Preface
Topoisomerases are complex molecular machines that modulate DNA topology to maintain chromosome superstructure and integrity. Although capable of stand-alone activity in vitro, topoisomerases frequently are linked to larger pathways and systems that resolve specific DNA superstructures and intermediates arising from cellular processes such as DNA repair, transcription, replication, and chromosome compaction. Topoisomerase activity is indispensible to cells, but requires the transient breakage of DNA strands. This property has been exploited, often for significant clinical benefit, by various exogenous agents that interfere with cell proliferation. Despite decades of study, surprising findings involving topoisomerases continue to emerge with respect to their cellular function, regulation, and utility as therapeutic targets.
Keywords: Topoisomerase, DNA replication, DNA repair, transcription, chromosome structure, topology, drug discovery
Introduction
The extended, double-helical structure of DNA poses a unique challenge to living organisms. Chromosomes must be folded and compacted in an orderly manner to fit within the confines of the cell. Natural nucleic-acid transactions such as transcription, replication, and repair – all of which require access to nucleotide sequence information – necessitate duplex melting events that overwind and underwind (or supercoil•) (Fig. 1A) nucleic acid segments, interfering with appropriate gene expression. DNA repair and replication further generate entanglements between chromosomal regions that, if left unresolved, can lead to potentially mutagenic or cytotoxic DNA strand breaks.
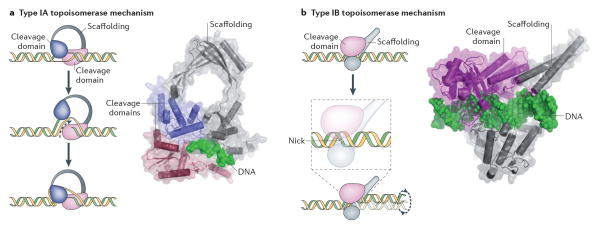
Fig 1. DNA cleavage and type I topoisomerase mechanisms.
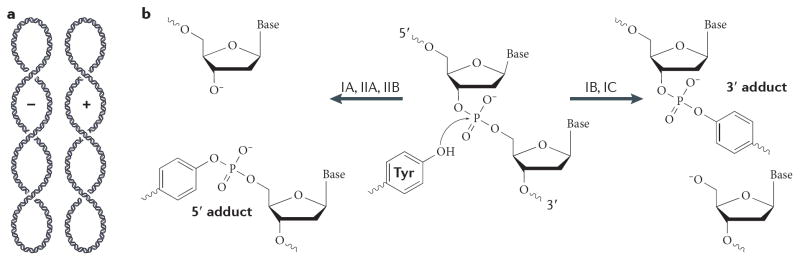
(A) Schematic of negative (−) and positive (+) plectonemic DNA supercoiling. The two forms can be distinguished by their right- and left-handed superhelical wrapping, respectively. (B) Schematic of the DNA cleavage reaction. Topoisomerases catalyze strand scission by forming a reversible, covalent enzyme–DNA adduct through their active site tyrosine. Type IB and IC topoisomerases become attached to 3′ DNA ends, and type IA and II topoisomerases attach to 5′ DNA ends. (C) Type IA topoisomerases pass a single-stranded DNA segment (yellow) through a transient break in a second, single DNA strand (green). The structure of E. coli topo III (a type IA topoisomerase) bound to single-stranded DNA is shown 210. (D) Type IB topoisomerases nick one DNA strand (yellow), allowing one duplex end (green strand) to rotate with respect to the other around the remaining phosphodiester bond. The structure of human topo IB bound to duplex DNA is shown 20.
Enzymes known as topoisomerases manage these transactions. Endowed with an ability to cut, shuffle, and religate DNA strands, topoisomerases can add or remove DNA supercoils, and disentangle snarled DNA segments. How topoisomerases carry out such complex functions has long been a significant question of both biochemical and biophysical interest. However, in recent years it has become evident that topoisomerases do not work at random; they can instead preferentially distinguish between different types of DNA juxtapositions and collaborate with disparate factors direct their action. It also has become clear that a rich and growing variety of natural and synthetic agents block topoisomerase function for the purposes of evolutionary competition or therapeutic gain.
In this Review, we touch upon a few of the many recent findings that have broadened our understanding of the roles played by topoisomerases in supporting chromosome biology and cell viability. Our goal is not to provide a comprehensive overview for those studying these complex enzymes, but rather to convey a sense of the vitality and richness of the topoisomerase field to the general biological community, particularly insofar as the evolutionary and functional diversity of these complex machines, and their roles in the flow of genetic information, genome maintenance, and human health. Given these limitations, we apologize to those colleagues whose work is only topically discussed, or excluded due to space limitations.
Topoisomerase families
At first glance, a bewildering number of different topoisomerases exist. However, nature’s topoisomerase collection is not as complicated as it first appears. For example, all topoisomerases contain a nucleophilic tyrosine, which they use to promote strand scission. This feature links DNA cleavage to the formation of a transient, covalent enzyme–DNA adduct, which in turn prevents the inadvertent release of nicked or broken DNA duplexes that might otherwise damage the chromosome (Fig. 1B). All topoisomerases also can be assigned into one of two primal classes, type I and type II, depending on whether they cleave one or two strands of DNA, respectively. Enzymes with an odd Roman numeral (for example, “I” or “V”) after their name fall into the type I class, whereas those with an even-numbered Roman numeral after their name are type II. Topoisomerase subtypes – “A,” “B,” or “C” – are then used to distinguish between enzyme families that have significantly different amino acid sequence relationships and/or structures. Some topoisomerase subtypes further encompass multiple paralogs1 that have diverged from one another in terms of their specific activities. A brief overview of these groupings follows to provide a context for subsequent discussion. For more extensive coverage of topoisomerase evolution, structure, and mechanism, the reader is invited to see 1,2.
Type IA: single-stranded ‘strand-passage’ enzymes
Type IA topoisomerases have a toroidal protein structure that bears a superficial resemblance to a padlock 3. These enzymes effect topological changes in DNA through a “strand-passage” mechanism 4,5, in which a single DNA strand is cleaved and physically opened, and a second DNA is navigated through the gap; following passage of the second DNA, the broken DNA is resealed (Fig. 1C). Type IA topoisomerases comprise three distinct subfamilies that are found throughout all three cellular domains of life2. These subfamilies are bacterial topo I (hereafter called topo IA), bacterial and eukaryotic topo III, and reverse gyrase 1. The primary activity of topo IA is to relax negatively-supercoiled DNA36,7. By contrast, topo III preferentially acts to resolve single-stranded DNA entanglements that can arise during DNA replication and repair 7–11. Reverse gyrase, found exclusively in thermophilic bacteria and archaea, positively supercoils DNA and renatures melted DNA strands through the action of an ATP-dependent superfamily-2 (SF-2) helicase4 domain fused to its type IA topoisomerase element12, 13.
Type IB: DNA “swivelases”
Type IB topoisomerases differ fundamentally in structure and mechanism from type IA enzymes 14. Rather than relying on strand passage, type IB topoisomerases effect supercoil relaxation by nicking a single strand of duplex DNA, and allowing one DNA end to rotate with respect to the other around the intact phosphodiester bond on the opposing strand 15. Rotation is controlled by friction between the DNA and the enzyme, which aids in aligning the broken ends for resealing 15,16 (Fig. 1D). Type IB proteins preferentially bind positively- or negatively- supercoiled substrates rather than relaxed substrates 17,18 using an interaction surface outside of its primary active site to bridge distal DNA segments 19. Some variants show a proclivity for positively-supercoiled DNA, which they can relax at a faster rate 18.
The active site architecture of type IB topoisomerases is evolutionarily related to that of tyrosine recombinases and integrases 20,21. Type IB enzymes are ubiquitous among eukaryotes, with some scattered homologs extant in certain viruses and bacteria 22. Type IB topoisomerases appear to be represented by a single family member (topo IB), although architectural differences that influence the rate and mechanism of supercoil relaxation are evident between various homologs in this group 15.
Type IC: a second class of swivelase
Type IC topoisomerases thus far have been found only in the archaeal genus Methanopyrus 23. Like topo IB, type IC topoisomerases relax positively- and negatively-supercoiled DNA through a nicking and rotation mechanism 24,25 (Fig 1D). However, the type IC active site shows little structural similarity to that of type IB enzymes, and appears to have a different evolutionary lineage 23,26. Type IC enzymes also retain functional elements that exhibit apurinic site lysase activity, which they can use for repairing abasic lesions5 in DNA 26,27. Only one topoisomerase variant, topo V, is presently known to comprise the type IC family 25.
Type IIA: duplex DNA ‘strand-passage’ enzymes
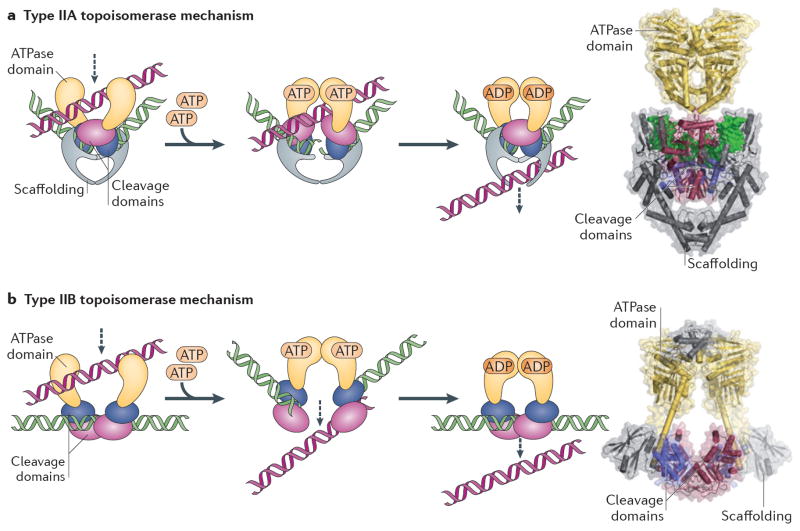
As with their type IA counterparts, type IIA topoisomerases employ an active strand-passage mechanism for effecting topological changes in DNA. Type IA and IIA topoisomerases also share certain catalytic domains used for DNA cleavage 28,29. However, type IIA enzymes differ in that they cleave both strands of a DNA duplex and pass a second intact duplex through the transient break 30–32 (Fig 2A), and they use ATP to power strand passage 31,33,34. These activities allow type IIA topoisomerases to resolve both positive and negative DNA supercoils, as well as disentangle long intertwined chromosomes and DNA catenanes632,35. Type IIA topoisomerases are found throughout all cellular organisms, as well as in some viruses and organelles, and can be partitioned into three paralogous subfamilies – eukaryotic topo II, bacterial topo IV, and prokaryotic gyrase – that exhibit distinct functional properties 1,2. For instance, gyrase actively adds negative supercoils to DNA and is only weakly able to unlink catenanes 33,36,37, while topo IV (and certain topo II isoforms) is a robust decatenase that preferentially relaxes positively-supercoiled substrates over negatively-supercoiled ones 38,39 (Box 1). Interestingly, type IIA topoisomerases show some ability to discriminate between, and preferentially act on, regions of high DNA curvature or DNA crossovers due to their ability to sharply bend DNA segments37,40–43. By contrast, yeast and Drosophila melanogaster topo II do not show any preference for one type of supercoiled DNA over another 44,45.
