Abstract
Purpose
Penile cancer is a rare malignancy in the developed world, with just over 1600 new cases diagnosed in the USA per year, however, the incidence is much higher in developing countries. Although HPV is known to contribute to tumourigenesis, little is known about the genetic or epigenetic alterations defining penile cancer (PeCa).
Experimental Design
Using high-density genome-wide methylation arrays we have identified epigenetic alterations associated with PeCa. Q-MSP was used to validate lymph node metastasis markers in 50 cases. 446 HNSCC and CESCC (head and neck squamous cell carcinoma and cervical squamous cell carcinoma) samples were used to validate HPV associated epigenetic alterations.
Results
We defined 6933 methylation variable positions (MVPs) between normal and tumour tissue, which include 997 hypermethylated differentially methylated regions associated with tumour supressor genes including CDO1, AR1 and WT1. Analysis of PeCa tumours identified a 4 gene epi-signature which accurately predicted lymph node metastasis in an independent cohort (AUC of 89%). Finally, we explored the epigenetic alterations associated with PeCa HPV infection and defined a 30 loci lineage independent HPV specific epi-signature which predicts HPV status and survival in independent HNSCC, CESC cohorts. Epi-signature negative patients have a significantly worse overall survival (HNSCC p=0.00073, CI 0.021-0.78, CESC p= 0.0094, HR=3.91, 95% CI =0.13-0.78), HPV epi-signature is a better predictor of survival than HPV status alone.
Conclusion
These data demonstrate for the first time genome-wide epigenetic events involved in an aggressive penile cancer phenotype and define the epigenetic alterations common across multiple HPV driven malignancies.
Introduction
Penile Cancer (PeCa) is relatively rare in the developed world, but represents a global health problem, showing high prevalence and posing significant morbidity and mortality in developing countries (1, 2). The age standardised incidence of PeCa is 0.3-1.0 per 100,000 men in European countries and the United States, equating to approximately 1600 new cases per annum in the USA (2). In contrast, the incidence in developing nations varies from 3 to 8.3 per 100,000 (3, 4).
The presence of inguinal lymph node involvement is at present the most important prognostic indicator of unfavourable prognosis in penile cancer (5). Although, histopathological factors including tumour subtype, grade, stage and the presence of lymphovascular and perineural invasion are useful predictors of inguinal lymph node metastases, they are still not accurate and if used exclusively would lead to overtreatment of a significant proportion of patients. The aetiology of PeCa, is multifactorial with smoking, phimosis, poor personal hygiene and low socioeconomic status all being risk factors for tumour development (6). Additionally, there is strong evidence linking development of PeCa to infection with high risk HPV (HPV 16, 18), suggesting that HPV plays a significant role in the pathogenesis of at least a subset of cases. High risk HPV infection is transformative in other tumour types including cervical squamous cell carcinoma and head and neck squamous cell carcinoma (CESC and HNSCC respectively) (7, 8). Contrary to cervical cancers, which appear to be almost exclusively (>90%) driven by HPV, only a proportion of penile, vulvar, anal, and oropharyngeal cancers appear to be HPV driven (9, 10). Interestingly, despite the clear oncogenic effects of HPV infection, HPV positivity appears to confer a survival benefit, this is particularly true for HNSCC, and also appears to be for PeCa, although as yet only limited data is available (11).
Changes in DNA methylation play a key role in malignant transformation, leading to the silencing of tumor-suppressor genes and overexpression of oncogenes(12). The ontogenic plasticity of DNA methylation makes epigenetic changes ideal biomarkers for diagnosis or as predictive and prognostic markers in cancer. However, little is known about the molecular genetics or epigenetics driving the development and progression of PeCa. Aberrant methylation of a handful of candidate genes has previously been identified, including CDKN2A and RASSF1A (13-16). Recently, epigenetic changes in both host and virus epigenomes have been reported in other HPV induced cancers (17-20). To date no substantial genome wide analysis has been performed in penile cancer and linkage between viral subtypes has not been elucidated. We have therefore sought to define the epigenetic alterations associated with penile carcinogenesis including a subset of cases associated with high risk HPV infection. Using high density genome-wide methylation array on a panel of PeCa and matched normal tissue we have annotated epigenetic alterations which define PeCa d, we also interrogated these data to reveal epigenetic changes associated with disease progression and HPV infection.
Materials and Methods
Ethics Approval
Ethics approval for this study was granted by the University College London (UCL) / University College London Hospital (UCLH) BioBank for Health and Human Disease (NC06.11). Informed consent was obtained.
Patient Samples and Clinical Data
Thirty-eight fresh penile cancers and 11 matched normal tissue samples (stored in RNAlater) from the UCL/UCLH Urology Biobank, and 50 formalin-fixed paraffin-embedded (FFPE) tissue blocks from the Department of Pathology (UCLH) with confirmed histopathological and clinical diagnosis of PeCa and with > 80% tumour cellularity were included and analysed. Normal samples taken adjacent from tumour tissue and confirmed to be histologically normal in pathological review (Supplementary Table 1 and 2).
DNA Extraction
DNA was extracted from RNA-later preserved frozen tissue using the QIAmp DNA MiniKit (Qiagen), and FFPE tissue using the QIAmp DNA FFPE Tissue Kit (Qiagen) according the manufacturer’s instructions.
HPV Assessment
All samples were assessed for the presence of low risk HPV 6 and 11 and high risk HPV 16, 18 and 31 viral DNA by qPCR with primers specific for each genotype (Supplementary Table 2A). The reference genes GAPDH and ACTB was used to normalise DNA input and calculate the number of HPV genomic copies present. HPV qPCR was carried out as previously described be Lechner et al (22). HPV type data for CESC and HNSCC TCGA samples were take from Tang et al., and based the expression of viral genes in RNA-seq data (21).
Methylation Analysis
500 ng of DNA from 38 tumour and 11 matched normal RNAlater-preserved samples from PeCa patients were bisulphite converted and hybridised to the Infinium 450K Human Methylation array, and processed in accordance with the manufacturer’s recommendations. DNA bisulphite conversion was carried out using the EZ DNA Methylation kit (Zymo Research) as per manufacturer’s instructions. Samples were processed in a single batch. R statistical software (version 2.14.0 (22)) was used for the subsequent data analysis. The ChAMP pipeline was used to extract and analyse data from iDat files, samples were normalised using BMIQ (23). Raw β values (methylation value) were subjected to a stringent quality-control analysis as follows: samples showing reduced coverage were removed and only probes with detection levels above background across all samples were retained (detection P < 0.01). DMRs (differentially methylated regions) were called using the Probe Lasso algorithm (implemented in ChAMP package; see Morris et al) with default parameters with the exception of applying a minimum DMR size of 100bp. As a result, all DMRs identified have a minimum of 3 significant probes, are at least 1Kb from a neighbouring DMR, and have a minimum size of 100bp. Maximum DMR size is effectively unbounded but is dependent the genomic separation between contiguous CpG probes, which itself is contingent on the local underlying genomic and epigenomic features with larger DMRs more likely to occur in probe-poor regions(Butcher et al., in press,(23))
The statistical significance of MVP enrichment in genomic and epigenomic features was calculated based on the random selection of equal numbers of probes (4935 for hypermethylated MVPs, 1998 hypomethylated MVPs), from the overall probe set (472,655 probes) used in the analysis and repeated 10,000 times(24).
