Abstract
Tumour heterogeneity is a major factor undermining the success of therapies targeting metastatic cancer. Two major theories are thought to explain the phenomenon of heterogeneity in cancer – clonal evolution and cell plasticity. In this review, we examine a growing body of work implicating the transcription factor FOS Related Antigen 1 (FRA-1) as a central node in tumour cell plasticity networks, and discuss mechanisms regulating its activity in cancer cells. We also discuss evidence from the FRA-1 perspective supporting the notion that clonal selection and cell plasticity represent two sides of the same coin. We propose that FRA-1-overexpressing clones featuring high plasticity undergo positive selection during consecutive stages of multistep tumour progression. This model underscores a potential mechanism through which tumour cells retaining elevated levels of plasticity acquire a selective advantage over other clonal populations within a tumour.
Keywords: FRA-1, EMT, plasticity, miRNA, tumour progression
INTRODUCTION
CELLULAR PLASTICITY AND CLONAL SELECTION AS DETERMINANTS OF TUMOUR HETEROGENEITY
The existence of heterogeneity between malignant cells in a tumour has long been recognised to pose an obstacle for successful cancer therapy. That tumours are highly heterogeneous was confirmed in recent years through massive efforts in whole genome sequencing of tumour DNA. Two theories, clonal evolution and cell plasticity, explain the heterogeneous nature of cancer.
According to the clonal evolution theory, mutations occur stochastically in cells within tumour tissue. Cancer cells in a growing tumour undergo Darwinian selection, and clones with competitive advantages survive and replace less aggressive cell populations, thereby driving tumour progression. On the other hand, subpopulations of tumour cells can cooperate and support growth and promote invasion of each other1-4. As mutation rate in cancer cells is generally high, different clones may coexist in any tumour.
The second theory of tumour heterogeneity is based on the view that cancer cells can reversibly change their phenotype, i.e. exhibit phenotypic plasticity5. This idea developed in parallel with the cancer stem cell (CSC) model, according to which tumours have a hierarchical organisation. CSCs represent a highly tumorigenic subpopulation that constantly re-establishes itself and produces less oncogenic progeny. The existence of CSCs has been demonstrated in different human tumour types and in mouse models. Though often discussed in separate contexts, clonal evolution and phenotypic plasticity are not by nature contradictory, but likely to be complementary concepts. In such a scenario, clones of cells with higher levels of plasticity would be predicted to undergo positive selection during tumour progression.
CSCs have been proposed as the source of metastases, and therefore, the necessity to refocus therapies on elimination of CSCs is broadly discussed. In this regard, understanding the molecular control of plasticity in tumour cells poses an important challenge. Two prominent studies have demonstrated that CSCs can be generated through epithelial-mesenchymal transitions (EMT), embryonic genetic programs aberrantly reactivated in cancer6, 7. This process is reversible, via mesenchymal-epithelial transitions (MET), which are considered critical for the establishment of metastases.
The AP-1 family member, FOS-related antigen 1 (FRA-1) is emerging as a key regulator of EMT/MET equilibrium in cancer cells. FRA-1 is an important downstream effector of signalling pathways activated by common human oncogenes, and tumors harbouring these “driver” mutations are likely to positively select clonal variants capable of expressing FRA-1. This would generate EMT-committed cells able to undergo EMT or MET depending on microenviromental cues.
EMT/MET, EMBRYONIC TRANSCRIPTION FACTORS (EMT-TFs) AND CANCER METASTASIS
EMT and MET are essential during embryonic development and for normal physiological responses to tissue injury in adult organisms. Cells undergoing EMT lose epithelial characteristics such as apicobasal polarity, experience massive cytoskeletal rearrangements and dissipate epithelial junctional complexes. At the same time, they acquire rear-to-front polarity, the ability to migrate individually and to invade surrounding tissues8. At different phases of embryonic development, such as neural crest delamination or organ formation, cells in a mesenchymal state migrate long distances and upon differentiation, give rise to new tissues. High level of phenotypic plasticity is a hallmark of embryonic cells, which undergo multiple consequent rounds of EMT and MET for final differentiation and organogenesis9. Several proteins belonging to the Zn finger (SNAIL1, SNAIL2/SLUG, ZEB1, ZEB2/SIP1), bHLH (TWIST1, TWIST2), forkhead (FOXC2) or homeobox (Goosecoid, SIX1, PRRX1) transcription factor families execute EMT programs in a developing embryo10-15. These EMT-TFs are also expressed in different solid tumours, and in some cases, as shown for ZEB1 in colorectal and pancreatic carcinoma, are highly enriched at the invasive front16, 17. EMT-TFs directly repress transcription of a number of epithelial genes, most notably CDH1, which encodes the major adherens junction component, E-cadherin.
The pattern of ZEB1 expression in cancer is compatible with the hypothesis that cancer cells recapitulate elements of physiological EMT programs to delaminate from the primary tumour mass. These cells may then form populations of circulating tumour cells (CTC). In breast cancer patients and in mouse models of pancreatic ductal adenocarcinoma (PDAC), these intermediates in the metastatic process have been shown to maintain mesenchymal characteristics18, 19. As most metastases are epithelial, a central role for MET in cancer spread has been proposed20-22 based on analogy with embryonic development – migrating embryonic or cancer cells undergo MET at destination sites in order to build new tissue or seed secondary tumours. This hypothesis was experimentally confirmed recently. Korpal et al. demonstrated that balancing EMT/MET equilibriums was important for growth and metastatic spread of breast cancer. While MET reduced the entry of cells from primary tumours into the circulation, it promoted formation of lung metastases. In contrast, EMT facilitated initial steps in metastasis associated with intravasation into thin blood vessels, but inhibited pulmonary colonisation23. In a mouse model of squamous cell carcinoma, induction of EMT by TWIST1 led to the formation of CTCs. However, TWIST1-expressing cells had low proliferative index, and switching off TWIST1 increased cell proliferation and enabled establishment of lung metastases24. Likewise, the EMT-TF PRRX1 induced EMT and invasiveness in breast carcinoma cells, but its loss was required for complete MET and lung colonisation in recipient mice14.