Fig 2. Type II topoisomerase mechanism.
(A) Type IIA topoisomerases cleave both strands of a duplex DNA (green) and pass another duplex DNA (purple) through the transient break in a reaction coupled to ATP turnover. The cleaved strands are religated, and the products of the reaction are released from the enzyme. The movement of the purple DNA duplex is indicated by a dashed arrow. The DNA cleavage domains are homologous to those of type IA topoisomerases. The structure of the yeast topo II ATPase and DNA cleavage domains are shown 187,211. (B) Type IIB topoisomerases use a duplex strand passage mechanism similar to that of type IIA enzymes, and share the same ATPase and cleavage domains but differ in overall tertiary structure. The structure of the topo VI holoenzyme is shown 51.
Box 1. Selectivity and function of type IIA DNA topoisomerase paralogs.
Vertebrates possess two topoisomerase II paralogs, topo IIα and topo IIβ, which share almost 77% sequence homology 198. However, the C-terminal regions of the two enzymes differ. In particular, the C-terminus of topo IIα shifts the activity of the enzyme toward the preferential relaxation of positive supercoils; by contrast, the equivalent region of topo IIβ does not appear to impart any supercoil preference 39. These differences may be linked to particular cellular functions 199, as topo IIα is essential in proliferating cells and assists in chromosome condensation, segregation and replication 200–202, while topo IIβ tends to be associated with DNA repair, transcription, and development 82,83,134.
Bacteria also often utilize two type IIA topoisomerase paralogs (gyrase and topo IV) with different activity profiles. Gyrase relaxes positive supercoils and actively adds negative supercoils to DNA, whereas topo IV relaxes both positive and negative supercoils and shows a heightened activity on positively-supercoiled and catenated substrates 33,38,203,204. An auxiliary domain appended to the primary strand-passage machinery of gyrase and topo IV appears to be responsible for controlling their functional distinctions 205,206, with sequence- and structure-based differences in this element permitting the wrapping of DNA by gyrase, but only DNA-binding by topo IV 206–209. The auxiliary DNA wrapping and binding domain of bacterial type IIA topoisomerases is not conserved in eukaryotic topo II, indicating that the improved action of topo IV and topo IIα on positively supercoiled DNA arose independently during their evolution.
Type IIB: a second class of duplex DNA strand-passage enzyme
Type IIB topoisomerases are found in archaea, plants, and a few bacterial, protist, and algal lineages 46,47. As with their type IIA cousins, type IIB topoisomerases unlink tangled DNA duplexes by strand passage (Fig 2B) and relax both negative and positive supercoils 48. Type IIB topoisomerases also possess ATPase and DNA-cleavage domains similar to those found in type IIA enzymes, although the relative arrangement of these elements in primary sequence space and their overall structural organization differs significantly 46,48–51. The principal DNA binding subunit of type IIB topoisomerases is noteworthy in that it is evolutionarily related to Spo11, the factor responsible for creating double-stranded DNA breaks that initiate meiotic recombination 46,52. Thus far, only one family member, topo VI, is known to comprise the type IIB topoisomerase clade 46.
Topoisomerase families – why so many?
Given this diversity of topoisomerases, and the seeming overlap in their basic function, how do cells decide which one to use for a particular purpose? This question has been difficult to address, in part because different organisms often possess markedly different topoisomerase repertoires 1,2. Moreover, the regulatory strategies for a given topoisomerase can vary widely among species. At present, there appears to be no single answer. Rather, the molecular logic behind topoisomerase choice appears to derive from a confluence of factors, including: the type of topological problem that needs to be addressed, which topoisomerase genes happen to be present in the resident genome, whether specific architectural elements have been acquired to modulate topoisomerase function, and if different accessory factors and/or modifications that alter topoisomerase localization and activity are in use (Table 1 and Supplementary information S1 (table)). Topoisomerases utilize a combination of these regulatory strategies in the majority of nucleic-acid transactions in which they participate. A discussion of the role of specific topoisomerases in different cellular processes follows.
DNA packaging
The length of chromosomal DNA far exceeds that of the cell in which it resides. Organisms thus employ a variety of mechanisms to assist with DNA compaction, including supercoiling and the use of chromosome organizing factors such as histones or nucleoid associated proteins 53. Plectonemic supercoiling7 alone has been estimated to condense DNA by two to three orders of magnitude (Figure 1A)54.
In bacteria, a need for compaction has been invoked as one of the principal reasons why chromosomal DNA is negatively supercoiled, and why gyrase, which adds those supercoils to DNA, is so ubiquitous throughout the bacterial kingdom 53,55. More recent studies, however, suggest that there are additional layers of complexity to this view. For example, two very closely related bacterial species, E. coli and S. typhimurium, share about 90% sequence identity across homologous genes 56, but have highly-dissimilar superhelical densities8, with E. coli DNA being significantly more underwound 57. Gyrase itself is not always necessary for DNA packaging, as the lone type IIA topoisomerase present in the hyperthermophilic bacterium Aquifex aeolicus is not a gyrase, as might be predicted on the basis of its sequence similarity with other gyrase N-terminal domains, but rather a topo IV 58. However, many thermophiles also possess reverse gyrase 59, which introduces positive supercoils that may aid compaction, as well as counteract thermal denaturation 60,61. Although the steady-state superhelical density of DNA in most prokaryotes has not been measured directly, multiple findings suggest that many species, particularly those that are adapted to growth at high temperatures, may have relatively relaxed chromosomes 62. How such organisms are able to sufficiently compact their chromosomes in the absence of supercoiling remains an open question, but the mechanism almost certainly relies on proteinaceous factors. Overall, the role of supercoiling in DNA compaction, and how topoisomerases collaborate to define the superhelical “setpoint” of the chromosome, is not fully understood.
Topoisomerases play another role in chromosome compaction by working with large condensation machineries (Table 1), principally a group of ATP binding cassette (ABC)-family ATPases9 known as Structural Maintenance of Chromosomes (SMC) proteins 63–67. SMCs, and their counterparts Rad50 (eukaryotes) and SbcC and RecF (E. coli), are conserved factors involved in multiple aspects of chromosome cohesion, condensation, and DNA repair 68. SMCs and type IIA topoisomerases indirectly co-localize as part of the protein network that helps stabilize long-range contacts between chromosomal segments 67,69. SMCs, or their affiliated accessory factors, also have been reported to directly associate with both topo II and topo IV. For example, the Drosophila melanogaster Barren subunit (an ortholog of the condensin H protein) co-immunoprecipiates with topo II in vitro 66, while a dimerization region of the E. coli SMC homolog MukB interacts with the C-terminal domain of the ParC subunit of topo IV 64,65. Interestingly, both interactions appear to potentiate the relaxation of negatively-supercoiled DNA by the associated topoisomerase, although how stimulation is achieved is unresolved. At present, little is known about the effect and role of these interactions on global DNA superstructure or about their utility to the cell, although the MukB–ParC interaction is important for cell viability 65. Whether these associations are preserved in other species is not clear, and constitutes a clear avenue for additional investigation.
Replication and chromosome segregation
Topoisomerases are required both for the successful management of DNA replication, and for specific developmental strategies that depend on replication. In the latter instance, the type IIB topoisomerase found in plants (topo VI) is required for a particular form of replication known as endoreduplication, a process by which chromosomes are copied multiple times in the absence of cell division 70–73. Endoreduplication gives rise to polyploid nuclei, which in turn can be used to regulate cell size 74. Why topo VI is needed specifically for endoreduplication, and why topo II cannot rescue a topo VI deficiency, is not clear 75. Similarly, replication of vertebrate mitochondrial DNA requires a special mitochondrial type IB topoisomerase, top1mt, a paralog of the nuclear type IB topoisomerase, top1 76,77. Mitochondrial DNA replication requires top1mt to form its regulatory displacement (D)-loop77, which in turn is thought to regulate DNA replication initiation and transcription 77. Surprisingly, when nuclear top1 is localized to the mitochondria, it cannot complement top1mt function and arrests the cell cycle 76. At present, it remains to be seen whether similar topoisomerase-dependent roles exist in other organisms. The role of topoisomerases in conventional DNA replication is better understood, with recent studies uncovering specific roles for topoisomerases in each of the three major replicative phases: initiation, fork progression, and termination.
Topoisomerases in replication initiation
In E. coli, replication initiation is dependent on local supercoiling at a lone origin10, oriC, that is regulated by the opposing activities of topo I and gyrase 78,79. During DNA replication initiation in eukaryotes, topoisomerases have been seen to associate directly with certain origins to aid in activation. For example, human topo IB and topo IIα co-localize with both the lamin B2 origin (an early firing, widely studied replication start site found on human chromosome 19) and the Orc2 subunit of the origin recognition complex11, while inhibition of topo IB interferes directly with origin firing 80 (Table 1). Formation of replication initiation complexes at the human Ors8 origin, (a late firing origin), as well as the lamin B2 origin, also have been reported to require DNA cleavage by topo IIβ – a topoisomerase associated with repair and development 81–83 – and the association of DNA repair proteins such as Ku70/80 and poly(adenosine diphosphate-ribose) polymerase-1 (PARP1) 84. Why topoisomerases are needed during these early stages of eukaryotic replication is not well understood, but they may be involved in the control of torsional stress or DNA deformations that could be used either to help clear DNA regions for replisome assembly, or to promote DNA unwinding in the early stages of replisome assembly.
Topoisomerases in replication fork progression
During strand synthesis, topoisomerases are required to relieve positive supercoiling that arises from DNA unwinding by replicative helicases (Fig 3A). In bacteria, this role is typically fulfilled by DNA gyrase and/or topo IV 37,85–87. By contrast, eukaryotes rely primarily on topo IB for positive supercoil relaxation, although topo II can assist or even substitute for topo IB in this capacity 88–91. Some type II topoisomerases (for example, topo IV and human topo IIα – see Box 1) are markedly more adept at removing positive supercoils than negative ones 38–40, suggesting that cells may be under evolutionary pressure to acquire particularly robust relaxases that can accommodate rapidly moving replication forks.