Gene set enrichment analysis (GSEA) was used to assess if gene associated DMRs are overrepresented in a particular gene set. Gene sets, categorised by gene ontology, molecular pathways, chromosomal locations, or targets of regulatory motifs and miRNAs, were derived from the Molecular Signatures Database (MSigDB). Enrichment was assessed by comparing the number of genes associated with DMRs belonging to the gene set with those that are not members. The significance of the over-representation was then assessed by a Fisher’s exact test and adjusted for false discovery by the Benjamini Hochberg procedure. Genes containing multiple DMRs were counted only once in order to remove any bias in gene set enrichment.
Motif analysis was performed using the MEME-ChIP tool of the MEME suite; parameters were set to default except for the number of repetitions (set to ‘Any number of repetitions’), motif width (min=4, max=15), and maximum number of motifs to find set to 20 (25).
Validation of methylation
Aberrant methylation was validated in the external cohort using Methylation Specific qPCR (MSP) (Supplementary Table 2).. Genomic DNA from FFPE samples was bisulphite converted as above. 10 ng of converted DNA was subjected to MSP. Briefly, all reactions were carried out in a 13 μL reaction volume containing 6.5 μL 2X SYBR Green reaction buffer, 0.3 μmol/L forward primer and 0.3 μmol/L reverse primer with 1 ng genomic DNA (RNAlater-preserved) or 10ng for FFPE samples. Reactions were run on an ABI 7300 RealTime PCR machine, denaturation for 10 minute at 95°C, with 40 cycles of 95 °C for 15 seconds and 60 °C for 60 seconds. All reactions were performed in triplicate. Sensitivity and specificity of all reactions was assessed using spiked dilutions of fully methylated DNA. The methylation state of individual samples was determined using a standard curve with a range of control methylation states (0% to 100%). The absolute methylation was subsequently used to determine the association with lymph node metastasis.
Integration of obtained methylation data with publicly available methylation data HNSCC data
R statistical software v2.15.1 [35] was used for pre-processing of data and for classic multidimensional scaling (MDS) using principal components analysis (PCA). HPV specific epigenetic signature and prediction of HPV infection was determined using the shrunken centroid method implemented through the pamr bioconductor package. Survival analysis was carried using the bioconductor package; Survival (26). MDS was used to visualize HPV+ve and HPV-ve PeCa methylation signatures within methylation datasets obtained from an HPV-induced head and neck squamous cell carcinomas ((20) GEO accession numbers: GSE38266, GSE38268, GSE38270 and GSE38271, and TCGA samples from HNSCC(27) and CESC(28). Raw iDAT files were processed and normalised in line with in house data as above.
RT-PCR
RNA was extracted from tissue, determined by H&E staining of frozen sections to be tumour or normal tissue from the same individuals, using an RNeasy kit according to the manufacturer’s instructions. RNA was quantified using a NanoDrop spectrophotomer and for each sample, 1 ug was reverse transcribed to cDNA in a 20 uL reaction using a Quantitect reverse transcription kit (QIAGEN) including a gDNA wipeout step. Completed reactions were diluted 10-fold with yeast tRNA 0.5 ug/mL and 2 ul were used for qPCR using Brilliant III SYBR Green UltraFast qPCR master mix (Agilent) and with primers at 500 nmol L-1 each in a final reaction volume of 10 uL. Standards (10^7-10^1 copies/rxn) were amplified together with samples in a Rotor-Gene Q (QIAGEN) using the following parameters: 95C for 3 minutes followed by 40 cycles of 95C for 5 sec and 57C for 10 sec. Melt curve data were collected to confirm product identity. For all assays efficiency was >95%, and reactions were linear over 7 log and sensitive to at least 10 copies and a single PCR product of the correct size was observed on a 2% agarose gel. Copy numbers/rxn were derived from the standard curves and normalized using the normalization factor for the three most stable reference genes identified by geNorm software: HPRT1, SDHA, YWHAZ. Data were analyzed using a paired Student’s t-test with alpha at 0.05
Results
Tumour specific methylation events
To investigate whether penile tumours are epigenetically distinct from normal tissue, we performed genome-wide DNA methylation profiling using the 450k Illumina Infinium platform (29) to interrogate the methylation state of over 485,000 cytosine residues.
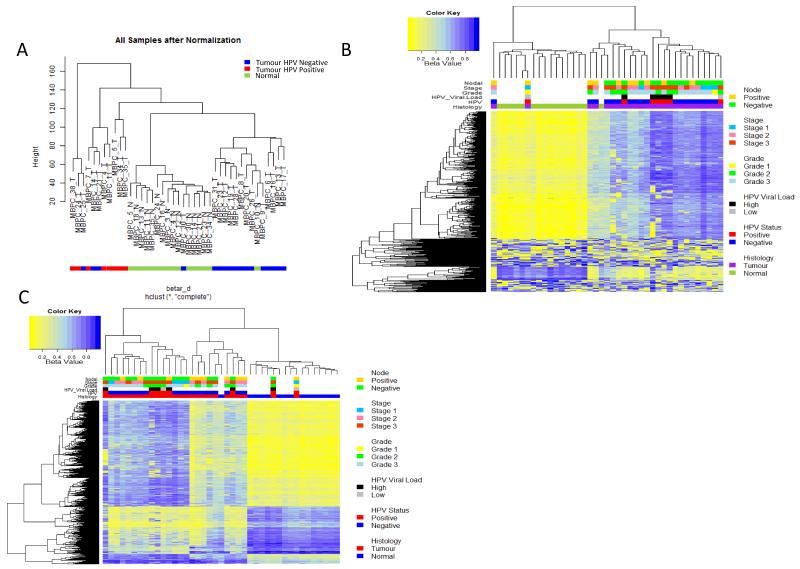
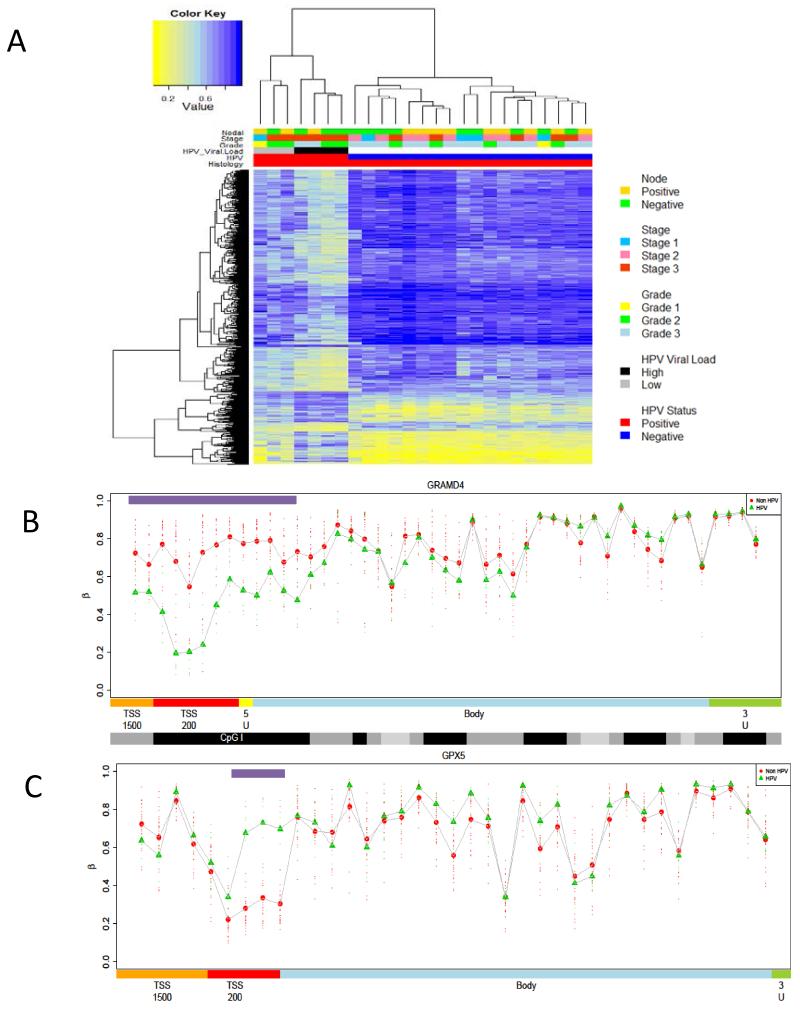
Unsupervised hierarchical clustering of beta values (methylation score) revealed three distinct clusters based on histological phenotype (Figure 1A). Clustering of the most variable probes (n=500) separated samples based on histopathology confirming that PeCa and normal penile tissue are epigenetically distinct, and pointing to a hypermethylation phenotype associated with malignant transformation (Figure 1B).
