Significance
The correct and regulated readout of epigenetic marks on chromatin is essential to modulate gene expression in living cells. The regulation of chromatin accessibility is ensured by such epigenetic tags, which form a platform for the binding of specific enzymatic modules. A clear example of this mechanism is represented by the histone demethylase LSD1-CoREST, which removes methylation marks from lysine 4 of histone protein H3. We developed a crosslinking technology to capture this histone demethylase in contact with the nucleosome and used this methodology to explore the structural and biophysical properties of this complex. This is one of the very few successful attempts to visualize the molecular mechanism underlying the recognition of the nucleosomal substrate by a histone-modifying enzyme complex.
Keywords: nucleosome, histone tails, molecular recognition, chromatin modification, small-angle X-ray scattering
Abstract
With its noncatalytic domains, DNA-binding regions, and a catalytic core targeting the histone tails, LSD1-CoREST (lysine-specific demethylase 1; REST corepressor) is an ideal model system to study the interplay between DNA binding and histone modification in nucleosome recognition. To this end, we covalently associated LSD1-CoREST to semisynthetic nucleosomal particles. This enabled biochemical and biophysical characterizations of nucleosome binding and structural elucidation by small-angle X-ray scattering, which was extensively validated through binding assays and site-directed mutagenesis of functional interfaces. Our results suggest that LSD1-CoREST functions as an ergonomic clamp that induces the detachment of the H3 histone tail from the nucleosomal DNA to make it available for capture by the enzyme active site. The key notion emerging from these studies is the inherently competitive nature of the binding interactions because nucleosome tails, chromatin modifiers, transcription factors, and DNA represent sites for multiple and often mutually exclusive interactions.
The mechanisms underlying the recruitment of histone-modifying complexes to specific chromatin regions, nucleosome components, or DNA sequences represent a challenge in the field of chromatin biology (1–3). Nucleosome recognition by histone-modifying enzymes is the result of two opposite requirements: binding to the target locus to locally shape and regulate chromatin accessibility and function, and nucleosome release to avoid the formation of a sticky complex bound too tightly to a chromatin region. A key feature is that it is essential to adopt a productive recognition mode so that the histone tails can be captured to undergo the necessary modifications (4, 5). To investigate these issues in nucleosome recognition, we have studied LSD1-CoREST, a flavin-dependent lysine 4 of histone protein H3 (H3-Lys4) demethylase (known also as KDM1A). This heterodimeric enzyme features several properties that make it an insightful model system for our purposes. First, the unique binding mode of the H3 tail to LSD1 has been largely studied by several biochemical and structural works (6–9). These data demonstrated that recognition by the enzyme active site requires a long stretch of 20 N-terminal amino acids and a specific pattern of posttranslational modifications. In addition, LSD1 alone is unable to recognize the nucleosomal substrate and needs the CoREST partner to effectively demethylate the nucleosomal H3 tail (10–12). Indeed, LSD1 has a characteristic “tower” domain, which protrudes from the catalytic core and represents the site for binding the C-terminal SANT2 (Swi3, Ada2, N-Cor, and TFIIIB) domain of CoREST corepressor (13–15). Furthermore, LSD1 typically acts in concert with histone deacetylases (HDAC 1/2) (9, 12). The stable and conserved LSD1-CoREST1-HDAC recognizes the nucleosomal substrate and exerts repressive activity through synergistic histone deacetylation and Lys4 demethylation (8, 9, 12).
In this work, we used biophysics to explore the properties of the single molecular components that build up the network of interactions of the LSD1-CoREST/nucleosome complex. The binding of the enzyme to DNA was investigated by fluorescent assays, mutagenesis, and structural modeling. These experiments were paralleled by the evaluation of the binding affinity of histone peptides to both the LSD1 active site and the nucleosomal DNA. We then combined these results with insights gained from small-angle X-ray scattering (SAXS) and biochemical approaches, based on designed semisynthetic nucleosomal particles, to unravel the structural and molecular mechanism of nucleosome recognition.
Results
LSD1-CoREST Binds DNA Unspecifically.