There is growing recognition that epithelial-mesenchymal plasticity contributes to tumour heterogeneity and is important for the accomplishment of early and late steps in metastatic process. Clearly, these findings have important clinical implications - co-targeting both epithelial and mesenchymal states in cancer might result in synergistic effects. Hence, recognition that epithelial and mesenchymal tumour cell populations are differentially susceptible to therapeutic agents may herald a new rationale for combinatorial cancer therapy. CSCs proliferate slowly and are capable of evading cell death25. As EMT induces stem-like traits, it is anticipated that the mesenchymal state is characterised by low proliferation rates and drug resistance. Indeed, some EMT-TFs can attenuate cell cycle progression and induce chemo- and radio-resistance in cell culture and in vivo24, 26-28. Differentiated epithelial cells in carcinomas proliferate actively and appear more vulnerable to conventional anti-proliferative treatments. The search for selective inhibitors of CSC/EMT cells has identified several compounds, including the anti-diabetic agent metformin29 and the potassium ionophore salinomycin30, which were found to synergise with conventional anti-proliferative agents in xenograft models29, 31. Treatment efficacy may also be improved through approaches that sensitise cancer cells to particular therapies by shifting the EMT/MET equilibrium. Efforts to characterise molecular pathways impacting on the EMT/MET balance thus have strong potential for the development of personalised medicine strategies.
EMT-TFs, microRNAs AND TGFβ SIGNALLING: FEEDBACK LOOPS DETERMINING EMT/MET BALANCE
At least two regulatory loops comprising EMT-TFs and several species of microRNA govern the equilibrium between epithelial and mesenchymal states. ZEB1 and ZEB2 are targeted by five members of the miR-200 family, which form two clusters – miR-200b/miR-200a/miR-429 and miR-200c/miR-141. These are grouped on human chromosomes 1 and 12, respectively, with each being expressed as a polycistronic transcript. Repression of ZEB1 and ZEB2 by ectopic expression of miR-200 family members is sufficient to restore E-cadherin expression and induce full MET in several cell line models32-34. Promoters driving transcription of both miR-200 clusters are in turn directly repressed by ZEB1 and ZEB2, thereby establishing a self-enforcing double negative feedback loop35,36. The circuit guiding expression of the EMT-TF SNAIL1 has a similar configuration. SNAIL1 is negatively regulated by miR-34a and miR-34b/c, both of which are in turn directly repressed by SNAIL137, 38. miR-34 also directly targets AXIN2, a Wnt pathway component that induces nuclear export of GSK3β thereby preventing phosphorylation of SNAIL1 and its subsequent degradation39. Thus, miR-34 controls SNAIL1 at two levels – mRNA abundance and protein stability. Of note, ZEB1 has also been reported to down-regulate miR-34a through δNTP6340, providing a potential mechanism for crosstalk between the miR-200/ZEB and miR-34/SNAIL1 loops.
The EMT/MET balance appears intricately linked with the occurrence of stem-like features in cells. TWIST1 directly activates transcription of the BMI1 gene encoding a polycomb group protein that maintains self-renewal in stem cells41. BMI1 and several other stemness-promoting genes, such as KLF4, SOX2, cMYC, TP63, CD44, CD133 and OLFM4, are also direct targets of miR-200 and miR-34 family members (and also ZEB1-controlled miR-203)38, 42, 43. Moreover, miR-200 directly targets Sec23a, an essential component of secreted COPII transport vesicles, and two components of the Sec23 secretome (Tinagl1 and Igfbp4) were shown to repress formation of macrometastases in lung colonisation assays23. Such findings present a plausible explanation for the high frequency of MET upon cancer dissemination.
EMT-TF/miR loops are also key orchestrators of cytoskeletal rearrangements during EMT. Ectopic miR-34a down-regulates the GTPase-activating protein ARHGAP1, which leads to increased levels of GTP-bound CDC42 and RAC1 and reduced actin dynamics40. Likewise, miR-200b can modify the cytoskeleton by repressing moesin, cofilin2 and WASF344. In addition, the TWIST1-inducible miR-10b indirectly promotes expression of RHOC45, a RHO family member that remodels cytoskeleton and is essential for cell locomotion and metastases46. The finding that regulators of stemness and cytoskeletal remodelling/cell motility are important target classes of EMT-TF/miR networks provides experimental support for a theory formulated by Thomas Brabletz in 2005, according to which, EMT produces migrating cancer stem cells that represent a source of metastases47. The rate at which these cells are generated depends both on intrinsic (gene mutations) and extrinsic (autocrine and microenvironmental) factors, collectively governing EMT/MET balance in cancer cells.
The TGFβ/SMAD pathway is a central contributor to tumour-associated EMT and metastasis48, 49. Paradoxically, despite its tumour suppressive action at early stages of cancer development, TGFβ can act as a potent driver of tumour progression. The latter is supported by studies in human cell lines and mouse models demonstrating that TGFβ signalling components are required for tumour cell invasion in vitro and metastasis in vivo50-56. Signalling through the pathway involves phosphorylation of the transcription factors SMAD2 and SMAD3 (receptor-regulated SMADs) in response to activation of type-I or type-II Ser/Thr kinase receptors by their ligands (TGFβ1-3). Phosphorylated SMAD2/3 bind SMAD4, facilitating translocation of the complex to the nucleus. SMADs regulate transcription of target genes in cooperation with other transcription factors and by recruiting co-activator or co-repressor complexes57. Amongst the extensive list of reported SMAD interactors are several key regulators of cell plasticity. SNAIL1 forms a complex with SMAD3/4, whose binding to adjacent E-boxes and SMAD-binding elements cooperatively down-regulates activation of target promoters such as CDH1 and CLDN358. SMADs also directly interact with ZEB1 and ZEB2 (though with different affinities) to mediate repression of BMP/TGFβ-regulated genes59, 60.
Autocrine or paracrine TGFβ signalling loops are important determinants of EMT/MET in carcinoma cells, acting to promote SMAD-dependent transcription of genes encoding EMT-TFs, including ZEB1, ZEB2, SNAIL1, SNAIL2 and TWIST161-65. While experimental manipulation of miR-200 levels can reverse the effects of TGFβ66, prolonged exposure of epithelial cells to TGFβ results in irreversible EMT sustained by autocrine TGFβ signalling. This can be explained by the finding that TGFβ ligands themselves are targets of the miR-200 family, and thus derepressed when paracrine signals shift the EMT/MET balance towards EMT35, 66. Adding to the complexity of TGFβ/ZEB/miR-200 networks, it was recently revealed that a long non-coding RNA (lncRNA) activated by TGFβ (lncRNA-ABT) induced EMT by competitively binding and sequestering miR-200 family members and enhancing ZEB expression67. It should however be emphasized that TGFβ control of tumour cell plasticity is not restricted to ZEB/miR-200 regulation. For example, in hepatocellular carcinoma cells, TGFβ down-regulates transcription of miR-34a68, though whether this effect is SNAIL1-dependent remains to be established.