Figure1. Unsupervised clustering of methylation variable positions (MVPs) in Penile cancer squamous cell carcinomas.
A) Hierarchical clustering of PeCa and normal tissue based on global epigenetic profiles, generates 3 groups: a normal (centre, green), non-HPV-associated group (right, blue) and HPV-associated group (left, red), B) Heat map of methylation values of the 500 most variable loci, showing clear separation of normal and malignant disease. The DNA-methylation (β) values are represented using a colour scale from yellow (low DNA methylation) to blue (high DNA methylation), C) Heatmap of beta values for significant MVPs (Methylation Variable Position) (n=6933) between normal and penile cancer tissue. The DNA-methylation (β) values are represented using a colour scale from yellow (low DNA methylation) to blue (high DNA methylation.
Supervised analysis, using a Wilcoxon rank-sum test to assign directionality, was used to identify MVPs (methylation variable positions) between PeCa versus normal tissue. MVPs were selected on the basis of statistical significance (Wilcoxon P-value>0.001), an additional filter of Δβ>0.30(+/−) was applied to compensate for not taking into account the absolute difference in methylation between the groups. The cut-off is empirically defined to result in a false discovery rate (FDR) of <2%. This allowed us to reduce our candidate loci to those with largest methylation differences and therefore greatest potential for functional effect. A total of 6933 MVPs met these requirements (4935 Hyper MVPs, 1998 Hypo MVPs), hierarchical clustering of the samples yielded three clusters 1) Normal, 2) Node positive and 3) Node negative (Figure 1C).
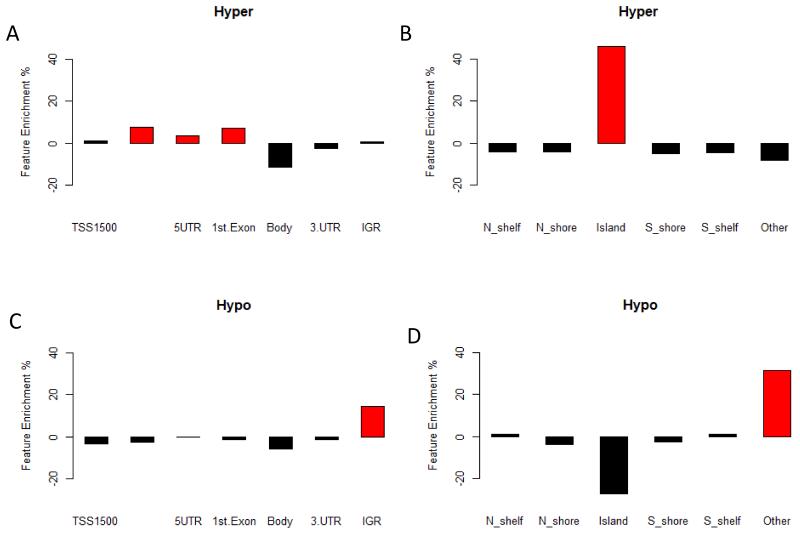
There is a clear hypermethylation profile associated with the cancer phenotype (Figure 1C), with over 71% of MVPs being hypermethylated in tumour tissue compared with matched normal tissue (Supplementary Figure 1). Mapping of the MVPs to gene features revealed a significant (random resampling p <0.0001) enrichment of hypermethylated CpG islands (CpGI), 44% enrichment (Figure 2A,B). To assess the potential functional impact of CpGI methylation on gene expression we tested the association with MVPs in either promoter associated or non-promoter associated CpGIs. This showed a enrichment (p<0.0001) of MVPs in promoter associated CpGIs, and is further supported by the enrichment (p<0.0001) of MVPs in regulatory regions including transcription start sites (TSS200), 1st exons, and 5′ UTRs, which show enrichments of 8%, 7% and 4% respectively (Figure 2A,B).
Figure 2. MVP canonical feature enrichment.
Assessment of MVP enrichment in canonical gene features, for both hyper- (A and B) and hypo- (C and D) methylated MVPs. Shows enrichment of hypermethylated MVP in promoter associated features (A) and CpGI (D), Hypomethylated MVP are enriched in inter-genic regions (IGR) (C). Genomic features with significant (P=<0.0001) enrichment are shown in red.
Analysis of hypomethylated MVPs showed enrichment (p=0.00101, 14%) of intergenic regions (IGR) (Figure 2C,D), potentially pointing to hypomethylation of repeats regions. This is confirmed by the enrichment of loci within ALU and SINE1 repeat elements.
As single MVPs are less likely to have functional effect on gene expression, we next sought to amalgamate individual MVPs into Differentially Methylated Regions (DMRs). The analysis defined 1255 significant DMRs (p<0.001) associated with the malignant phenotype (997 hyper DMRs and 258 hypo DMRs). The DMRs were associated with 367 genes, CpGIs were the predominant genomic feature associated DMRs.
