecific band in all IPs also seen in (b). (e) IP of mutant p53 with DO-1. HCT116 p53−/− cells were transfected with 0.75 μg HA-tagged TAp73α, TAp73β, TAp73γ or TAp73α ΔDBD and 1.5 μg mutant p53 (R175H). Input for p53 and input and co-precipitated p73 was detected by western blotting with DO-1 and HA antibodies.
As p53R175H was the better binding partner, we used this mutant to investigate which domains in p63 and p73 were required to interact with mutant p53. Previous studies have shown that the DBD of p53 is required for the interaction of mutant p53 with p63 or p73.19, 20 Interestingly, while deletion of the DBD of TAp73α (Δ131–307) or ΔNp73α (Δ82–258) reduced the interaction with p53R175H, deletion of this domain in TAp63α (Δ138–319) or ΔNp63α (Δ84–265) somewhat enhanced binding (Figures 1b and c and see below). The contribution of the DBD of TAp73α to the interaction with mutant p53 was confirmed in a reciprocal immunoprecipitation of p73 (using a hemagglutinin (HA) antibody) to identify co-precipitating p53R175H (Supplementary Figure S1A). However, although deletion of the DBD of p73α reduced binding to p53, we consistently saw that this interaction was not completely abolished by this deletion. We therefore explored further the possibility that binding of p53R175H involved regions distinct from the DBD in both p63 and p73.
As both ΔNp63 and ΔNp73 retained p53R175H binding (Figure 1a), suggesting that the N terminus was not necessary for the interaction, we examined the contribution of the C terminus of each protein using deletion mutants and the C-terminal truncated isoforms (Figure 1c). In the case of p63, the shorter isoforms TAp63β and TAp63γ were consistently expressed at lower levels compared with TAp63α (Figure 1d), an observation previously noted with other TAp63α proteins mutated in the transactivation inhibitory domain.12 However, we consistently found that both TAp63β and TAp63γ were significantly impaired for p53R175H binding compared with TAp63α (Figure 1, long exposure and Supplementary Figure S1B). Analysis of further TAp63α mutants indicated that deletion of amino acids 462–561 also perturbed the interaction with p53R175H (Figure 1d and Supplementary Figures 1c and d).
During the course of these studies, we noted that removal of the DBD of TAp63α led to somewhat increased binding to p53R175H (Figure 1d and Supplementary Figure S1C). Previous studies have shown that TAp63α can be held in an inactive compact dimer through an interaction between the C- and N-terminal domains of the protein,13, 37 and we considered that removal of the entire DBD may prevent this intramolecular interaction and inhibit the formation of closed dimers. To test directly the effect of inhibiting the dimer formation in TAp63α, we used a mutant within the N terminus of TAp63α (TAp63αFWL) that creates an open and tetrameric state that is also transcriptionally inactive.13 Interestingly, this mutant also showed enhanced p53R175H binding (Supplementary Figure S1D), supporting the suggestion that mutant p53 binds preferentially to the tetrameric form of TAp63.
Turning to TAp73, we noted that TAp73β clearly showed loss of binding to p53R175H (Figure 1e), indicating that similar to p63, the C terminus of p73 has a role in mediating this interaction. Surprisingly, however, TAp73γ regained the ability to bind to p53R175H (Figure 1e). Despite the loss of the C-terminal sequences present in TAp73α, TAp73γ contains 77 additional unique amino acids at the C terminus (Figure 1c), which appear to confer binding competence.
A previously proposed aggregation domain of mutant p53 is not required to bind p63 or promote invasion
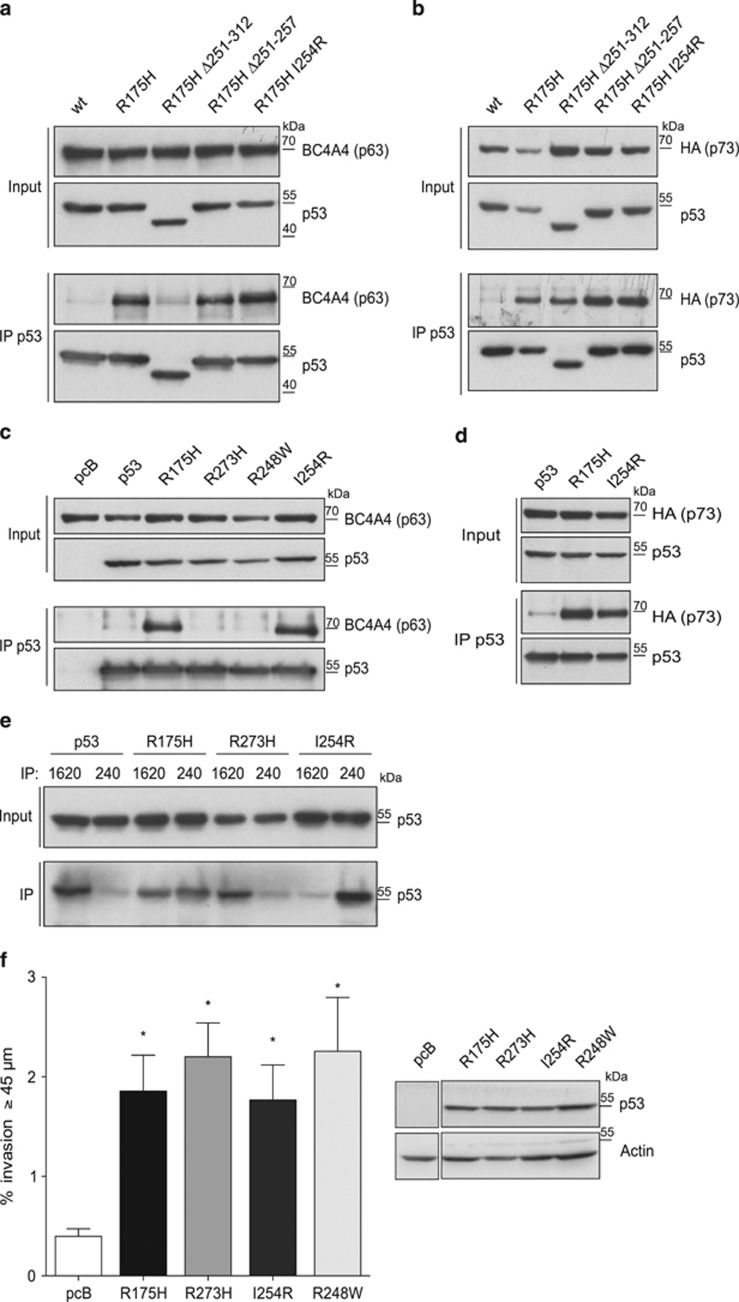
Although p63 and p73 can form mixed tetramers through their C-terminal tetramerization domains,15 the interaction of mutant p53 with p63/p73 has been shown to depend on the DBD of mutant p53.19, 20 While examining the effects of small deletions in the DBD of p53, which would be expected to destabilize p53, we found that the binding of p53R175H Δ251–312 to TAp63α appeared somewhat reduced (Figure 2a), although this was less apparent when looking at the p53R175H Δ251–312 interaction with TAp73α (Figure 2b). This observation was of interest in light of a recent publication reporting the formation of aggregates by mutant p53 that can include p63 and p73.38 This study identified an aggregation domain within mutant p53 spanning residues 251–258, with an absolute requirement for isoleucine 254. We therefore sought to determine whether aggregation was necessary for p53 to bind p63 or p73. A mutant in which the complete aggregation domain had been removed (R175H