Abstract
The PIK3CA gene encodes for the p110 alpha isoform of PI3 kinase and is one of the most frequently mutated oncogenes in human cancers. However, the mechanisms by which PIK3CA mutations activate cell signaling are not fully understood. Here we used a phosphoproteomic approach to compare differential phosphorylation patterns between human breast epithelial cells and two isogenic somatic cell knock in derivatives, each harboring a distinct PIK3CA mutation. We demonstrated differential phosphorylation patterns between isogenic cell lines containing a PIK3CA helical domain mutation (E545K) compared to cells with a PIK3CA kinase domain mutation (H1047R). In particular, the receptor tyrosine kinase, HER3, showed increased phosphorylation at tyrosine 1328 in H1047R cells versus E545K cells. Genetic studies using shRNA demonstrated that H1047R cells have a profound decrease in growth factor independent proliferation upon HER3 knock down, but this effect was attenuated in E545K cells. In addition, HER3 knock down led to reductions in both PI3 kinase and MAP kinase pathway activation in H1047R cells, but in E545K cells only PI3 kinase pathway diminution was observed. These studies demonstrate the power of using paired isogenic cell lines for proteomic analysis to gain new insights into oncogenic signal transduction pathways.
Keywords: Biomedicine, HER3, MAP kinase, PIK3CA, PI3 kinase
1 Introduction
Human cancers originate from the stepwise accumulation of genetic and epigenetic alterations. Recent human genome sequencing efforts have identified genes that are often somatically mutated in many human malignancies. The PIK3CA gene encodes the catalytic subunit of PI3 kinase, p110 alpha and is one of the most frequently mutated oncogenes in breast cancers [1]. Previously, our group demonstrated that PIK3CA is mutated in approximately 25% of breast cancers, and further studies demonstrated a higher percentage of 40 to 50% mutational frequency in estrogen receptor—alpha (ER) positive disease [2]. However, it has become increasingly clear that not all mutations are “drivers” and that “passenger” mutations can occur in any given oncogene [3, 4]. Adding to this complexity, it is now appreciated that oncogenic mutations within the same gene may impart their transformative phenotypes via different mechanisms [5], thus making therapeutic targeting distinct for a given mutation, rather than a specific gene. For example, common kinase domain mutations in EGFR found in nonsmall lung cancers predict for sensitivity to the small molecule inhibitors gefinitib and erlotinib. However, certain oncogenic mutations such as EGFR Exon 20 insertion mutations, do not confer increased sensitivity to these agents [6]. As more data surfaces through the sequencing of cancer genomes, it is becoming clear that specific mutations can affect protein function in distinct manners. Elucidating how this occurs would provide mechanistic insights into signaling pathways that are aberrantly activated by cancer mutations and potentially provide opportunities for more effective targeted therapies.
In this study, we used a phosphoproteomic approach to identify unique mediators of oncogenic PIK3CA signaling. Although somatic mutations of PIK3CA in human cancers were discovered over a decade ago [7], targeted therapies specific for mutant PIK3CA have been elusive. Our study demonstrates that the use of isogenic cell lines carrying common oncogenic PIK3CA mutations coupled with a phosphoproteomic screen can identify unique pathways that are “rewired” leading to aberrant signaling and pathway activation. This knowledge presents opportunities for targeting pathways that are mutation, rather than gene, specific.
2 Materials and methods
2.1 Cell culture
The nontransformed human breast epithelial cell line MCF-10A [8] and its derivatives were grown in DMEM/F12 (1:1) supplemented with 5% horse serum (Hyclone), EGF at 20 ng/mL, insulin at 10 μg/mL, hydrocortisone at 0.5 μg/mL, and cholera toxin at 0.1 μg/mL (hereafter denoted as “growth medium”). Targeted knock in PIK3CA exon 9 (E545K) and exon 20 (H1047R) mutations into MCF-10A cells have been previously described [9]. Cell lines with gene targeted PIK3CA mutations were grown in growth media without EGF. HER3 knock down cell lines derived from PIK3CA mutant cell lines were maintained in EGF containing media due to the effects of HER3 knock down. MCF-7 cells and T47D cells were grown in DMEM media with 5% FBS. All supplements were purchased from Sigma-Aldrich unless otherwise noted. All cell lines were purchased from ATCC and verified by DNA STR fingerprint analysis.
2.2 Cell line labeling
MCF-10A and derivative cell lines were propagated in DMEM/F12 SILAC media with corresponding complete supplements but deficient in both L-lysine and L-arginine (Thermo Fisher Scientific) and supplemented with light lysine (K) and arginine (R) for light, 2H4-K and 13C6-R for medium state and 13C615N2-K and 13C615N4-R for heavy state labeling (Cambridge Isotope Laboratories). Cells were seeded at 80% confluence in 0.2 ng/mL EGF complete medium overnight and followed by serum starvation with corresponding SILAC basal DMEM/F12 medium for 16 h prior to harvest with urea lysis buffer containing 20 mM HEPES pH 8.0, 9 M urea, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, and 5 mM sodium fluoride.