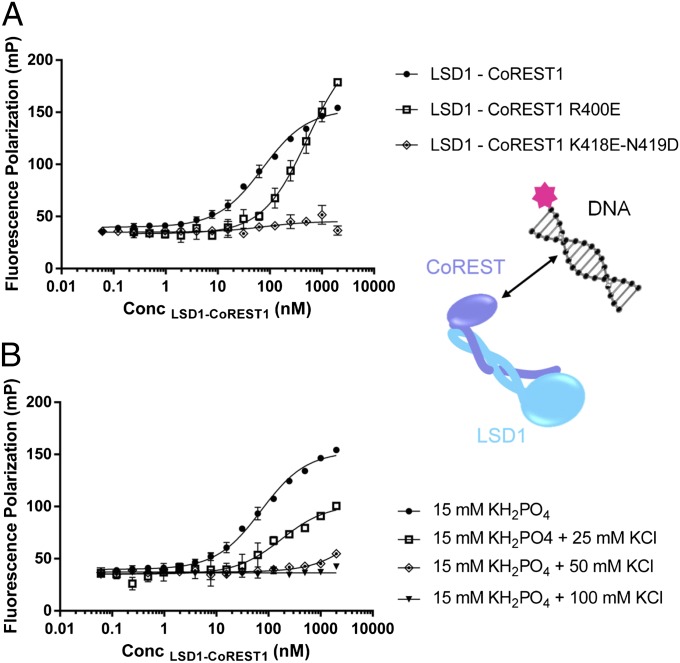
Our analysis started from the published observation that the SANT2 domain of CoREST1 binds to DNA (14, 16). With the goal of defining a biochemical model and the experimental assays for nucleosome recognition by LSD1-CoREST, we initially sought to determine the affinity for DNA and the factors that modulate it. We set up a fluorescence polarization binding assay using 21-mer oligonucleotides conjugated with a fluorescent label. One of the sequences contained a mismatch that slightly distorts the double-stranded helix, which was used to probe the effect of alterations in secondary structure. We observed interaction of LSD1-CoREST with the DNA oligos, although its strength is strongly salt-dependent (Fig. 1 and SI Appendix, Table S1). These experiments collectively confirmed that LSD1-CoREST1 is a nonspecific DNA-binding enzyme and that the nature of this interaction is mainly electrostatic.
Fig. 1.
Binding affinities of LSD1-CoREST to DNA, measured by fluorescence polarization. (A) Increasing concentrations of purified LSD1-CoREST1 were incubated with 5(6)-carboxytetramethylrhodamine-conjugated 21-bp DNA (final DNA concentration of 1 nM). Changes in polarization were measured in millipolarization (mP) units and plotted against the concentration of LSD1-CoREST1. Error bars correspond to SDs for all measurements (n ≥ 3). Binding curves of wild-type and two representative LSD1-CoREST1 mutants are shown. The Kd values are reported in SI Appendix, Table S1. (B) Effect of ionic strength on the binding affinities. The increasing additions of KCl reduce both the amplitudes of the curve and the Kd values.
An Assay for H3 Tail Binding to LSD1-CoREST.
We next probed the binding of H3 N-terminal peptides to LSD1-CoREST1. The rationale for these experiments was evaluating peptide binding, using the same methodology employed for studying the interactions with DNA (SI Appendix, Table S2). The observed tight affinity (Kd of 56 nM) was found to be significantly affected by increasing ionic strength, following the same trend observed for the enzymatic activity (SI Appendix, Fig. S1) (8). As a further check, a competition assay was performed. In this case, unlabeled Lys4-H3 peptide was used to competitively displace the fluorescently labeled peptide, resulting in a concentration-dependent decrease of polarization signal with a calculated Kd value of 115 nM (SI Appendix, Fig. S2A). The similarity between the Kd values measured in the direct and competitive assays proved that the hydrophobic fluorescent label does not alter the binding of the histone tail to the active site pocket, validating our approach.
Measuring the Binding of H3 Tails to Nucleosomes.
Having reliable assays in hand to quantitatively evaluate protein–protein and protein–DNA binding, we decided to use the same approaches to probe nucleosome binding to the demethylase. To that aim, native nucleosomes (either extracted from chicken erythrocytes or reconstituted from recombinant proteins) were titrated as competitors of the fluorescent histone-tail. The outcome of the experiment did not show at all the expected result: binding of nucleosomes to LSD1 was expected to outcompete the labeled peptide, resulting in a decreased polarization signal. Instead, we observed a “direct-binding” type of curve (i.e., an increase in the signal), indicating that the fluorescent peptide was associating to the nucleosomal particle (SI Appendix, Fig. S2B). On this basis, we next directly measured the affinity of free H3 tail peptide for nucleosomes, using both chicken and recombinant particles. The tight affinity observed in both cases (Kd ∼2 nM) was found to be strongly affected by either salt concentration or acetylation of Lys residues, consistent with the notion that these interactions mostly involve charged groups (SI Appendix, Table S3 and Fig. S3). It has been described in the literature that H3 N-terminal tails specifically mediate inter- and intranucleosome contacts in vitro and in vivo (17, 18), but to our knowledge, this is the first time such an interaction has been described in quantitative terms.
