Abstract
p73 is a structural and functional homologue of the p53 tumor suppressor protein. Like p53, p73 induces apoptosis and cell cycle arrest and transactivates p53-responsive genes, conferring its tumor suppressive activity. In addition, p73 has unique roles in neuronal development and differentiation. The importance of p73-induced apoptosis lies in its capability to substitute the pro-apoptotic activity of p53 in various human cancer cells in which p53 is mutated or inactive. Despite the great importance of p73-induced apoptosis in cancer therapy, little is known about the molecular basis of p73-induced apoptosis. In this review, we discuss the p73 structures reported to date, detailed structural comparisons between p73 and p53, and current understanding of the transcription-dependent and -independent mechanisms of p73-induced apoptosis. [BMB Reports 2015; 48(2): 81-90]
Keywords: Apoptosis, Cancer therapy, p53 protein family, p73, Structure
INTRODUCTION
Since its first discovery in 1997, p73, a member of the p53 family of transcription factors, has become one of the most extensively studied proteins because of the possibility of replacing p53 with p73 in p53-defective cells due to its structural and functional homologies with p53. p73 has cellular activities similar to those of p53, including binding and transactivation of p53-responsive genes and induction of apoptosis and cell cycle arrest, which confer the tumor-suppressive activities of p73 (1). In addition to its p53-like functions, p73 fulfills its own unique roles including roles in neuronal development and differentiation (1), metabolic control (2), and spermatogenesis and the maintenance of male fertility (3). The observation that p73 is rarely mutated in human cancers (< 1%), unlike p53 which is mutated in more than 50% of human cancers (1), and the functional overlaps of p73 and p53, suggest that p73 can substitute the pro-apoptotic roles of p53 in the majority of cancers in which p53 is mutated or inactive.
More than 10 p73 variants have been identified, generated using two promoters at the N-terminus and alternative splicing at the C-terminus (4). Among these, however, p73 exists as two major isoforms with opposing activities; the pro-apoptotic transcriptionally active (TA) isoform and the anti-apoptotic N-terminal transactivation domain (TAD)-truncated delta-N (DN) isoform (5). The DNp73 isoform is dominant negative toward TAp73 and wild-type p53, thus inhibiting pro-apoptotic activities.
The stability and activity of p73 are primarily regulated by post-translational modifications (PTMs). In healthy cells, both the TAp73 and DNp73 isoforms are degraded by either calpains (6) or in an ubiquitin-dependent or -independent manner (7, 8), which keeps their steady-state levels low. p73 is degraded by NAD(P)H quinone oxidoreductase 1 and NADH through the ubiquitin-independent 20S proteasome (7), and is also subject to ubiquitin-dependent 26S proteasomal degradation mediated by the HECT-domain E3 ubiquitin ligase Itch (8). Additional negative control occurs through competitive binding between p73 and Nedd4-binding protein 1 (N4BP1) and yes-associated protein (YAP) (9, 10). In response to genotoxic stress such as DNA damage, however, TAp73 rapidly accumulates due to tyrosine phosphorylation by c-Abl, acetylation by p300 (11, 12), and Itch down-regulation (8). In addition to these non-selective degradations, selective degradation of the DNp73 isoform by RING finger-domain E3 ubiquitin ligase p73-induced ring finger protein 2 (PIR2) (13) and other molecules (14-16) leads to greater accumulation of TAp73 than the DNp73 isoform, allowing TAp73 to induce apoptosis. Thus, the TA/DNp73 isoform ratio controls apoptotic activity and consequently, cell fate and oncogenesis following DNA damage (17-19). Indeed, DNp73 levels are elevated in several human cancers (5, 18-21).
The capability of both p53 and TAp73 isoforms to induce apoptosis is central to their tumor suppressive activities (22). While p53 induces apoptosis by transactivating pro-apoptotic genes such as Bax (23), p53 up-regulated modulator of apoptosis (PUMA) (24), and Noxa (25) and death receptor CD95 (26), it can also induce apoptosis by acting directly in the cytoplasm and mitochondria (27-30). p53 induces apoptosis both by inhibiting anti-apoptotic Bcl-XL to release pro-apoptotic Bcl-2 homology 3 (BH3)-only proteins sequestered by Bcl-XL and by directly activating the Bak effector in the cytoplasm (27, 28, 31-34). It has also been suggested that when cytoplasmic p53 is sequestered by Bcl-XL , PUMA, induced by p53, in turn, releases p53 from Bcl-XL to trigger Bax oligomerization and thus cytochrome c release from mitochondria (29).
In contrast to the well-known mechanisms of p53-induced apoptosis, our understanding of the mechanism of p73-induced apoptosis is incomplete, despite several studies to date. p73 induces apoptosis in both transcription-dependent and -independent manners. Although the pro-apoptotic activity of p73 as a transactivator is relatively well understood, its transcription-independent apoptosis mechanism is less understood. This transcription-independent mechanism is of importance because it is responsible for the rapid induction of apoptosis (27). In addition, despite several domain structures of p73 reported to date, transcription-independent apoptosis of p73 has never been understood in terms of protein structure. In this review, we discuss the p73 structures and molecular mechanisms of p73-induced apoptosis reported to date and compare them with those of p53 to provide mechanistic insights into apoptosis mediated by non-canonical p53 family members.