2.3 In-solution trypsin digestion
Cell lysates were sonicated and cleared by centrifugation at 3000 × g at 4°C for 10 min. Protein estimation was carried out using BCA protein assay and 8 mg protein from each SILAC labeled cell line was equally mixed and reduced with 5 mM dithiothreitol and alkylated with 10 mM iodoacetamide. Proteins were then diluted to less than 2 M urea final concentration using 20 mM HEPES (pH 8.0) and in-solution digestion was carried out using TPCK-treated trypsin on an orbital shaker at 25°C overnight. The reaction was quenched using 1% TFA and the digest was cleared by centrifugation. The protein digest was desalted using SepPak C18 cartridge and eluted peptides were lyophilized and subjected to phosphotyrosine peptide enrichment.
2.4 Immunoaffinity purification of phosphotyrosine peptides
The phosphotyrosine peptide enrichment was performed according to the manufacturer’s protocol (Cell Signaling Technology). Briefly, lyophilized peptides were reconstituted in immunoaffinity purification buffer containing 50 mM MOPS pH 7.2, 10 mM sodium phosphate, 50 mM NaCl. Antiphosphotyrosine antibody (pY100, Cell Signaling Technology) was added to the peptide solution and rotated at 4°C for 30 min. Postincubation, antibody and peptide complexes were washed twice with immunoaffinity purification buffer and water. The phosphopeptides were eluted using 0.1% TFA. The eluted phosphopeptides were desalted using C18 STAGE tips, vacuum dried, and kept at −80°C before LC–MS analysis.
2.5 Liquid chromatography tandem mass spectrometry
LC–MS/MS analysis of enriched phosphopeptides was carried out using a RPLC system interfaced with an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific) essentially as previously described [10, 11]. The peptides were loaded onto an enrichment column (2 cm × 75 μm, Magic C18 AQ 5 μm, 120 A)° with a flow rate of 3 μL/min using 0.1% formic acid in water. The peptides were separated on an analytical column (10 cm × 75 μm, Magic C18 AQ 5 μm, 120 A) by 0.1% formic acid and eluted with a linear gradient with 180 min running time. Precursor scans (FTMS) from 350–1800 m/z at 60 000 resolution followed by MS2 scan (FTMS) of HCD fragmentation of the 20 most abundant ions (isolation width: 1.90 m/z; normalized collision energy: 39%) at 7500 resolution.
2.6 MS data analysis
The MS/MS data were searched using SEQUEST search algorithm against a human RefSeq database (version 59) supplemented with frequently observed contaminants through the Proteome Discoverer platform (version 1.4, Thermo Scientific). The search parameters included a maximum of one missed cleavage; carbamidomethylation at cysteine as a fixed modification; N-terminal acetylation, oxidation at methionine, phosphorylation at serine, threonine and tyrosine, and SILAC labeling 13C615N2-lysine; 13C615N4-arginine as variable modifications. The MS tolerance was set at 10 ppm and MS/MS tolerance to 0.05 Da. The false discovery rate was set to 0.01 at the peptide level. The quantitation ratio for each phosphopeptide-spectrum match (phosphoPSM) was calculated by the quantitation node and the probability of phosphorylation for each Ser/Thr/Tyr site on each peptide was calculated by the PhosphoRS node (Version 3.0) in the Proteome Discoverer. Phosphorylation sites were assigned based on the phosphoRS probability ≥75% threshold. In this study, phosphotyrosine peptides were specifically enriched for MS analysis. Thus, if the phosphoRS probabilities of ambiguous phosphorylation sites are same for tyrosine or serine/ threonine residues, we assigned phosphorylation onto the tyrosine residue.
2.7 Cell proliferation assays
For cell growth assays with MCF-10A and derivative cell lines, exponentially growing cells were washed with HBSS twice and seeded in growth media as well as “assay media” consisting of DMEM/F12 medium, 1% charcoal dextran-treated FBS, insulin at 10 μg/mL, hydrocortisone at 0.5 μg/mL, and cholera toxin at 0.1 μg/mL, with 0.2 ng/mL EGF. For MCF-7 and T47D and derivative cell lines, identical conditions were used except media was phenol red free DMEM, 1% charcoal dextran-treated FBS, and estrogen (beta-estradiol; Sigma-Aldrich) at the specified concentration as previously described [12]. Cells were seeded at a density of 2 × 104 cells/well of a 6-well tissue culture dish on day 0. Medium was changed every third day. Cells were harvested on days 0, 2, 4, and 6, and cell numbers were counted using a Beckman Coulter counter. In some assays, cells were also stained with crystal violet (Sigma-Aldrich) after 3 days, or when cells in control flasks were ~90–95% confluent, diluted in formalin at a concentration of 2 mg/mL. All assays were performed in triplicate and repeated at least three times.