Gene set enrichment analysis
GO analysis of genes associated with DMRs identified genes involved in DNA binding (GO:0003677), Signal Transduction (GO:0007165) and Receptor activity (GO:0004872) pathways. We also performed gene set enrichment analysis, assigning MVPs to their closest gene, to assess whether specific classes of genes are enriched. Interrogation of the PeCa-associated hypermethylated genes showed significant enrichment (P=0.000106) of genes which are targets of the PRC2 complex, including TBX5, GATA4, CDH7 and SOX14. Motif analysis of PRC2 target DMRs showed enrichment for PBX1, KLF4 and HIF1A transcription factor binding sites. Interestingly, we also see an increase in the expression of PRC2 complex members SUZ12 and EZH2 in tumours compared to normal tissue (Supplementary Figure 2).
The high rate of CpGI methylation would suggest the potential for frequent inactivation of tumour suppressor genes (TSGs). We therefore compared genes associated with both MVPs and DMRs with a list of 712 known TSGs. This revealed the enrichment of hyper-MVPs in TSGs (p=0.0019), with 52 TSGs showing CpGI hypermethylation, these include RASSF2, WT1 & CDO1.
We also identified aberrant methylation of several potential therapeutic targets, including tyrosine kinases, EPHA5, EPHA6 along with FLT1 (VEGFR1), FLT3 and FLT4 (VEGFR3), and aberrant methylation of the androgen receptor (AR) and programmed cell death receptor 1 (PDCD1) the gene which encodes PD1, highlighting potential therapeutic targets for the treatment of PeCa (Supplementary Figure 3, Supplementary Figure 4A-B).
To confirm the functional relevance of methylation we assessed the expression of two candidate genes (CDO1 and AR) in an independent cohort of matched PeCa and normal tissues. This showed a significant reduction of expression in PeCa compared with matched normal tissue (Supplementary Figure 4C-D).
Epigenetic markers of lymph node metastasis
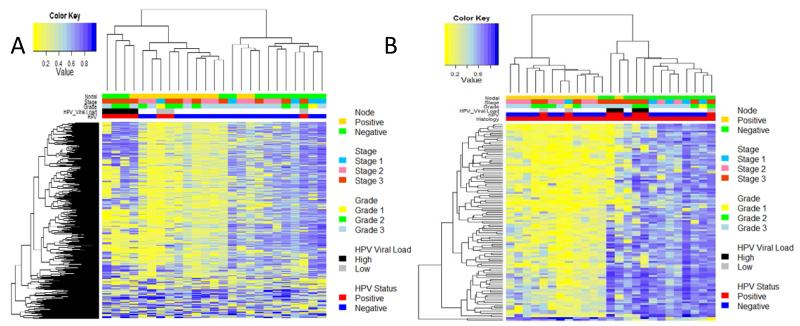
Unsupervised clustering of the top 500 most variable (tumour only) probes was performed to assess the association of aberrant epigenetic events with pathological factors. This defined two clusters (Figure 3A), which showed a significant correlation with lymph node status (P=000017), with a hypermethylated lymph node positive cluster and hypomethylated lymph node negative cluster. No correlation was found between these clusters and tumour grade or stage (P>0.05).
Figure 3. Epigenetic signature of local lymphatic metastasis.
Heat map for the top 500 most variably methylated loci in penile cancers shows three pathologically defined clusters, a hypermethylated lymph node negative (right (upper green bar)), lymph node positive hypomethylated group (centre (upper gold bar)) and a HPV associated cluster (left (lower two bars (red = 1st-HPV positive, blue = HPV negative, black = 2nd HPV Viral Load, High HPV (>1 copy/cell), white = low (<1copy/cell) grey, no HPV detectable). B) Heatmap of methylation values for 962 significant MVP between node negative (green upper bar), and node positive (gold upper bar).
In order to more clearly define the epigenetic alterations associated with local metastatic spread we carried out a supervised analysis utilising all 48577 informative loci (Figure 3B). This defined a small number of MVPs (n=112), which separate samples into two main groups, a hypomethylated lymph node positive group and a hypermethylated lymph node negative disease group. Analysis of the enriched MVPs in canonical gene features, shows enrichment of hypomethylated MVPs within CpGIs (P<0.0001), with 72% of MVPs located in CpGIs. These data suggest that CpGI hypermethylation is associated with lower metastatic potential.
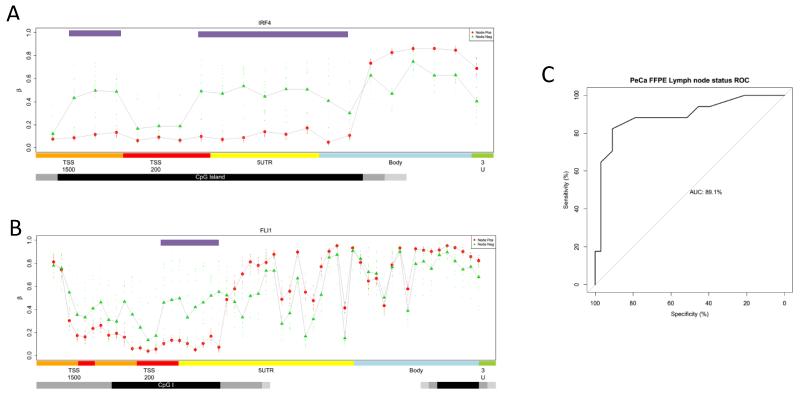
The ability to predict lymph node metastasis may have potential utility in the clinical management of patients by identifying which patients with clinically impalpable inguinal lymph nodes require an inguinal lymphadenectomy. To explore this we sought to define a minimal epigenetic signature, which could be used to predict lymph node metastasis. Using a shrunken centroids approach, we identified a minimum 54 CpG signature which in cross validation, could predict the lymphatic metastases with an accuracy of 93%. When individual MVPs were coalesced into potentially functional DMRs, we identified DMRs in four genes, HMX3, IRF4, FLI1 and PPP2R5C, to be predictive of lymph node positive disease (Figure 4 A-B, Supplementary Figure 5). These DMRs were combined to define a final predictive methylation index for each sample (mean methylation state across DMRs). This predictive index reached an ROC of 98% (specificity 100%, sensitivity 92%,) (Supplementary Figure 5). We then tested the association of this gene panel in a validation cohort of a further 50 patients with FFPE DNA using qMSP for each DMR. In the validation cohort the predictive lymph node metastasis signature reached an AUC of 0.89 (specificity 80%, sensitivity 93%)(Figure 4C).
Figure 4. Epigenetic genomic profiles of DMRs associated with lymph node metastasis.
Methylation profiles of candidate genes associated with local lymphatic metastases, for A) IRF4 and B) FLI1. Feature annotation are taken form the Infinium methylation arrays, methylation values are color-coded accordingly: TSS1500, orange (1500 bp to 200 bp upstream of the transcription start site (TSS)); TSS200, red (200 bp upstream of the TSS); 5′ untranslated region (UTR), yellow; gene body, blue; CpGI, black; CpGI shores, grey; and CpGI shelves, light grey. Regions defined as Differentially Methylated Regions (DMRs) are highlighted by upper purple bars. Intermarker distances are not to genomic scale. C) ROC curve for the accuracy of lymph node metastasis using the QMSP epi-signature in a 50 case validation cohort.
Multivariable analysis showed this minimal signature to be an independent predictor of lymph node metastasis (P=0.0053), a surrogate for disease-specific survival, there was no significant association with age, stage or grade (p=1, p=0.98, p=0.76) in multivariable analysis.