p73 STRUCTURE
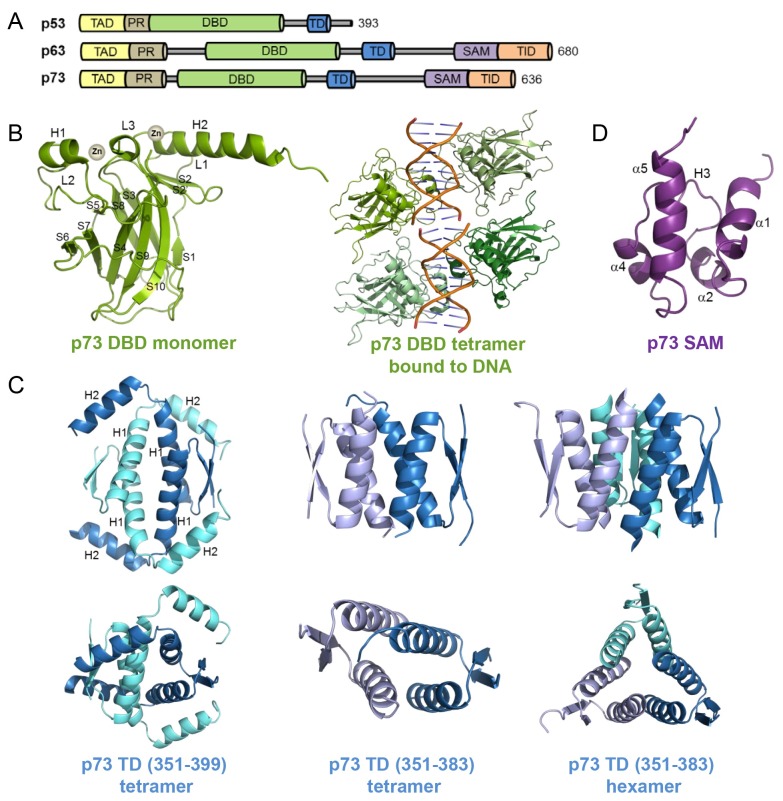
p53 family members, including p53, p63, and p73, share high sequence homology as well as domain organization. They consist of an intrinsically disordered transactivation domain (TAD) followed by a proline-rich domain (PR) at their N-termini. They also contain a DNA-binding domain (DBD) and a tetramerization domain (TD) at the center (Fig. 1A). An additional sterile alpha motif (SAM) domain and a transcription-inhibition domain (TID) are present only in the C-termini of p63 and p73, but not in p53 (Fig. 1A).
Fig. 1. Structures of p73. (A) Domain organization of p53 family members, p53, p63, and p73. TAD, transactivation domain; PR, proline-rich domain; DBD, DNA-binding domain; TD, tetramerization domain; SAM, sterile alpha motif, TID transcription-inhibition domain (TID) (B) Crystal structures of p73 DBD in its free (left, PDB code: 2XWC) and response elements (REs)-bound (right, PDB code: 2VD0) states. (C) Three different p73 TD crystal structures: long form tetramer (left, PDB code: 2WQI), short form tetramer (middle, PDB code: 2WQJ), and short form hexamer (right, PDB code: 2WQJ). Structures shown at the bottom are rotated views of those at the top. (D) Solution structure of p73 SAM domain (PDB code: 1COK).
Transactivation domain (TAD)
p73 TAD consists of two distinctive subdomains, TAD1 and TAD2, corresponding to residues 1-31 and 46-67, respectively, that are connected by a linker encompassing residues 33-45. Like p53 (35), p73 TAD is intrinsically disordered under physiological conditions (36) and serves as a promiscuous binding site for a variety of interacting partners. In 2009, Burge et al. reported that p73 TAD could bind to four different domains of transcriptional coactivator p300 by biophysical methods (36). While the two subdomains of p73 TAD are involved in binding to the Taz1 and Taz2 domains of p300, only the p73 TAD1 subdomain is necessary for binding to the Kix and IBiD domains of p300. Dissociation constants for these bindings between p73 TAD and p300 range from 0.89 to 25.5 μM, which is weaker than those between p53 TAD and p300, as determined by fluorescence anisotropy. For Taz1 and Taz2, both p73 TAD subdomains are required for maximum binding to p73, as each TAD subdomain alone has weaker affinity for Taz1 or Taz2 than full TAD, suggesting cooperative roles of p73 TAD subdomains for binding to Taz1 or Taz2 domains.
The hydrophobic motif within TAD is known to be critical for protein-protein interaction. p53 QS mutants (L22Q/W23S and W53Q/F54S) have been reported to impair transcriptional activity due to reduced binding to p300 (37). Similarly, p73 QS mutation (L18Q/W19S) within TAD abrogates binding to p300 domains, suggesting the importance of the motif in binding to p300 (36). Consistent with this, compared with wildtype p73, the p73 QS mutant shows a marked reduction in transactivation of pro-apoptotic Bax expression, as evidenced by a luciferase assay using H1299 cells. This suggests the critical role of hydrophobic residues within TAD in its pro-apoptotic function.
In contrast to the disorder of p73 TAD upon binding to p300 domains, as evidenced by lack of amide chemical shift dispersion (36), eight residues within p73 TAD adopt an α-helical conformation when bound to the E3 ubiquitin ligase Mdm2 in our recently determined structure (38). The helical conformation of p73 TAD when bound to its binding partner corresponds to the previously reported helical conformation of a fragment within the p53 TAD1 upon binding to Mdm2 (39). This suggests that the intrinsically disordered p73 TAD is highly versatile in structure and probably capable of adopting multiple conformations depending on its binding targets.
DNA-binding domain (DBD)
In contrast to the low sequence identity of TAD between p53 family members (22% identity between p63 and p53 and 30% identity between p73 and p53), their DBDs share high sequence identity (60% identity between p63 and p53 and 63% identity between p73 and p53) (1). Among the DBDs of p53 family members, p73 DBD was the last domain to be determined structurally. In 2012, structures of p73 DBD in both its free and DNA-bound states were determined by X-ray crystallography (40, 41) (Fig. 1B).
As expected from the high sequence homology of p73 DBD with p53 and p63 DBDs, the free p73 DBD structure (Fig. 1B, left) is highly similar to those of p53 and p63 (40). It consists of a β-sandwich fold with two anti-parallel β-sheets, one containing four β-strands (S1, S3, S5, and S8) and the other containing five β-strands (S4, S6, S7, S9, and S10), and three long loops emerging from the β-sandwich fold. There are two coordinated Zn2+ ions that are critical for dimerization and DNA binding. One is coordinated by Cys194 and His197 from the H1 helix and Cys258 and Cys262 from the L3 loop, while the other is trapped at the interface of the crystallographic dimer formed by the H2 helix and the C-terminal tail (40).