2.8 Immunoblotting
MCF-10A cells and derivatives were seeded in media using EGF-free or EGF containing conditions. MCF-7, T47D cells and derivatives were seeded in media using no or supplemented estrogen conditions for proliferation assays. Cells were harvested after 48 h for protein lysates and immunoblotting was performed as previously described [13]. Briefly, whole cell protein extracts prepared in Laemmli sample buffer were resolved by SDS-PAGE using NuPAGE gels (Invitrogen), transferred to Invitrolon PVDF membranes (Invitrogen) and probed with primary antibody followed by incubation with HRP-conjugated secondary antibodies. The primary antibodies used in this study include anti-p44/p42 MAP kinase rabbit antibody (9102; Cell Signaling Technology), anti-phospho p44/p42 MAP kinase (Thr 202/Tyr 204) mouse antibody (9106; Cell Signaling Technology), anti-AKT rabbit antibody (9272; Cell Signaling Technology), anti-phospho AKT (Ser 473) rabbit antibody (9271; Cell Signaling Technology), anti-HER3 rabbit antibody (4754S; Cell Signaling Technology), anti-phospho HER3 (Tyr 1289) rabbit antibody (4791S; Cell Signaling Technology), and anti-GAPDH mouse antibody (6C5) (ab8245; Abcam).
2.9 RNA interference
Short-hairpin RNA constructs were purchased from Origene (TR320343) to target ERBB3 mRNA and were used to create stable shRNA expressing cell lines as previously described [14].
Cell lines were transfected and selected with G418 (Life Technologies) as previously described [15]. Due to potential deleterious effects of HER3 knock down, MCF-10A cell line derivatives were grown in successively increasing concentrations of EGF (0.2 and 20 ng/mL).
2.10 Statistical analysis
All statistical analyses were performed using GraphPad InStat software (La Jolla, CA). A p-value of less than 0.05 was considered significant.
3 Results
3.1 Phosphoproteomic profiles differ between PIK3CA E545K and H1047R isogenic cell lines
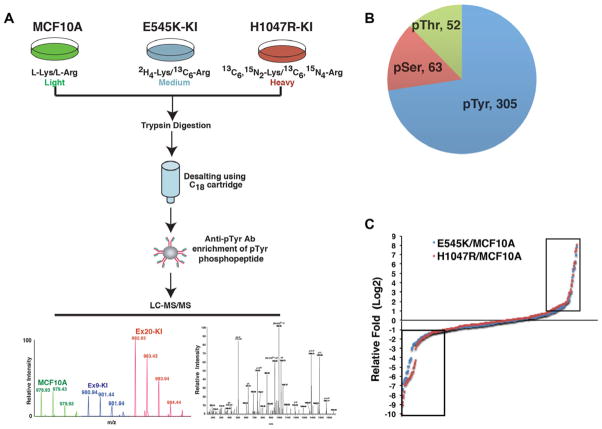
We have previously described human breast epithelial cell lines that were gene targeted to create isogenic derivatives harboring an E545K or H1047R PIK3CA mutation [9]. These frequently recurring “hotspot” mutations imparted features of cellular transformation including EGF independent growth, activation of the PI3 kinase pathway, and unexpectedly, increased MAP kinase pathway signaling. Further work demonstrated that PI3 kinase signaling was dependent on Ras/p110 alpha binding, but disrupting this interaction did not abrogate MAP kinase signaling induced by PIK3CA mutations [16]. In order to further elucidate the mechanism of oncogenic mutant PIK3CA signaling responsible for MAP kinase pathway activation, we chose to characterize the phosphoproteomic differences resulting from E545K and H1047R PIK3CA mutations. We therefore subjected parental MCF-10A cell lines and their E545K and H1047R derivatives to MS-based phosphotyrosine proteomic analysis in conjunction with stable isotope labeling by amino acids in cell culture (SILAC) as shown in Fig. 1A. We performed one round of MS analysis on the enriched tyrosine phosphorylated peptides and the data were processed and searched against a database with the SEQUEST algorithm through the Proteome Discoverer platform. Using a false discovery rate cutoff of 1% and a target-decoy analysis, this generated 2611 phosphopeptide-spectrum-matches (PhosphoPSM). We set the phosphoRS possibility of 75% as the cutoff and identified 2594 Phospho-PSM. After consolidating redundant phosphopeptides, 412 unique phosphopeptides derived from 209 proteins were identified (Supporting Information Table 1). Among them, 305 peptides contain tyrosine phosphorylation, 63 peptides have serine phosphorylation, and 52 peptides have threonine phosphorylation (Fig. 1B). In comparison to MCF-10A parental cells, 49 peptides derived from 39 proteins and 62 peptides derived from 51 proteins were hyperphosphorylated (> twofold) in E545K or H1047R knock in cells, respectively (Supporting Information Table 2). On the other hand, we also found 108 peptides derived from 77 proteins and 106 peptides derived from 69 proteins that were hypophosphorylated (< 0.5 fold) in E545K or H1047R knock in cells, respectively (Fig. 1C and Supporting Information Table 2). As shown in Fig. 2A, the majority of the hyperphosphorylated peptides overlapped between E545K and H1047R knock in cells, whereas ~60% of the hypophosphorylated peptides overlapped between the two knock in cell lines. This suggested that E545K and H1047R mutations have not only largely similar downstream signaling effects but also have some signaling effects that are unique to individual mutations.