THE EMERGING ROLE OF FRA-1 IN CONTROL OF TUMOUR CELL PLASTICITY
EMT-TF/miR feedback loops are an integral feature of cancer signalling networks69, with new players in these circuits constantly being identified. Recent research has highlighted a critical role for FRA-1 in configuring EMT-TF/miR loops by linking them with intrinsically or extrinsically activated pathways. FRA-1 belongs to the Activator Protein-1 (AP-1) family of transcription factors, which provide a dynamic platform for integrating multi-pathway signalling events. AP-1 complexes comprise homo/hetero-dimers of mainly FOS, JUN, ATF or MAF family members, each of which contains an evolutionarily conserved bZIP domain, and is expressed and/or activated by specific physiological or pathological signals70-73. They can also physically and functionally interact with other factors on chromatin, underscoring their versatility in signal integration during cell fate control.
Early studies demonstrated that c-FOS and c-JUN could malignantly transform fibroblasts, while their dominant negative variants inhibited transformation induced by a various oncogenes. These functional characteristics were linked to the potent activities of these proteins as transcription factors. By contrast, the FRA proteins (FRA-1, FRA-2) lacked this ability and were unable to transform fibroblasts, in some cases behaving in an inhibitory manner70-74. Though it has been known for decades that enforced expression of c-FOS in murine epithelial cells induces fibroblastoid phenotype75, endogenous levels of the protein in mesenchymal cell lines of epithelial origin are low. By contrast, FRA-1 is upregulated in the same context76-78, generating much interest in understanding how it accumulates in tumour cells.
CONTROL OF FRA-1 EXPRESSION IN CANCER CELLS: TRANSCRIPTION, TRANSLATION AND PROTEIN STABILITY
FRA-1 accumulation in cells is regulated on two major levels – transcription of the FRA-1 gene (FOSL1) and post-translational modifications affecting protein half-life. The primary modification that FRA-1 undergoes is phosphorylation, which has been reported to affect both its transcriptional activity and stability. However, only the latter is well characterized. A C-terminal destabilizer domain (DEST) comprising 30-40 residues is critical for conferring instability to FRA-1, but its influence is largely neutralized upon phosphorylation on Ser-252 (by RSK) and Ser-265 (by ERK2) upon MEK pathway activation79, thereby prolonging half-life of the protein.
FRA-1 is also one of an expanding class of proteins that can be targeted to the proteasome for destruction without requiring ubiquitylation. Its susceptibility to proteolysis was unperturbed when all its lysine residues were replaced with arginines and its N-terminus fused to a myc epitope-tag79, 80. Targeting of FRA-1 for destruction may instead involve direct interaction with components of the proteasome such as TBP1, whose deficiency increased FRA-1 levels in a variety of cell types81. In contrast to FRA-1, c-FOS does not appear to associate with TBP1, indicating that proteasomal routing of these FOS family members involves distinct mechanisms.
Recently, two additional strategies that cells can employ to control FRA-1 protein levels were identified. A pathway involving sequential activation of the kinases PKCθ and SPAK1 was found to promote FRA-1 stability in subtypes of breast cancer cells where the contribution of ERK1/2 was weak82. Here, PKCθ-dependent FRA-1 stabilisation involved phosphorylation on Ser-265, Thr-223 and Thr-230. By contrast, in tumour cells bearing RAS/RAF mutations, FRA-1 stabilisation appears entirely dependent on ERK-induced phosphorylation83. Evidence has also emerged that mTORC1/S6K1 signalling can regulate FRA-1 expression. In cases of pulmonary lymphangioleomyomatosis, mTORC1 activation enhanced FRA-1 translation efficiency in a subset of smooth muscle-like cells through S6K1-dependent phosphorylation of the eukaryotic translation initiation factor 4B, eIF4B, at Ser-42284.
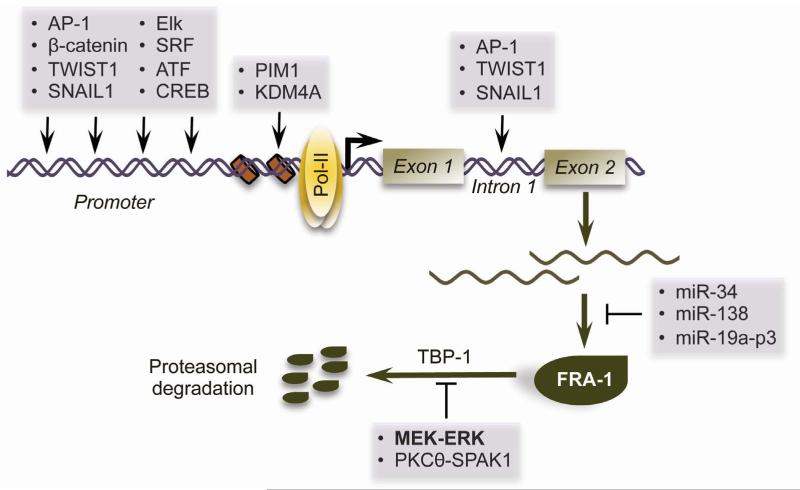
Transcription of FOSL1 is induced immediately after treatment with various growth factors and cytokines85, 86. The FOSL1 promoter can be occupied by a multitude of transcription factors, including AP-1, Elk1, SRF and ATF/CREB87, thus serving as a hub at which multiple pathways converge to modulate FRA-1 expression. Signal-induced activation of FOSL1 also requires extensive chromatin remodelling, including PIM1-mediated phosphorylation of Ser-10 on histone H3, which facilitates sequential recruitment of 14-3-3, BRD4 and p-TEFb to release paused Pol-II at the promoter88. FOSL1 transcription is also strongly regulated by an enhancer in its first intron, which contains AP-1-binding elements. This region is required for positive auto-regulation of FOSL1 by the RAS-ERK pathway, and for FOSL1 induction upon c-JUN activation89-91. As c-Jun appears to be the major dimerization partner for FRA-1 in the context of EMT92, 93, it is likely that the activation of both proteins must be coordinated during EMT programming. This notion is supported by recent work demonstrating that loss of the KDM4 demethylase induces both FOSL1 and JUN in SCC cells, resulting in an EMT-like phenotypic switch94. Collectively, the studies highlighted in this section suggest a model wherein FRA-1 accumulates in tumour cells through MEK/ERK, PKCθ/SPAK1 or mTOR/S6K1-dependent protein stabilisation, which perpetuates increased FOSL1 transcription via a positive feedback mechanism (Figure 1).