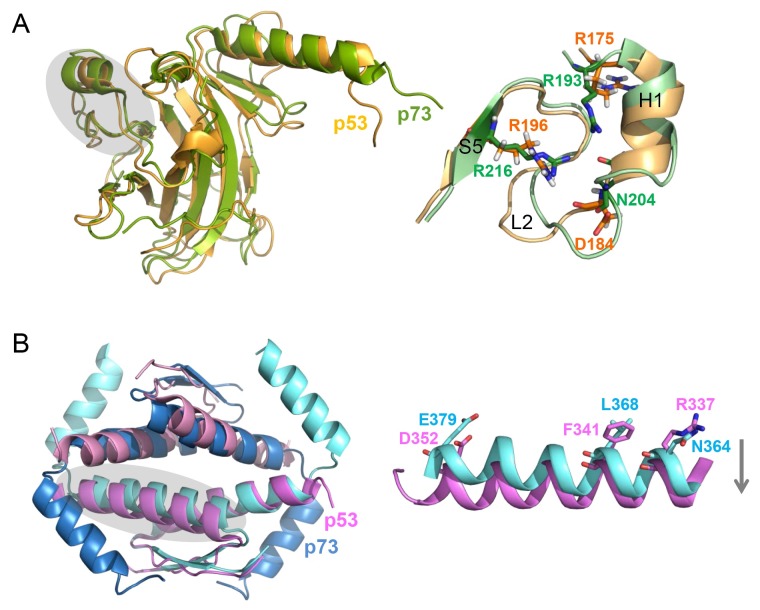
p73 DBD forms a tetramer in the presence of DNA, following dimerization on the DNA, whereas it remains a monomer in the absence of DNA (41). Despite the highly homologous structure of p53 and p73 DBDs (the overall Cα r.m.s. deviation of 1.7 Å), the most divergent region in the structure is identified as the L2 loop, encompassing p73 residues 182-212 (40) (Fig. 2A). In contrast to the high sequence homology in the H1 helix of p53 family members, sequence homology in the L2 loop is relatively low. p73 and p63 contain two more residues than p53 in this region. This leads to a significantly different conformation at p73 residues R201-A209 compared to the corresponding p53 residues C182-A189 (Fig. 2A, right). This L2 loop is anchored to the core β-sandwich fold by two arginine residues, R193 and R216. Although the conformation of these p73 residues is similar to the corresponding p53 residues, R175 and R196, their interaction with aspartate residues is different. p53 D184 forms a salt bridge and a hydrogen bond with R175 and R196, respectively, which are critical for p53 folding. Thus, oncogenic p53 mutation at R175H is regarded as the most disruptive factor for p53 activity (42). On the contrary, p73 D202, equivalent to D184 in p53, is not involved in any interactions with aspartates. Instead, a loose helical turn of p73, induced by the two extra residues, places N204 in the position to interact with R193 and R216.
Fig. 2. Structural comparison between p73 and p53. (A) Superimposed structure of p73 (green, PDB code: 2XWC) and p53 DBD (orange, PDB code: 2FEJ) (left). The L2 loop region of p73 and p53 (right), shaded in the structures on the left, shows the most distinct structural difference between them. (B) Superimposed structure of p73 (blue, PDB code: 2WQI) and p53 TD (pink, PDB code: 1C26) tetramers (left). The p53 helix is farther apart than the p73 helix, shaded on the left, as shown in the monomer (right).
In addition to the free structure, crystal structures of the p73 DBD tetramer bound to response elements (REs) (Fig. 1B, right) with spacers of different lengths have been determined (41). The overall fold between free and DNA-bound p73 DBD is very similar to each other (r.m.s. deviation of 0.8 Å). DNA is bound to a p73 DBD via a loop-sheet-helix motif formed by L1 and L3 loops, S10 strand and H2 helix. Interestingly, when the p73 DBD bound to the DNA with different spacer lengths between the RE half site, their quaternary structures changed significantly. While the structures bound to the DNA with zero or one base pair spacers resulted in a compact p73 DBD structure and a large tetramerization interface, those with two base -pair spacers led to DNA unwinding and a smaller interface. In addition, the transcriptional activity of p73 decreased dramatically with any insertion, whereas p53 tolerated up to 4 base pairs of spacers in a yeast-based functional assay. Thus, the spacer between half site REs determines the p73 DBD quaternary structure and subsequent transcriptional activity of p73.
Tetramerization domain (TD)
The main difference between the p63 and p73 TDs and that of p53 is the presence of an additional C-terminal helix in p63 and p73. The TDs of p63 and p73 share ∼40% sequence identity with that of p53. In 2009, Joerger et al. solved the crystal structures of p73 TD using two different versions: a short form (residues 351-383) corresponding to p53 TD and a long form (residues 351-399) containing an additional C-terminal helix (43).
The overall fold of the long form of p73 TD is that of a dimer of dimers (Fig. 1C, left). Each monomer consists of a short β-strand, an α-helix with a C-terminal 310-helical segment (H1) and the additional C-terminal helix (H2) unique to p63 and p73. Two monomers form a symmetrical primary dimer via an anti-parallel intermolecular β-sheet and an antiparallel helical orientation. Two such primary dimers then form a tetramer with a 2-fold symmetry via helix-helix packing interactions. At the core of the tetramer interface, H1 helices are arranged in a four-helical bundle. Additional dimer-dimer contact occurs at the H2 helices, which clasp the second dimer, packing against H1 helices. In the tetramer structure of the p73 TD short form (Fig. 1C, middle), despite the similar overall fold between the long and short forms, the packing angles of their H1 helices are significantly different: 160o in the long form and ∼135o in the short form. This packing angle difference is possibly due to the stabilizing effect of the H2 helix on the overall fold of p73 TDs. In addition, the hexamer structure of the p73 TD short form shows that three dimers are aligned along the 3-fold axis, with each helix packed against a helix from a neighboring dimer (Fig. 1C, right).
There are two significant structural differences between p53 and p73 TDs. The crystal structure of p53 TD shows nearly orthogonal packing of two dimers with an angle of 81o, however, this dimer-dimer packing angle is twisted by ∼16o in the p73 structure. Despite this distortion in the long form of p73 TD, the overall packing of the p73 long form is still closer to that of p53 TD than the p73 TD short form (Fig. 2B). Due to the stabilizing effect of the H2 helix of p73, the total buried surface area of p73 TD (8,420 Å2) is found to be larger than that of p53 TD (6.720 Å2). An additional structural difference between p53 and p73 can be found in the H1 helical packing. The H1 helices pack farther apart in p53 than in p73. This is attributed to large-to-small substitutions of the two critical residues at the dimer interface, for example, Phe341 in p53 and Leu368 in p73.
Sterile alpha motif (SAM)
An additional C-terminal extension is found in both p73 and p63 at their C-termini, but not in p53. We determined the solution structure of the C-terminal extension of p73 and identified it as a SAM domain (44). In the solution structure of the C-terminal region of p73 encompassing amino acids 487-554, there are four α-helices (α1, α2, α4, and α5) and a short 310 helix (H3) between the α2 and α4 helices, thus forming a five helical bundle (Fig. 1D). The α1 and α5 helices run in an anti-parallel orientation with α2, H3, and α4. Notably, DALI structural analysis reveals that this p73 domain has high structural similarities with the SAM domains of two ephrin receptor tyrosine kinases, EphB2 (45) and EphA4 (46). The SAM domain mediates a protein-protein interaction that forms both homo- and hetero-oligomers. However, such oligomers are not formed with either p63 or p73; they remain monomeric. This observation indicates the possibility that they may interact with other proteins that have unidentified yet.