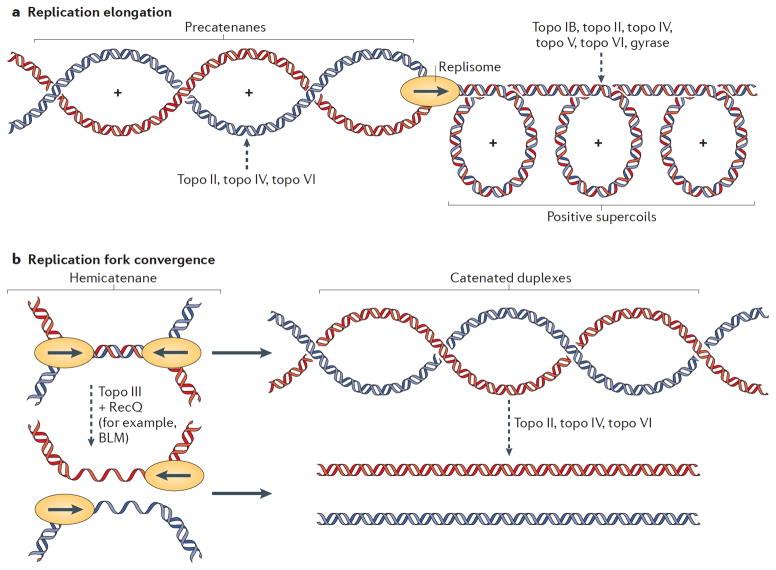
Fig 3. Topoisomerase functions during DNA replication.
Schematic of topological problems that arise during DNA replication. The names of the topoisomerases that resolve these superstructures are listed. Topoisomerase action is indicated by a dashed arrow. (A) Replication elongation. As a replisome progresses, positive supercoils form ahead of the fork, and newly-replicated precatenanes behind it. If unresolved, precatenanes can give rise to tangled or catenated DNAs that can lead to abnormal DNA segregation upon entry into cell division. Unresolved positive supercoils can impede fork progression and terminate DNA replication prematurely. (B) Replication termination. Hemicatenanes are formed as two forks converge, and these must be resolved before chromosome segregation can occur. The unreplicated mother duplex can be resolved by topo III, together with a RecQ-family helicase (for example, the BLM helicase in eukaryotes), after which the single-stranded gaps are filled in (1). Alternatively, the unreplicated mother duplex can be replicated to form duplex linkages, which are then removed by a type II topoisomerase (2).
A second consequence of a progressing fork is the formation of daughter duplex intertwinings (termed precatenanes12 in circular chromosomes) behind it (Fig. 3A). Such structures are produced as a natural byproduct of replicating the double helix of the mother chromosome and, if left unchecked, give rise to tangled or catenated DNAs that can lead to abnormal DNA segregation upon entry into cell division 92–96. The duplex strand-passage activity of type II topoisomerases plays a key role in resolving these topological linkages.
As two forks near one another, the unreplicated region between them represents a topological linkage, known as a hemicatenane13, which must be resolved before chromosome segregation can occur (Fig. 3B). If the two strands of the hemicatenane can be fully replicated prior to resolution, the resulting duplex DNA segments become a natural substrate for type II topoisomerases. Consistent with a need for these enzymes in resolving inter-chromosome crossovers, the removal or inactivation of topo II or topo IV can lead to hyper-catenated DNAs and the production of broken chromosomes during cell division 96–101.
However, there also is mounting evidence that type IA topoisomerases, particularly topo III, can participate directly in hemicatenane resolution before forks converge. For example, in bacteria, topo IV temperature-sensitive alleles can be rescued by overexpression of topo III 102. Topo III, which decatenates single-stranded DNA in vitro, has been shown to collaborate with RecQ-family helicases in disentangling hemicatenated structures (Table 1); in this partnership, the helicase may help generate single-stranded DNA to aid topo III function. The connection between these two enzyme classes is sufficiently entwined that they frequently form stable or co-localized complexes, along with auxiliary single-stranded DNA binding proteins such as SSB (in bacteria), or replication protein A (RPA)-like factors (Rmi1 in eukaryotes and Rmi2 in metazoans) 9,103–105. Consistent with the collaboration between these proteins as part of a functional “resolvosome”, ablation of the RecQ-partner of topo III in eukaryotes, the BLM helicase, leads to the enhanced formation of ultrafine DNA tangles, termed “microbridges,” between condensed chromosomes 106. The natural fusion of a RecQ-like (SF-2) helicase domain with a type IA topoisomerase module in reverse gyrase – which likewise can resolve hemicatenanes 13,107,108 – further highlights the frequent need for paired, RecQ-family helicase–topoisomerase activities in the cell.
Topoisomerases in replication termination
With respect to replication termination, topoisomerases have been found to play a role in at least two instances. In bacteria, the completion of DNA synthesis can be assisted by chromosomally-encoded termination regions (Ters), which bind dedicated factors that help arrest replicative helicases. In E. coli, the absence of topo IA diminishes the ability of the bacterium’s cognate termination factor (Tus) to block progression of the replicative DnaB helicase 109. This effect may arise as a consequence of the increased levels of negative supercoiling left by topo IA’s absence, which can stimulate DNA unwinding by the replicative helicase, DnaB, and may also serve to reduce the duration of the Tus–DnaB interaction. In yeast, topo II appears to facilitate fork progression at termination elements by localizing to such sites and working with the Rrm3 helicase to resolve the torsional stress arising from converging forks 110. This controlled pausing at termination elements ensures complete replication and counteracts abnormal genome rearrangements and breaks arising from fork convergence at replication termination (Table 1). Whether there exist other, more specific connections between topoisomerases and replication termination remains to be determined.
Topoisomerases in chromosome segregation
Once replication is complete, daughter chromosomes must be pulled apart and separated from each other. Topoisomerases promote these events by facilitating DNA compaction and disentangling (as described above) and by working directly with chromosome partitioning machineries and/or cytoskeletal elements. For instance, many bacteria use a dedicated motor protein, FtsK or SpoIIIE, to move newly replicated chromosomes into different cellular locales to aid segregation 111. In E. coli, topo IV physically associates with, and is stimulated by, FtsK, possibly as a means to both counteract the supercoils created by the translocating motor and to help unlink tangled DNA regions 112,113 (Table 1). A physical association between E. coli topo IV and the actin-like protein MreB also has been observed, and this pairing stimulates DNA decatenation as a possible additional force promoting chromosome segregation 114. Although no eukaryotic topoisomerase has been reported to bind to cytoskeletal elements, topo II does associate with factors responsible for centromere formation and chromosome segregation, including the aurora B kinase and the polo-like kinase (Plk)-1 115,116 (Table 1). The biological rationale for these interactions is not fully understood; however, the colocalization of topo II with aurora B and Plk-1 may be important for centromere resolution between sister chromatids. Additional links between topoisomerases and chromosome segregation appear likely to exist, but have yet to be discovered.
What attracts a given topoisomerase to a particular point in the replicative process? As the aforementioned examples highlight, this control can be achieved not only by the types of DNA intermediates that derive from a particular process (for example, hemicatenanes, pre-catenanes and supercoils), but also by topoisomerase-associated partner proteins. Other intriguing interactions have been implicated in topoisomerase function during replication. For example, topo IV can associate with both the polymerase processivity clamp-loader assembly, which may localize the enzyme to oriC to decatenate newly synthesized chromosomes 117, and the SeqA factor, a protein that prevents replication reinitiation by sequestering hemimethylated DNA within oriC, and that stimulates topo IV decatenation and supercoil relaxation activity 118 (Table 1). These connections further highlight cellular needs for particular topoisomerase activities during elongation and initiation, respectively.
Transcription and gene regulation
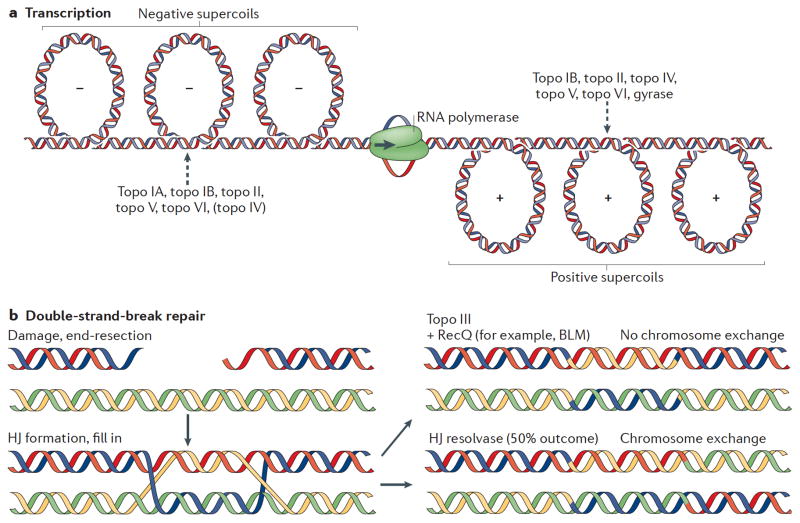
As with replication, transcription induces changes in DNA topology. A moving RNA polymerase produces localized positive supercoiling ahead of the transcription bubble and negative supercoiling in its wake 119,120 (Fig. 4A). Left unchecked, these supercoiling alterations can lead to significant changes in gene expression 121. Underwound DNA also has a higher propensity to form stable RNA–DNA hybrids (R-loops) than relaxed duplexes 122. Such structures can block cell growth and may contribute to genomic instability by stalling replication forks and promoting DNA breaks at highly transcribed genes 123,124.
Fig 4. Topoisomerase functions during transcription and DNA repair.
(A) Schematic of topological problems that arise during transcription. As RNA polymerase progresses, positive and negative supercoils form ahead and behind it, respectively. The names of the topoisomerases that act on these superstructures are listed; topoisomerase action is indicated by a dashed arrow. Topo IV can remove negative supercoils; however, it is listed in brackets because it is not the primary enzyme used by cells to resolve these superstructures. (B) Double-strand break repair through homologous recombination. The repair of broken DNA ends can proceed through a pathway that leads to the formation of a double-Holliday junction (HJ). Topo III, together with a RecQ-type helicase (such as BLM in eukaryotes) can resolve these junctions, generating disentangled chromosomes that have no crossovers between DNA ends (1). By contrast, the resolution of Holliday junctions by branch-endonucleases have a 50% chance of giving rise to repair chromosomes, the arms of which are now swapped (2).
Both type IA and type IB topoisomerases have been implicated in the removal of negative supercoils and the resulting suppression of R-loop formation 123,125. Indeed, the E. coli topo IA C-terminus interacts directly with RNA polymerase, and this interaction may help recruit the topoisomerase to negative supercoils 55,126. Type IB and type II topoisomerases further help relax positive supercoils in front of transcribing polymerases, with topo IB playing a more significant role in eukaryotic transcription and topo IV and gyrase handling these substrates in bacteria 37,55,127–129.