Δ251–257) still showed interaction with TAp63α (Figure 2a) and TAp73α (Figure 2b), although we considered that this may reflect a misfolding of the protein caused by the partial deletion of the domain. Surprisingly, however, even the point mutation I254R that has previously been shown to prevent aggregation38 did not inhibit binding of the R175H p53 mutant to p63 or p73 in our hands (Figures 2a and b). Furthermore, introduction of the I254R point mutant alone into p53 (without the R175H mutation) induced enhanced binding to TAp63α to a similar extent as R175H, under conditions where DNA contact mutants such as R273H or R248W showed very low binding (Figure 2c). The efficiency of mutant p53 binding to p63 and p73 correlated with a misfolded conformation of the p53 protein, as assessed by a change in reactivity to antibodies 1620 (which recognizes folded wild-type p53) and 240 (which recognizes misfolded p53 associated with mutations such as R175H, which cause structural instability of the p53 protein).39 Using this approach, we found that, similar to R175H, introduction of the I254R mutation resulted in clear misfolding of p53, which correlated with the acquisition of reactivity with Ab240 and TAp63α or TAp73α binding (Figures 2c–e). Although these results differ somewhat from those previously published,38 it is well established that even wild-type p53 is structurally very unstable,40 and small differences in extraction procedures may result in differences in the extent of unfolding of p53 proteins.41 To assess the functional consequence of the I254R mutation, we examined whether this aggregation mutant acquired the gain of function that is characteristic of mutant p53s in cells. Several studies have shown that mutant p53s (such as R175H and R273H) can promote the invasive behavior of p53-null cells in Matrigel invasion assays22, 23 and that this correlates with the ability to inactivate TAp63α. Interestingly, a p53 protein carrying only the I254R mutation promoted invasion in this assay as efficiently as a series of tumor-derived p53 mutants that have previously been shown to acquire this gain of function (Figure 2f) and was similarly able to inhibit TAp63α transcriptional activity (Supplementary Figure 1E). Taken together, these results suggest that mutations in p53 that have been reported by others to prevent aggregation do not necessarily impede the acquisition of p63-binding activity or the ability to promote invasive behavior in cells.
Figure 2.
The aggregation domain in p53 is not required for p63/p73 binding or the ability to promote invasion. (a) Immunoprecipitation of p53 with DO-1 in HCT116 p53−/− cells transfected with 3 μg TAp63α in combination with 3 μg p53wt, p53R175H, p53R175H Δ251–312, p53R175H Δ251–257 or p53R175H I254R. p53 and p63 expression was determined in the inputs and in precipitated material by western blot using DO-1 and BC464 (p63) antibodies. (b) Immunoprecipitation of p53 with DO-1 in HCT116 p53−/− cells transfected with 3 μg HA-tagged TAp73α in combination with 3 μg p53wt, p53R175H, p53R175H Δ251–312, p53R175H Δ251–257 or p53R175H I254R. p53 and p73 expression was determined in the inputs and in precipitated material by western blot using DO-1 and HA antibodies. (c) Immunoprecipitation of p53 with DO-1 in HCT116 p53−/− cells transfected with 3 μg TAp63α in combination with 2 μg p53wt, p53R175H, p53R273H, p53R248W or p53I254R. p53 and p63 expression was determined in the inputs and in precipitated material by western blot using DO-1 and BC464 (p63) antibodies. (d) Immunoprecipitation of p53 in HCT116 p53−/− cells transfected with 3 μg HA-tagged TAp73α in combination with 3 μg p53wt, p53R175H or p53I254R. p53 and p73 expression was determined in the inputs and in precipitated material by western blot using DO-1 and HA antibodies. (e) Immunoprecipitation of p53 (folded: 1620; unfolded: 240) in HCT116−/− cells transfected with 3μg p53, p53R175H, p53R273H or p53I254R. p53 expression was determined in the inputs and in precipitated material by western blot using a DO-1 antibody. (f) H1299 cells were retrovirally infected with an empty vector or the indicated p53 mutants (in a pWZL blast vector). Blasticidin-resistant cells were selected and their invasion towards hepatocyte growth factor (HGF) over 72h was measured in inverted transwell invasion assays. Bars represent the mean of invasion beyond 45 μm in four sets of triplicates. Error bars as standard error of the mean. *A P-value <0.02 compared with the empty vector-expressing cells. The expression of the constructs is shown by western blot and actin was used as a loading control (right). Panels form part of the same gel.