TRANSCRIPTION-DEPENDENT p73-INDUCED APOPTOSIS
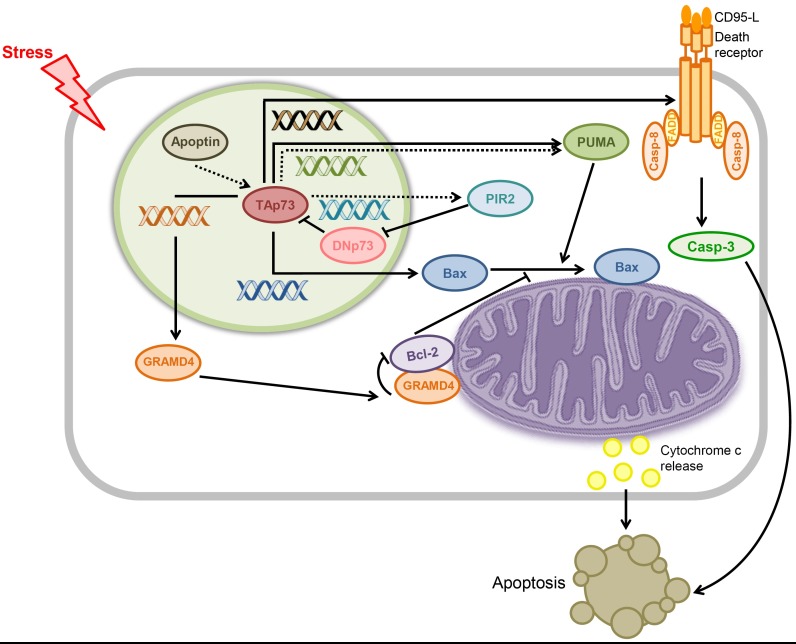
The capability of p73 to induce apoptosis is generally attributed to the transactivation of pro-apoptotic genes by p73. p73-responsive target genes include PUMA and Bax (classical pro-apoptotic Bcl-2 family members) as well as the recently identified GRAMD4 and viral protein apoptin in an intrinsic apoptotic pathway and the death receptor CD95 in an extrinsic apoptotic pathway. In these mechanisms, p73 indirectly induces apoptosis within the nucleus by transactivating the genes, enabling them to act in the mitochondria or cytoplasm (Fig. 3).
Fig. 3. Proposed mechanism for transcription-dependent p73-induced apoptosis. In response to cellular stress, the pro-apoptotic, transcriptionally active p73 (TAp73) isoform transactivates pro-apoptotic genes such as Bax and GRAMD4 and apoptin, whereas the anti-apoptotic transcriptionally inactive p73 (DNp73) isoform inhibits the pro-apoptotic activity of TAp73. In turn, overexpressed pro-apoptotic proteins act directly on the mitochondria to trigger apoptosis. For the extrinsic apoptotic pathway, apoptosis is initiated upon binding of CD95-L to the CD95 death receptor, induced by p73. The apoptotic activity of nuclear p73 is indirect via these mechanisms. Dotted arrows indicate the activation pathway for the viral protein apoptin and the solid arrows designate the pathways for other human proteins.
PUMA and Bax
Bcl-2 family members are critical regulators of apoptosis. The pore-forming activities of Bax/Bak, which are critical in apoptosis, are tightly regulated by complex interactions between pro- and anti-apoptotic members of the Bcl-2 family (47). The Bcl-2 family is categorized into three subfamilies: anti-apoptotic proteins, pro-apoptotic effectors, and pro-apoptotic BH3-only proteins (47). Anti-apoptotic Bcl-2 family proteins such as Bcl-XL , Bcl-2, and Bcl-w preserve outer mitochondrial membrane (OMM) integrity by directly inhibiting pro-apoptotic Bcl-2 proteins. The pro-apoptotic effectors Bax and Bak homo-oligomerize in the OMM to promote mitochondrial outer membrane permeabilization (MOMP). The pro-apoptotic BH3-only proteins are further divided functionally into two groups: sensitizers/de-repressors and direct activators. Where sensitizer/derepressors bind only to the anti-apoptotic Bcl-2 family proteins to inhibit their anti-apoptotic functions but do not cause apoptosis directly, direct activators such as Bim and Bid interact with anti-apoptotic Bcl-2 family proteins as well as effectors, resulting in the direct activation of the effectors to promote oligomerization and MOMP.
In 2004, Melino et al. demonstrated a p73-induced apoptosis mechanism that involved the pro-apoptotic BH3 only proteins, PUMA and Bax, as mediators. PUMA is induced by direct transactivation of p73 in response to DNA damage and subsequently translocates the pro-apoptotic effector molecule Bax to the mitochondria and activates it to trigger cytochrome c release (48). Although p73 transactivates both PUMA and Bax, the kinetics of p73-mediated apoptosis is incompatible with increased protein levels of Bax but is compatible with its mitochondrial translocation. This suggests that the effect of Bax overexpression initiated by p73 on p73-induced apoptosis is indirect - Bax induction is insufficient to trigger apoptosis. Instead, PUMA, overexpressed by p73, causes mitochondrial translocation of Bax, thereby releasing mitochondrial cytochrome c and resulting in apoptotic cell death. Thus, p73-induced apoptosis is correlated with PUMA-mediated mitochondrial translocation of Bax, rather than with direct transactivation of Bax by p73. In addition, DNp73 isoforms inhibit apoptosis mediated by TAp73 or p53, thus acting as dominant negatives toward their pro-apoptotic activities. During p73-dependent apoptosis, p73 is localized and remains within the nucleus, whereas Bax translocates from the cytosol to the mitochondria to trigger cytochrome c release, indicating an indirect interaction between p73 and Bax.