Figure 1. Mechanisms regulating FRA-1 accumulation in tumour cells.
Transcription of the FRA-1 gene, FOSL1, is regulated by various transcription factors that bind to its promoter and/or intron 1, as well as by chromatin remodelling factors. Once transcribed, FOSL1 transcripts can be targeted for degradation by several species of miRNAs. The FRA-1 protein itself is highly labile and is targeted to the proteasome for destruction through an ubiquitin-independent route. This process is antagonized by phosphorylation of FRA-1 by MEK-ERK-RSK or PKCθ-SPAK1 induced.
FRA-1 DIRECTLY REGULATES TRANSCRIPTION OF EMT-TFs
Early studies into the functions of FRA-1 in tumour cells established a link between its expression levels, cytoskeletal dynamics and cell locomotion. In colon carcinoma cells, FRA-1 stimulates cell motility by uncoupling RAS-activated RHO GTPase from stress fibre formation95, 96, resulting in more dynamic cell-matrix adhesion, reduced integrin β1 function and increased cell migration in vitro. FRA-1 was also identified as an important downstream effector of RSK-induced cell motility programs through its ability to control a number of genes encoding extracellular matrix-degrading enzymes, integrin subunits and cell-cell adhesion proteins97. To our knowledge, there are currently no published cell models in which ectopic FRA-1 expression alone affects epithelial differentiation and EMT-TF expression. However, FRA-1 activity is an essential requirement for establishment of EMT in carcinoma cells. This conclusion stems from work addressing the mechanism of oncogenic RAS-induced EMT in mammary epithelial cells, which revealed that RAS/ERK2 driven FRA-1 upregulation was necessary for the increased expression of ZEB1 and ZEB2, which led to a full EMT98. These data are in line with the results of genome-wide ChIP-seq analyses of FRA-1 targets in cancer cells, which identified several EMT-TF genes including SNAI2 and ZEB1 in mesenchymal colon cancer cells and ZEB2 in triple-negative breast carcinoma cells as direct FRA-1-induced targets93, 99.
FRA-1 has also been shown to directly bind AP-1 binding elements in the vicinity of the ZEB1, ZEB2, TWIST1 and SNAI2 genes in malignant melanoma cells. However, the effects of FRA-1 on EMT-TF expression differ in the carcinoma and melanoma contexts – whereas ZEB1 and TWIST1 were upregulated by FRA-1 in melanoma, ZEB2 and SNAI2 were repressed. The outcome of this differential regulation is a switch in the expression of EMT-TFs that demarcates the balance between melanocytic differentiation (high SNAIL2 and ZEB2) and tumorigenicity (high ZEB1 and TWIST1)100.
Further evidence supporting the centrality of FRA-1 in EMT regulation recently emerged for the Weinberg laboratory, which found that FOSL1 was directly activated by TWIST1 and SNAIL1 in immortalised human mammary epithelial cells, thus serving as effector of EMT pathways. Moreover, activation of ZEB1 and ZEB2 by TWIST1 and SNAIL2 required FRA-1 expression, but whether FRA-1 was present on the chromatin of ZEB genes was not examined76. Nevertheless, in this model, FRA-1 was integral for operation of EMT-TF networks and was a key factor driving EMT and stem cell functions. Together, these data suggest that FRA-1 directly regulates EMT-TF expression and is essential for establishing and maintaining EMT programs in different cancer types. Additionally, several indirect links between FRA-1 and pathways controlling EMT-TF/miR feedback loops further implicate FRA-1 in regulation of tumour cell plasticity.
FRA-1 IN THE CONTEXT OF EMT-TF/miRNA/TGFβ AND TUMOUR CELL PLASTICITY NETWORKS
RAS signalling provides one avenue through which tumor cells can overcome TGFβ-mediated growth inhibition, and the two pathways strongly cooperate in promoting EMT and metastasis in several systems, particularly in breast cancer models101-103. The use of effector-specific RAS mutants showed that RAF rather than PI3K activation was required for induction of an EMT program, which was then reinforced by induction of autocrine TGFβ signalling104-106. As autocrine TGFβ signalling is indicative of activated EMT-TF/miR/TGFβ loops, and transcriptional activation of EMT-TF genes in different cell models requires FRA-1, RAS-ERK induction of FRA-1 is likely to be a key determinant of TGFβ-driven EMT in cancers. This suggestion is supported by results of global analyses of FRA-1-regulated genes in colon and breast cancer cells, which identified genes implicated in autocrine TGFβ signalling as FRA-1 targets99, 107. In addition to the classical pathway, TGF-β may induce so-called SMAD-independent signalling leading to the activation of MAPK and mTOR108, 109, which may then contribute to the accumulation of FRA-1. In breast cancer cells, TGFβ treatment was recently found to promote formation of SMAD2/3-FRA-1 complexes, which appear essential for the induction of SNAIL2 and other mesenchymal genes102. Thus, FRA-1 can act at multiple levels to connect EMT-TF/miR/TGFβ loops with different pathways activated in cancer.
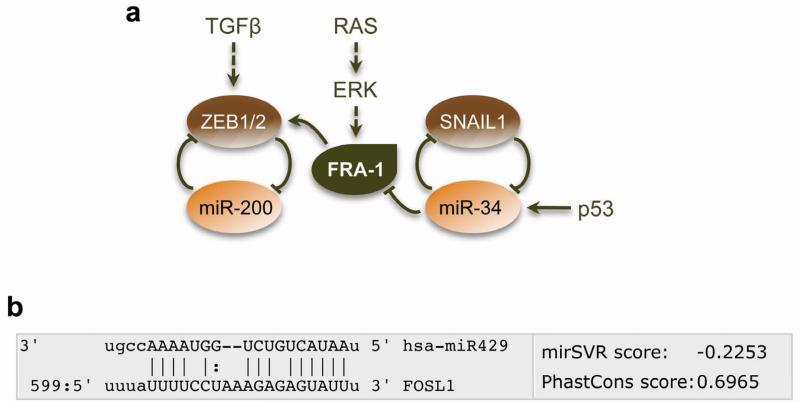
miR-34a and miR-200 are transcriptional targets of p53 family members37, 110, 111. Consequently, their expression is reduced when p53 is mutated or lost in cancers, thus shifting the EMT/MET equilibrium towards a mesenchymal/stem cell state. Interestingly, miR-34a directly targets FOSL1112, 113, whose 3′-UTR also contains a miR-200 recognition element that has not been characterised (Figure 2). Loss of p53 control over miR-34a (and possibly miR-200) would thus be expected to increase FOSL1 expression and amplify the effect of mutant p53 on EMT-TFs through FRA-1. This regulatory circuit may be further reinforced via direct transcriptional repression of miR-200 by ZEB proteins and indirect δNTP63-dependent down-regulation of miR-34a by ZEB140. These important findings give rise to a model in which p53 controls tumour cell plasticity through FRA-1/miR/EMT-TF regulatory modules (Figure 2). However, as differentiated tumours and epithelial carcinoma cell lines often contain mutant p53 or are p53 deficient, it is highly likely that p53 acts in combination with other pathways to determine differentiation status of cancer cells.