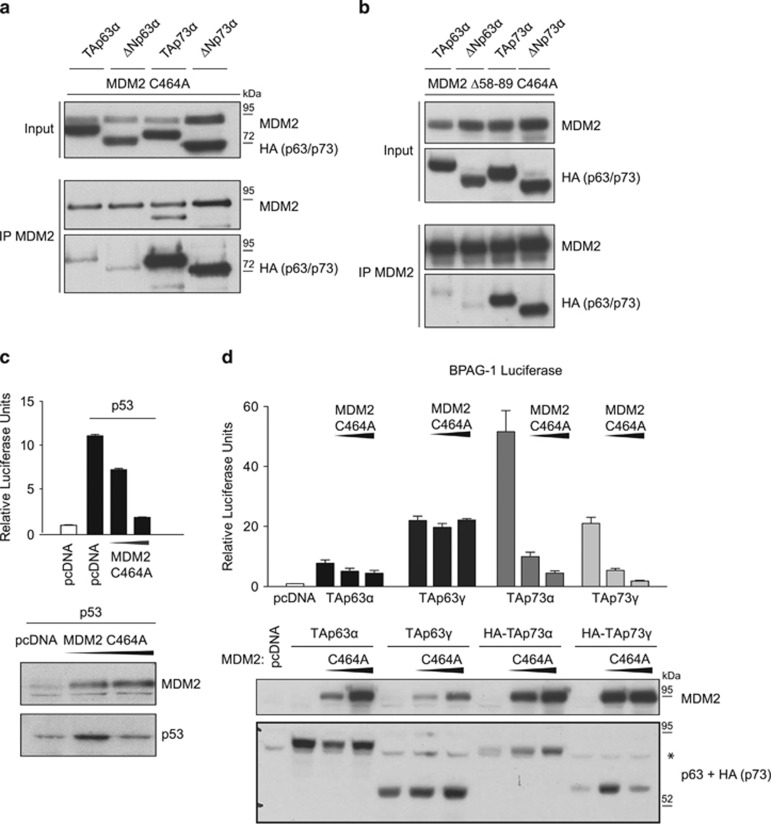
MDM2 binds p73 more strongly than p63
MDM2 is a key regulator of p53 activity, and functions through various mechanisms to inhibit p53. Previous studies have shown that MDM2 also binds p73, but interacts much more weakly, if at all, with p63.34, 42 We were also able to confirm that MDM2 bound well to TAp73α, TAp73γ and ΔNp73α (Figure 3a and Supplementary Figure 1F), but showed a much weaker ability to interact with either TAp63α or ΔNp63α, using the same antibody and similar immunoprecipitation conditions as for the p73 isoforms (Figure 3a). The principal binding site for p53 on MDM2 is within the N-terminal binding pocket on MDM2, which is disrupted by deleting residues 58–89. Interestingly, loss of this region did not reduce the binding of MDM2 to TAp73 or ΔNp73α (Figure 3b), suggesting that p73 and p53 bind to different regions on MDM2. Although MDM2 controls p53 to a large extent through ubiquitination and degradation, interaction with MDM2 can also directly inhibit the transcriptional activity of p53, as seen by the effect of coexpression of an MDM2 mutant that lacks E3 ligase activity (MDM2 C464A) with p53 (Figure 3c). MDM2 does not target p73 for degradation,29 but the E3 mutant MDM2 C464A also inhibited the transcriptional activity of TAp73α and TAp73γ (Figure 3d). However, coexpression of this MDM2 mutant did not significantly affect the transcriptional activity of either TAp63α or TAp63γ (Figure 3d), consistent with the much weaker binding of MDM2 to TAp63. We note that, as shown previously, TAp63α shows relatively weak transcriptional activity in these assays,6, 37 most likely reflecting the adoption of the closed dimeric and therefore inactive conformation of this transfected protein.13
Figure 3.
MDM2 binds and inhibits TA and ΔNp73 but not p63. (a) Immunoprecipitation of MDM2 (Ab1) in HCT116 p53−/− cells transfected with 3 μg HA-tagged TAp63α, ΔNp63α, TAp73α (Simian) or ΔNp73α in combination with 2 μg MDM2 C464A. MDM2 and p63 or p73 expression was determined in the inputs and in precipitated material by western blot using the Ab1 (MDM2) and HA antibodies. (b) Immunoprecipitation of MDM2 (Ab1) in HCT116 p53−/− cells transfected with 3 μg HA-tagged TAp63α, ΔNp63α, TAp73α or ΔNp73α in combination with 2 μg MDM2 Δ58–89 C464A. MDM2 and p63 or p73 expression was determined in the inputs and in precipitated material by western blot using the Ab1 (MDM2) and HA antibodies. (c) HCT116 p53−/− cells were transfected with 300 ng PG13 luciferase, 100 ng TK Renilla and either 300 ng empty vector or p53 together with increasing amounts of MDM2 C464A (25 and100 ng). Activation of the promoters was assayed using the Promega Luciferase System. Data are plotted as relative luciferase units (RLU) (firefly luciferase readings divided by Renilla luciferase readings) fold change relative to p53-null control. The diagram represents the mean of triplicates with error bars as s.e.m. The expression of MDM2 and p53 was determined by western blot using Ab1 or DO-1 antibodies. (d) HCT116 p53−/− cells were transfected with 300 ng BPAG luciferase, 100 ng TK Renilla in combination with 300 ng TAp63α, TAp63γ, HA-TAp73α or HA-TAp73γ and 0.2 or 0.5 μg of MDM2 C464A and luciferase activity determined. Data are plotted as RLUs (firefly luciferase readings divided by Renilla luciferase readings) fold change relative to p53-null control. The diagram represents the mean of triplicates with error bars as s.e.m. The expression of MDM2 and p63 or p73 was determined by western blot using an Ab1 (MDM2) or BC4A4 (p63)+HA (p73). *A nonspecific band.