Topoisomerases have been linked to specific transcriptionally-related events, such as the activation or repression of particular promoters or nucleosome remodeling. For example, inactivation of topo IB in S. cerevisiae leads to histone-specific acetylation and methylation events that increase the transcription of telomere-proximal genes 130. In S. pombe, there is evidence that the presence or absence of topo IB activity can influence nucleosome disassembly and assembly, respectively, at certain promoter regions 129. Eukaryotic topo IB has been implicated in the control of gene expression through an associated kinase activity, which is reported to phosphorylate splicing factors such as SR (serine/arginine-rich) proteins131, regulating splicing factor localization and enhancing their activity132,133; topo IB-mediated phosphorylation of SR proteins in turn can be negatively regulated by the presence of poly(ADP-ribose) 133 (Table 1 and Supplementary information S1 (table)).
Remarkably, the transient, site-specific cleavage of DNA by human topo IIβ has been reported to be required to activate the transcription of genes regulated by certain nuclear receptors, a process that necessitates the activity of PARP14 and Ku80/Ku70, as well as DNA repair proteins such as DNA-PK (DNA-dependent protein kinase) 81,83. DNA cleavage and the subsequent recruitment of PARP and associated repair proteins appears necessary to exchange histone-1 for the high mobility group (HMG)B-1/2 factor, and to stimulate transcription 83. Similarly, topo IIβ associates with the promoter region of genes in human acute promyelocytic leukemia cell lines that are regulated by retinoic acid receptor elements, initially stimulating transcription; sustained transcriptional activation then leads to decreased transcript levels and the inhibition of granulocytic differentiation in a topo IIβ-dependent manner 134.
Some of these findings, such as those suggesting that topo IB might directly phosphorylate target proteins, or that cells might rely on a potentially mutagenic topo IIβ-meditated DNA-cleavage event to control gene expression, raise as many questions as they answer. Nonetheless, even though it can be difficult to definitively determine whether the effects of topoisomerases on promoter function and structure are direct or indirect, it seems that these enzymes can influence transcriptional events in a manner that may not depend solely on their ability to control supercoiling. This complex area of investigation is likely to yield many more surprises.
Recombination and repair
Yet another area where topoisomerases play a critical role is in forming and managing double-stranded DNA (dsDNA) breaks. As noted earlier, DNA cleavage by at least one topoisomerase, topo IIβ, has been implicated in the control of promoter activity 81,83,134. However, double-stranded DNA-break formation through a variety of topoisomerases and topoisomerase-like proteins also impacts processes such as meiotic recombination, cell cycle checkpoint activation, and DNA repair. For instance, during meiotic recombination, the type IIB topoisomerase A-subunit homolog, Spo11, creates double-strand breaks that allow chromosomes to exchange segments through homologous recombination 46,52,135. Although there is no evidence presently linking Spo11 to a bona fide topoisomerase activity (in the sense that it would be capable of DNA strand passage) 136, its DNA-cleavage activity during meiosis is clearly controlled, as only a fraction of the total Spo11 pool bound to chromosomal loci is used to form DNA breaks and recombinogenic sites 137. Eukaryotes (with the exception of plants) do not appear to possess any clear homologs of the ATP-binding B-subunit of topo VI, and how Spo11 is activated is unknown at present 136.
The creation of nicks and double-strand DNA breaks are potentially deleterious events for the cell. Hence, DNA cleavage by topoisomerases or topoisomerase-like factors has the capacity to elicit DNA damage responses. For example, following DNA-breakage by Spo11, the protein remains covalently attached to the DNA and must be removed 137. The removal of Spo11 and downstream DNA-end processing events appear to be carried out by double-strand break repair proteins such as the Rad50–Mre11–Nbs1 (MRN) complex 136,137, Rad51 and Dmc1-like factors 138,139, and the nuclease Sae2/CtIP 140. Meiotic recombination elicits a p53 damage response, which appears to be independent of the ATM and ATR (ataxia telangiectasia mutated and ATM Rad3-related) kinases and their associated repair pathways 141. Similar responses are seen when topoisomerases become inactivated 96, or are aberrantly linked to DNA through the action of DNA lesions or small-molecule inhibitors that promote the formation of topoisomerase-DNA cleavage complexes. However, additional systems also can be activated in these instances (for example, the ATM and ATR pathways, and BRCA1 or BRCA2 proteins 142,143) that induce cell cycle arrest and DNA repair. When topoisomerases become inadvertently trapped in a covalent complex with DNA, they further can be targeted for destruction by SUMOylation- and ubiquitinylation-mediated mechanisms, and the covalent tyrosine-DNA adducts can be repaired by enzymes such as Tdp1 and Tdp2 143–152 (Supplementary information S1 (table)). Some of these latter events may be promoted by collision encounters with proteins such as RNA polymerase153–155.
Other evidence also exists linking topoisomerase activity to mutagenesis. For instance, highly transcribed genes in eukaryotes are sometimes associated with enhanced levels of genetic instability and spontaneous mutation 156. Several recent studies have found that 2–3 nucleotide deletions commonly observed in these regions are the result of topo IB activity 157,158. The mechanism behind these alterations has not been fully elucidated; topo IB may generate lesions by binding and cleaving within and adjacent to tandem dinucleotide repeats, forming a stable covalent DNA link that is then processed in part by endonucleases such as Mus81 and Rad1 157,158. Alternatively, topo IB may cleave at the scissile phosphate 15 of a misincorporated ribonucleotide within or adjacent to the tandem dinucleotide repeats. The misincorporated ribonucleotide would then undergo an internal 2′-3′ cyclization event to release the enzyme and leave a small nick or gap in the DNA 159, giving rise to microdeletions from DNA misalignments during repair. In this regard, it is interesting to note that type II and type IB topoisomerases form stable adducts at sites of ribonucleotide incorporation in DNA 160, as well as at DNA lesions such as nicks 161 and abasic sites 162,163,164. Thus, both classes of enzymes may contribute to a steady-state background of damage events with mutagenic potential.
Topoisomerases further play an important role during and after the repair of damaged DNA strands. This is particularly true during homologous recombination (HR), which generates interlinked intermediates that bear some similarities to those occurring during replication fork convergence (Fig. 4B). Recent work has found ample evidence for the participation of topo III in HR where, as with hemicatenane resolution during replication, it breaks up double-Holliday junctions in concert with both a RecQ-type helicase (Sgs1 in yeast 165, BLM in metazoans 166,167) and single-stranded DNA binding proteins such as SSB168, Rmi1 104 and Rmi2 105,169 (Table 1). Notably, the cooperative activity of this resolvosome allows HR to proceed without the generation of crossover products that exchange large chromosomal segments, which occurs when branch-endonucleases resolve double-Holliday junctions, and may therefore constitute the dominant means for resolving Holliday junctions in mitotic cells 104,166,167. Topo III also may help recruit other HR proteins to broken DNA ends. For example, Sgs1, Dna2, and RPA derived from budding yeast can resect DSBs in a topo III-independent manner in vitro; however, topo III stimulates Sgs1 helicase activity and is probably required at physiological concentrations to recruit Sgs1 to broken DNA ends 170,171. How disparate topoisomerase, helicase, and SSB protein activities collaborate to disentangle hemicatenated substrates is not known; studies of reverse gyrase may provide some mechanistic insights into this problem, since reverse gyrase is composed of an Sgs1-like (SF-2) helicase domain fused to a type IA topoisomerase element 107,108.
PTM of eukaryotic topoisomerases
Eukaryotic topoisomerases are subject to a number of post-translational modifications (PTMs) that can alter their localization and activities at various stages in the cell cycle. These modifications may also influence the types of DNA structures on which topoisomerases preferentially act. Thus far, four types of topoisomerase PTM have been observed: phosphorylation 172, acetylation 173, SUMOylation 145,151, and ubiquitinylation146.
The modification of poisoned topoisomerases trapped on DNA by SUMOylation and ubiquitinylation, in response to DNA damage, appears to target these enzymes for processing and/or degradation 145,146,151–155. However, apart from this function, the physiological role of topoisomerase PTMs has been somewhat controversial. Indeed, it is not uncommon to find studies that link the same type of modification to the regulation of different protein–protein partnering events, the activation or repression of specific topoisomerase activities, and/or proper and improper topoisomerase localization, sometimes in a mutually-exclusive manner. An example of this complexity is the role of SUMOylation in the temporal activation and localization of topo IIα through two E3 ligases16, RanBP2 and protein-inhibitor-of-activated-STAT-1 γ (PIASγ). In Xenopus laevis, PIASγ has been linked to the SUMOylation of topo IIα, a modification that appears to reduce topo IIα’s ability to decatenate DNA in vitro 174. By contrast, PIASγ is reported to have no effect on topo IIα in mouse embryonic fibroblasts; instead, these cells appear to SUMOylate topo IIα through RanBP2, which enhances topo IIα localization and activation at centromeres during sister chromosome resolution in anaphase 175,176. The role of phosphorylation has been similarly difficult to tease apart, with studies variously reporting that this modification can potentiate, depress, or exert no effect on topoisomerase function 116,177–179. Nonetheless, some of these differences are almost certainly due to the particular site on the topoisomerase that is being modified. The regulation of eukaryotic topoisomerases through PTMs is a highly-interesting, but still relatively murky, area of research that undoubtedly holds significant surprises.
Topoisomerase inhibition
Although indispensible for cell viability, it is clear that topoisomerases also present a fundamental peril: they can be inadvertently inactivated, or their DNA cleavage activity can be corrupted, resulting in cytotoxic or mutagenic DNA breaks, which leads to cell death. The evolution of general repair systems, as well as more specific ones such as the tyrosyl-DNA phosphodiesterases (Tdps) 147–150, help cells to deal with these problems. However, nature and humans also have been able to exploit topoisomerase inactivity and stalling through diverse means to deliberately kill cells, at times for tangible therapeutic benefit.
A number of proteins and protein-like factors have been found that specifically inhibit topoisomerase activity, particularly in bacteria (Table 1 and Supplementary information S1 (table)). These proteins range from toxins that control plasmid stability (CcdB) 180,181 to agents that are involved in microbial competition (microcin B17) 182 and antibiotic resistance (Qnr and MfpA) 183. Such factors have varied modes of action that include attenuating the DNA binding activity of topoisomerases, or promoting their ability to cleave DNA. In considering protein-based anti-topoisomerase factors, it is interesting to note that their identification has come not through directed screens, but from basic research and serendipity. The continual discovery of new proteinaceous inhibitors points to the utility – and potentially relative ease – by which topoisomerase activity can be subverted, and strongly suggests that more anti-topoisomerase systems remain to be discovered.