GRAMD4
Another p73 target gene during p73-induced apoptosis is GRAMD4. Recently, John et al. reported that p73-induced apoptosis is mediated by the transactivation and mitochondrial translocation of GRAMD4 by p73. GRAMD4 facilitates the mitochondrial translocation and oligomerization of Bax in the mitochondria, resulting in apoptosis (49). GRAMD4 consists of a nuclear localization signal in the N-terminus, two transmembrane domains in the middle, and a GRAM domain at the C-terminus. GRAMD4 was previously reported to enhance apoptosis when up-regulated by transcription factor E2F1 or overexpressed in various cancer cells (50). GRAMD4 is responsive to p73, but not to p53, and p73-induced apoptosis is mediated by mitochondrial translocation of GRAMD4 from the nucleus. Interestingly, the ectopically expressed GRAMD4 up-regulates Bax and down-regulates Bcl-2, decreasing the Bcl-2/Bax ratio, which is a signature of apoptosis that is also seen with p53. Because GRAMD4 lacks a DNA-binding or transactivation domain, this influence of p73 on the Bax and Bcl-2 levels might be indirect, probably at the PTM level.
Notably, GRAMD4 mimics the transcription-independent apoptotic function of p53 in the mitochondria. Like p53, GRAMD4 physically interacts with anti-apoptotic Bcl-2 and facilitates mitochondrial translocation and oligomerization of Bax in the mitochondria, resulting in MOMP. Despite the absence of a BH3 domain that mediates protein-protein interaction in the Bcl-2 family of proteins, GRAMD4 binds to Bcl-2 via a Bcl-2-like domain located between its N-terminal domain and the first TM domain. It has been suggested that the inhibitory complex formed between GRAMD4 and Bcl-2 releases Bax sequestered by Bcl-2, activating the released Bax to cause MOMP. In this p73-induced apoptosis mechanism, the apoptotic activity of p73 at the mitochondria is indirect and transcription-dependent. It requires GRAMD4, which acts directly in the mitochondria in a manner similar to that of p53.
Apoptin
Apoptin is a chicken anemia virus-derived protein that induces apoptosis selectively in cancer cells but not in normal cells. Taebunpakul et al. proposed a cellular mechanism for apoptin-induced apoptosis mediated by two molecules: pro-apoptotic BH3 protein PUMA and RING finger-domain E3 ubiquitin ligase PIR2 (51). The overexpression of apoptin increases the stability of TAp73 but has no effect on the anti-apoptotic DNp73 isoform, suggesting the pro-apoptotic activity of apoptin. In turn, p73 transactivates PUMA to trigger apoptosis; PUMA knock-down reduces cell death in apoptin-induced apoptosis. The increased stability of TAp73 by apoptin overexpression is attributed to the expression of PIR2, which selectively degrades the DNp73 isoform. Once apoptin stabilizes p73 in tumor cells, p73, in turn, transactivates PUMA and PIR2 to trigger apoptosis and degrade the anti-apoptotic DNp73 isoform, respectively. Thus, apoptin-induced cell death depends on the transcriptional activation of TAp73 in the nucleus.
CD95
In addition to the intrinsic apoptotic pathway, TAp73 also induces apoptosis via the extrinsic pathway. CD95 (FAS, TNFRSF6, APO-1) is a cell surface receptor that induces apoptosis when bound to its natural ligand, CD95L, a member of the tumor necrosis factor (TNF) cytokine family. Like p53, p73 as well as p63 transactivates the CD95 gene by binding to the p53-binding site within the gene promoter, causing cell death through the death receptor via a caspase-dependent pathway (52, 53). Cooperation among the p53 family members in transactivating the CD95 target gene results in maximal transcriptional activation of the gene. When p73 was activated by either cisplatin or ectopic expression, CD95 was up-regulated, whereas death-inducing signaling complex (DISC) proteins including FADD, FLICE, and c-FLIP were unaffected (53). This finding suggests that p73 induces apoptosis as a transactivator via the extrinsic pathway.
TRANSCRIPTION-INDEPENDENT p73-INDUCED APOPTOSIS
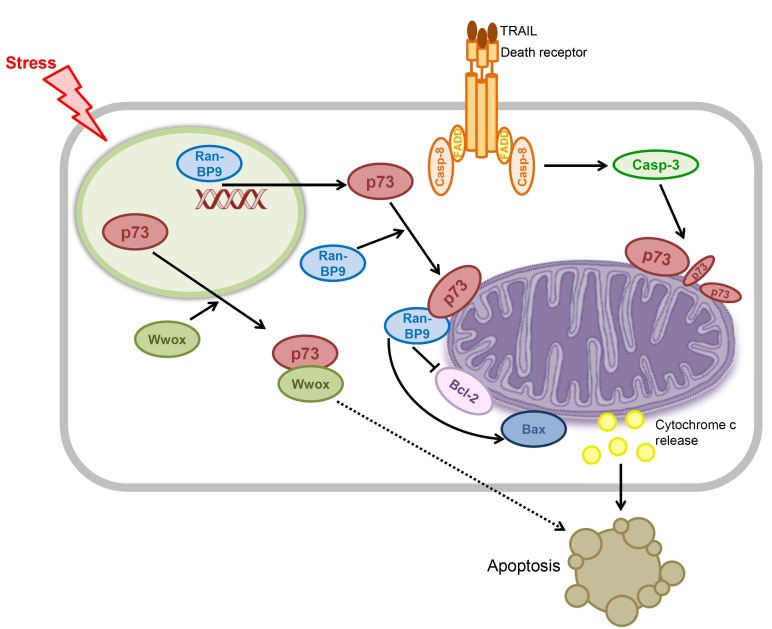
Contrary to the observation that p73 is localized and remains within the nucleus during apoptosis (48), accumulating evidence indicate that it is also localized in the cytosol and mitochondria during apoptosis, suggesting direct, exonuclear apoptotic roles of p73, independent of its transcriptional activities. The following sections describe several mediators of transcription-independent apoptosis induced by p73 (Fig. 4).
Fig. 4. Proposed mechanism for transcription-independent p73-induced apoptosis. p73 induces apoptosis by acting directly within the cytoplasm and mitochondria, independent of its transcriptional activity. p73 requires a mediator to trigger apoptosis in some mechanisms but not in others. It is unclear whether Wwox proceeds via the mitochondrial apoptotic pathway (dotted arrow).
WW domain-containing oxidoreductase (Wwox)
Wwox is a 46 kDa protein that has biological implications in tumor suppression. The WW domain contains ∼40 amino acids that recognize proline-containing ligands in protein-protein interactions that can be regulated by tyrosine phosphorylation. The first WW domain of Wwox interacts directly with the PPxY motif of p73 in the cytoplasm both in vivo and in vitro (54). Whereas p73 and Wwox are separately localized in the nucleus and cytoplasm, respectively, co-transfection of p73 and Wwox sequesters p73 in the cytoplasm, where the two interact with each other. Cytoplasmic localization of p73 is thought to be mediated by the association of the p73 C-terminal nuclear export signal with exportin-1 (55). In addition, phosphorylation of Wwox at tyrosine 33 (within the first WW domain) by Src, a non-receptor tyrosine kinase, enhances binding to p73.