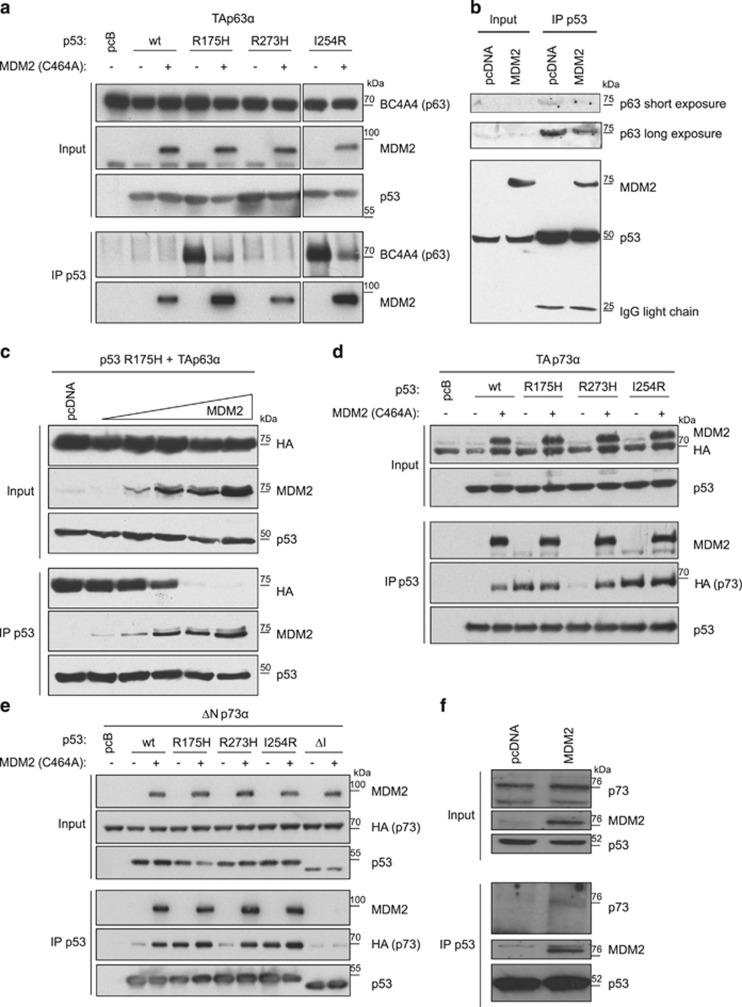
Having established that TAp73α forms an interaction with both p53R175H and MDM2, whereas TAp63α bound p53R175H well but MDM2 only very weakly, we examined the effect of MDM2 expression on the mutant p53/p63 or p73 interaction. Intriguingly, the interaction of mutant p53 (either R175H or I254R) with TAp63α was substantially reduced following coexpression of an E3-inactive MDM2 without apparently affecting the weak interaction of p53R273H or wild-type p53 to TAp63α in HCT116−/− cells (which have lost full-length p53 expression but retain the expression of some N-terminally truncated p53 isoforms) (Figure 4a) or H1299 cells (which are null for all forms of p53) (Supplementary Figure 2A). Overexpression of MDM2 C464A in BXPC3 cells expressing p53Y220C (a conformational mutant) (Figure 4b) or SKBR3 cells expressing p53R175H (Supplementary Figure 2B) further demonstrated a decrease in the binding capacity of endogenous mutant p53 to endogenous p63 (most likely the TAp63α variant based on size) in the presence of MDM2. Consistent with these observations, titrating MDM2 expression resulted in a dose-dependent decrease in the interaction between p53R175H and TAp63α, indicating that MDM2 competes with TAp63α for binding to mutant p53 (Figure 4c).
Figure 4.
MDM2 competes for binding of mutant p53 with p63, but promotes the binding of mutant p53 with p73. (a) Immunoprecipitation of p53 with DO-1 in HCT116 p53−/− cells transfected with 2μg TAp63α in combination with 2 μg p53wt, p53R175H, p53R273H, p53I254R and MDM2 C464A. P53, MDM2 and p63 expression was determined in the inputs and p63 and MDM2 expression was determined in precipitated material by western blot using DO-1, Ab1 (MDM2) and BC4A4 (p63) antibodies. (b) Immunoprecipitation of p53 with DO-1 in BXPC3 cells (three 10 cm plates) that were transfected with 3 μg MDM2 C464A or pcDNA. MDM2, p63 and p53 expression was determined in inputs and immunoprecipitations by western blot using p53 DO-1, MDM2 SMP14 and p63 4A4 antibodies. (c) Immunoprecipitation of p53 using DO-1 in HCT116 −/− cells transfected with 3 μg HA-TAp63α and p53R175H each in combination with 0, 0.5, 1.0, 2.0, 4.0 or 6.0 μg MDM2 C464A or compensatory amounts of pcDNA. MDM2, p63 and p53 expression levels were determined in the input and precipitated material using western blot and SMP14, HA and DO-1 antibodies, respectively. (d and e) Immunoprecipitation of p53 with DO-1 (d) or 1801 (e) in HCT116 p53−/− cells transfected with 2 μg HA-TAp73α (d) or HA-ΔNp73α (e) in combination with 2 μg p53wt, p53R175H, p53R273H, p53I254R, p53ΔI (only in e) and MDM2 C464A. P53, MDM2 and p73 (HA) expression was determined in the inputs and in precipitated material by western blot using DO-1 (d), 1801 (e), Ab1 (MDM2) and HA antibodies. (f) Immunoprecipitation of p53 with DO-1 in U251 cells that were transfected with 3 μg MDM2 C464A or pcDNA. MDM2, p73 and p53 expression was determined in inputs and immunoprecipitations by western blot using p53 DO-1, MDM2 SMP14 and a p73 antibody.
In contrast to the behavior of p63, coexpression of MDM2 C464A did not clearly affect the binding of R175H or I254R to TAp73α or ΔNp73α and markedly enhanced the interaction of these p73 isoforms with p53R273H in HCT116−/− cells (Figures 4d and e) or H1299 cells (Supplementary Figure 2C). In these assays, MDM2 even promoted an interaction between wild-type p53 and TAp73α or ΔNp73α (Figure 4e and Supplementary Figure 2C). However, immunoprecipitation of the complex through MDM2 instead of p53 showed that while p53R273H bound MDM2, it did not increase the binding between MDM2 and TAp73α, compared with the interaction seen in the absence of mutant p53 (Supplementary Figure 2D). These data suggest that MDM2 facilitates the binding of TAp73α to p53R273H, but that p53R273H does not govern the interaction between MDM2 and TAp73α. An increase in binding of p53R248W (in HCT116 cells) and p53R273H (in U251 cells) to p73 (most likely TAp73α based on size) following MDM2 expression demonstrated that the effect of MDM2 is also apparent with endogenously expressed mutant p53 and TAp73α (Figure 4f and Supplementary Figure 2E). Importantly, the ability of MDM2 to enhance the p53R273H/p73 interaction was dependent on the N-terminal MDM2-binding domain on p53, as shown by a p53 protein deleted for this domain (ΔI). The p53 ΔI protein did not co-precipitate MDM2 (as expected) and did not gain the ability to interact with ΔNp73α in the presence of MDM2 (Figure 4e). As MDM2 can bind both mutant p53 and TA/ΔNp73α, these results indicate that p53/MDM2/p73α can form a trimeric complex, and that the p53/TAp73α or p53/ΔNp73α interaction is enhanced by MDM2.
Various reports show that mutant p53s can be stabilized by chaperones that impair MDM2-mediated degradation of p53.43, 44 Furthermore, heat-shock protein 70 (HSP70) can have a role in stabilizing the p53R175H/TAp73α interaction and destabilizing the p53R175H/TAp63α interaction,45 suggesting that there may be a similarity to our observations with MDM2. However, under our experimental conditions, HSP70 did not alter the R175H/TAp63α or R175H/TAp73α interaction (Supplementary Figures 3A and B), indicating that the effect of MDM2 on these interactions is not mediated entirely through HSP70.