Immunohistochemical analysis for FLI1 and IRF4 (available antibodies) was carried out on a tissue microarray containing the 50 PeCa tumours. Although we observe a reduction in protein expression in samples with corresponding hypermethylation, the relationship with lymph node metastasis was not statistically significant (Supplementary Figure 5B).
HPV-driven tumourigenesis
Unsupervised clustering points to the presence of a potential HPV related epigenetic component (Figure 5A). To define a HPV induced epigenetic signature we performed a supervised analysis and ranked probes using a Bayesian regularised t-statistics model. We identified a significant association between DNA methylation and HPV status, with 960 significant MVPs at an FDR of less than 0.01, and 5037 at an FDR of < 0.05. Of the 960 MVPs, the overwhelming majority (747, 77%) were hypo-MVPs in HPV positive samples, compared with HPV negative, indicating that HPV infection is associated with widespread loss of DNA methylation (Figure 5A). Analysis of the canonical gene features in which these MVPs reside showed that over 67% are located with CpGI’s, shores and shelves, with a significant enrichment (p<0.001) of MVPs in CpGI shores. When individual MVPs were coalesced into potentially functional DMRs, we identified DMRs in several candidate genes including GRAMD4 and GPX5 (Figure 5B-C). GO analysis of analysis of genes associated with PeCa HPV DMRs identified genes involved in WNT signaling, DNA binding, Signal Transduction and Receptor activity pathways. They also showed significant overlap with genes shown to be up-regulated in nasopharyngeal tumours, which are also frequently driven by HPV. Motif analysis of PeCa HPV DMRs showed enrichment for TCF3, MAZ, JUN, PAX4 and MYC transcription factor binding sites.
Figure 5. PeCa HPV induced epigenetic signature.
A) Heat map of significant MVP (p<0.01) between HPV positive and HPV negative PeCa. HPV positive samples show a significantly lower methylation profile than HPV negative disease. B,C) Methylation profiles of candidate genes epigenetically deregulated during HPV tumorigenic transformation. Comparison of DMR profiles across canonical features for HPV associated PeCa (green) and non-HPV associated PeCa (red), for candidate epigenetically regulated genes involved in the HPV driven penile cancer ((B)GPX5 and (C)GRAMD4). Profiles show Feature annotation is as provided by BeadChip, and methylation values are colour-coded accordingly: orange = TSS1500, (1500 bp to 200 bp upstream of the transcription start site (TSS)); red = TSS200 (200 bp upstream of the TSS,); yellow = 5′ untranslated region (UTR), blue = gene body; black = CpG islands,; darker grey = CpG shores,; and light grey = CpG shelves
Lineage independent HPV signature
We sought to assess if the effect of HPV infection on DNA methylation is lineage dependent by evaluating the methylation state of PeCa HPV MVPs in HNSCC and CESC. Using all PeCa HPV MVPs we were able to accurately define HPV positive from HPV negative disease in 42 HNSCC (Data not shown). We subsequently identified the overlapping loci between these two data sets, in order to define a lineage independent HPV signature. Despite the apparent strong association of our PeCa HPV epigenetic signature across different tissue lineages, there is little overlap in epigenetically altered loci, with only 30 overlapping loci MVPs in both tissue types. Analysis of the methylation state of these loci reveals a distinct hypomethylated signature associated with HPV-positive disease (Data not shown). For cross validation we performed a shrunken centroid class prediction used the 30 MVPs and were able to accurately predict the HPV status of 27/28 HPV positive and 57/58 HPV negative samples from the combined PeCa-HNSCC training cohort. We were also able to accurately predict the HPV status of a panel of HPV positive and HPV negative HNSCC cell lines (n=6) (Supplementary Figure 6).
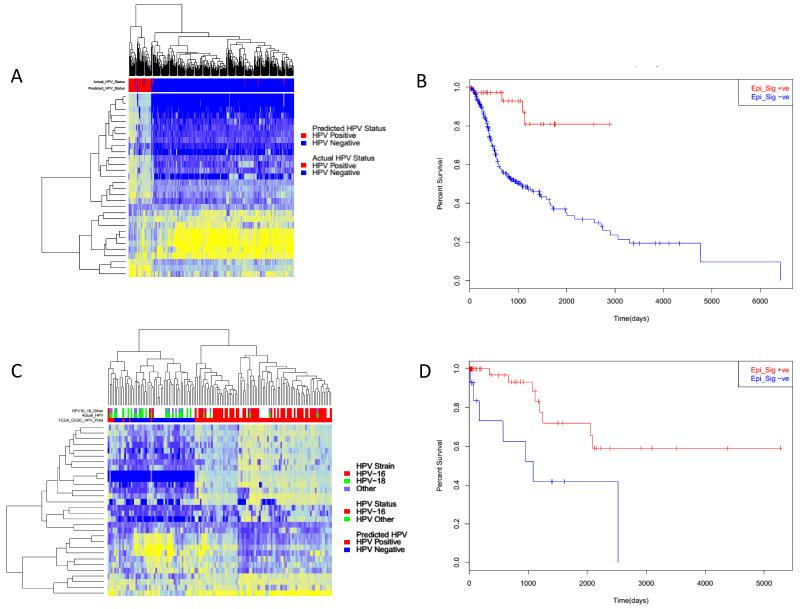
We subsequently applied this HPV epi-signature to an independent set of HNSCC (n=310) and CESC (n=136) samples. When applied to HNSCC the HPV epi-signature predicated 40 HPV positive and 290 HPV negative (Figure 6). When comparing those samples with a known HPV status this accurately predicted the HPV status of 299/310 HNSCC samples (4 false positives, 7 false negatives), giving an overall misclassification rate of 3.5% (Figure 6A).
Figure 6. Analysis of HPV epi-signature in independent HNSC and CESC.
A) Heatmap of 310 TCGA HNSCC samples showing the methylation of the 30 probe set classifier. Showing the epi-signature predicted HPV status (Positive – red, Negative –blue), Actual HPV status, HPV 16 positive (red), HPV negative (blue). B) Kaplan–Meier curve showing for HNSC epi-signature positive (red) and epi-sganture negative (Blue). C) Heatmap of 136 CESC samples showing the methylation of the epi-siganutre loci. Showing the epi-signature predicted HPV status (Positive – red, Negative –blue), Actual HPV status, HPV 16 positive (red) samples containing other HPV subtypes (green), comparison of HPV sub type, HPV 16 (red), HPV18 (green) and other HPV (purple). D) Kaplan–Meier curve showing for CESC epi-signature positive (red) and epi-sganture negative (blue) patients.
When comparing the predicted HPV status of all 310 HNSCC compared with pathological features, there was a significant association with patient overall survival, with a 5-year survival for signature negative patients of 38% compared to 81% for signature positive patients (p=0.00073, HR = 5.6, 95% CI 0.021-0.78) (Figure 6B) although not independent of HPV status. There was no significant association of our HPV epigenetic signature with stage, age or gender.
