Abstract
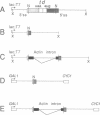
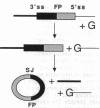
Studies on the function of circular RNA and RNA topology in vivo have been limited by the difficulty in expressing circular RNA of desired sequence. To overcome this, the group I intron from the phage T4 td gene was split in a peripheral loop (L6a) and rearranged so that the 3' half intron and 3' splice site are upstream and a 5' splice site and 5' half intron are downstream of a single exon. The group I splicing reactions excise the internal exon RNA as a circle (RNA cyclase ribozyme activity). We show that foreign sequences can be placed in the exon and made circular in vitro. Expression of such constructs (RNA cyclase ribozymes) in Escherichia coli and yeast results in the accumulation of circular RNA in these organisms. In yeast, RNA cyclase ribozymes can be expressed from a regulated promoter like an mRNA, containing 5' leader and 3' trailer regions, and a nuclear pre-mRNA intron. RNA cyclase ribozymes have broad application to questions of RNA structure and function including end requirements for RNA transport or function, RNA topology, efficacy of antisense or ribozyme gene control elements, and the biosynthesis of extremely long polypeptides.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993 Jun 4;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- Chan W. K., Belfort G., Belfort M. Stability of group I intron RNA in Escherichia coli and its potential application in a novel expression vector. Gene. 1988 Dec 20;73(2):295–304. doi: 10.1016/0378-1119(88)90494-5. [DOI] [PubMed] [Google Scholar]
- Chandry P. S., Belfort M. Activation of a cryptic 5' splice site in the upstream exon of the phage T4 td transcript: exon context, missplicing, and mRNA deletion in a fidelity mutant. Genes Dev. 1987 Nov;1(9):1028–1037. doi: 10.1101/gad.1.9.1028. [DOI] [PubMed] [Google Scholar]
- Cocquerelle C., Daubersies P., Majérus M. A., Kerckaert J. P., Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992 Mar;11(3):1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Couture S., Szostak J. W. A multisubunit ribozyme that is a catalyst of and template for complementary strand RNA synthesis. Science. 1991 Mar 29;251(5001):1605–1608. doi: 10.1126/science.1707185. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Galloway Salvo J. L., Coetzee T., Belfort M. Deletion-tolerance and trans-splicing of the bacteriophage T4 td intron. Analysis of the P6-L6a region. J Mol Biol. 1990 Feb 5;211(3):537–549. doi: 10.1016/0022-2836(90)90264-m. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Zaug A. J., Cech T. R. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena. Cell. 1981 Feb;23(2):467–476. doi: 10.1016/0092-8674(81)90142-2. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Harland R., Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988 Apr;102(4):837–852. doi: 10.1242/dev.102.4.837. [DOI] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J Mol Biol. 1986 May 5;189(1):143–165. doi: 10.1016/0022-2836(86)90387-6. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Rosbash M. Efficient trans-splicing of a yeast mitochondrial RNA group II intron implicates a strong 5' exon-intron interaction. Science. 1986 Nov 28;234(4780):1099–1104. doi: 10.1126/science.2430332. [DOI] [PubMed] [Google Scholar]
- Jarrell K. A., Dietrich R. C., Perlman P. S. Group II intron domain 5 facilitates a trans-splicing reaction. Mol Cell Biol. 1988 Jun;8(6):2361–2366. doi: 10.1128/mcb.8.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. A. Inverse splicing of a group II intron. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8624–8627. doi: 10.1073/pnas.90.18.8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems J., Garrett R. A. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell. 1988 Aug 26;54(5):693–703. doi: 10.1016/s0092-8674(88)80014-x. [DOI] [PubMed] [Google Scholar]
- Macejak D. G., Sarnow P. Internal initiation of translation mediated by the 5' leader of a cellular mRNA. Nature. 1991 Sep 5;353(6339):90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Nigro J. M., Cho K. R., Fearon E. R., Kern S. E., Ruppert J. M., Oliner J. D., Kinzler K. W., Vogelstein B. Scrambled exons. Cell. 1991 Feb 8;64(3):607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- Pan T., Gutell R. R., Uhlenbeck O. C. Folding of circularly permuted transfer RNAs. Science. 1991 Nov 29;254(5036):1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- Pan T., Uhlenbeck O. C. Circularly permuted DNA, RNA and proteins--a review. Gene. 1993 Mar 30;125(2):111–114. doi: 10.1016/0378-1119(93)90317-v. [DOI] [PubMed] [Google Scholar]
- Price J. V., Engberg J., Cech T. R. 5' exon requirement for self-splicing of the Tetrahymena thermophila pre-ribosomal RNA and identification of a cryptic 5' splice site in the 3' exon. J Mol Biol. 1987 Jul 5;196(1):49–60. doi: 10.1016/0022-2836(87)90510-9. [DOI] [PubMed] [Google Scholar]
- Puttaraju M., Been M. D. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 1992 Oct 25;20(20):5357–5364. doi: 10.1093/nar/20.20.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaraju M., Perrotta A. T., Been M. D. A circular trans-acting hepatitis delta virus ribozyme. Nucleic Acids Res. 1993 Sep 11;21(18):4253–4258. doi: 10.1093/nar/21.18.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D. Replication and evolution of viroid-like pathogens. Curr Top Microbiol Immunol. 1992;176:213–219. doi: 10.1007/978-3-642-77011-1_14. [DOI] [PubMed] [Google Scholar]
- Szostak J. W. Enzymatic activity of the conserved core of a group I self-splicing intron. Nature. 1986 Jul 3;322(6074):83–86. doi: 10.1038/322083a0. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Van der Horst G., Kamps A. M., Arnberg A. C. Interlocked RNA circle formation by a self-splicing yeast mitochondrial group I intron. Cell. 1987 Jan 16;48(1):101–110. doi: 10.1016/0092-8674(87)90360-6. [DOI] [PubMed] [Google Scholar]
- Waugh D. S., Pace N. R. Gap-scan deletion analysis of Bacillus subtilis RNase P RNA. FASEB J. 1993 Jan;7(1):188–195. doi: 10.1096/fasebj.7.1.7678561. [DOI] [PubMed] [Google Scholar]
- Woodson S. A., Cech T. R. Alternative secondary structures in the 5' exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991 Feb 26;30(8):2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- Zavanelli M. I., Ares M., Jr Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes Dev. 1991 Dec;5(12B):2521–2533. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]