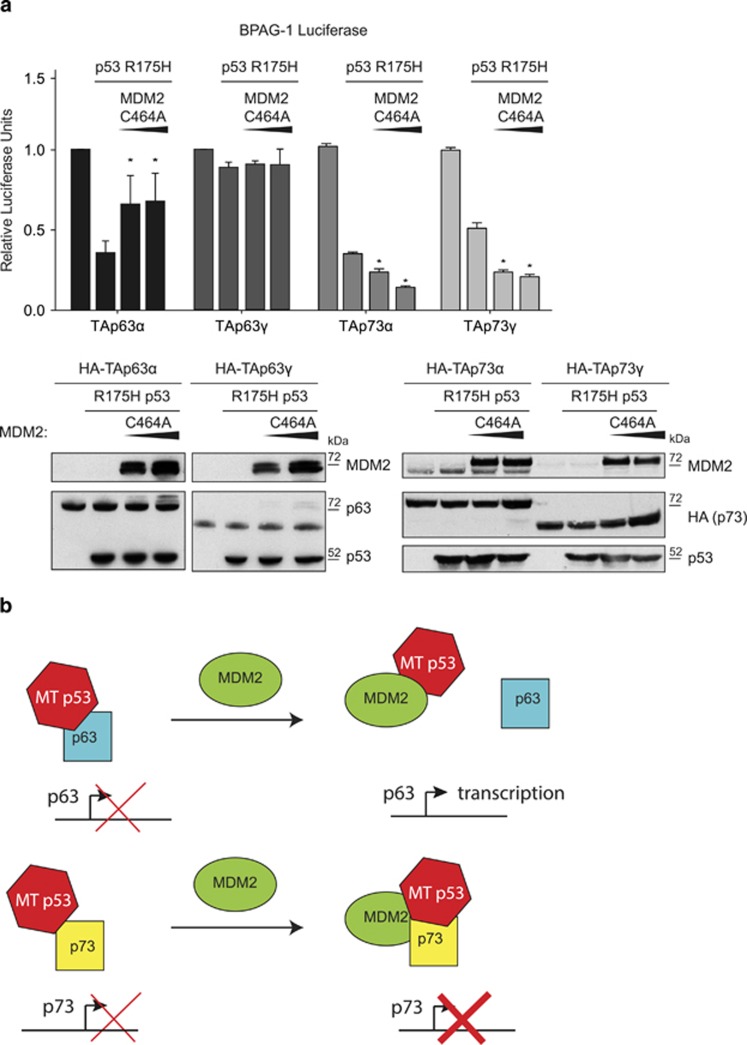
To test the consequences of the different effects of MDM2 on the interaction of mutant p53 with p63 and p73, we looked at the impact on TAp63α/γ and TAp73α/γ transcriptional activity (Figure 5a). As expected, expression of p53R175H reduced the transcriptional activity of TAp63α, but not TAp63γ (Figure 5a), reflecting the capacity of mutant p53 to interact with the different p63 isoforms (Figures 1d and 5a). Interestingly, titration of increasing amounts of MDM2, which was shown to dissociate binding of p53R175H to TAp63α (Figure 4a), significantly alleviated the inhibition of p53R175H on TAp63α, while having no effect on TAp63γ (Figure 5a). In contrast, p53R175H inhibited the transcriptional activity of both TAp73α and TAp73γ (reflecting the ability to bind both isoforms; Figure 1) and a titration of MDM2 even slightly increased this inhibition (Figure 5a), correlating with the formation of the mutant p53/TAp73α/MDM2 complex (Figure 4b). Previous studies have shown that p53R273H can inhibit the transcriptional activity of TAp73α, despite showing only weak binding.23 Additional expression of MDM2 further decreased TAp73α luciferase activity (Supplementary Figure 3C), consistent with the concurrent binding of p53R273H, TAp73α and MDM2.
Figure 5.
MDM2 prevents mutant p53 from inhibiting TAp63α, but potentiates the inhibition of TAp73. (a) HCT116 p53−/− cells were transfected with 300 ng BPAG luciferase, 100 ng TK Renilla, 2 μg p53R175H and increasing concentrations (0.2, 0.4 and 0.6 μg) of MDM2 C464A in combination with 2 μg HA-TAp63α, HA-TAp63γ, HA-TAp73α or HA-TAp73γ, and luciferase activity was determined. Data are plotted as relative luciferase units (RLUs) (firefly luciferase readings divided by Renilla luciferase readings) fold change relative to p53-null control. The diagram represents the mean of triplicates with error bars as s.e.m. The expression of MDM2, p63, p73 and p53 was determined by western blot using Ab1 (MDM2), BC4A4 (p63), HA or DO-1 (p53). (b) Schematic representation of the consequences of MDM2 expression on the inhibitory function of mutant p53 on p63 or p73. Expression of MDM2 alleviates the inhibitory role of mutant p53 on p63, but potentiates the inhibition of p73 by mutant p53.
Taken together, these results show that the binding of mutant p53 to TAp63α and TAp73α is differentially modulated by MDM2, which attenuates mutant p53 binding and inhibition of TAp63α, while potentiating mutant p53 binding and inhibition of TAp73α (Figure 5b).
Discussion
p53, p63 and p73 are a family of related proteins with distinct but overlapping functions. Although each of these proteins can homo-oligomerize, wild-type p53 does not interact efficiently with either p63 or p73. By contrast, tumor-associated point mutations in the DBD of p53 can induce an ability of the mutant p53 to bind both p63 and p73, an activity that correlates with the acquisition of a misfolded conformation of the mutant p53 protein. Our results are in agreement with previous reports that have shown a contribution of the DBD of p73 to the interaction with mutant p53.20 However, we also show that the C termini of p63 and p73 have an important role in permitting this interaction. Intriguingly, although binding is decreased in TAp63β, TAp63γ and TAp73β, the novel amino acids at the C terminus of TAp73γ reinstate the ability to bind mutant p53. These results present the interesting possibility that mutant p53 may differentially inhibit the C-terminal isoforms of TAp63 and TAp73. Indeed, our data support this suggestion, as we show that mutant p53R175H inhibits the transcriptional activity of TAp63α, but is unable to inhibit this function of TAp63γ. However, the transcriptional activity of both TAp73α and TAp73γ is efficiently inhibited by mutant p53R175H (Figure 5). Given the pro-oncogenic role of mutant p53, this differential activity of mutant p53 may be important in tumor development.