Design of Semisynthetic Nucleosomes to Generate Covalent LSD1-CoREST/Nucleosome Complexes.
Fluorescence polarization allowed a direct comparison of the binding affinity of histone tails to both nucleosomal DNA and LSD1 active sites but, as previously described, was not suitable to monitor nucleosome/LSD1-CoREST interactions. To address this issue, we resorted to other biophysical methodologies. Surface plasmon resonance produced incomplete and ambiguous results because of the multiplicity and complexity of the mutually binding elements (histone tails, DNA, and LSD1-CoREST). All efforts to copurify the LSD1-CoREST1/nucleosome assembly were unsuccessful, likely because this complex is characterized by fast association and dissociation, leading to separation of the interacting species in diluted conditions.
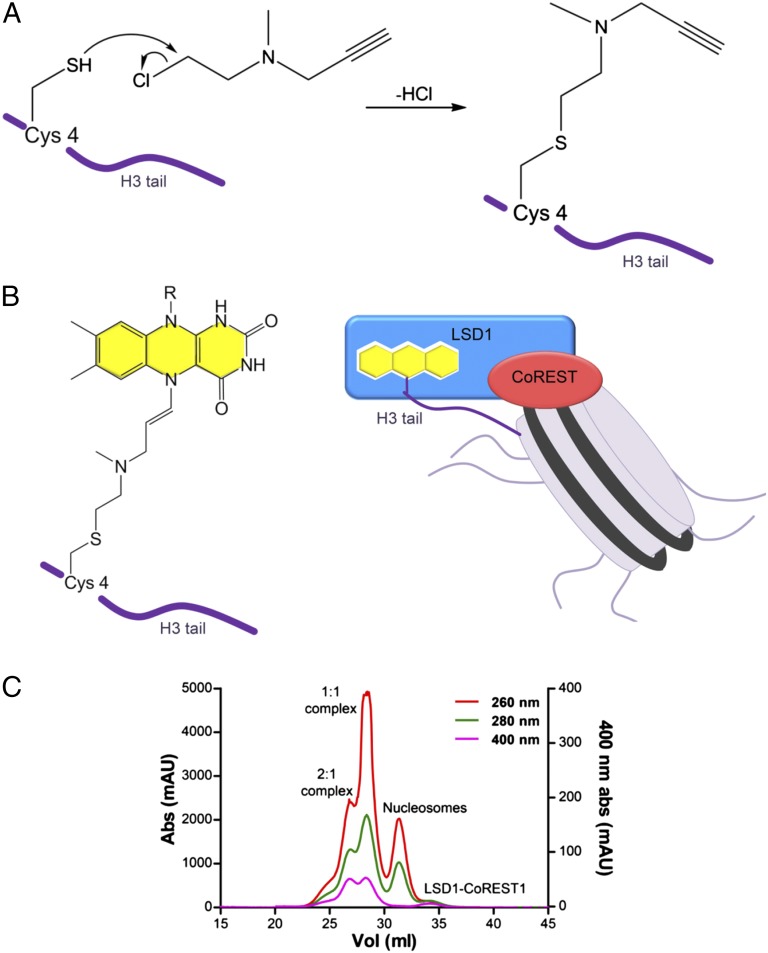
Given these difficulties, we sought to develop a chemical method to stabilize the complex. In 2007, Yang and coworkers published the crystal structure of LSD1-CoREST1 in complex with the 21-amino acid N-terminal tail of H3 covalently bound to the active site (19). In that work, the peptide was chemically modified on the side chain of Lys4 with a propargylic moiety able to react with the flavin cofactor of LSD1, forming a stable adduct. We decided to take advantage of this work, introducing the propargylic group on the recombinant H3 protein used for nucleosome reconstitution. We created a H3 mutant containing a single Cys residue located in position 4 and installed a propargylic group onto the Cys4 side chain, creating a dimethyl-Lys analog (Fig. 2) (20). The added moiety did not hamper octamer refolding and nucleosome reconstitution. Most important, both semisynthetic histone H3 and nucleosome turned out to covalently react with FAD cofactor, forming a stable covalent complex with LSD1-CoREST. The covalent adduct formation can be followed by monitoring the changes of the absorbance spectrum in the UV region (SI Appendix, Fig. S4). The semisynthetic nucleosomes proved to be a most powerful experimental tool, as described here.
Fig. 2.