Figure 1.
Phosphoproteomic analysis of PIK3CA E545K and H1047R isogenic knock in cell lines. (A) A schematic depicting the strategy used for quantitative phosphoproteomic profiling of MCF10A parental, E545K, and H1047R knock in cells. (B) Number of phosphotyrosine (pTyr) phosphoserine (pSer) and phosphothreonine (pThr) sites identified in the study. (C) Distribution of log 2 ratio of phosphopeptide intensities identified in E545K (blue) or H1047R (red) cells vs. MCF10A cells. The dots inside the two boxes represent peptides with phosphorylation altered more than twofold in E545K or H1047R cells comparing to MCF10A cells. Left: hypophosphorylation, right: hyperphosphorylation.
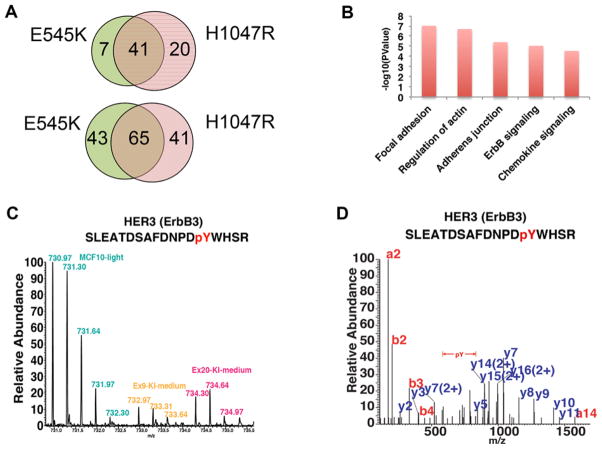
Figure 2.
Differential peptide phosphorylation induced by PIK3CA E545K or H1047R mutations. (A) Hyperphosphorylated (top panel) and hypophosphorylated (bottom panel) peptides identified in E545K or H1047R knock in cells. (B) Top signaling pathways enriched in differentially phosphorylated proteins between E545K and H1047R knock in cells. (C) MS spectrum shows the relative abundance of the HER3 phosphopeptide (SLEATDSAFD-NPDpYWHSR) in MCF10A parental, E545K or H1047R knock in cells. (D) MS/MS spectrum of HER3 phosphopeptide (SLEATDSAFDNPDpYWHSR).
In order to focus on the unique signaling alterations induced by E545K or H1047R mutations, we compared quantitative differences between mutant E545K and H1047R knock in cell lines. We found 42 phosphopeptides from 39 proteins were hyperphosphorylated in H1047R knock in cells (Supporting Information Table 3), whereas 39 phosphopeptides from 32 proteins were hypophosphorylated in H1047R knock in cells compared to E545K knock in cells (Supporting Information Table 4). To understand the phosphorylation signaling alterations induced by E545K or H1047R mutations, we performed a KEGG pathway analysis using an integrated online functional annotation tool, DAVID [17] for the differentially phosphorylated proteins. Interestingly, we noticed that the EGFR/HER signaling pathway was among the top five enriched signaling pathways (Fig. 2B). The most intriguing protein identified was HER3, as prior reports suggested that H1047R, but not E545K, mutations could enhance HER2-mediated transformation via activation of HER3 [18]. Our data predicted a differential degree of tyrosine phosphorylation at codon 1328 (Y1328) between E545K and H1047R isogenic cell lines (Fig. 2C, D). Based on this finding and relevant past literature, this protein was selected for further analysis.
3.2 HER3 knock down reveals differential effects in signaling between oncogenic PIK3CA mutants
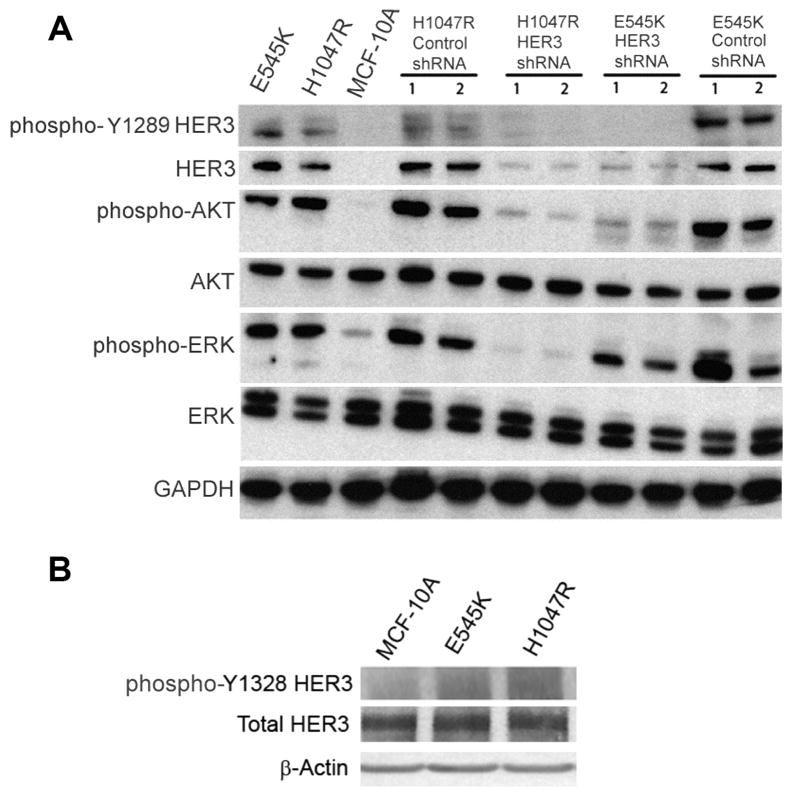
To functionally validate HER3’s role in oncogenic PIK3CA signaling, we transfected E545K and H1047R cells with two distinct shRNA constructs against HER3 as well as a control shRNA. As shown in Fig. 3A, we were able to achieve significant knock down of HER3 in the E545K and H1047R cell lines using these shRNA constructs, while scrambled control shRNA did not show any appreciable knock down. Interestingly, MCF-10A cells did not have appreciable levels of HER3 by Western blot relative to isogenic mutant PIK3CA knock in cells. The reasons for this are unclear, though increased expression of HER3 after mutant PIK3CA overexpression has been previously reported [19], and may be regulated by FoxO1 and FoxO3a transcription factors [20]. However, we were unable to confirm differential phosphorylation via Western blotting using a phosphospecific