On the biomedical side, topoisomerase inhibition has proven to be a highly versatile and useful approach for therapeutic intervention. Small-molecule agents targeting topoisomerases fall into two broad classes based on their mode of action: “inhibitors,” which attenuate enzyme activity, and “poisons”, which stabilize DNA-cleavage complexes (see Fig. 5 for a partial list of type II topoisomerase antagonists and their modes of action). Although many of the most significant therapeutics (for example, epipodophyllotoxins, quinolones, and camptothecins) have been used for decades, only relatively recently have their mechanisms of action been understood in molecular detail. For instance, camptothecin, and its derivatives such as Topotecan, poison topo IB by intercalating between cleaved DNA ends in the enzyme active site 184, impeding DNA religation and the relaxation of supercoils. Fluoroquinolones, which are used primarily as antibiotics that target gyrase and topo IV, as well as topo II poisons such as etoposide, act in a similar manner, binding between the 5′ and 3′ end of a broken DNA strand to prevent resealing and release 42. Other compounds that act in a very different manner, blocking either ATPase activity (for example, novo- and cholorobiocins 185,186, bisdioxopiperazines 187, and radicicol 188), DNA binding (simocyclinone D8) 189, or DNA cleavage (NTBI GSK299423)190. These compounds have been imaged in the presence of their respective targets, yielding insights into their function. Together, these findings have not only highlighted the molecular determinants that enable drug binding and explained their inhibitory effects, but further demonstrated that there is a rich abundance of molecular scaffolds capable of inhibiting topoisomerases (Supplementary information S2 (figure)). At present, no small-molecule agent is known to target type IA topoisomerases, although biochemical studies have suggested that such compounds likely would be cytotoxic 191. A hopeful, but presently unrealized, goal for these combined efforts is to point the way toward design of new inhibitors that show improved patient tolerance and that can circumvent emerging problems with drug-resistance.
Fig 5. Inhibition or poisoning points for type II topoisomerases.
Detailed schematic of the type II topoisomerase reaction cycle, showing all of the points where exogenous agents can disrupt function. During its reaction cycle, the topoisomerase initially binds one duplex DNA segment. Following binding, the topoisomerase can then associate with a second duplex DNA segment. ATP binding stimulates cleavage and opening of the first DNA, and passage of the second through the opening. Agents have been identified that interfere with: (1) DNA binding, (2) ATP binding, (3) DNA cleavage, (4) strand passage, (5) religation, and (6) product release. Only a partial list of all inhibitors and poisons is shown.
How cells respond to topoisomerase poisons has been difficult to tease out. In bacteria, it was discovered relatively recently that fluoroquinolones help promote cell death by inducing the formation of reactive oxygen species (ROS) that further damage DNA 192,193. Interestingly, however, some fluoroquinolones (moxifloxacin and PD161144) also kill cells even when the ROS cascade is blocked (or when oxygen is absent), suggesting that there exists a second pathway for killing by some anti-topoisomerase agents 194. In eukaryotes, treatment of cells with poisons leads to the ubiquitinylation and SUMOylation of both topo I and topo II 145,146,151–155. These modifications are necessary in part for initiating the repair responses described earlier. However, the need for active repair mechanisms also sensitizes cells to agents that block such pathways. In this regard, inhibitors of PARP have shown some promise when administered with topoisomerase poisons in promoting tumor cell killing. Initially, the PARP-1 inhibitor NU1025 was reported to potentiate damage mediated by the topo I inhibitor camptothecin in mouse lymphocytic cells, but not to promote further damage in conjunction with the topo II-targeting agent, etoposide 195. However, more recent work has shown that mouse fibroblasts lacking PARP-1 are more sensitive to the topo II inhibitor C-1305 196, and that the PARP inhibitor ANI enhances the activity of the topo II inhibitor doxorubicin against liver cells 197. Fully mapping the pathways and mechanisms by which cells contend with anti-topoisomerase agents, and looking for synergy between such compounds and other drugs, promises to be a rich vein of research, with powerful implications for the treatment of bacterial infections and cancer.
Concluding Remarks
Although first discovered over 40 years ago, the study of topoisomerases remains highly vigorous. Our knowledge of the role of cellular topoisomerases and their regulation by external factors continues to expand, but many pressing questions remain. For instance, there remain many significant gaps in describing the molecular operating principles that define where, when, and how a particular topoisomerase will act in the cell. The role of higher-order topoisomerase complexes (many of which are transient), the control of topoisomerases by post-translational modifications, and the mechanisms by which topoisomerases are directly involved in processes such as gene expression or the formation of DNA damage, similarly are poorly understood. At the same time, while studies are at long last explaining how certain classes of anti-topoisomerase agents function – particularly for small-molecule compounds – many of these insights have yet to be translated into the development of more efficacious drugs. Moreover, the available evidence suggests novel inhibitors and poisons remain to be identified, indicating that the therapeutic potential of topoisomerases is far from exhausted. Future cellular, biochemical, and structural efforts in this area will no doubt continue to offer up new surprises and insights into the action and use of these essential and complex molecular machines.
Supplementary Material
Supplementary information S1 (table): A partial list of the topoisomerase interactome
Acknowledgments
The authors would like to thank Karl Drlica for thoughtful discussions and editing. This work was supported by an NSF pre-doctoral fellowship (to SMV) and the NCI (CA077373, to JMB).
Footnotes
Glossary term: Paralogs. Homologous genes separated by a gene duplication event, which can evolve new functions.
Glossary term: Domains of life. Refers to the three major evolutionary branches – bacteria, archaea, and eukaryotes – of modern day cellular lineages.
Glossary Term: Negatively-supercoiled DNA. DNA becomes negatively supercoiled when undertwisted (that is, when wound to < 10.5 bp/turn). This destabilizes DNA, allowing complementary strands to be more easily denatured. In contrast, DNA becomes positively supercoiled and more stabilized when over-twisted (that is, wound to > 10.5 bp/turn).
Glossary Term: Superfamily-2 (SF-2) helicase. A diverse group of ATP-dependent nucleic-acid helicases and translocases that share a conserved domain architecture. Many members of this family can locally melt or unwind short duplex regions.
Glossary term: Abasic lesions. A form of DNA damage in which the nucleobase has been excised from the sugar backbone.
Glossary Term: Catenanes. Topologically interlinked duplex DNA rings.
Glossary term: Plectonemic supercoiling. The natural tendency of supercoiled DNA is to wrap back on itself, forming intra-molecularly wound structures known as plectonemes (Figure 1A). Because DNA is itself a chiral molecule, negatively- and positively-supercoiled states cause the DNA to coil in opposite directions (in the form of right- and left-handed supertwists, respectively).
Glossary Term: Superhelical density. A measurement of the over- or under-twistedness of DNA, which is generally expressed as the ratio to which the twist of the supercoiled state differs from that of the relaxed state.
Glossary Term: ATP Binding Cassette (ABC) ATPases. A family of proteins that include membrane bound transporters, repair proteins, and SMCs. ABC ATPases possess a conserved ATP binding domain that is often formed at dimer interfaces. Binding of ATP results in conformational changes, often dimerization, that affect associated binding partners and substrates.
Glossary Term: Origin. A chromosomal site for replisome assembly.
Glossary Term: Origin recognition complex. A multi-subunit protein complex that localizes to specific DNA origins to initiate DNA replication in eukaryotes.
Glossary term: Precatenanes. Entangled daughter duplexes formed behind a replication fork during strand synthesis.
Glossary Term: Hemicatenane. A junction between two double-stranded DNA molecules where one strand of one DNA forms a duplex with a complementary strand on the other duplex.
Glossary Term: PARP. Poly (ADP ribose) polymerase, an enzyme that adds chains of ADP to proteins as part of DNA damage and cell death responses.
Glossary Term : Scissile phosphate. The phosphate at which a nucleic acid backbone is broken by nucleophilic attack.
Glossary term: E3 ligase. An enzyme that attaches ubiquitin or ubiquitin like proteins to a target protein.
Glossary Term: Supercoil. The result of duplex DNA twisting upon itself in 3-dimensional space.
The authors declare no competing financial interests.