We also assessed an independent cohort of 136 cervical cancer samples, using the same 30 loci HPV epi-signature 66% (90) were predicted to be signature positive compared to 34% (46) predicted to be signature negative. Epi-signature negative samples had a significantly (p=0.05) worse overall survival than signature positive samples, with a 5 year overall survival for signature positive patients of 77% compared to 50% for signature negative patients. Age (p=0052) and stage (p=0.035) were also significant in multivariate analysis.
As >90% of CESCC are a result of HPV infection, using only those samples with a known HPV status (n=84) we compared the predicted and actual HPV status (Figure 6C). Of those 62 epi-signature positive samples 53 (85%) were HPV16 positive, compared to 9/62 (15%) which contained other high risk HPV subtypes, including HPV18. Of those epi-signature negative samples only 2 out of 22 (9%) contained HPV16, suggesting the possibility of a HPV16 specific epigenetic alteration signature. Of the 84 patients with a confirmed HPV genotype, 73 had confirmed outcome data. Signature positive patients had a significantly better overall survival than signature negative (Figure 6D) (p= 0.0094, HR=3.91, 95% CI =0.13-0.78) (adjusted for age, grade and stage). Despite correlating strongly with HPV genotype, the HPV epi-signature appears a stronger predictor of CESC patient survival than HPV genotype alone (p=0.07, HR=2.56, 95% CI=0.14129-1.083) (adjusted for age, grade and stage).
Discussion
Penile cancer is a rare disease in the developed world, however represents a significant source of patient morbidity and mortality in developing nations. The results reported here represent the most comprehensive epigenetic study of penile squamous cell carcinoma to date and shed light on to the epigenetic alterations involved in penile cancer. Using high density genome-wide methylation arrays we have revealed distinct PeCa associated epigenetic signatures and define an epigenetic signature which can predict local lymph node metastasis, one of the most important prognostic indicators for PeCa survival, and, to our knowledge, this is the first study to demonstrate the existence of an HPV-mediated DNA-methylation signature in HPV positive PeCa.
Previous studies have identified differentially methylated genes in PeCa(14, 15). These have been targeted studies in which candidate epigenetic regulated genes have been identified including RAS and THBS1. Using the Illumina Infinium Human Methylation arrays, we defined over 1,200 DMRs associated with the malignant phenotype and CIMP relating to 367 genes. Supervised analysis of PeCa versus normal tissue identified PeCa-associated hypermethylated genes with significant enrichment of genes which are targets of PRC2 complex, these include TBX5, GATA4, CDH7 and SOX14. Aberrant methylation of genes regulated by the PRC2 complex has been observed in many cancer types, including head and neck, cervical and prostate cancer but not previously in penile cancer. However, changes in the epigenetic regulation of PRC2 target genes has been noted during the HPV16 transformation of normal foreskin keratinocytes, with HPV16 infection resulting in the increased EZH2 expression and decreased global H3K27me3 (30). Furthermore, we also see overexpression of the members of the PRC2 complex (EZH2 and SUZ12) in PeCas. This has been reported in other tumour types and shown to result in loss of PRC2 target gene expression(31). These data would suggest that deregulation [through either aberrant methylation, altered histone code or increased PRC2 complex expression] of PRC2 regulated genes is an essential part of the oncogenic transformation of both HPV and non-HPV related PeCa and warrants further investigation.
The hypermethylation of tumour suppressor genes (TSGs) is a key feature of tumourigenesis. To identify key TSGs regulated by methylation we compared with both MVP and DMR with a list of 712 known TSGs. This included CDO1, which we also show to be differentially expressed between PeCa and normal tissue. The inactivation of CDO1 by DNA methylation has recently been implicated in many cancers including bladder, breast cancer colon and lung cancer (32-36). Cysteine deoxygenase 1 (CDO1) is integral to the biodegradation of toxic cysteine, and reduced CDO1 expression has been shown to increase cell proliferation in vitro, whereas over expression resulted in decreased tumour growth both in vitro and in vivo (33).
We also identified aberrant methylation of several potential therapeutic targets, including the hypermethylation and epigenetic regulation of the androgen receptor (AR). The aberrant methylation of the AR is particularly intriguing. Increased AR signalling is important in hormonally driven tumours including prostate and breast cancers. Although it is assumed increased AR expression is oncogenic in hormonally driven cancers, it has recently been shown that loss of AR in hormone refractory prostate cancer results in the activation of STAT3 (37). STAT3 regulates gene involved in the control of cellular processes including proliferation, survival and immune responses (38). Persistent activation of STAT3 is oncogenic and has been implicated in the development of a wide variety of human malignancies including leukaemia and lymphoma and solid tumours including head and neck cancer, prostate, breast and colon cancers (39-41). Although still to be functionally validated, these data would suggest the potential for a pivotal role for loss of the androgen receptor in the development of penile cancer.
The presence of metastatic disease in the inguinal lymph nodes is one of the most important prognostic factors in penile cancer (42). Occult nodal metastasis are present in 20 - 25% of cases at presentation (43, 44) and inguinal lymph node dissection is largely directed by clinical examination and the histopathological features of the primary lesion. Due to the lack of biomarkers which can accurately identify or predict lymph node metastasis, all patients with ≥T1G2 disease and impalpable inguinal lymph nodes undergo inguinal lymphadenectomy (removal of the inguinal lymph nodes), which is unnecessary in 75 - 80% of patients. Lymph node metastasis is an independent predictor of survival in penile cancer and therefore may be used as a surrogate disease-specific survival (45).
Methylome analysis identified a distinct epigenetic signature associated with lymph node metastasis. This 122 CpG classifier, which in cross validation, could predict the lymphatic metastases with an accuracy of 93%. The majority of MVP were located within DMRs in 4 genes, HMX3, IRF4, FLI1 and PPP2R5C and DMR methylation was also predictive of lymph node positive disease. When combined as predictive methylation index for each sample, the predictive accuracy of this signature (90% methylation array and 89% for qMSP) to identify the presence of lymph node metastasis is at least comparable to if not better than the sensitivity of sentinel lymph node biopsy. We are currently assessing the feasibility of using the methylation state of these loci as biomarkers in ‘liquid’ biopsy, using plasma cell free DNA to detect metastasis specific methylation events.