Figure 2. Interactions between FRA-1 and EMT-TF/miR loops.
(a) FRA-1 can directly regulate transcription of ZEB proteins, which control cell plasticity by operating in a double-negative feedback loop with miR-200 family members. Expression of FRA-1 is repressed by p53-inducible miR-34, which acts in a double-negative feedback loop with SNAIL1. (b) Identification of a potential miR-200 recognition element in the 3′-UTR of FOSL1.
Canonical WNT signalling is critical during embryogenesis and cooperates with loss of p53 to induce EMT in cancer cells. Mutations that activate the WNT pathway occur in a range of cancers, most notably in colorectal cancers, where they suppress APC function (90% of cases) or stabilize β-catenin114. Ultimately these lesions inhibit the APC/AXIN/GSK-3β destruction complex, leading to the nuclear import of β-catenin. β-catenin in complex with TCF/LEF family transcription factors controls transcription of many genes, including key modulators of cell plasticity – FOSL1, c-JUN115 and ZEB1116.
Though β-catenin is expected to accumulate in the nucleus of tumour cells where the WNT pathway is constitutively active, this staining pattern is often detected only at the tumour/stroma interphase, but not within the central region of tumours (so called β-catenin paradox)117. Of note, expression of ZEB1 and FRA-1, as well as ZEB2 and SNAIL1, has also been detected at the invasive front in CRC specimens, but not in the tumour centre99, 112, 116, 118. This pattern indicates that APC mutations are not sufficient for full-scale activation of the pathway, and other factors, such as stromal cues or additional mutations facilitate β-catenin signalling and localised invasion.
TP53 gene mutations represent one class of candidate intrinsic lesions stimulating WNT pathway and consequent EMT. Several critical components of the WNT pathway, including β-catenin, LEF1, WNT1, WNT3, LPR6 and AXIN2 are directly targeted by p53-regulated miR-34, and loss of p53 activity or experimental manipulation of miR-34 expression affects WNT signalling119, 120. Increased AXIN2 expression invoked by loss of miR-34 may have a dual effect on the EMT/MET equilibrium. As part of the β-catenin destruction complex, AXIN2 may repress WNT signalling and consequently reduce transcription of FOSL1 and ZEB1 to maintain a differentiated state of tumour cells. On the other hand, AXIN2 stabilises SNAIL1 by controlling nucleocytoplasmic shuttling of GSK-3ß, thereby promoting EMT39. Although crosstalk between the WNT, p53 and EMT/MET pathways has been studied predominantly in CRC, it may also exist in other cancer types. Indeed, a correlation between β-catenin/TCF transcriptional activity, p53 and miR-34 functional status has also been documented in breast cancer and paediatric neuroblastoma patients119.
FRA-1 LINKS TWO CONCEPTS UNDERLYING TUMOUR HETEROGENEITY – CLONAL EVOLUTION AND CELL PLASTICITY
It is becoming increasingly evident that EMT can occur at early stages of tumorigenesis, and EMT-TFs play an early role in cancer. This notion is supported by histological analyses of early stage tumours and premalignant lesions in human samples and mouse models121. Mechanistically, TWIST1 and ZEB1 have been shown to cooperate with classical oncoproteins by promoting escape from oncogene-induced failsafe programs of premature senescence and apoptosis. The dual function of EMT-TFs in oncogenic transformation and cell invasion suggests their crucial role in early metastatic dissemination. The existence of early metastatic spread (model of parallel progression of primary tumours and metastases) and implication of EMT-TFs in this process has been experimentally demonstrated in patient samples and in mouse models of breast and pancreatic carcinoma19, 122-124. A role for FRA-1 in transformation and metastases was not examined in these studies. However, in immortalised melanocytes, FRA-1 was rapidly induced by the BRAFV600E mutation, leading to an EMT-TF switch involving up-regulation of ZEB1 and TWIST1 and down-regulation of SNAIL2 and ZEB2. This reprogramming was required for oncogenic BRAF to induce both transformation and invasion of melanoma cells. Upon melanoma development, different negative feedback loops are activated to repress MEK-ERK pathway and revert EMT-TF expression patterns. In late-stage melanoma, accumulating mutations override negative feedback regulation, restore the activity of MEK-ERK module and bring the expression of EMT-TFs to the “transformed/invasive pattern” (high FRA-1, high ZEB1 and TWIST1). This late switch is associated with metastatic disease and predicts cancer-related death100.
Akin to melanocytes, FOSL1 is also rapidly induced by dominant active components of the RAS pathway in epithelial and fibroblastoid cell backgrounds83, 125. In immortalised fibroblasts, FRA-1 has been characterised as a factor essential for transformation by RAS126. Whether EMT-TFs have any role downstream of FRA-1 has not been addressed in fibroblast models. Although an early role for FRA-1 in epithelial tumorigenesis has not been addressed, one can speculate that, by analogy with melanomagenesis, carcinoma-initiating mutations may lead to the activation of FRA-1/EMT-TF pathways contributing to the transformation and early metastases.