References
- 1.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. A review that extensively covers the evolution of topoisomerases. [DOI] [PubMed] [Google Scholar]
- 2.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. A review that extensively covers the biochemical and structural nature of topoisomerases. [DOI] [PubMed] [Google Scholar]
- 3.Lima CD, Wang JC, Mondragón A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. First crystal structure of a type IA topoisomerase. [DOI] [PubMed] [Google Scholar]
- 4.Tse YC, Kirkegaard K, Wang JC. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J Biol Chem. 1980;255:5560–5565. [PubMed] [Google Scholar]
- 5.Brown PO, Cozzarelli NR. Catenation and knotting of duplex DNA by type 1 topoisomerases: a mechanistic parallel with type 2 topoisomerases. Proc Natl Acad Sci USA. 1981;78:843–847. doi: 10.1073/pnas.78.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JC. Interaction between DNA and an Escherichia coli protein omega. Journal of Molecular Biology. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. The first report of a purified topoisomerase activity. [DOI] [PubMed] [Google Scholar]
- 7.Hiasa H, DiGate RJ, Marians KJ. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J Biol Chem. 1994;269:2093–2099. [PubMed] [Google Scholar]
- 8.Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. Discovery of eukaryotic topo III and its possible role in homologous recombination. [DOI] [PubMed] [Google Scholar]
- 9.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. The first paper to link topo III and RecQ function. [DOI] [PubMed] [Google Scholar]
- 10.DiGate RJ, Marians KJ. Identification of a potent decatenating enzyme from Escherichia coli. J Biol Chem. 1988;263:13366–13373. This is the first paper to identify a decatenase activity for Topo III. [PubMed] [Google Scholar]
- 11.Lopez CR, et al. A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol Microbiol. 2005;58:80–101. doi: 10.1111/j.1365-2958.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi A, Asai K. Reverse gyrase-a topoisomerase which introduces positive superhelical turns into DNA. Nature. 1984;309:677–681. doi: 10.1038/309677a0. Identified and charachterized reverse gyrase, a novel type IA topoisomerase that is found in thermophiles. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh T, Plank JL. Reverse gyrase functions as a DNA renaturase. Journal of Biological Chemistry. 2006;281:5640. doi: 10.1074/jbc.M513252200. [DOI] [PubMed] [Google Scholar]
- 14.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. Reports first eukaryotic type IB topoiosmerase structure. This enzyme was crystallized with and without DNA. [DOI] [PubMed] [Google Scholar]
- 15.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. Paper describing the detailed mechanism of topo IB through elegant single molecule experiments. [DOI] [PubMed] [Google Scholar]
- 16.Champoux JJ, Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay) Proc Natl Acad Sci USA. 1972;69:143–146. doi: 10.1073/pnas.69.1.143. The first discovery of topo IB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madden KR, Stewart L, Champoux JJ. Preferential binding of human topoisomerase I to superhelical DNA. EMBO J. 1995;14:5399–5409. doi: 10.1002/j.1460-2075.1995.tb00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frohlich RF, et al. Tryptophan-205 of human topoisomerase I is essential for camptothecin inhibition of negative but not positive supercoil removal. Nucleic Acids Res. 2007;35:6170–6180. doi: 10.1093/nar/gkm669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel A, Yakovleva L, Shuman S, Mondragón A. Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site. Structure. 2010;18:725–733. doi: 10.1016/j.str.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. Using structural information, the authors propose a detailed mechanism for type IB topoisomerase activity. [DOI] [PubMed] [Google Scholar]
- 21.Cheng C, Kussie P, Pavletich N, Shuman S. Conservation of Structure and Mechanism between Eukaryotic Topoisomerase I and Site-Specific Recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 22.Krogh BO, Shuman S. A poxvirus-like type IB topoisomerase family in bacteria. Proc Natl Acad Sci USA. 2002;99:1853–1858. doi: 10.1073/pnas.032613199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forterre P. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Taneja B, Schnurr B, Slesarev A, Marko JF, Mondragón A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc Natl Acad Sci USA. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slesarev AI, et al. DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature. 1993;364:735–737. doi: 10.1038/364735a0. First discovery of topo IC. [DOI] [PubMed] [Google Scholar]
- 26.Taneja B, Patel A, Slesarev A, Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belova GI, et al. A type IB topoisomerase with DNA repair activities. Proc Natl Acad Sci USA. 2001;98:6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aravind L, Leipe DD, Koonin EV. Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. This paper showed that many nucleotidyl transferases, topoisomerases, and nucleases share a catalytic domain known as the Toprim domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger JM, Fass D, Wang JC, Harrison SC. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc Natl Acad Sci U S A. 1998;95:7876–7881. doi: 10.1073/pnas.95.14.7876. This paper identifies similarities between type IA and type II topoisomerase DNA interaction regions, which suggest the enzymes share a common DNA cleavage mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu LF, Liu CC, Alberts BM. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980;19:697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- 31.Brown PO, Cozzarelli NR. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979;206:1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 32.Mizuuchi K, Fisher LM, O’Dea MH, Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci USA. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. The first discovery of a type II topoisomerase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goto T, Wang JC. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J Biol Chem. 1982;257:5866–5872. [PubMed] [Google Scholar]
- 35.Hsieh T, Brutlag D. ATP-dependent DNA topoisonmerase from D. melanogaster reversibly catenates duplex DNA rings. Cell. 1980;21:115–125. doi: 10.1016/0092-8674(80)90119-1. [DOI] [PubMed] [Google Scholar]
- 36.Peng H, Marians KJ. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc Natl Acad Sci USA. 1993;90:8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zechiedrich E, Cozzarelli N. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes & Development. 1995;9:2859. doi: 10.1101/gad.9.22.2859. Defines distinct roles of topo IV and gyrase during bacterial DNA replication. [DOI] [PubMed] [Google Scholar]
- 38.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase II alpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J Biol Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 40.Baxter J, et al. Positive Supercoiling of Mitotic DNA Drives Decatenation by Topoisomerase II in Eukaryotes. Science. 2011;331:1328–1332. doi: 10.1126/science.1201538. This paper suggests that positive supercoiling may drive topoisomerase II decatenation activity. [DOI] [PubMed] [Google Scholar]
- 41.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. First DNA bound type IIA topoisomerase structure shows bent DNA conformation confirming previous models. [DOI] [PubMed] [Google Scholar]
- 42.Laponogov I, et al. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010;5:e11338. doi: 10.1371/journal.pone.0011338. This paper reported the structure of a ternary topo:DNA:quinolone complex, showing that the quinolone intercalates between the +1/−1 bases at each of two active sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vologodskii AV, et al. Mechanism of topology simplification by type II DNA topoisomerases. Proc Natl Acad Sci USA. 2001;98:3045. doi: 10.1073/pnas.061029098. This paper proposed a model for DNA bending by type II topoisomerases to account for topology simplification below the expected thermodynamic equilibrium value. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charvin G, Bensimon D, Croquette V. Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9820–9825. doi: 10.1073/pnas.1631550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roca J, Wang JC. The probabilities of supercoil removal and decatenation by yeast DNA topoisomerase II. Genes to Cells. 1996;1:17–27. doi: 10.1046/j.1365-2443.1996.01001.x. [DOI] [PubMed] [Google Scholar]
- 46.Bergerat A, et al. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. One of two papers linking a topo II-like protein, Spo11, to the formation of double stranded DNA breaks during meiosis. [DOI] [PubMed] [Google Scholar]
- 47.Malik SB, Ramesh MA, Hulstrand AM, Logsdon JM., Jr Protist homologs of the meiotic Spo11 gene and topoisomerase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol Biol Evol. 2007;24:2827–2841. doi: 10.1093/molbev/msm217. [DOI] [PubMed] [Google Scholar]
- 48.Bergerat A, Gadelle D, Forterre P. Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J Biol Chem. 1994;269:27663–27669. Reports the discovery of the first type IIB topoisomerase. [PubMed] [Google Scholar]
- 49.Corbett KD, Berger JM. Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J. 2003;22:151–163. doi: 10.1093/emboj/cdg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat Struct Mol Biol. 2007;14:611–619. doi: 10.1038/nsmb1264. This paper was the first to describe a full-length type IIB topoisomerase structure and utilized SAXS to image enzyme nucleotide-mediated conformational changes. [DOI] [PubMed] [Google Scholar]
- 52.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. One of two papers linking a topo II-like protein, Spo11, to the formation of double stranded DNA breaks during meiosis. [DOI] [PubMed] [Google Scholar]
- 53.Luijsterburg MS, White MF, van Driel R, Dame RT. The Major Architects of Chromatin: Architectural Proteins in Bacteria, Archaea and Eukaryotes. Critical Reviews in Biochemistry and Molecular Biology. 2008;43:393–418. doi: 10.1080/10409230802528488. [DOI] [PubMed] [Google Scholar]
- 54.Boles TC, White JH, Cozzarelli NR. Structure of plectonemically supercoiled DNA. J Mol Biol. 1990;213:931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- 55.Zechiedrich EL, et al. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 56.McClelland M, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 57.Champion K, Higgins NP. Growth rate toxicity phenotypes and homeostatic supercoil control differentiate Escherichia coli from Salmonella enterica serovar Typhimurium. The Journal of Bacteriology. 2007;189:5839–5849. doi: 10.1128/JB.00083-07. This paper shows that distinct differences in overall supercoiling densities between two closely related species is directly linked to small differences in gyrase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tretter EM, Lerman JC, Berger JM. A naturally chimeric type IIA topoisomerase in Aquifex aeolicus highlights an evolutionary path for the emergence of functional paralogs. Proc Natl Acad Sci U S A. 2010;107:22055–22059. doi: 10.1073/pnas.1012938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brochier-Armanet C, Forterre P. Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers. Archaea. 2007;2:83–93. doi: 10.1155/2006/582916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forterre P, Mirambeau G, Jaxel C, Nadal M, Duguet M. High positive supercoiling in vitro catalyzed by an ATP and polyethylene glycol-stimulated topoisomerase from Sulfolobus acidocaldarius. The EMBO Journal. 1985;4:2123. doi: 10.1002/j.1460-2075.1985.tb03902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perugino G, Valenti A, D’Amaro A, Rossi M, Ciaramella M. Reverse gyrase and genome stability in hyperthermophilic organisms. Biochem Soc Trans. 2009;37:69–73. doi: 10.1042/BST0370069. [DOI] [PubMed] [Google Scholar]
- 62.Charbonnier F, Forterre P. Comparison of plasmid DNA topology among mesophilic and thermophilic eubacteria and archaebacteria. J Bacteriol. 1994;176:1251–1259. doi: 10.1128/jb.176.5.1251-1259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tadesse S, Mascarenhas J, Koesters B, Hasilik A, Graumann PL. Genetic interaction of the SMC complex with topoisomerase IV in Bacillus subtilis. Microbiology. 2005;151:1–9. doi: 10.1099/mic.0.28234-0. [DOI] [PubMed] [Google Scholar]
- 64.Hayama R, Marians KJ. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:18826–18831. doi: 10.1073/pnas.1008140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, et al. Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc Natl Acad Sci USA. 2010;107:18832–18837. doi: 10.1073/pnas.1008678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- 67.Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Dev Cell. 2003;4:467–480. doi: 10.1016/s1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 68.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 69.Bhalla N, Biggins S, Murray AW. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Molecular Biology of the Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartung F, et al. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr Biol. 2002;12:1787–1791. doi: 10.1016/s0960-9822(02)01218-6. [DOI] [PubMed] [Google Scholar]