Semisynthetic nucleosomes as a tool to probe recognition by LSD1-CoREST. (A) Chemical reaction between the Cys4 thiol group and 1-methyl-1-(prop-2-ynyl)aziridinium chloride, leading to a propargylamine-containing mimic of dimethyl Lys4. The propargylic reagent was installed onto the Cys4 side chain, creating a dimethyl-Lys analog using an H3 double mutant that does not contain any other Cys (Lys4Cys-Cys110Ala). (B) Propargylamine-derivative H3 covalently tethered to FAD (Left) and schematics of the covalent complex formed by LSD1-CoREST and the semisynthetic nucleosome (Right). (C) Elution profile of the covalent complex on a Superdex 200 column. Purification was obtained by serially connecting three 10/300 columns (GE Healthcare). Absorbance at 260, 280, and 400 nm is used to monitor DNA, protein, and FAD, respectively. Peaks for LSD1-CoREST1/nucleosome and (LSD1-CoREST1)2/nucleosome complexes are labeled by “1:1” and “2:1.”
A Chromatography-Based Assay for LSD1-CoREST/Nucleosome Complex Formation.
We devised size-exclusion chromatography protocols to achieve, for the first time to our knowledge, the purification of the LSD1-CoREST/nucleosome complex as well as a semiquantitative evaluation of the efficiency of its formation. Fig. 2C and SI Appendix illustrate the logic and outcome of this type of experiments. LSD1-CoREST1 and semisynthetic nucleosomes were incubated for variable times (e.g., 2 h) and subsequently loaded on both preparative and analytical gel filtration columns. The ratio between the UV absorbances was used to quantify the relative protein/DNA content of each of the four elution peaks typically observed in all these experiments. We were able to assign the four peaks as follows: the LSD1-CoREST1/nucleosome complex with 2:1 stoichiometry (i.e., both nucleosomal H3s engage one LSD1 each), the LSD1-CoREST1/nucleosome with 1:1 stoichiometry (i.e., only one nucleosomal H3 engages LSD1), free nucleosomes, and free LSD1-CoREST1, as verified by SDS and native electrophoretic shift assay (Fig. 2C and SI Appendix, Figs. S4B and S5). From the analysis of the relative intensities of each peak in the experiments performed with varying sample ratios (SI Appendix, Fig. S6), we could make two key observations. First, significant amounts of unreacted nucleosomes are always present. Second, the 2:1 complex represents a small fraction compared with the 1:1 complex, even in the presence of a large excess of LSD1-CoREST1. These findings both suggest that some of the nucleosomal particles are possibly engaged in catalytically nonproductive modes of interaction and dissociate during elution, and demonstrate that the predominant stoichiometry of the LSD1-CoREST1/nucleosome complex is 1:1.
Mapping the Protein Regions Involved in Nucleosome Recognition.
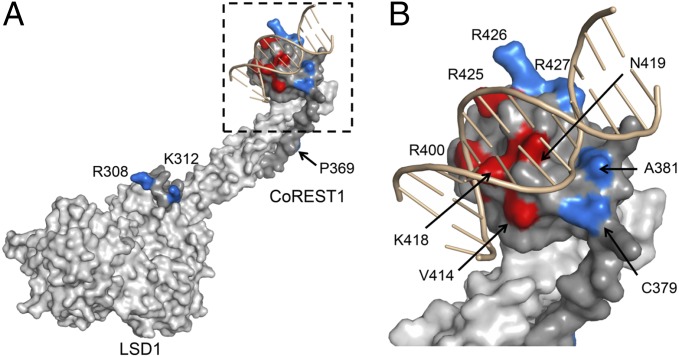
To dissect the structural elements involved in nucleosome substrate recognition, we built a structural model for LSD1-CoREST1 binding to DNA by comparison with the 3D structure of the SANT domain of c-Myb in complex with a short DNA duplex (PDB ID code 1H88) (21) (Fig. 3). To validate this model and support further structural studies, we mutated CoREST1 at 12 different sites, located on the SANT2 domain as well on other regions of the protein (i.e., the helical linker and the portion of CoREST1 close to the cleft of the LSD1 catalytic core). These experiments revealed that the mutation sites affecting DNA binding are all clustered in the region of the SANT2 domain predicted by the structural model to be directly engaged in DNA recognition (Fig. 1A, red-colored in Fig. 3 and SI Appendix, Table S1).
Fig. 3.
Structural model for DNA-binding to LSD1-CoREST. (A) Surface representation of the structure of LSD1 (light gray) and CoREST (gray) modeled with double-stranded DNA (pink; 13 nucleotides). (B) Details of the SANT2 domain of CoREST1 facing the DNA major groove. Residues that show no/little effect on DNA binding and those that instead affect it are highlighted in blue and red, respectively (SI Appendix, Table S1). To create the model, LSD1-CoREST SANT2 domain (PDB ID code 2V1D) was superposed to a c-Myb protein (PDB ID code 1H88), the SANT domain of which had been crystallized with a DNA oligo.