Previous studies have shown that most mutant p53s can form aggregates in vitro and in cells,38, 46 and that these can also include full-length and N-terminal truncated p63 and p73 isoforms.38 A single point mutation of isoleucine 254 in p53 was shown to prevent aggregation and interaction of p53R175H with TAp63 or TAp73. We show here, however, that the interaction of mutant p53 with TAp63 and TAp73 is not dependent on isoleucine 254. Instead, p53 carrying a substitution of this residue to arginine, which would disrupt aggregation, behaves like a tumor-derived DBD point mutant. Specifically, p53I254R acquires the ability to react with a mutant-specific antibody, bind to TAp63α/TAp73α, inhibit TAp63α and promote invasion in a three-dimensional cell culture system. Indeed, I245R is found in the p53 database of somatic tumor mutations, albeit rarely (0.2% of all recorded somatic p53 mutations), suggesting that this mutation can be advantageous during cancer development.47 Although the ability to form aggregates may contribute to the oncogenic activity of mutant p53, our data show that aggregation mediated through the previously identified domain within amino acids 251–257 of p53 is not required for mutant p53 to show oncogenic activities. Indeed, the ability of mutant p53 to regain wild-type p53 activity (e.g. with temperature-sensitive mutant p5348) suggests that these mutants are not necessarily locked into irreversible aggregates. It seems that multiple mechanisms through which mutant p53 can inhibit p63/p73 functions exist, including the direct binding (as described here) and mechanisms that do not depend on tight and stable complex formation, as exhibited by p53 mutants such as R273H.23
One of the key regulators of p53 function is MDM2, which also binds to mutant p53 through the N-terminal binding regions in both proteins. MDM2 can promote the degradation of mutant p53,49, 50 although in tumor cells this regulation is disrupted, for example, through the expression of MDM2 isoforms that inhibit full-length MDM2 E3 ligase activity,51 and mutant p53 protein accumulates. In agreement with previous results,34, 42 we can detect MDM2 binding to TAp73α and ΔNp73α, but to a much lesser extent to TAp63α and ΔNp63α in cells. This differential binding is reflected by a far superior inhibition of TAp73α and ΔNp73α transcriptional activity by MDM2, which has almost no effect on TAp63α and ΔNp63α transcriptional activity in these assays. These data support in vitro studies showing that TAp63 can bind MDM2 with a much lower affinity than p53 or TAp73.52 As with the binding of mutant p53, these observations suggest that there are significant differences in the ability of both N-terminal variants of p63 and p73 to interact with protein binding partners. Interestingly, this differential ability to bind MDM2 is reflected by different consequences of MDM2 on the mutant p53 interaction with p63 or p73. We find that while MDM2 can inhibit the p53R175H/TAp63α interaction—suggesting competition between MDM2 and TAp63α for binding to mutant p53—MDM2 can enhance the interaction of p53R273H with TAp73α or ΔNp73α. This latter effect is seen most clearly using mutants of p53 that do not strongly activate p73 binding (such as R273H), whereas the interaction of strongly binding mutant p53 (such as R175H) are unaffected by MDM2. MDM2 is not the only protein that can form a complex with p53 family members, and a recent publication has suggested that HSP70 shows a similar differential effect of reactivating p63, but not p73, in the presence of mutant p53.45 In addition to HSP70 and MDM2, a number of other proteins could affect mutant p53/p63/p73 interactions, including HSP90, the CCT (chaperonin containing tailless complex polypeptide 1) complex, HAUSP (herpesvirus-associated ubiquitin-specific protease) or CHIP (C-terminus of Hsc70-interacting protein). Additional complexity is provided by the potential for expression of many isoforms of all the p53 family proteins.9, 53
In this study, we have focused on the consequences of MDM2 expression on mutant p53/p63 and p73 interactions. We showed that MDM2 relieves the inhibition of p63 activity by mutant p53, but can enhance the inhibition of p73 by mutant p53. Although the impact of other interacting proteins remains to be addressed, our observations point to an interesting differential ability of MDM2 to regulate the activity of mutant p53.
Materials and methods
Cells
H1299 and HEK293T cells (obtained from American Type Culture Collection, Teddington, UK) and HCT116 p53-null cells (a gift from Dr Bert Vogelstein) were maintained in DMEM (Life Technologies, Paisley, UK) supplemented with 10% fetal bovine serum, 1% pen/strep and 1% glutamine at 37 °C in 5% CO2.
Plasmids
The following plasmids have been previously described: pcDNA constructs—HA-TAp63α, HA-ΔNp63α, HA-TAp73α, HA-ΔNp73α, TAp63α, TAp63β, TAp63γ, ΔNp63α, ΔNp63γ, TAp63αFWL, Myc-p63α and Myc-p63αFWL;13, 54, 55, 56, 57 pcB6 constructs—p53 wild type, p53R175H, p53R273H amd p53ΔI;58 and PCHDM1A constructs—MDM2, MDM2 Δ58–89 and MDM2 C464,59, 60 BPAG luciferase and PG13 luciferase,61, 62 Myc-TAp63α and Myc-TAp63α FWL13 and pWZL blast p53.63
The following constructs were generated by site-directed mutagenesis: pcDNA3 constructs—p53R175H, p53I254R, p53R248W, p53R273H, p53R175H Δ251–312, p53R175H Δ251–257, p53R175H I254R, HA-TAp63α Δ263–361, HA-TAp63α Δ362–461, HA-TAp63α Δ462–561, HA-TAp63α Δ138–319 (ΔDBD), HA-ΔNp63α Δ84–265 (ΔDBD), HA-TAp73α Δ131–307 (ΔDBD), HA-TAp73α Δ308–386 (ΔDBD), pWZL blast p53R175H, pWZL blast p53R273H, pWZL blast p53I254R, pWZL blast p53R248W and pCHDM1A MDM2 Δ58–89 C464A. Oligonucleotides used for site-directed mutagenesis are listed in the Supplementary Information. HA-tagged p63β and p63γ were generated using site-directed mutagenesis in the non-tagged constructs provided by Dr Caron de Fromentel.