Y1328 antibody due to the lack of sensitivity and specificity of this antibody. As shown in Fig. 3B, this antibody did not provide adequate resolution to demonstrate differences between our isogenic cell lines. Interestingly, using another HER3 phosphospecific antibody (Y1289), no appreciable difference in phosphorylation was seen via Western blot as shown in Fig. 3A. In fact, at this particular residue, Y1289, there was a slightly increased phosphorylation of HER3 for E545K cells compared to H1047R cells, which was in contrast to the HER3 Y1328 phosphorylation data from MS. These results underscore that phosphorylation of specific residues of the same protein may be altered differentially during the process of pathway activation, and that commercially available antibodies may not provide sufficient sensitivity and/or specificity for detection, reaffirming the power of using a relatively global phosphoproteomic approach.
Figure 3.
HER3 knock down results in decreased Akt phosphorylation in MCF-10A PIK3CA mutant E545K and H1047R knock in cell lines, and decreased Erk phosphorylation in H1047R cells. (A) Western blot demonstrating levels of phosphorylated HER3 (Y1289), total HER3, phosphorylated Akt (Ser-473), total Akt, phosphorylated Erk (Thr-202/Tyr-204), total Erk, in MCF-10A cell lines, derivatives and controls. GAPDH is shown as a loading control. (B) Western blot demonstrating levels of phosphorylated HER3 (Y1328) and total HER3 in MCF-10A cells and PIK3CA E545K and H1047R knock in cell lines. GAPDH is shown as a loading control.
We next investigated PI3 kinase and MAP kinase activation by examining phosphorylation of AKT and ERK1/2, respectively, as described in prior studies [9]. As shown in Fig. 3A, knock down of HER3 led to significant decreases in AKT phosphorylation for both E545K and H1047R cell lines. In contrast, decreased ERK phosphorylation was evident only in H1047R HER3 knock down cell lines, while E545K HER3 knock down cells continued to have increased ERK phosphorylation relative to parental MCF-10A cells. Given that HER3 lies proximal to p110 alpha in the PI3 kinase pathway, these data lend further credence to the rewiring that can occur with oncogenic mutations. They also support studies suggesting that oncogenic PIK3CA signaling may be distinct between E545K and H1047R mutations [5, 21], with each mutation relying upon specific molecules for full pathway signaling.
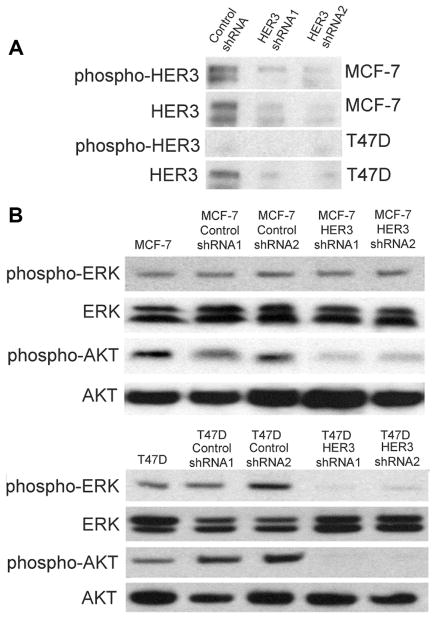
In order to determine if our findings were applicable to cancer cells, we next created stable HER3 knock down via shRNA in two human breast cancer cell lines, MCF-7 and T47D. MCF-7 has a helical E545K PIK3CA mutation, while T47D has a PIK3CA kinase domain H1047R mutation [1]. As seen in Fig. 4A, we generated two stable knock down clones for each cell line, noting significant knock down relative to parental counterparts. Interestingly, MCF-7 (E545K) cells appeared to have increased HER3 phosphorylation of Y1289 compared to T47D (H1047R) cells relative to total HER3, consistent with our isogenic cell line results. Similarly, HER3 knock down had a dramatic effect of reducing AKT phosphorylation in both MCF-7 and T47D cells, yet ERK phosphorylation was reduced only in T47D cells (Fig. 4B). Together, these data suggest that E545K and H1047R PIK3CA mutations rely upon HER3 for PI3 kinase pathway activation as measured by phosphorylated AKT, but that H1047R mutations also use HER3 for MAP kinase activation in stark contrast to E545K mutants.