Two Wwox mutants, WW-Y33R and WW-Y61R, mutated within the first and second WW domains, respectively, were generated to map the region of Wwox responsible for p73 binding and to determine the effect of the tyrosine residue, which is critical for protein-protein interaction. Although p73 binding still occurred with the WW-61R mutant, it was nonexistent in the WW-33R mutant, as evidenced by co-immunoprecipitation in 293 cells. Consistent with this finding, co-transfection with the WW-Y61R mutant and p73 caused cytoplasmic translocation of p73 in NIH 3T3 cells. However, this translocation did not occur after co-transfection with the WWY33R mutant and p73 as evidenced by confocal microscopy. These findings suggest that the first WW domain of Wwox is indispensable for direct binding to p73 and for cytoplasmic localization of nuclear p73.
Cytoplasmic localization of p73 suppresses its transcriptional activity, as evidenced by a luciferase assay. However, fluorescence-activated cell sorting analysis (FACS) for DNA content showed increases in apoptotic activity, suggesting a transcription-independent apoptosis. This effect has been suggested to be due to the interaction between p73 and Wwox in the cytoplasm, with p73 enhancing the pro-apoptotic activity of Wwox. This is further supported by the observation that anti-apoptotic DNp73 also interacts with Wwox in the cytoplasm and induces apoptosis. Thus, the transcription-independent apoptotic activity of p73 is thought to be mediated by Wwox, not caused by direct involvement of p73.
RanBP9
Recently, Liu et al. demonstrated that p73 induces apoptosis by cooperating with RanBP9 in the mitochondria (56). RanBP9 is a scaffolding protein that brings together low density lipoprotein receptor-related protein (LRP), amyloid precursor protein (APP), and site app cleaving enzyme 1 (BACE1) via interactions with their cytoplasmic tails. RanBP9 is increased in Alzheimer’s disease, induces neurodegeneration, and mediates amyloid-induced neurotoxicity. In addition to its roles in neurodegenerative processes, RanBP9 reportedly exhibits proapoptotic activity in response to DNA damage (57). RanBP9 also interacts directly with the C-terminal region of p73 and the resulting complex regulates p73 levels as well as the nuclear translocation of RanBP9 (58). Liu et al. have also discovered that the RanBP9/p73 complex participates in the induction of apoptosis. This finding is supported by the observation that knockdown of endogenous p73 by siRNA inhibits RanBP9-induced apoptosis. Conversely, siRNA knockdown of RanBP9 blocks p73-induced apoptosis. RanBP9 or p73 expressed ectopically either alone or together in HT22 cells induces mitochondrial dysfunction and apoptosis by increasing mitochondrial reactive oxygen species (ROS) and the Bax/Bcl-2 ratio, promoting Bax oligomerization. The expression also induces mitochondrial dysfunction and apoptosis by releasing cytochrome c, which is suppressed by anti-apoptotic proteins Bcl-2, Bcl-XL, and XIAP.
RanBP9 increases p73 levels at both transcriptional and post-translational levels. The observation that RanBP9 increases the half-life of endogenous p73 suggests the stabilization of p73 by RanBP9 at the post-translational level. Furthermore, as a transcriptional co-regulator, RanBP9 increases the mRNA levels of p73 and PUMA, but not p21, as shown by quantitative real time reverse transcription PCR analysis (59). This also suggests that RanBP9 specifically stabilizes p73, but not p53, because PUMA is overexpressed by both p53 and p73 whereas p21 is only overexpressed by p53.
Bcl-2 family proteins
Although mechanistic overlap occurs between p73 and p53 in apoptosis, the underlying mechanism for transcription-independent p73-induced apoptosis has not been investigated in the context of the Bcl-2 family of proteins. We recently identified a novel pathway of p73-induced apoptosis in which direct interaction occurs between p73 and Bcl-2 family proteins in the mitochondria. Our studies showed that p73 induces apoptosis in the absence of transcription and translation by triggering cytochrome c release, but the pro-apoptotic activity of p73 is inhibited by the direct binding of Bcl-2 family proteins in the mitochondria. This revealed a molecular basis for a novel apoptotic pathway of p73 in the mitochondria.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)
Apart from its intrinsic apoptotic pathway, p73 is also involved in an extrinsic pathway. TAp73 is reportedly cleaved by caspase-3 and -8 both in vitro and in vivo during TRAIL-induced apoptosis, and TAp73 and its cleavage products have been detected in the mitochondria using subcellular fractionation and electron microscopy (EM) (60). This apoptosis also proceeds via the mitochondrial apoptotic pathway, as recombinant p73 triggers mitochondrial release of cytochrome c (60). These observations suggest a direct pro-apoptotic role of p73 in the mitochondria via an extrinsic apoptotic pathway, independent of its transcriptional activity in the nucleus.
COMPARISON OF THE APOPTOSIS MECHANISMS OF p73 AND p53
Structural and functional homologies between p73 and p53 allow p73 to undertake p53-like functions, including induction of apoptosis and cell cycle arrest in response to DNA damage, which are critical for the tumor-suppressive activities of these proteins. As expected from the functional homologies, the apoptotic mechanisms of p73 are very similar to those of p53. Like p53, p73 recognizes, binds, and transactivates p53-responsive genes encoding pro-apoptotic proteins such as PUMA and Bax and the death receptor CD95, suggesting pro-apoptotic activities as a transactivator in both intrinsic and extrinsic apoptotic pathways. In addition, we have recently identified a novel apoptosis mechanism by which p73 induces apoptosis at the mitochondria independent of transcription, similar to p53. During apoptosis through an extrinsic pathway, as full length p53 and its fragments generated by caspases were found in the cytoplasm and mitochondria, p73 induced apoptosis by acting directly on the mitochondria following cleavage by caspases in a transcription-independent manner through TRAIL-mediated apoptosis (60). Despite their apparent redundancies, these functional, mechanistic overlaps between p53 and p73 are of great importance because they allow p73 to substitute the apoptotic functions of p53 in cancer cells in which p53 is mutated or inactive.