Finally, we also sought to understand the relationship between epigenetic alterations and HPV and clinical pathological factors. High risk HPV infection is a key oncogenic driver in several different tumour types, including, cervical cancer, head and neck squamous cell cancers along with PeCa. It is well documented in HNSCC and cervical cancers that HPV infection results in the epigenetic reprogramming of the host cell during malignant transformation resulting in a distinct HPV-induced epigenetic phenotype (20, 46). In this cohort, we found HPV infection in 23% of samples which was lower than expected although the incidence of HPV positive penile cancer ranges from 14%-100% and is also dependent on prior circumcision which was not recorded in our cohort (47). Only HPV 16 was detected in our cohort and HPV 16 represents the predominant subtype in PeCa and head and neck cancers (20, 45, 48). We defined a distinct, predominately hypomethylated, HPV 16-associated epigenetic signature. This large probe set was able to accurately separate an independent cohort of HNSCC cases, suggesting a lineage independent HPV specific epigenetic phenotype (20). However, despite the apparent synergy in epigenetic alterations associated with HPV infection, only 30 HPV specific MVPs were found to be overlapping between the two cohorts. We validated this minimal HPV signature, in independent HNSCC and CESC cohorts, and show it to be predictive of disease free survival in both HNSCC and CESC, and predictive of HPV infection in HNSCC. Interestingly when applied to CESC, this signature appeared to separate by HPV subtype, specifically HPV16 v HPV18/other HPV, supporting the postulate that we have defined a HPV16 signature. While 50% to 60% of CESC are associated with HPV16 infection, a further 20% are associated with HPV18 (6, 8, 49), this contrasts with HNSCC and PeCa in which >90% of HPV infection is HPV16. We found only HPV 16 in each of the two training cohorts. Although only a single CESC cohort, these data suggest the presence of specific HPV subtype epigenetic alterations, and further suggest a distinct survival advantage to HPV 16 driven tumours compared to those associated with other high risk HPVs, such as HPV 18 (50). In future studies it will be important to elucidate the functional impact of differential methylation of these genes and their role in HPV subtype specific driven cancer development. In terms of clinical utility, this novel methylation signature can be tested as a strategy to stratify cases at high risk with the potential to direct multimodal therapy. Moreover, the encoded proteins affected by aberrant methylation may represent promising drug targets for innovative and more efficient cancer therapy.
In summary, this work shows that changes in DNA methylation are a key components in penile cancer. We show the utility of an epigenetic signature, which has been validated on an independent cohort, to identify occult lymph node metastasis in PeCa with equivalent or greater sensitivity to methods in current clinical practice. In addition we define a PeCa specific HPV signature and a HPV associated host epigenetic signature which is a lineage independent predictor of disease free survival and suggests distinct HPV sub-type specific epigenetic alterations.
Supplementary Material
Translational Statement.
Penile Cancer (PeCa) is rare in the developed world, but represents a global health problem, with an incidence of up to 8.3:100,00 in developing nations. The most important predictive factor of an unfavourable prognosis in PeCa is the presence of regional inguinal lymph node involvement. Currently, no molecular markers exist that can accurately predict the presence of lymph node metastases. Using genome wide DNA methylation profiling, we defined the epigenetic alterations involved in PeCa and validated an epigenetic signature which is predictive of lymph node metastasis. HPV represents a major oncogenic driver in PeCa, we identify HPV induced epigenetic alterations, from these we define an epigenetic signature that is predictive of survival across multiple HPV driven cancers. The identification of epigenetic biomarkers of metastasis and survival may play a significant role in improving the management, treatment and survival of penile cancer and also other HPV driven cancers.
Acknowledgements
The Authors would like to thank Jenny Patterson and UCL Advanced Diagnostics for the assistance with immunohistochemical analysis. Along with Kerra Peirce from UCL genomics for processing the Illumina methylation arrays.
Grant Support
AF is supported by the UCL/UCLH Comprehensive Biomedical Research Centre, the Rosetrees Trust and research involved in this project was supported by Orchid. MA is supported by Orchid. Research in the Beck lab was supported by the Wellcome Trust (WT084071, WT093855), Royal Society Wolfson Research Merit Award (WM100023), MRC (G100041), IMI-JU OncoTrack (115234) and EU-FP7 projects EPIGENESYS (257082), IDEAL (259679) and BLUEPRINT (282510). JK is supported by the UCLH/UCL Comprehensive Biomedical Research Programme.
Footnotes
There is no conflict of interest from any of the authors.
References
- 1.Arya M, Li R, Pegler K, Sangar V, Kelly JD, Minhas S, et al. Long-term trends in incidence, survival and mortality of primary penile cancer in England. Cancer causes & control: CCC. 2013;24:2169–76. doi: 10.1007/s10552-013-0293-y. [DOI] [PubMed] [Google Scholar]
- 2.Morris BJ, Gray RH, Castellsague X, Bosch FX, Halperin DT, Waskett JH, et al. The Strong Protective Effect of Circumcision against Cancer of the Penis. Advances in urology. 2011;2011:812368. doi: 10.1155/2011/812368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misra S, Chaturvedi A, Misra NC. Penile carcinoma: a challenge for the developing world. The lancet oncology. 2004;5:240–7. doi: 10.1016/S1470-2045(04)01427-5. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez BY, Barnholtz-Sloan J, German RR, Giuliano A, Goodman MT, King JB, et al. Burden of invasive squamous cell carcinoma of the penis in the United States, 1998-2003. Cancer. 2008;113:2883–91. doi: 10.1002/cncr.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ornellas AA, Nobrega BL, Wei Kin Chin E, Wisnescky A, da Silva PC, de Santos Schwindt AB. Prognostic factors in invasive squamous cell carcinoma of the penis: analysis of 196 patients treated at the Brazilian National Cancer Institute. The Journal of urology. 2008;180:1354–9. doi: 10.1016/j.juro.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 6.Dillner J, von Krogh G, Horenblas S, Meijer CJ. Etiology of squamous cell carcinoma of the penis. Scandinavian journal of urology and nephrology Supplementum. 2000:189–93. doi: 10.1080/00365590050509913. [DOI] [PubMed] [Google Scholar]
- 7.Tran N, Rose B, O’Brien C. Role of human papillomavirus in the etiology of head and neck cancer. Head Neck. 2007;29:64–70. doi: 10.1002/hed.20460. [DOI] [PubMed] [Google Scholar]
- 8.Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–65. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 10.Watson R. European centre urges vaccination of girls against HPV. Bmj. 2008;336:241. doi: 10.1136/bmj.39472.537697.DB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lont AP, Kroon BK, Horenblas S, Gallee MP, Berkhof J, Meijer CJ, et al. Presence of high-risk human papillomavirus DNA in penile carcinoma predicts favorable outcome in survival. Int J Cancer. 2006;119:1078–81. doi: 10.1002/ijc.21961. [DOI] [PubMed] [Google Scholar]
- 12.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 13.Ferreux E, Lont AP, Horenblas S, Gallee MP, Raaphorst FM, von Knebel Doeberitz M, et al. Evidence for at least three alternative mechanisms targeting the p16INK4A/cyclin D/Rb pathway in penile carcinoma, one of which is mediated by high-risk human papillomavirus. J Pathol. 2003;201:109–18. doi: 10.1002/path.1394. [DOI] [PubMed] [Google Scholar]