Figure 4.
HER3 knock down results in decreased Akt phosphorylation in PIK3CA mutant E545K and H1047R breast cancer cells, and decreased Erk phosphorylation in H1047R breast cancer cells. (A) Western blot demonstrating levels of phosphorylated HER3 (Y1289) and total HER3 in MCF-7 (E545K) and T47D (H1047R) breast cancer cell lines. (B) Western blot demonstrating levels of phosphorylated Erk (Thr-202/Tyr-204), total Erk, phosphorylated Akt (Ser-473), and total Akt in MCF-7 (top panel) and T47D (bottom panel) cell lines, knock down derivatives and controls.
3.3 HER3 knock down preferentially affects H1047R proliferation
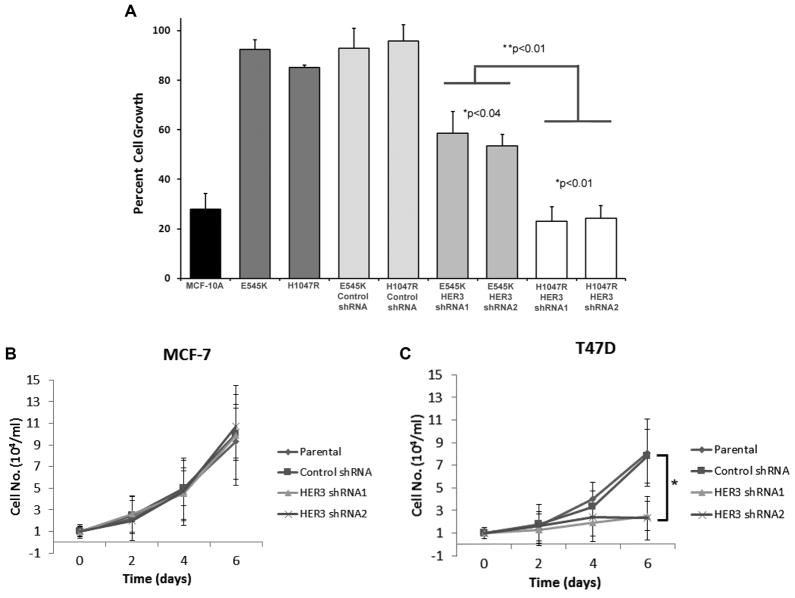
In order to determine whether HER3 knock down led to any phenotypic changes, we then subjected MCF-10A, E545K, H1047R, and HER3 knock down derivatives to cell proliferation assays. Because parental MCF-10A cells require exogenous growth factors, cell lines were tested in growth media, with 20 ng/mL EGF and compared to proliferation in assay conditions, using physiologic EGF concentrations of 0.2 ng/mL. This was necessary due to differences in basal proliferation rates between parental MCF-10A cells and mutant PIK3CA knock in cells as we have previously described [9]. As seen in Fig. 5A, the percentage of cells in assay media relative to growth media on day 6 of the proliferation assay demonstrated that E545K and H1047R HER3 knock down cells had significantly decreased cell proliferation compared to E545K and H1047R cells and shRNA controls. H1047R cells had the most pronounced effect and in EGF free conditions did not proliferate, similar to parental MCF-10A cells (data not shown). Next, we extended these observations to our cancer cell lines: MCF-7 and T47D HER3 knock down cells. Because these are breast cancer cell lines, they are not dependent upon EGF for cell proliferation. However, they are both ER positive cell lines and have a relative dependence on estrogens for growth. Thus, we analyzed whether HER3 knock down could have effects on estrogen dependence in these cell lines. Using assay condition with low estrogen conditions as previously described [12], we showed that estrogen dependence was minimally affected in MCF-7 (E545K) cells with HER3 knock down (Fig. 5B). However, T47D cells demonstrated significantly decreased growth in response to estrogen relative to parental controls (Fig. 5C). These findings corroborate with our results in isogenic MCF-10A cells and together demonstrate that HER3 mediated knock down in H1047R PIK3CA mutation positive cells leads to a decreased proliferative response to growth factors/hormones that is associated with diminished MAPK kinase signaling.
Figure 5.