- 71.Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC. DNA Topoisomerase VI Is Essential for Endoreduplication in Arabidopsis. Current Biology. 2002;12:1782–1786. doi: 10.1016/s0960-9822(02)01198-3. [DOI] [PubMed] [Google Scholar]
- 72.Yin Y, et al. A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc Natl Acad Sci USA. 2002;99:10191. doi: 10.1073/pnas.152337599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Breuer C, et al. BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell. 2007;19:3655–3668. doi: 10.1105/tpc.107.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes & Development. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sugimoto-Shirasu K, et al. RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. PNAS. 2005;102:18736–18741. doi: 10.1073/pnas.0505883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosa ID, et al. Adaptation of topoisomerase I paralogs to nuclear and mitochondrial DNA. Nucleic acids research. 2009;37:6414–6428. doi: 10.1093/nar/gkp708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang H, Pommier Y. Mitochondrial Topoisomerase I Sites in the Regulatory D-Loop Region of Mitochondrial DNA. Biochemistry. 2008;47:11196–11203. doi: 10.1021/bi800774b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaguni JM, Kornberg A. Topoisomerase I confers specificity in enzymatic replication of the Escherichia coli chromosomal origin. J Biol Chem. 1984;259:8578–8583. [PubMed] [Google Scholar]
- 79.Hiasa H, Marians KJ. Topoisomerase IV can support oriC DNA replication in vitro. The Journal of biological chemistry. 1994;269:16371–16375. [PubMed] [Google Scholar]
- 80.Abdurashidova G, et al. Functional interactions of DNA topoisomerases with a human replication origin. EMBO J. 2007;26:998–1009. doi: 10.1038/sj.emboj.7601578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lyu YL, et al. Role of Topoisomerase IIβ in the Expression of Developmentally Regulated Genes. Mol Cell Biol. 2006;26:7929. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 83.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. This paper links topo II beta-dependent dsDNA breaks with the DNA damage and repair enzyme PARP-1 in transcription regulation. [DOI] [PubMed] [Google Scholar]
- 84.Rampakakis E, Zannis-Hadjopoulos M. Transient dsDNA breaks during pre-replication complex assembly. Nucleic Acids Res. 2009;37:5714. doi: 10.1093/nar/gkp617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Lesterlin C, Reyes-Lamothe R, Ball G, Sherratt DJ. Replication and segregation of an Escherichia coli chromosome with two replication origins. PNAS. 2011;108:E243–250. doi: 10.1073/pnas.1100874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khodursky AB, et al. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc Natl Acad Sci USA. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hiasa H, Marians KJ. Two distinct modes of strand unlinking during θ-type DNA replication. Journal of Biological Chemistry. 1996;271:21529. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- 88.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 89.Kim RA, Wang JC. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. Journal of Molecular Biology. 1989;208:257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- 90.Yang L, Wold MS, Li JJ, Kelly TJ, Liu LF. Roles of DNA topoisomerases in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1987;84:950–954. doi: 10.1073/pnas.84.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kegel A, et al. Chromosome length influences replication-induced topological stress. Nature. 2011;471:392–396. doi: 10.1038/nature09791. This paper presents the finding that chromosome length in eukaryotes determines the need for release and development of superhelical tension. [DOI] [PubMed] [Google Scholar]
- 92.Champoux J, Been M. Topoisomerases and the swivel problem. Mechanistic studies of DNA replication and recombination: ICN-UCLA symposia on molecular and cellular biology. 1980:809–815. [Google Scholar]
- 93.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94:819–827. doi: 10.1016/s0092-8674(00)81740-7. This paper showed that replication dependent supercoiling is spread throughout the nascent and unreplicated DNA, and supercoiling is not localized. [DOI] [PubMed] [Google Scholar]
- 94.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 95.Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. Shows that Topo II depletion and topo II inhibition kill eukaryotic cells by two distinct mechanisms. [DOI] [PubMed] [Google Scholar]
- 97.Uemura T, et al. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. In addition to their role in chromosome decatenation, this paper discovered that type II topoisomerases are required for condensation of chromosomes prior to segregation. [DOI] [PubMed] [Google Scholar]
- 98.Adams DE, Shekhtman EM, Zechiedrich EL, Schmid MB, Cozzarelli NR. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. This paper was the first to show that topo IV is responsible for unlinking of daughter chromosomes prior to cell division in bacteria. [DOI] [PubMed] [Google Scholar]
- 99.Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989;9:159. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. Identified essential role of type II topoisomerases for resolving sister chromosmes after DNA replication, prior to cell segregation in eukaryotes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Uemura T, Yanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. The EMBO Journal. 1986;5:1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nurse P, Levine C, Hassing H, Marians KJ. Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J Biol Chem. 2003;278:8653–8660. doi: 10.1074/jbc.M211211200. In this paper, overexpression of topo III compensates for the loss of topo IV, and thus can act as the main cellular decatenase. [DOI] [PubMed] [Google Scholar]
- 103.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. This paper provides the first evidence that RecQ helicases and topo III collaborate to resolve converging replication forks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang M, et al. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu D, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes & Development. 2008;22:2843. doi: 10.1101/gad.1708608. Identifies another single-stranded binding protein, RMI1/NCE4, that associates with the topo III-RecQ helicase complex. The interaction of RMI1/NCE4 with the topo III-RecQ helicase complex diminishes and regulates sister chromosome exchange during homologous recombination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. In this study, ultrafine anphase bridges formed at a higher frequency when BLM helicase was absent, suggesting a role for BLM dependent topo III localization for appropriate segregation of sister chromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Confalonieri F, et al. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc Natl Acad Sci USA. 1993;90:4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Déclais AC, Marsault J, Confalonieri F, de La Tour CB, Duguet M. Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J Biol Chem. 2000;275:19498–19504. doi: 10.1074/jbc.M910091199. [DOI] [PubMed] [Google Scholar]
- 109.Valjavec-Gratian M, Henderson TA, Hill TM. Tus-mediated arrest of DNA replication in Escherichia coli is modulated by DNA supercoiling. Mol Microbiol. 2005;58:758–773. doi: 10.1111/j.1365-2958.2005.04860.x. [DOI] [PubMed] [Google Scholar]
- 110.Fachinetti D, et al. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Crozat E, Grainge I. FtsK DNA translocase: the fast motor that knows where it’s going. Chembiochem. 2010;11:2232–2243. doi: 10.1002/cbic.201000347. [DOI] [PubMed] [Google Scholar]
- 112.Bigot S, Marians KJ. DNA chirality-dependent stimulation of topoisomerase IV activity by the C-terminal AAA+ domain of FtsK. Nucleic Acids Res. 2010;38:3031–3040. doi: 10.1093/nar/gkp1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Espeli O, Lee C, Marians KJ. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J Biol Chem. 2003;278:44639–44644. doi: 10.1074/jbc.M308926200. [DOI] [PubMed] [Google Scholar]
- 114.Madabhushi R, Marians KJ. Actin homolog MreB affects chromosome segregation by regulating topoisomerase IV in Escherichia coli. Mol Cell. 2009;33:171–180. doi: 10.1016/j.molcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Coelho PA, et al. Dual role of topoisomerase II in centromere resolution and aurora B activity. PLoS Biol. 2008;6:e207. doi: 10.1371/journal.pbio.0060207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li H, Wang Y, Liu X. Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIα in cell cycle progression. Journal of Biological Chemistry. 2008;283:6209. doi: 10.1074/jbc.M709007200. [DOI] [PubMed] [Google Scholar]
- 117.Espeli O, Levine C, Hassing H, Marians KJ. Temporal regulation of topoisomerase IV activity in E. coli. Mol Cell. 2003;11:189–201. doi: 10.1016/s1097-2765(03)00013-3. [DOI] [PubMed] [Google Scholar]
- 118.Kang S, Han J, Park J, Skarstad K, Wang DH. SeqA protein stimulates the relaxing and decatenating activities of topoisomerase IV. J Biol Chem. 2003;278:48779–48785. doi: 10.1074/jbc.M308843200. [DOI] [PubMed] [Google Scholar]
- 119.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. This paper presented the first concept of DNA supercoiling induced by a translocating motor protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. This paper provided experimental evidence that transcription results in the formation of positive supercoils ahead of the RNA polymerase and negative supercoils behind. [DOI] [PubMed] [Google Scholar]
- 121.Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Drolet M, Bi X, Liu LF. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. Journal of Biological Chemistry. 1994;269:2068. [PubMed] [Google Scholar]
- 123.Tuduri S, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Drolet M, et al. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA. 1995;92:3526. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Massé E, Drolet M. Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J Biol Chem. 1999;274:16654–16658. doi: 10.1074/jbc.274.23.16654. [DOI] [PubMed] [Google Scholar]
- 126.Cheng B, Zhu CX, Ji C, Ahumada A, Tse-Dinh YC. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J Biol Chem. 2003;278:30705–30710. doi: 10.1074/jbc.M303403200. [DOI] [PubMed] [Google Scholar]
- 127.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 128.Mondal N, et al. Elongation by RNA polymerase II on chromatin templates requires topoisomerase activity. Nucleic Acids Res. 2003;31:5016–5024. doi: 10.1093/nar/gkg705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Durand-Dubief M, Persson J, Norman U, Hartsuiker E, Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010;29:2126–2134. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lotito L, et al. Global transcription regulation by DNA topoisomerase I in exponentially growing Saccharomyces cerevisiae cells: activation of telomere-proximal genes by TOP1 deletion. J Mol Biol. 2008;377:311–322. doi: 10.1016/j.jmb.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 131.Rossi F, et al. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80–82. doi: 10.1038/381080a0. First paper to show a topo IB associated kinase activity for SR proteins. [DOI] [PubMed] [Google Scholar]
- 132.Juge F, Fernando C, Fic W, Tazi J. The SR protein B52/SRp55 is required for DNA topoisomerase I recruitment to chromatin, mRNA release and transcription shutdown. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J Biol Chem. 2008;283:19991–19998. doi: 10.1074/jbc.M709495200. [DOI] [PubMed] [Google Scholar]
- 134.Mcnamara S, Wang H, Hanna N, Miller W. Topoisomerase IIb Negatively Modulates Retinoic Acid Receptor a Function: a Novel Mechanism of Retinoic Acid Resistance. Mol Cell Biol. 2008;28:2066–2077. doi: 10.1128/MCB.01576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Peciña A, et al. Targeted stimulation of meiotic recombination. Cell. 2002;111:173–184. doi: 10.1016/s0092-8674(02)01002-4. [DOI] [PubMed] [Google Scholar]
- 136.Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. Describes how SpoII is removed from chromosome ends to permit homologous recombination to occur. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li W, Ma H. Double-stranded DNA breaks and gene functions in recombination and meiosis. Cell Res. 2006;16:402–412. doi: 10.1038/sj.cr.7310052. [DOI] [PubMed] [Google Scholar]
- 139.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 140.Hartsuiker E, et al. Ctp1 CtIP and Rad32 Mre11 nuclease activity are required for Rec12 Spo11 removal, but Rec12 Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol. 2009;29:1671–1681. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lu WJ, Chapo J, Roig I, Abrams JM. Meiotic recombination provokes functional activation of the p53 regulatory network. Science. 2010;328:1278–1281. doi: 10.1126/science.1185640. Recent paper that highlights the role of p53 in SpoII mediated homologous recombination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cliby WA. S Phase and G2 Arrests Induced by Topoisomerase I Poisons Are Dependent on ATR Kinase Function. Journal of Biological Chemistry. 2002;277:1599–1606. doi: 10.1074/jbc.M106287200. [DOI] [PubMed] [Google Scholar]
- 143.Treszezamsky AD, et al. BRCA1- and BRCA2-Deficient Cells Are Sensitive to Etoposide-Induced DNA Double-Strand Breaks via Topoisomerase II. Cancer Research. 2007;67:7078–7081. doi: 10.1158/0008-5472.CAN-07-0601. [DOI] [PubMed] [Google Scholar]