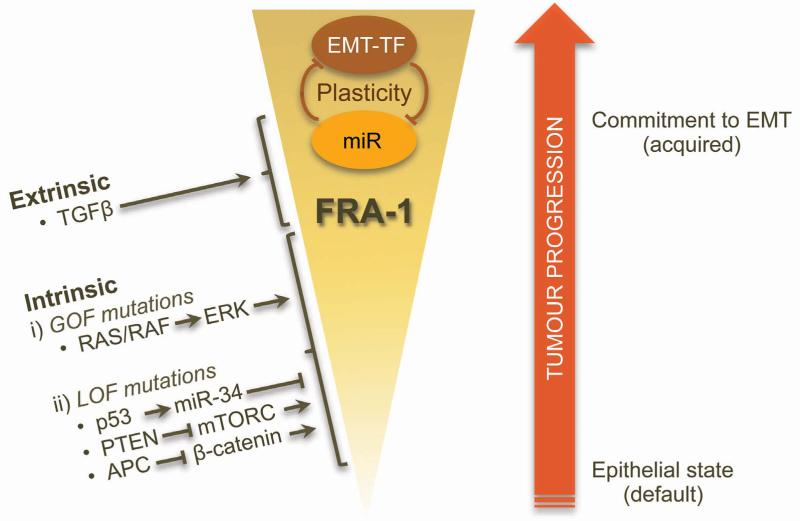
In most carcinoma samples the expression of FRA-1 is not uniform, and higher FRA-1 immunopositivity is observed at the invasive front in the areas of tumour/stroma interphase99, 127, 128. As the epithelial phenotype has been proposed to be the default state of carcinoma cells21, 129, overriding this condition during carcinoma progression by intrinsic and/or extrinsic factors leads to enhanced cell plasticity. As discussed in earlier sections, FRA-1 is emerging as a central hub at which these factors, which include tumour suppressors (p53, APC, PTEN) and oncogenes of the MEK-ERK module, intersect with cell plasticity regulators. FRA-1 may trigger and maintain positive signalling loops that affect EMT-TF/miR regulation, the net outcome being production of EMT-committed cells. However, though a necessary prerequisite, the presence of FRA-1 alone is insufficient for EMT induction. This is likely to require additional extrinsic inputs, such as availability of TGFβ. Dependence of EMT-committed cells on the availability of extrinsic factors is thus a major determinant of tumour cell plasticity.
According to the clonal evolution model, successive cycles of mutations, clonal selection and expansion drive tumour progression1 (Figure 3). In this context, pathways driving up-regulation of FRA-1 will have pleiotropic effects on tumour cells, and the corresponding mutations will be subject to positive selection. Consequently, tumour evolution will lead to the expansion of EMT-committed cell populations. Therefore, both clonal evolution and phenotypic plasticity interact to generate tumour heterogeneity through the selection of FRA-1-expressing variants, in which epithelial or mesenchymal phenotypes are adopted in response to micro-environmental cues.
Figure 3. Contribution of FRA-1 to tumour cell plasticity control during cancer progression.
In this model, positive selection of tumour cell clones overexpressing FRA-1 during cancer progression is predicted to generate EMT-committed cell populations. The enhanced plasticity of these populations may then contribute to tumour heterogeneity, by enabling epithelial or mesenchymal phenotypes to be adopted in response to micro-environmental cues.
ACKNOWLEDGMENTS
We apologize to the colleagues whose relevant work we were unable to cite due to space limitations. This work was supported by project grants from the National Health and Medical Research Council of Australia (to ASD), CRUK and AICR (to ET).
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463:545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celia-Terrassa T, Meca-Cortes O, Mateo F, de Paz AM, Rubio N, Arnal-Estape A, et al. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. The Journal of clinical investigation. 2012;122:1849–1868. doi: 10.1172/JCI59218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014 doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PloS one. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nature reviews Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 9.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene. 2014;33:1755–1763. doi: 10.1038/onc.2013.128. [DOI] [PubMed] [Google Scholar]
- 11.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartwell KA, Muir B, Reinhardt F, Carpenter AE, Sgroi DC, Weinberg RA. The Spemann organizer gene, Goosecoid, promotes tumor metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18969–18974. doi: 10.1073/pnas.0608636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCoy EL, Iwanaga R, Jedlicka P, Abbey NS, Chodosh LA, Heichman KA, et al. Six1 expands the mouse mammary epithelial stem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119:2663–2677. doi: 10.1172/JCI37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 15.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nature reviews Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 16.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30:770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brabletz T. To differentiate or not--routes towards metastasis. Nature reviews Cancer. 2012;12:425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 22.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 23.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nature medicine. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Signore M, Ricci-Vitiani L, De Maria R. Targeting apoptosis pathways in cancer stem cells. Cancer letters. 2013;332:374–382. doi: 10.1016/j.canlet.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes and development. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mejlvang J, Kriajevska M, Vandewalle C, Chernova T, Sayan AE, Berx G, et al. Direct repression of cyclin D1 by SIP1 attenuates cell cycle progression in cells undergoing an epithelial mesenchymal transition. Molecular biology of the cell. 2007;18:4615–4624. doi: 10.1091/mbc.E07-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer research. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang GN, Liang Y, Zhou LJ, Chen SP, Chen G, Zhang TP, et al. Combination of salinomycin and gemcitabine eliminates pancreatic cancer cells. Cancer letters. 2011;313:137–144. doi: 10.1016/j.canlet.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 32.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 33.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. The Journal of biological chemistry. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes and development. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO reports. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer research. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 37.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. The Journal of cell biology. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 39.Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nature cell biology. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 40.Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, et al. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. The Journal of clinical investigation. 2012;122:3170–3183. doi: 10.1172/JCI63608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nature cell biology. 2010;12:982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 42.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature cell biology. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 43.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Roslan S, Johnstone CN, Wright JA, Bracken CP, Anderson M, et al. MiR-200 can repress breast cancer metastasis through ZEB1-independent but moesin-dependent pathways. Oncogene. 2014;33:4077–4088. doi: 10.1038/onc.2013.370. [DOI] [PubMed] [Google Scholar]
- 45.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 46.Hakem A, Sanchez-Sweatman O, You-Ten A, Duncan G, Wakeham A, Khokha R, et al. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes and development. 2005;19:1974–1979. doi: 10.1101/gad.1310805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nature reviews Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 48.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell research. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drabsch Y, ten Dijke P. TGF-beta signalling and its role in cancer progression and metastasis. Cancer Metastasis Rev. 2012;31:553–568. doi: 10.1007/s10555-012-9375-7. [DOI] [PubMed] [Google Scholar]