HER3 knock down decreases cell proliferation preferentially in PIK3CA mutant H1047R cells. (A) Cell proliferation assays were performed as described in Materials and Methods, with MCF-10A cells, knock in derivatives, HER3 knock down and controls, under growth media and assay media conditions. Data are shown as the percentage of cell proliferation under assay conditions (0.2 ng/mL EGF) relative to full growth media conditions at day 6. Error bars represent the standard error of the mean from triplicate samples and data are representative of three independent experiments. * is comparison between HER3 shRNA cells and their parental counterparts, while ** represents comparison between HER3 shRNA E545K versus HER3 shRNA H1047R cells. (B) Cell proliferation assays were performed as described in Materials and Methods, with MCF-7 (E545K), T47D (H1047R) cells, HER3 knock down and controls, with 1.25 nM 17-β-estradiol. Error bars represent the standard deviation from triplicate samples and data are representative of three independent experiments. *p < 0.01.
4 Discussion
Somatic alterations in kinases and phosphatases are found in many human cancers. As such, there has been great enthusiasm for developing small molecular inhibitors against mutant kinases, and the successes of imatinib, gefinitib, erlotinib, crizotinib, vemurafenib, and many other kinase inhibitors speak to the feasibility of this approach. However, despite these achievements, many other kinases have been recalcitrant to targeted therapies. A prime example is the fact that PI3 kinase inhibitors have had limited success in clinical trials for various malignancies and correlating responses with PIK3CA mutations has been inconsistent. Although part of this may be due to differences in PIK3CA mutation detection technologies, passenger mutations reported as PIK3CA mutation positive tumors, and tumor heterogeneity, the fact that most PI3 kinase inhibitors have significant side effects accentuates the point that these inhibitors have off target effects and are not mutation specific. In an effort to define pathways that are distinctly activated or dependent upon specific PIK3CA mutations, we used a global phosphoproteomic approach to define proteins that might be targets for therapy in cancers harboring oncogenic PIK3CA mutations.
Our results demonstrate that both E545K and H1047R PIK3CA mutations rely upon HER3 for full oncogenic pathway activation, though notably, more so for H1047R mutants. Importantly, our SILAC based approach identified differences between E545K and H1047R mutations in terms of phosphorylation status that could not be detected using conventional antibody detection via Western blot. However, our genetic approach using shRNA confirms that knock down of HER3 has a much more pronounced effect on cell growth in breast cells carrying a H1047R mutation versus cells harboring an E545K mutation. Mechanistically, this appeared to be due to the dependence of the H1047R mutation, but not the E545K mutation, on HER3 for MAP kinase signaling. Although the precise manner in which oncogenic PIK3CA mutations can activate HER3 is not yet known, it is tempting to speculate that differences in how these mutant p110 alpha proteins function could lead to preferential phosphorylation of HER3 at critical residues. For example, Huang et al. demonstrated that H1047R mutations may structurally alter p110 alpha allowing the kinase domain to be more accessible to substrates at the cell membrane [22]. In contrast, Wang and colleagues recently showed that E545K mutations rely upon IRS1 for full oncogenic pathway activation [21]. Our use of isogenic cell lines and confirmation in other cancer cell types, strongly support these two studies and demonstrate that both mutations require HER3 for PI3 kinase pathway activation, yet surprisingly, only H1047R mutations depend upon HER3 for MAP kinase activation. Equally intriguing, the mechanism of how oncogenic PIK3CA mutations activate the MAP kinase pathway has not been fully elucidated. The discovery that these mutations can rewire pathways and utilize upstream receptor tyrosine kinases such as HER3 provides a mechanistic explanation of how this may occur, since many receptor tyrosine kinases including HER2/HER3 heterodimers typically activate both MAP kinase and PI3 kinase pathways. Work by the Arteaga laboratory has demonstrated that upregulation of HER3 by mutant PI3 kinase may be mediated by heregulin and its transcriptional activation induced by FoxO family members [18–20]. However, in contrast to their models of overexpression, in our hands, physiologic expression of mutant PIK3CA by gene targeting did not lead to differential upregulation of the erbB ligands heregulin, amphiregulin or TGF-α (data not shown). This would suggest that our isogenic system can be further explored to determine the mechanism of how specific PIK3CA mutations selectively alter signaling pathways. Interestingly, a recent study using the EGFR/HER2 inhibitor lapatinib in ER positive, HER2 negative breast cancer patients demonstrated that tumors with PIK3CA mutations responded to this drug, whereas those without these mutations generally did not [23, 24]. Taken together, these results suggest that targeting pathways activated by specific oncogenic PIK3CA mutations may be possible.
In sum, we have demonstrated the power of combining a phosphoproteomic screen using isogenic cell lines to discover subtle yet distinct changes in phosphorylation patterns used by common oncogenic mutation within the same gene. This approach has the potential for uncovering new signal transduction pathways utilized by cancer mutations that may lend themselves for therapeutic targeting.
Supplementary Material
Acknowledgments
We apologize to our colleagues whose important work could not be cited due to space constraints.