- 14.Yanagawa N, Osakabe M, Hayashi M, Tamura G, Motoyama T. Detection of HPV-DNA, p53 alterations, and methylation in penile squamous cell carcinoma in Japanese men. Pathology international. 2008;58:477–82. doi: 10.1111/j.1440-1827.2008.02259.x. [DOI] [PubMed] [Google Scholar]
- 15.Yanagawa N, Osakabe M, Hayashi M, Tamura G, Motoyama T. Frequent epigenetic silencing of the FHIT gene in penile squamous cell carcinomas. Virchows Archiv: an international journal of pathology. 2008;452:377–82. doi: 10.1007/s00428-008-0597-6. [DOI] [PubMed] [Google Scholar]
- 16.Guerrero D, Guarch R, Ojer A, Casas JM, Ropero S, Mancha A, et al. Hypermethylation of the thrombospondin-1 gene is associated with poor prognosis in penile squamous cell carcinoma. BJU international. 2008;102:747–55. doi: 10.1111/j.1464-410X.2008.07603.x. [DOI] [PubMed] [Google Scholar]
- 17.Paschos K, Allday M. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 2010;18:439–47. doi: 10.1016/j.tim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez A, Esteller M. Viral epigenomes in human tumorigenesis. Oncogene. 2010;29:1405–20. doi: 10.1038/onc.2009.517. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez A, Rosales C, Lopez-Nieva P, Grana O, Ballestar E, Ropero S, et al. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome research. 2009;19:438–51. doi: 10.1101/gr.083550.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lechner M, Fenton T, West J, Wilson G, Feber A, Henderson S, et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome medicine. 2013;5:15. doi: 10.1186/gm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang KW, Alaei-Mahabadi B, Samuelsson T, Lindh M, Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nature communications. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R: A language and environment for statistical computing [Google Scholar]
- 23.Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, et al. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014;30:428–30. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guilhamon P, Eskandarpour M, Halai D, Wilson GA, Feber A, Teschendorff AE, et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nature communications. 2013;4:2166. doi: 10.1038/ncomms3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.TCGA . The Cancer Genome Project. HSNCC; 2014. Available from: https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm?mode=ApplyFilter&showMatrix=true&diseaseType=HNSC&tumorNormal=TN&tumorNormal=T&tumorNormal=NT&platformType=2&platformType=42. [Google Scholar]
- 28.TCGA . The Cancer Genome Project. CESC; 2014. Available from: https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm?mode=ApplyFilter&showMatrix=true&diseaseType=CESC&tumorNormal=TN&tumorNormal=T&tumorNormal=NT&platformType=2&platformType=42. [Google Scholar]
- 29.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–95. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Hyland PL, McDade SS, McCloskey R, Dickson GJ, Arthur K, McCance DJ, et al. Evidence for alteration of EZH2, BMI1, and KDM6A and epigenetic reprogramming in human papillomavirus type 16 E6/E7-expressing keratinocytes. Journal of virology. 2011;85:10999–1006. doi: 10.1128/JVI.00160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato T, Kaneda A, Tsuji S, Isagawa T, Yamamoto S, Fujita T, et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Scientific reports. 2013;3:1911. doi: 10.1038/srep01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Booken N, Gratchev A, Utikal J, Weiss C, Yu X, Qadoumi M, et al. Sezary syndrome is a unique cutaneous T-cell lymphoma as identified by an expanded gene signature including diagnostic marker molecules CDO1 and DNM3. Leukemia. 2008;22:393–9. doi: 10.1038/sj.leu.2405044. [DOI] [PubMed] [Google Scholar]
- 33.Brait M, Ling S, Nagpal JK, Chang X, Park HL, Lee J, et al. Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PloS one. 2012;7:e44951. doi: 10.1371/journal.pone.0044951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietrich D, Krispin M, Dietrich J, Fassbender A, Lewin J, Harbeck N, et al. CDO1 promoter methylation is a biomarker for outcome prediction of anthracycline treated, estrogen receptor-positive, lymph node-positive breast cancer patients. BMC cancer. 2010;10:247. doi: 10.1186/1471-2407-10-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeschke J, O’Hagan HM, Zhang W, Vatapalli R, Calmon MF, Danilova L, et al. Frequent inactivation of cysteine dioxygenase type 1 contributes to survival of breast cancer cells and resistance to anthracyclines. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:3201–11. doi: 10.1158/1078-0432.CCR-12-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon YJ, Lee SJ, Koh JS, Kim SH, Lee HW, Kang MC, et al. Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2012;7:20–33. doi: 10.1097/JTO.0b013e3182307f62. [DOI] [PubMed] [Google Scholar]
- 37.Schroeder A, Herrmann A, Cherryholmes G, Kowolik C, Buettner R, Pal S, et al. Loss of androgen receptor expression promotes a stem-like cell phenotype in prostate cancer through STAT3 signaling. Cancer research. 2014;74:1227–37. doi: 10.1158/0008-5472.CAN-13-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature reviews Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, Johnson JT, et al. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer discovery. 2012;2:694–705. doi: 10.1158/2159-8290.CD-12-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu ZB, Bai L, Qian P, Xiao YB, Wang GS, Qian GS, et al. Restoration of SOCS3 suppresses human lung adenocarcinoma cell growth by downregulating activation of Erk1/2, Akt apart from STAT3. Cell biology international. 2009;33:995–1001. doi: 10.1016/j.cellbi.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–15. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 42.Horenblas S, van Tinteren H. Squamous cell carcinoma of the penis. IV. Prognostic factors of survival: analysis of tumor, nodes and metastasis classification system. The Journal of urology. 1994;151:1239–43. doi: 10.1016/s0022-5347(17)35221-7. [DOI] [PubMed] [Google Scholar]
- 43.Ornellas AA, Kinchin EW, Nobrega BL, Wisnescky A, Koifman N, Quirino R. Surgical treatment of invasive squamous cell carcinoma of the penis: Brazilian National Cancer Institute long-term experience. Journal of surgical oncology. 2008;97:487–95. doi: 10.1002/jso.20980. [DOI] [PubMed] [Google Scholar]
- 44.Horenblas S, van Tinteren H, Delemarre JF, Moonen LM, Lustig V, van Waardenburg EW. Squamous cell carcinoma of the penis. III. Treatment of regional lymph nodes. The Journal of urology. 1993;149:492–7. doi: 10.1016/s0022-5347(17)36126-8. [DOI] [PubMed] [Google Scholar]
- 45.Sonpavde G, Pagliaro LC, Buonerba C, Dorff TB, Lee RJ, Di Lorenzo G. Penile cancer: current therapy and future directions. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2013;24:1179–89. doi: 10.1093/annonc/mds635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henken F, Wilting S, Overmeer R, van Rietschoten J, Nygren A, Errami A, et al. Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. British journal of cancer. 2007;97:1457–64. doi: 10.1038/sj.bjc.6604055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flaherty A, Kim T, Giuliano A, Magliocco A, Hakky TS, Pagliaro LC, et al. Implications for human papillomavirus in penile cancer. Urologic oncology. 2014;32:53, e1–8. doi: 10.1016/j.urolonc.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 48.Miralles-Guri C, Bruni L, Cubilla AL, Castellsague X, Bosch FX, de Sanjose S. Human papillomavirus prevalence and type distribution in penile carcinoma. J Clin Pathol. 2009;62:870–8. doi: 10.1136/jcp.2008.063149. [DOI] [PubMed] [Google Scholar]
- 49.Burd EM. Human papillomavirus and cervical cancer. Clinical microbiology reviews. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz SM, Daling JR, Shera KA, Madeleine MM, McKnight B, Galloway DA, et al. Human papillomavirus and prognosis of invasive cervical cancer: a population-based study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2001;19:1906–15. doi: 10.1200/JCO.2001.19.7.1906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.