- 50.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nature genetics. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 51.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 52.Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, et al. TGFβ1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 53.Oft M, Heider KH, Beug H. TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 54.Kang Y, He W, Tulley S, Gupta GP, Serganova I, Chen CR, et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci U S A. 2005;102:13909–13914. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welch DR, Fabra A, Nakajima M. Transforming growth factor beta stimulates mammary adenocarcinoma cell invasion and metastatic potential. Proc Natl Acad Sci U S A. 1990;87:7678–7682. doi: 10.1073/pnas.87.19.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muraoka-Cook RS, Kurokawa H, Koh Y, Forbes JT, Roebuck LR, Barcellos-Hoff MH, et al. Conditional overexpression of active transforming growth factor beta1 in vivo accelerates metastases of transgenic mammary tumors. Cancer Res. 2004;64:9002–9011. doi: 10.1158/0008-5472.CAN-04-2111. [DOI] [PubMed] [Google Scholar]
- 57.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nature cell biology. 2009;11:943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Grunsven LA, Taelman V, Michiels C, Opdecamp K, Huylebroeck D, Bellefroid EJ. deltaEF1 and SIP1 are differentially expressed and have overlapping activities during Xenopus embryogenesis. Developmental dynamics. 2006;235:1491–1500. doi: 10.1002/dvdy.20727. [DOI] [PubMed] [Google Scholar]
- 60.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. The Journal of cell biology. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishimura G, Manabe I, Tsushima K, Fujiu K, Oishi Y, Imai Y, et al. DeltaEF1 mediates TGF-beta signaling in vascular smooth muscle cell differentiation. Developmental cell. 2006;11:93–104. doi: 10.1016/j.devcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 63.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature reviews Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 64.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer science. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu J, Guo H, Treekitkarnmongkol W, Li P, Zhang J, Shi B, et al. 14-3-3zeta Cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer cell. 2009;16:195–207. doi: 10.1016/j.ccr.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Molecular biology of the cell. 2011;22:1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 68.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. International journal of cancer Journal international du cancer. 2013;132:745–754. doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 70.Lloyd A, Yancheva N, Wasylyk B. Transformation suppressor activity of a Jun transcription factor lacking its activation domain. Nature. 1991;352:635–638. doi: 10.1038/352635a0. [DOI] [PubMed] [Google Scholar]
- 71.Tulchinsky E. Fos family members: regulation, structure and role in oncogenic transformation. Histology and histopathology. 2000;15:921–928. doi: 10.14670/HH-15.921. [DOI] [PubMed] [Google Scholar]
- 72.Ransone LJ, Verma IM. Nuclear proto-oncogenes fos and jun. Annual review of cell biology. 1990;6:539–557. doi: 10.1146/annurev.cb.06.110190.002543. [DOI] [PubMed] [Google Scholar]
- 73.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nature reviews Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 74.Matthews CP, Colburn NH, Young MR. AP-1 a target for cancer prevention. Current cancer drug targets. 2007;7:317–324. doi: 10.2174/156800907780809723. [DOI] [PubMed] [Google Scholar]
- 75.Reichmann E, Schwarz H, Deiner EM, Leitner I, Eilers M, Berger J, et al. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial-fibroblastoid cell conversion. Cell. 1992;71:1103–1116. doi: 10.1016/s0092-8674(05)80060-1. [DOI] [PubMed] [Google Scholar]
- 76.Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, et al. Protein kinase C alpha is a central signaling node and therapeutic target for breast cancer stem cells. Cancer cell. 2013;24:347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kustikova O, Kramerov D, Grigorian M, Berezin V, Bock E, Lukanidin E, et al. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Molecular and cellular biology. 1998;18:7095–7105. doi: 10.1128/mcb.18.12.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Belguise K, Kersual N, Galtier F, Chalbos D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene. 2005;24:1434–1444. doi: 10.1038/sj.onc.1208312. [DOI] [PubMed] [Google Scholar]
- 79.Basbous J, Chalbos D, Hipskind R, Jariel-Encontre I, Piechaczyk M. Ubiquitin-independent proteasomal degradation of Fra-1 is antagonized by Erk1/2 pathway-mediated phosphorylation of a unique C-terminal destabilizer. Molecular and cellular biology. 2007;27:3936–3950. doi: 10.1128/MCB.01776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Basbous J, Jariel-Encontre I, Gomard T, Bossis G, Piechaczyk M. Ubiquitin-independent-versus ubiquitin-dependent proteasomal degradation of the c-Fos and Fra-1 transcription factors: is there a unique answer? Biochimie. 2008;90:296–305. doi: 10.1016/j.biochi.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 81.Pakay JL, Diesch J, Gilan O, Yip YY, Sayan E, Kolch W, et al. A 19S proteasomal subunit cooperates with an ERK MAPK-regulated degron to regulate accumulation of Fra-1 in tumour cells. Oncogene. 2012;31:1817–1824. doi: 10.1038/onc.2011.375. [DOI] [PubMed] [Google Scholar]
- 82.Belguise K, Milord S, Galtier F, Moquet-Torcy G, Piechaczyk M, Chalbos D. The PKCtheta pathway participates in the aberrant accumulation of Fra-1 protein in invasive ER-negative breast cancer cells. Oncogene. 2012;31:4889–4897. doi: 10.1038/onc.2011.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vial E, Marshall CJ. Elevated ERK-MAP kinase activity protects the FOS family member FRA-1 against proteasomal degradation in colon carcinoma cells. J Cell Sci. 2003;116:4957–4963. doi: 10.1242/jcs.00812. [DOI] [PubMed] [Google Scholar]
- 84.Gu X, Yu JJ, Ilter D, Blenis N, Henske EP, Blenis J. Integration of mTOR and estrogen-ERK2 signaling in lymphangioleiomyomatosis pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14960–14965. doi: 10.1073/pnas.1309110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murphy LO, MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Molecular and cellular biology. 2004;24:144–153. doi: 10.1128/MCB.24.1.144-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nature cell biology. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 87.Adiseshaiah P, Papaiahgari SR, Vuong H, Kalvakolanu DV, Reddy SP. Multiple cis-elements mediate the transcriptional activation of human fra-1 by 12-O-tetradecanoylphorbol-13-acetate in bronchial epithelial cells. The Journal of biological chemistry. 2003;278:47423–47433. doi: 10.1074/jbc.M303505200. [DOI] [PubMed] [Google Scholar]
- 88.Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 89.Bergers G, Graninger P, Braselmann S, Wrighton C, Busslinger M. Transcriptional activation of the fra-1 gene by AP-1 is mediated by regulatory sequences in the first intron. Molecular and cellular biology. 1995;15:3748–3758. doi: 10.1128/mcb.15.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Casalino L, De Cesare D, Verde P. Accumulation of Fra-1 in ras-transformed cells depends on both transcriptional autoregulation and MEK-dependent posttranslational stabilization. Molecular and cellular biology. 2003;23:4401–4415. doi: 10.1128/MCB.23.12.4401-4415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vikhanskaya F, Toh WH, Dulloo I, Wu Q, Boominathan L, Ng HH, et al. p73 supports cellular growth through c-Jun-dependent AP-1 transactivation. Nature cell biology. 2007;9:698–705. doi: 10.1038/ncb1598. [DOI] [PubMed] [Google Scholar]