- 144.Lin CP, Ban Y, Lyu YL, Liu LF. Proteasome-dependent Processing of Topoisomerase I-DNA Adducts into DNA Double Strand Breaks at Arrested Replication Forks. Journal of Biological Chemistry. 2009;284:28084–28092. doi: 10.1074/jbc.M109.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc Natl Acad Sci USA. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276:40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- 147.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. This paper reports the discovery of TDP, a repair protein that removes covalently attached topo IB from DNA ends. [DOI] [PubMed] [Google Scholar]
- 148.Nitiss KC, Malik M, He X, White SW, Nitiss JL. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc Natl Acad Sci U S A. 2006;103:8953–8958. doi: 10.1073/pnas.0603455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 150.Zeng Z, Cortes-Ledesma F, El Khamisy SF, Caldecott KW. TDP2/TTRAP is the major 5′-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA Damage. J Biol Chem. 286:403–409. doi: 10.1074/jbc.M110.181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mao Y, Desai SD, Liu LF. SUMO-1 Conjugation to Human DNA Topoisomerase II Isozymes. J Biol Chem. 2000;275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 152.Desai SD, Liu LF, Vazquez-Abad D, D’Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- 153.Sordet O, et al. Hyperphosphorylation of RNA Polymerase II in Response to Topoisomerase I Cleavage Complexes and Its Association with Transcription- and BRCA1-dependent Degradation of Topoisomerase I. Journal of Molecular Biology. 2008;381:540–549. doi: 10.1016/j.jmb.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Desai SD, et al. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Molecular and cellular biology. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xiao H, et al. The topoisomerase IIbeta circular clamp arrests transcription and signals a 26S proteasome pathway. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3239–3244. doi: 10.1073/pnas.0736401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- 157.Lippert MJ, et al. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci U S A. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2011;108:692–697. doi: 10.1073/pnas.1012582108. This paper and reference 157 are the first to report on transcription mediated genomic deletions that are a result of topo IB action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. This recent paper is the first to report topo IB’s mutagenic role in the removal of genomically misincorpated ribonucleotides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang Y, Knudsen BR, Bjergbaek L, Westergaard O, Andersen AH. Stimulated activity of human topoisomerases IIalpha and IIbeta on RNA-containing substrates. The Journal of biological chemistry. 1999;274:22839–22846. doi: 10.1074/jbc.274.32.22839. [DOI] [PubMed] [Google Scholar]
- 161.Deweese JE, Osheroff N. Coordinating the Two Protomer Active Sites of Human Topoisomerase IIα: Nicks as Topoisomerase II Poisons. Biochemistry. 2009;48:1439–1441. doi: 10.1021/bi8021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sayer J, Jerina D, Shuman S. Individual nucleotide bases, not base pairs, are critical for triggering site-specific DNA cleavage by vaccinia topoisomerase. Journal of Biological Chemistry. 2004 doi: 10.1074/jbc.M407376200. [DOI] [PubMed] [Google Scholar]
- 163.Kingma PS, Osheroff N. Apurinic sites are position-specific topoisomerase II poisons. The Journal of biological chemistry. 1997;272:1148–1155. doi: 10.1074/jbc.272.2.1148. [DOI] [PubMed] [Google Scholar]
- 164.Kingma PS, Osheroff N. Spontaneous DNA damage stimulates topoisomerase II-mediated DNA cleavage. The Journal of biological chemistry. 1997;272:7488–7493. doi: 10.1074/jbc.272.11.7488. [DOI] [PubMed] [Google Scholar]
- 165.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. This paper shows that interactions between RecQ helicases and topo III restrict crossover events during DSB repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. Shows that topo III and a RecQ helicase interact to resolve double Holliday junctions, which results in limited crossover during homologous recombination. [DOI] [PubMed] [Google Scholar]
- 167.Plank JL, Wu J, Hsieh TS. Topoisomerase IIIalpha and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci U S A. 2006;103:11118–11123. doi: 10.1073/pnas.0604873103. Shows that Topo III collaberates with a RecQ like helicase and RPA to remove double holliday junctions(DHJs), using a convergent migration mechanism to collapse DHJs producing noncrossover products. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Harmon FG, Brockman JP, Kowalczykowski SC. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. Journal of Biological Chemistry. 2003;278:42668–42678. doi: 10.1074/jbc.M302994200. First paper to show a RecQ helicase’s ability to stimulate topo III, and the interaction is suggested to be important for recombination. [DOI] [PubMed] [Google Scholar]
- 169.Singh TR, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase–double Holliday junction dissolvasome. Genes & Development. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. In this paper, it was reported that nucleases required for end resectioning may be recruited to broken ends by topo III in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wells NJ, Addison CM, Fry AM, Ganapathi R, Hickson ID. Serine 1524 is a major site of phosphorylation on human topoisomerase II alpha protein in vivo and is a substrate for casein kinase II in vitro. J Biol Chem. 1994;269:29746–29751. [PubMed] [Google Scholar]
- 173.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 174.Ryu H, Furuta M, Kirkpatrick D, Gygi SP, Azuma Y. PIASy-dependent SUMOylation regulates DNA topoisomerase II alpha activity. J Cell Biol. 2010;191:783–794. doi: 10.1083/jcb.201004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dawlaty MM, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 177.Shapiro P, et al. Extracellular Signal-Regulated Kinase Activates Topoisomerase IIα through a Mechanism Independent of Phosphorylation. Mol Cell Biol. 1999;19:3551–3560. doi: 10.1128/mcb.19.5.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Plo I, et al. Overexpression of the Atypical Protein Kinase C zeta Reduces Topoisomerase II Catalytic Activity, Cleavable Complexes Formation, and Drug-induced Cytotoxicity in Monocytic U937 Leukemia Cells. Journal of Biological Chemistry. 2002;277:31407–31415. doi: 10.1074/jbc.M204654200. [DOI] [PubMed] [Google Scholar]
- 179.Kimura K, Saijo M, Tanaka M, Enomoto T. Phosphorylation-independent stimulation of DNA topoisomerase II alpha activity. J Biol Chem. 1996;271:10990–10995. doi: 10.1074/jbc.271.18.10990. [DOI] [PubMed] [Google Scholar]
- 180.Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 181.Smith AB, Maxwell A. A strand-passage conformation of DNA gyrase is required to allow the bacterial toxin, CcdB, to access its binding site. Nucleic Acids Res. 2006;34:4667–4676. doi: 10.1093/nar/gkl636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Vizán J, Hernández-Chico C, del Castillo I, Moreno F. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J. 1991;10:467–476. doi: 10.1002/j.1460-2075.1991.tb07969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hegde S, et al. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480. doi: 10.1126/science.1110699. This paper identified and structurally characterized a DNA-mimic protein in M. tuberculosis that provides quinolone resistance to gyrase. [DOI] [PubMed] [Google Scholar]
- 184.Staker BL, et al. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci U S A. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lewis RJ, et al. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 1996;15:1412–1420. [PMC free article] [PubMed] [Google Scholar]
- 186.Tsai FT, et al. The high-resolution crystal structure of a 24-kDa gyrase B fragment from E. coli complexed with one of the most potent coumarin inhibitors, clorobiocin. Proteins. 1997;28:41–52. [PubMed] [Google Scholar]
- 187.Classen S, Olland S, Berger JM. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc Natl Acad Sci U S A. 2003;100:10629–10634. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Corbett KD, Berger JM. Structural basis for topoisomerase VI inhibition by the anti-Hsp90 drug radicicol. Nucleic Acids Res. 2006;34:4269–4277. doi: 10.1093/nar/gkl567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Edwards MJ, et al. A crystal structure of the bifunctional antibiotic simocyclinone D8, bound to DNA gyrase. Science. 2009;326:1415–1418. doi: 10.1126/science.1179123. [DOI] [PubMed] [Google Scholar]
- 190.Bax BD, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. Bacterial type IIA topoisomerases are inhibited by a novel compound, which binds across the dimer interface and intercalates DNA. [DOI] [PubMed] [Google Scholar]
- 191.Cheng B, et al. Asp-to-Asn substitution at the first position of the DxD TOPRIM motif of recombinant bacterial topoisomerase I is extremely lethal to E. coli. J Mol Biol. 2009;385:558–567. doi: 10.1016/j.jmb.2008.10.073. This paper reports how a single active-site mutation turns a type IA topo into a highly lethal machine, and suggests this previously unrealized drug target may become a viable future target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 194.Wang X, Zhao X, Malik M, Drlica K. Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J Antimicrob Chemother. 65:520–524. doi: 10.1093/jac/dkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bowman KJ, Newell DR, Calvert AH, Curtin NJ. Differential effects of the poly (ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br J Cancer. 2001;84:106–112. doi: 10.1054/bjoc.2000.1555. This paper shows that the PARP-1 inhibitor NU1025 enhances the toxicity of the topo I inhibitor camptothecin, suggesting that this kind of synergy with topo inhibitors may be a promising path forward in the treatment of cancer and microbial infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wesierska-Gadek J, Schloffer D, Gueorguieva M, Uhl M, Skladanowski A. Increased susceptibility of poly(ADP-ribose) polymerase-1 knockout cells to antitumor triazoloacridone C-1305 is associated with permanent G2 cell cycle arrest. Cancer Res. 2004;64:4487–4497. doi: 10.1158/0008-5472.CAN-03-3410. [DOI] [PubMed] [Google Scholar]
- 197.Munoz-Gamez JA, et al. Inhibition of poly (ADP-ribose) polymerase-1 enhances doxorubicin activity against liver cancer cells. Cancer Lett. 301:47–56. doi: 10.1016/j.canlet.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 198.Austin C, Sng J, Patel S, Fisher L. Novel HeLa topoisomerase II is the II [beta] isoform: complete coding sequence and homology with other type II topoisomerases. Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression. 1993;1172:283–291. doi: 10.1016/0167-4781(93)90215-y. [DOI] [PubMed] [Google Scholar]
- 199.Linka RM, et al. C-terminal regions of topoisomerase IIα and IIβ determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007;35:3810–3822. doi: 10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ye J, et al. TRF2 and apollo cooperate with topoisomerase 2 alpha to protect human telomeres from replicative damage. Cell. 2010;142:230–242. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 201.Niimi A, Suka N, Harata M, Kikuchi A, Mizuno S. Co-localization of chicken DNA topoisomerase IIa, but not β, with sites of DNA replication and possible involvement of a C-terminal region of a through its binding to PCNA. Chromosoma. 2001;110:102–114. doi: 10.1007/s004120100140. [DOI] [PubMed] [Google Scholar]
- 202.Grue P, et al. Essential mitotic functions of DNA topoisomerase IIalpha are not adopted by topoisomerase IIbeta in human H69 cells. J Biol Chem. 1998;273:33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 203.Peng H, Marians KJ. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 204.Kato J, et al. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. Discovery of topo IV and its importance for chromosome segregation in E. coli. [DOI] [PubMed] [Google Scholar]
- 205.Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc Natl Acad Sci U S A. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J Mol Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 207.Hsieh TJ, Farh L, Huang WM, Chan NL. Structure of the topoisomerase IV C-terminal domain: a broken beta-propeller implies a role as geometry facilitator in catalysis. J Biol Chem. 2004;279:55587–55593. doi: 10.1074/jbc.M408934200. [DOI] [PubMed] [Google Scholar]
- 208.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc Natl Acad Sci U S A. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. Presents the first high-resolution structure of the bacterial type IIA topoisomerase C-terminal domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ruthenburg AJ, Graybosch DM, Huetsch JC, Verdine GL. A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J Biol Chem. 2005;280:26177–26184. doi: 10.1074/jbc.M502838200. [DOI] [PubMed] [Google Scholar]
- 210.Changela A, DiGate RJ, Mondragón A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 211.Schmidt B, Burgin A, Deweese J, Osheroff N, Berger J. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information S1 (table): A partial list of the topoisomerase interactome