Immunoprecipitation and immunoblot
A total of 7.5 × 105 cells were seeded in 10 cm plates and transfected with 3–6 μg DNA the following day using 15–18 μl GeneJuice in 300 μl Opti-Mem (Life Technologies). The transfection mix was added to the plates dropwise and cells were harvested 20–24 h later in RIPA buffer (50mM Tris, pH 7.4, 150 mM NaCl, 1% Triton, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate). Lysates were incubated for 15 min on ice and the supernatant was taken for immunoprecipitation with the indicated antibodies (0.5–2 μg per sample) overnight at 4 °C using protein G magnetic beads (Dynabeads; Invitrogen, Life Technologies). The following antibodies were used for immunoprecipitation: p53 DO-1 (Santa Cruz, Santa Cruz, CA, USA), p53 pAb1620 (Calbiochem, Nottingham, UK), p53 pAb240 (Calbiochem), HA (Covance, Leeds, UK) and MDM2 Ab1 (Calbiochem). Input and precipitates were run on western page and the following first antibodies were used: p53 DO-1, p63 BC4A4 (Santa Cruz), p63 4A4 (Santa Cruz), p73,29 p73 Ab 7824 (Millipore, Watford, UK), HA (Covance), actin C4 (Chemicon), HSP70 (Enzo, Life Sciences, Exeter, UK), Ab1 MDM2 (Calbiochem), MDM2 SMP14 (Santa Cruz) or Myc (9E10; Covance). For immunoprecipitations, a rat anti-mouse IgK light-chain horse radish peroxidase (BD Biosciences, Oxford, UK) was used and all other detections were carried out with regular mouse or rabbit horse radish peroxidase-coupled Amersham secondary antibodies (GE Healthcare Life Sciences, Little Chalfont, UK).
Luciferase assays
Cells were seeded to 70% confluency into 24 wells for transfection using GeneJuice (25 or 100 ng p53 constructs, 50 ng p63 or p73, 50 ng MDM2, 25 ng Renilla, 100 ng BPAG luciferase). At 24 h after transfection, cells were lysed in 100 μl lysis buffer using a Renilla Luciferase Kit according to the manufacturer's protocol (Promega, Southampton, UK). Luciferase and Renilla activity was measured using a Veritas Microplate luminometer (Turner Biosystems, Promega) and the Glomax software from Promega. Relative luciferase units were determined by correcting luciferase readings for Renilla luciferase expression. Error bars represent the s.e.m. for three independent experiments. Of each lysate, 20μl was run on the immunoblot to test the expression of the transfected constructs.
Matrigel invasion
H1299 cells were retrovirally infected with empty vector or p53 mutants using HEK293T cells as packaging cells and were subsequently selected with 7.5 μg/ml blasticidin for 3–4 days. In Transwell inserts (6.5 mm, 8 μm pore size; Corning Life Sciences, High Wycombe, UK) in a 24-well plate 60 μl Matrigel batch A6520 (BD Bioscience) diluted 1:1 in phosphate-buffered saline, containing 25 ng/ml fibronectin was pipetted carefully onto the center of the membrane and incubated 45 min at 37 ºC. The inserts were inverted on the lid of a 24-well plate and 100 μl of a 2.5 × 105 cells per ml cell suspension was pipetted onto the membrane, whereafter the 24-well plate was fitted over the inserts to create ‘hanging droplets' of cells. Cells were allowed to settle for 5 h at 37 ºC, whereafter the transwells were washed in serum-free medium and transferred (upright) to a 24-well plate containing 1 ml serum-free medium. One hundred microliters of medium containing serum and 10 ng/ml hepatocyte growth factor was pipetted into the center of the insert on top of the polymerized Matrigel to attract cells from above the Matrigel plug. Cells were allowed to migrate for 3 days and stained in the medium containing 4 nM calcein (Life Technologies) for 1 h. Migration was visualized on a Leica TCS SP2 laser scanning confocal microscope (Leica, Peterborough, UK), taking images in serial sections every 15 μm starting at the membrane and moving up into the Matrigel plug until no more cells could be detected. The number of pixels with intensity of 100 or above was quantified in each binary confocal image using ImageJ.64 The percentage of invading cells was calculated by dividing the sum of pixels at or beyond 45 μm by the sum of pixels in all images.
Acknowledgments
We thank Drs Gerry Melino, Caron de Fromentel and Kevin Ryan for plasmids and are grateful to Cancer Research UK, the DFG (DO 545/8-1), The Wellcome Trust/the Royal Society (Sir Henry Dale Fellowship) and the Cluster of Excellence Frankfurt (Macromolecular Complexes) for funding.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. P53 mutations in human cancers. Science (New York) NY. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, et al. Understanding the function–structure and function–mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. P53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. P63, a p53 homolog at 3q27–29, encodes multiple products withtransactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;3:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J-C, Valent A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Ishimoto O, Kawahara C, Enjo K, Obinata M, Nukiwa T, Ikawa S. Possible oncogenic potential of DeltaNp73: a newly identified isoform of human p73. Cancer Res. 2002;62:636–641. [PubMed] [Google Scholar]
- Khoury MP, Bourdon JC. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol. 2010;2:a000927. doi: 10.1101/cshperspect.a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natan E, Joerger AC. Structure and kinetic stability of the p63 tetramerization domain. J Mol Biol. 2012;415:503–513. doi: 10.1016/j.jmb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethayathulla AS, Tse PW, Monti P, Nguyen S, Inga A, Fronza G, et al. Structure of p73 DNA-binding domain tetramer modulates p73 transactivation. Proc Natl Acad Sci USA. 2012;109:6066–6071. doi: 10.1073/pnas.1115463109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub WE, Weber TA, Schafer B, Candi E, Durst F, Ou HD, et al. The C-terminus of p63 contains multiple regulatory elements with different functions. Cell Death Dis. 2010;1:e5. doi: 10.1038/cddis.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch GB, Zielonka EM, Coutandin D, Weber TA, Schafer B, Hannewald J, et al. DNA damage in oocytes induces a switch of the quality control factor TAp63alpha from dimer to tetramer. Cell. 2011;144:566–576. doi: 10.1016/j.cell.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luh LM, Kehrloesser S, Deutsch GB, Gebel J, Coutandin D, Schafer B, et al. Analysis of the oligomeric state and transactivation potential of TAp73alpha. Cell Death Differ. 2013;20:1008–1016. doi: 10.1038/cdd.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutandin D, Lohr F, Niesen FH, Ikeya T, Weber TA, Schafer B, et al. Conformational stability and activity of p73 require a second helix in the tetramerization domain. Cell Death Differ. 2009;16:1582–1589. doi: 10.1038/cdd.2009.139. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Rajagopalan S, Natan E, Veprintsev DB, Robinson CV, Fersht AR. Structural evolution of p53, p63, and p73: implication for heterotetramer formation. Proc Natl Acad Sci USA. 2009;106:17705–17710. doi: 10.1073/pnas.0905867106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Gaiddon C, Prives C. P73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiddon C, Lokshn M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Fontemaggi G, Costanzo A, Rizzo MG, Monti O, Baccarini A, et al. Physical interaction with human tumor derived p53 mutants inhibits p63 activities. J Biol Chem. 2002;277:18817–18826. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Cristofanellu B, Shaul Y, Castagnoli L, et al. Physical and functional interaction between p53 mutants and difference isoforms of p73. J Biol Chem. 2000;275:29503–29512. doi: 10.1074/jbc.M003360200. [DOI] [PubMed] [Google Scholar]