This work was supported by: Career Catalyst Award from the Susan G. Komen for the Cure foundation KG100459 (X.W.), DOD Breast Cancer Research Program BC083057 (M.M.), BC051652 (A.P.), BC100972 (B.G.B.); Flight Attendant Medical Research Institute (FAMRI) (B.H.P.); The Avon Foundation (B.H.P.); The Stetler Fund (J.A.B.); and NIH CA088843 (B.H.P.), U54GM103520 (A.P.), U24CA160036 (A.P.), GM007309 (D.J.Z.), CA168180 (R.L.C.), CA167939 (S.C.), CA62924 (A.P.) and NHLBI contract HHSN268201000032C (A.P.). We thank the Majlis Amanah Rakyat (MARA) of the Government of Malaysia for the research fellowship to M.S.Z. We would also like to thank and acknowledge the support of the Sandy Garcia Charitable Foundation, the Commonwealth Foundation, the Santa Fe Foundation, the Breast Cancer Research Foundation, the Health Network Foundation, the ME Foundation and The Robin Page / Lebor Foundation. None of the funding sources influenced the design, interpretation or submission of this manuscript.
Footnotes
Potential conflict of interest: B.H.P. is a paid member of the scientific advisory boards of Horizon Discovery, Ltd. and Loxo Oncology and has a research contract with Genomic Health, Inc. Under separate licensing agreements between Horizon Discovery, Ltd. and The Johns Hopkins University, B.H.P. is entitled to a share of royalties received by the University on sales of products. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. All other authors have declared no conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher’s web-site
References
- 1.Bachman KE, Argani P, Samuels Y, Silliman N, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sjoblom T, Jones S, Wood LD, Parsons DW, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 4.Yi KH, Axtmayer J, Gustin JP, Rajpurohit A, Lauring J. Functional analysis of non-hotspot AKT1 mutants found in human breast cancers identifies novel driver mutations: implications for personalized medicine. Oncotarget. 2013;4:29–34. doi: 10.18632/oncotarget.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasuda H, Park E, Yun CH, Sng NJ, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuels Y, Wang Z, Bardelli A, Silliman N, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 8.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 9.Gustin JP, Karakas B, Weiss MB, Abukhdeir AM, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci USA. 2009;106:2835–2840. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong J, Kim MS, Chaerkady R, Wu X, et al. TSLP signaling network revealed by SILAC-based phosphoproteomics. Mol Cell Proteomics. 2012;11:M112.017764. doi: 10.1074/mcp.M112.017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim MS, Zhong Y, Yachida S, Rajeshkumar NV, et al. Heterogeneity of pancreatic cancer metastases in a single patient revealed by quantitative proteomics. Mol Cell Proteomics. 2014;13:2803–2811. doi: 10.1074/mcp.M114.038547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abukhdeir AM, Vitolo MI, Argani P, De Marzo AM, et al. Tamoxifen-stimulated growth of breast cancer due to p21 loss. Proc Natl Acad Sci USA. 2008;105:288–293. doi: 10.1073/pnas.0710887105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konishi H, Karakas B, Abukhdeir AM, Lauring J, et al. Knock-in of mutant K-ras in nontumorigenic human epithelial cells as a new model for studying K-ras mediated transformation. Cancer Res. 2007;67:8460–8467. doi: 10.1158/0008-5472.CAN-07-0108. [DOI] [PubMed] [Google Scholar]
- 14.Lauring J, Abukhdeir AM, Konishi H, Garay JP, et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111:856–864. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karakas B, Weeraratna AT, Abukhdeir AM, Konishi H, et al. p21 Gene knock down does not identify genetic effectors seen with gene knock out. Cancer Biol Ther. 2007;6:1025–1030. doi: 10.4161/cbt.6.7.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang GM, Wong HY, Konishi H, Blair BG, et al. Single copies of mutant KRAS and mutant PIK3CA cooperate in immortalized human epithelial cells to induce tumor formation. Cancer Res. 2013;73:3248–3261. doi: 10.1158/0008-5472.CAN-12-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakrabarty A, Rexer BN, Wang SE, Cook RS, et al. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29:5193–5203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakrabarty A, Sanchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci USA. 2012;109:2718–2723. doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanker AB, Pfefferle AD, Balko JM, Kuba MG, et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci USA. 2013;110:14372–14377. doi: 10.1073/pnas.1303204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao Y, Wang C, Cao B, Hirsch BM, et al. Gain of interaction with IRS1 by p110alpha-helical domain mutants is crucial for their oncogenic functions. Cancer Cell. 2013;23:583–593. doi: 10.1016/j.ccr.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 23.Guarneri V, Generali DG, Frassoldati A, Artioli F, et al. Double-blind, placebo-controlled, multicenter, randomized, phase IIb neoadjuvant study of letrozole-lapatinib in post-menopausal hormone receptor-positive, human epidermal growth factor receptor 2-negative, operable breast cancer. J Clin Oncol. 2014;32:1050–1057. doi: 10.1200/JCO.2013.51.4737. [DOI] [PubMed] [Google Scholar]
- 24.Zabransky DJ, Park BH. Estrogen receptor and receptor tyrosine kinase signaling: use of combinatorial hormone and epidermal growth factor receptor/human epidermal growth factor receptor 2-targeted therapies for breast cancer. J Clin Oncol. 2014;32:1084–1086. doi: 10.1200/JCO.2013.53.5070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.