- 92.Gilan O, Diesch J, Amalia M, Jastrzebski K, Chueh A, Verrills NM, et al. PR55alpha-containing protein phosphatase 2A complexes promote cancer cell migration and invasion through regulation of AP-1 transcriptional activity. Oncogene. 2014 doi: 10.1038/onc.2014.26. [DOI] [PubMed] [Google Scholar]
- 93.Zhao C, Qiao Y, Jonsson P, Wang J, Xu L, Rouhi P, et al. Genome-wide Profiling of AP-1-Regulated Transcription Provides Insights into the Invasiveness of Triple-Negative Breast Cancer. Cancer research. 2014;74:3983–3994. doi: 10.1158/0008-5472.CAN-13-3396. [DOI] [PubMed] [Google Scholar]
- 94.Ding X, Pan H, Li J, Zhong Q, Chen X, Dry SM, et al. Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis. Science signaling. 2013;6:ra28.1–13. doi: 10.1126/scisignal.2003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pollock CB, Shirasawa S, Sasazuki T, Kolch W, Dhillon AS. Oncogenic K-RAS is required to maintain changes in cytoskeletal organization, adhesion, and motility in colon cancer cells. Cancer research. 2005;65:1244–1250. doi: 10.1158/0008-5472.CAN-04-1911. [DOI] [PubMed] [Google Scholar]
- 96.Vial E, Sahai E, Marshall CJ. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer cell. 2003;4:67–79. doi: 10.1016/s1535-6108(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 97.Doehn U, Hauge C, Frank SR, Jensen CJ, Duda K, Nielsen JV, et al. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Molecular cell. 2009;35:511–522. doi: 10.1016/j.molcel.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Molecular cell. 2010;38:114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Diesch J, Sanij E, Gilan O, Love C, Tran H, Fleming NI, et al. Widespread FRA1-dependent control of mesenchymal transdifferentiation programs in colorectal cancer cells. PloS one. 2014;9:e88950. doi: 10.1371/journal.pone.0088950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer cell. 2013;24:466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 101.Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, et al. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. The Journal of cell biology. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sundqvist A, Zieba A, Vasilaki E, Herrera Hidalgo C, Soderberg O, Koinuma D, et al. Specific interactions between Smad proteins and AP-1 components determine TGFbeta-induced breast cancer cell invasion. Oncogene. 2012 doi: 10.1038/onc.2012.370. [DOI] [PubMed] [Google Scholar]
- 103.Lehmann K, Janda E, Pierreux CE, Rytomaa M, Schulze A, McMahon M, et al. Raf induces TGFbeta production while blocking its apoptotic but not invasive responses: a mechanism leading to increased malignancy in epithelial cells. Genes and development. 2000;14:2610–2622. doi: 10.1101/gad.181700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lehmann K, Janda E, Pierreux CE, Rytomaa M, Schulze A, McMahon M, et al. Raf induces TGF-beta production while blocking its apoptotic but not invasive responses: a mechanism leading to increased malignancy in epithelial cells. Genes and development. 2000;14:2610–2622. doi: 10.1101/gad.181700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, et al. Ras and TGF-beta cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. The Journal of cell biology. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Janda E, Nevolo M, Lehmann K, Downward J, Beug H, Grieco M. Raf plus TGF-beta-dependent EMT is initiated by endocytosis and lysosomal degradation of E-cadherin. Oncogene. 2006;25:7117–7130. doi: 10.1038/sj.onc.1209701. [DOI] [PubMed] [Google Scholar]
- 107.Desmet CJ, Gallenne T, Prieur A, Reyal F, Visser NL, Wittner BS, et al. Identification of a pharmacologically tractable Fra-1/ADORA2B axis promoting breast cancer metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5139–5144. doi: 10.1073/pnas.1222085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 109.Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R. TGF-beta-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J Cell Sci. 2012;125:1259–1273. doi: 10.1242/jcs.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nature cell biology. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. The Journal of experimental medicine. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue YF, et al. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis. 2012;33:519–528. doi: 10.1093/carcin/bgr304. [DOI] [PubMed] [Google Scholar]
- 113.Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2013;32:4294–4303. doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 114.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochimica et biophysica acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 115.Mann B, Gelos M, Siedow A, Hanski ML, Gratchev A, Ilyas M, et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sanchez-Tillo E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A. beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19204–19209. doi: 10.1073/pnas.1108977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Current opinion in cell biology. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 118.Kahlert C, Lahes S, Radhakrishnan P, Dutta S, Mogler C, Herpel E, et al. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clinical cancer research. 2011;17:7654–7663. doi: 10.1158/1078-0432.CCR-10-2816. [DOI] [PubMed] [Google Scholar]
- 119.Kim NH, Kim HS, Kim NG, Lee I, Choi HS, Li XY, et al. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Science signaling. 2011;4:ra71. doi: 10.1126/scisignal.2001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cha YH, Kim NH, Park C, Lee I, Kim HS, Yook JI. MiRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell cycle. 2012;11:1273–1281. doi: 10.4161/cc.19618. [DOI] [PubMed] [Google Scholar]
- 121.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nature cell biology. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 122.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 123.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 124.Klein CA. Parallel progression of primary tumours and metastases. Nature reviews Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 125.Treinies I, Paterson HF, Hooper S, Wilson R, Marshall CJ. Activated MEK stimulates expression of AP-1 components independently of phosphatidylinositol 3-kinase (PI3-kinase) but requires a PI3-kinase signal To stimulate DNA synthesis. Molecular and cellular biology. 1999;19:321–329. doi: 10.1128/mcb.19.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kakumoto K, Sasai K, Sukezane T, Oneyama C, Ishimaru S, Shibutani K, et al. FRA1 is a determinant for the difference in RAS-induced transformation between human and rat fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5490–5495. doi: 10.1073/pnas.0601222103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sayan AE, Stanford R, Vickery R, Grigorenko E, Diesch J, Kulbicki K, et al. Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL. Oncogene. 2012;31:1493–1503. doi: 10.1038/onc.2011.336. [DOI] [PubMed] [Google Scholar]
- 128.Usui A, Hoshino I, Akutsu Y, Sakata H, Nishimori T, Murakami K, et al. The molecular role of Fra-1 and its prognostic significance in human esophageal squamous cell carcinoma. Cancer. 2012;118:3387–3396. doi: 10.1002/cncr.26652. [DOI] [PubMed] [Google Scholar]
- 129.Frisch SM. The epithelial cell default-phenotype hypothesis and its implications for cancer. BioEssays. 1997;19:705–709. doi: 10.1002/bies.950190811. [DOI] [PubMed] [Google Scholar]