- Santini S, Di Agostino S, Coppari E, Bizzarri AR, Blandino G, Cannistraro S. Interaction of mutant p53 with p73: asurface plasmon resonance and atomic force spectroscopy study. Biochim Biophys Acta. 2014;1840:1958–1964. doi: 10.1016/j.bbagen.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Neilsen PM, Noll JE, Suetani RJ, Schulz RB, Al-Ejeh F, Evdokiou A, et al. Mutant p53 uses p63 as a molecular chaperone to alter gene expression and induce a pro-invasive secretome. Oncotarget. 2011;2:1203–1217. doi: 10.18632/oncotarget.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, George DL, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Balint E, Bates S, Vousden KH. Mdm2 binds p73a without targeting degradation. Oncogene. 1999;18:3923–3929. doi: 10.1038/sj.onc.1202781. [DOI] [PubMed] [Google Scholar]
- Zeng X, Chen L, Jost CA, Maya R, Keller D, Wang X, et al. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M, Wienzek S, Koenig S, Roth J. Inactivation of the p53-homologue p73 by the mdm2-oncoprotein. Oncogene. 1999;18:2101–2106. doi: 10.1038/sj.onc.1202512. [DOI] [PubMed] [Google Scholar]
- Kadakia M, Slader C, Berberich SJ. Regulation of p63 function by Mdm2 and MdmX. DNA Cell Biol. 2001;20:321–330. doi: 10.1089/10445490152122433. [DOI] [PubMed] [Google Scholar]
- Calabro V, Mansueto G, Parisi T, Vivo M, Calogero RA, La Mantia G. The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53 homolog p63. J Biol Chem. 2002;277:2674–2681. doi: 10.1074/jbc.M107173200. [DOI] [PubMed] [Google Scholar]
- Little NA, Jochemsen AG. Hdmx and Mdm2 can repress transcription activation by p53 but not by p63. Oncogene. 2001;20:4576–4580. doi: 10.1038/sj.onc.1204615. [DOI] [PubMed] [Google Scholar]
- Kojima T, Ikawa Y, Katoh I. Analysis of molecular interactions of the p53-family p51(p63) gene products in a yeast two-hybrid system: homotypic and heterotypic interactions and association with p53-regulatory factors. Biochem Biophys Res Commun. 2001;281:1170–1175. doi: 10.1006/bbrc.2001.4486. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. Structure–function-rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- Serber Z, Lai HC, Yang A, Ou HD, Sigal MS, Kelly AE, et al. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol Cell Biol. 2002;22:8601–8611. doi: 10.1128/MCB.22.24.8601-8611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- Gannon JV, Greaves R, Iggo R, Lane DP. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 1990;9:1595–1602. doi: 10.1002/j.1460-2075.1990.tb08279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53 and cancer-associated mutants. Adv Cancer Res. 2007;97:1–23. doi: 10.1016/S0065-230X(06)97001-8. [DOI] [PubMed] [Google Scholar]
- Trinidad AG, Muller PA, Cuellar J, Klejnot M, Nobis M, Valpuesta JM, et al. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell. 2013;50:805–817. doi: 10.1016/j.molcel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arooz T, Siu WY, Chiu CH, Lau A, Yamashita K, et al. MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett. 2001;490:202–208. doi: 10.1016/s0014-5793(01)02124-x. [DOI] [PubMed] [Google Scholar]
- Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F, et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Hrstka R, Coomber D, Lane DP, Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene. 2008;27:3371–3383. doi: 10.1038/sj.onc.1211010. [DOI] [PubMed] [Google Scholar]
- Wiech M, Olszewski MB, Tracz-Gaszewska Z, Wawrzynow B, Zylicz M, Zylicz A. Molecular mechanism of mutant p53 stabilization: the role of HSP70 and MDM2. PLoS One. 2012;7:e51426. doi: 10.1371/journal.pone.0051426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano Bom AP, Rangel LP, Costa DC, de Oliveira GA, Sanches D, Braga CA, et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: implications for cancer. J Biol Chem. 2012;287:28152–28162. doi: 10.1074/jbc.M112.340638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- Midgley CA, Lane DP. P53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X, et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun. 2013;4:2996. doi: 10.1038/ncomms3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdzalik M, Pustelny K, Kedracka-Krok S, Huben K, Pecak A, Wladyka B, et al. Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle (Georgetown, TX. 2010;9:4584–4591. doi: 10.4161/cc.9.22.13871. [DOI] [PubMed] [Google Scholar]
- Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. P63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni M, Falco M, Annicchiarico-Petruzzelli M, et al. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Ruptier C, Tribollet V, Hautefeuille A, Chardon F, Cavard C, et al. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis. 2008;29:273–281. doi: 10.1093/carcin/bgm258. [DOI] [PubMed] [Google Scholar]
- De Laurenzi V, Rossi A, Terrinoni A, Barcaroli D, Levrero M, Costanzo A, et al. P63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem Biophys Res Commun. 2000;273:342–346. doi: 10.1006/bbrc.2000.2932. [DOI] [PubMed] [Google Scholar]
- Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differen. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol. 2007;27:8284–8295. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MHG, Ludwig RL, Levine AJ, Vousden KH. Analysis of the degradation function of Mdm2. Cell Growth Differ. 1999;10:87–92. [PubMed] [Google Scholar]
- Osada M, Nagakawa Y, Park HL, Yamashita K, Wu G, Kim MS, et al. P63-specific activation of the BPAG-1e promoter. J Invest Dermatol. 2005;125:52–60. doi: 10.1111/j.0022-202X.2005.23801.x. [DOI] [PubMed] [Google Scholar]
- El-Deiry W, Tokino T, Velculescu VE, Levy DB, Parson VE, Trent JM, et al. WAF1, a potential mediator of p53 tumour suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Stindt MH, Carter S, Vigneron AM, Ryan KM, Vousden KH. MDM2 promotes SUMO-2/3 modification of p53 to modulate transcriptional activity. Cell Cycle (Georgetown, TX) 2011;10:3176–3188. doi: 10.4161/cc.10.18.17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins TJ. Image J for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.