In addition to the apoptotic mechanisms shared by p53 and p73, other mechanisms act specifically on p73. The GRAMD4 gene responds specifically to p73 but not to p53 (49). RanBP9, a transcriptional co-regulator, selectively increases the transcription of p73 but not that of p53 (56). Although mouse Wox1 protein binds to p53 (61), human Wwox does not, nor does it induce cytoplasmic localization of p53. In contrast, human Wwox causes cytoplasmic localization of p73 and also interacts with p73 in the cytoplasm (54). The unique apoptotic mechanisms specific to p73 suggest that it induces apoptosis in a p53-independent manner. They also suggest that p73 may increase the sensitivity of cells to apoptotic stimuli by participating in other mechanisms shared by both proteins, whether the mechanism is transcription-dependent or not.
CONCLUSION
In summary, we presented a detailed comparison between p73 and p53 in terms of structure and apoptotic function. Apart from its unique function in neuronal development and differentiation, p73 closely resembles p53 in terms of apoptotic function. In response to apoptotic stimuli, p73 transactivates p53-responsive genes, inducing transcription-dependent apoptosis. In addition, cytosolic p73 can move to the mitochondria, leading to transcription-independent apoptosis. Understanding the molecular basis for the similarities and differences between p73 and p53 functions may explain why p73 can function as a substitute for p53 in p53-deficient cancer cells, which will be useful for establishing a novel p73-based cancer therapy. Because the mechanisms of p73-induced apoptosis remain controversial, with conflicting activities depending on cellular context, further studies are needed for complete understanding of the multifunctional role of p73.
Acknowledgments
This work was supported by a grant from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1220150).
References
- 1.Dötsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. (2010);2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutruzzola F, Avigliano L, Candi E. p73 keeps metabolic control in balance. Cell Cycle. (2014);13:179–180. doi: 10.4161/cc.27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoue S, Tomasini R, Rufini A, et al. TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc Natl Acad Sci U S A. (2014);111:1843–1848. doi: 10.1073/pnas.1323416111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. (2006);13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 5.Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. (2002);2:605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 6.Munarriz E, Bano D, Sayan AE, Rossi M, Melino G, Nicotera P. Calpain cleavage regulates the protein stability of p73. Biochem Biophys Res Commun. (2005);333:954–960. doi: 10.1016/j.bbrc.2005.05.188. [DOI] [PubMed] [Google Scholar]
- 7.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. (2005);19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossi M, De Laurenzi V, Munarriz E, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. (2005);24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oberst A, Malatesta M, Aqeilan RI, et al. The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc Natl Acad Sci U S A. (2007);104:11280–11285. doi: 10.1073/pnas.0701773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy D, Adamovich Y, Reuven N, Shaul Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. (2007);14:743–751. doi: 10.1038/sj.cdd.4402063. [DOI] [PubMed] [Google Scholar]
- 11.Costanzo A, Merlo P, Pediconi N, et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol Cell. (2002);9:175–186. doi: 10.1016/S1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani F, Piazza S, Gostissa M, et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell. (2004);14:625–636. doi: 10.1016/j.molcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Sayan BS, Yang AL, Conforti F, et al. Differential control of TAp73 and DeltaNp73 protein stability by the ring finger ubiquitin ligase PIR2. Proc Natl Acad Sci U S A. (2010);107:12877–12882. doi: 10.1073/pnas.0911828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toh WH, Siddique MM, Boominathan L, Lin KW, Sabapathy K. c-Jun regulates the stability and activity of the p53 homologue, p73. J Biol Chem. (2004);279:44713–44722. doi: 10.1074/jbc.M407672200. [DOI] [PubMed] [Google Scholar]
- 15.Danovi SA, Rossi M, Gudmundsdottir K, Yuan M, Melino G, Basu S. Yes-associated protein (YAP) is a critical mediator of c-Jun-dependent apoptosis. Cell Death Differ. (2008);15:217–219. doi: 10.1038/sj.cdd.4402226. [DOI] [PubMed] [Google Scholar]
- 16.Dulloo I, Gopalan G, Melino G, Sabapathy K. The antiapoptotic DeltaNp73 is degraded in a c-Jun-dependent manner upon genotoxic stress through the antizyme-mediated pathway. Proc Natl Acad Sci U S A. (2010);107:4902–4907. doi: 10.1073/pnas.0906782107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramadan S, Terrinoni A, Catani MV, et al. p73 induces apoptosis by different mechanisms. Biochem Biophys Res Commun. (2005);331:713–717. doi: 10.1016/j.bbrc.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 18.Zaika AI, Slade N, Erster SH, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. (2002);196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi SG, Chang SG, Lee SJ, Lee CH, Kim JI, Park JH. Elevated and biallelic expression of p73 is associated withprogression of human bladder cancer. Cancer Res. (1999);59:2791–2793. [PubMed] [Google Scholar]
- 20.Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. (2004);2:371–386. [PubMed] [Google Scholar]
- 21.Casciano I, Mazzocco K, Boni L, et al. Expression of DeltaNp73 is a molecular marker for adverse outcome in neuroblastoma patients. Cell Death Differ. (2002);9:246–251. doi: 10.1038/sj.cdd.4400993. [DOI] [PubMed] [Google Scholar]
- 22.Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. (1997);389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 23.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. (1995);80:293–299. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 24.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. (2001);7:683–694. doi: 10.1016/S1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 25.Oda E, Ohki R, Murasawa H, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. (2000);288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 26.M#252;ller M, Wilder S, Bannasch D, et al. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med. (1998);188:2033–2045. doi: 10.1084/jem.188.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mihara M, Erster S, Zaika A, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. (2003);11:577–590. doi: 10.1016/S1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 28.Chipuk JE, Kuwana T, Bouchier-Hayes L, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. (2004);303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 29.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. (2005);309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 30.Chi SW. Structural insights into the transcription-independent apoptotic pathway of p53. BMB Rep. (2014);47:167–172. doi: 10.5483/BMBRep.2014.47.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chipuk JE, Bouchier-Hayes L, Green DR. Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario. Cell Death Differ. (2006);13:1396–1402. doi: 10.1038/sj.cdd.4401963. [DOI] [PubMed] [Google Scholar]
- 32.Ha JH, Shin JS, Yoon MK, et al. Dual-site interactions of p53 protein transactivation domain with anti-apoptotic Bcl-2 family proteins reveal a highly convergent mechanism of divergent p53 pathways. J Biol Chem. (2013);288:7387–7398. doi: 10.1074/jbc.M112.400754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ha JH, Won EY, Shin JS, et al. Molecular mimicry-based repositioning of nutlin-3 to anti-apoptotic Bcl-2 family proteins. J Am Chem Soc. (2011);133:1244–1247. doi: 10.1021/ja109521f. [DOI] [PubMed] [Google Scholar]
- 34.Lee DH, Ha JH, Kim Y, et al. A Conserved Mechanism for Binding of p53 DNA-Binding Domain and Anti-Apoptotic Bcl-2 Family Proteins. Mol Cells. (2014);37:264–269. doi: 10.14348/molcells.2014.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell S, Klein C, Muller L, Hansen S, Buchner J. p53 contains large unstructured regions in its native state. J Mol Biol. (2002);322:917–927. doi: 10.1016/S0022-2836(02)00848-3. [DOI] [PubMed] [Google Scholar]
- 36.Burge S, Teufel DP, Townsley FM, Freund SM, Bycroft M, Fersht AR. Molecular basis of the interactions between the p73 N terminus and p300: effects on transactivation and modulation by phosphorylation. Proc Natl Acad Sci U S A. (2009);106:3142–3147. doi: 10.1073/pnas.0900383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu G, Xia T, Chen X. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREBbinding protein. J Biol Chem. (2003);278:17557–17565. doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]
- 38.Shin JS, Ha JH, Lee DH, et al. Structural convergence of unstructured p53 family transactivation domains in MDM2 recognition. Cell Cycle. (2015);14:533–543. doi: 10.1080/15384101.2014.998056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kussie PH, Gorina S, Marechal V, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. (1996);274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 40.Canning P, von Delft F, Bullock AN. Structural basis for ASPP2 recognition by the tumor suppressor p73. J Mol Biol. (2012);423:515–527. doi: 10.1016/j.jmb.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ethayathulla AS, Tse PW, Monti P. Structure of p73 DNA-binding domain tetramer modulates p73 transactivation. Proc Natl Acad Sci U S A. (2012);109:6066–6071. doi: 10.1073/pnas.1115463109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bullock AN, Henckel J, Fersht AR. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene. (2000);19:1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 43.Joerger AC, Rajagopalan S, Natan E, Veprintsev DB, Robinson CV, Fersht AR. Structural evolution of p53, p63, and p73: implication for heterotetramer formation. Proc Natl Acad Sci U S A. (2009);106:17705–17710. doi: 10.1073/pnas.0905867106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chi SW, Ayed A, Arrowsmith CH. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. (1999);18:4438–4445. doi: 10.1093/emboj/18.16.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thanos CD, Goodwill KE, Bowie JU. Oligomeric structure of the human EphB2 receptor SAM domain. Science. (1999);283:833–836. doi: 10.1126/science.283.5403.833. [DOI] [PubMed] [Google Scholar]
- 46.Stapleton D, Balan I, Pawson T, Sicheri F. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat Struct Biol. (1999);6:44–49. doi: 10.1038/4917. [DOI] [PubMed] [Google Scholar]
- 47.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. (2010);37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melino G, Bernassola F, Ranalli M, et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem. (2004);279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- 49.John K, Alla V, Meier C, P#252;tzer BM. GRAMD4 mimics p53 and mediates the apoptotic function of p73 at mitochondria. Cell Death Differ. (2011);18:874–886. doi: 10.1038/cdd.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng Y, Wu X. Peg3/Pw1 promotes p53-mediated apoptosis by inducing Bax translocation from cytosol to mitochondria. Proc Natl Acad Sci U S A. (2000);97:12050–12055. doi: 10.1073/pnas.97.22.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taebunpakul P, Sayan BS, Flinterman M, et al. Apoptin induces apoptosis by changing the equilibrium between the stability of TAp73 and DeltaNp73 isoforms through ubiquitin ligase PIR2. Apoptosis. (2012);17:762–776. doi: 10.1007/s10495-012-0720-7. [DOI] [PubMed] [Google Scholar]
- 52.Schilling T, Schleithoff ES, Kairat A, et al. Active transcription of the human FAS/CD95/TNFRSF6 gene involves the p53 family. Biochem Biophys Res Commun. (2009);387:399–404. doi: 10.1016/j.bbrc.2009.07.063. [DOI] [PubMed] [Google Scholar]
- 53.Terrasson J, Allart S, Martin H, et al. p73-dependent apoptosis through death receptor: impairment by human cytomegalovirus infection. Cancer Res. (2005);65:2787–2794. doi: 10.1158/0008-5472.CAN-04-2019. [DOI] [PubMed] [Google Scholar]
- 54.Aqeilan RI, Pekarsky Y, Herrero JJ, et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci U S A. (2004);101:4401–4406. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inoue T, Stuart J, Leno R, Maki CG. Nuclear import and export signals in control of the p53-related protein p73. J Biol Chem. (2002);277:15053–15060. doi: 10.1074/jbc.M200248200. [DOI] [PubMed] [Google Scholar]
- 56.Liu T, Roh SE, Woo JA, Ryu H, Kang DE. Cooperative role of RanBP9 and P73 in mitochondriamediated apoptosis. Cell Death Dis. (2013);4:e476. doi: 10.1038/cddis.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Atabakhsh E, Bryce DM, Lefebvre KJ, Schild-Poulter C. RanBPM has proapoptotic activities that regulate cell death pathways in response to DNA damage. Mol Cancer Res. (2009);7:1962–1972. doi: 10.1158/1541-7786.MCR-09-0098. [DOI] [PubMed] [Google Scholar]
- 58.Kramer S, Ozaki T, Miyazaki K, et al. Protein stability and function of p73 are modulated by a physical interaction with RanBPM in mammalian cultured cells. Oncogene. (2005);24:938–944. doi: 10.1038/sj.onc.1208257. [DOI] [PubMed] [Google Scholar]
- 59.Rao MA, Cheng H, Quayle AN, Nishitani H, Nelson CC, Rennie PS. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J Biol Chem. (2002);277:48020–48027. doi: 10.1074/jbc.M209741200. [DOI] [PubMed] [Google Scholar]
- 60.Sayan AE, Sayan BS, Gogvadze V, et al. P73 and caspase-cleaved p73 fragments localize to mitochondria and augment TRAIL-induced apoptosis. Oncogene. (2008);27:4363–4372. doi: 10.1038/onc.2008.64. [DOI] [PubMed] [Google Scholar]
- 61.Chang NS, Pratt N, Heath J, et al. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity. J Biol Chem. (2001);276:3361–3370. doi: 10.1074/jbc.M007140200. [DOI] [PubMed] [Google Scholar]