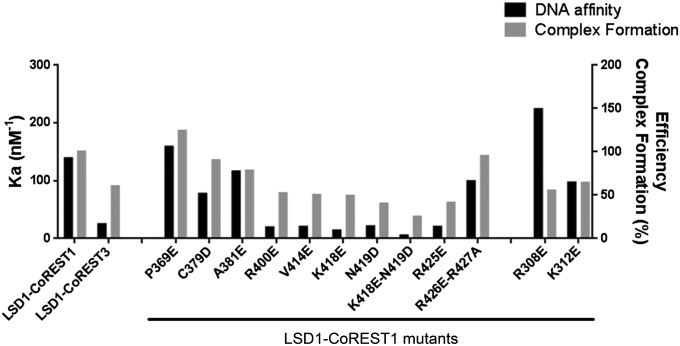
On the basis of these data, we subsequently addressed the question of the role of DNA binding in the actual process of nucleosome recognition and demethylation. Each of the 12 mutants was tested with the chromatographic assay, based on the semisynthetic nucleosomes. As shown in Fig. 4, mutants featuring weaker DNA binding were proportionally impaired in their ability to productively recognize the nucleosome. Such a correlation demonstrated that nucleosome recognition requires a DNA binding step. This concept is further highlighted by the properties of human CoREST3, a CoREST1 homolog with a lower repressive activity (SI Appendix, Fig. S7). LSD1-CoREST3 features both lower affinity for DNA and lower capacity to form the covalent complex with the nucleosome.
Fig. 4.
Studies of LSD1-CoREST1/nucleosome binding by mutagenesis. For each LSD1-CoREST mutant or variant, the affinity constant Ka (calculated from the Kd values listed in SI Appendix, Table S1) for DNA and a qualitative indicator of complex formation with semisynthetic nucleosomes are represented as black and gray bars, respectively. Efficiency of complex formation is defined as the ratio between the heights of the 1:1 complex and free nucleosome elution peaks (Fig. 2C). The values are expressed as percentage, using as reference (100%) the value observed for the wild-type LSD1-CoREST1.
Structural Studies with SAXS.
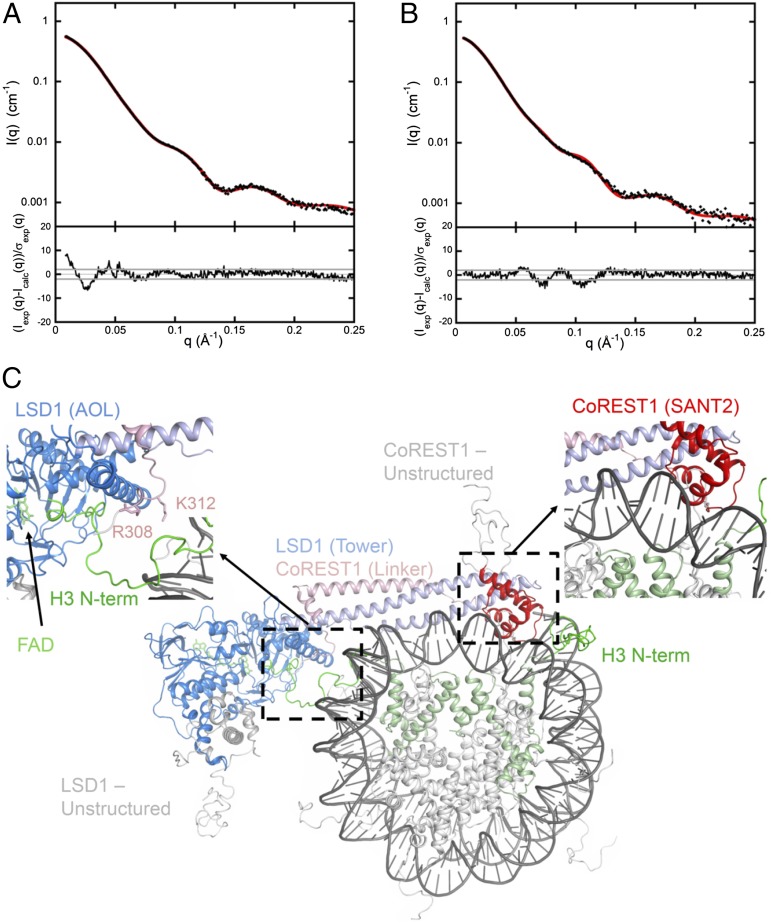
To gain insights into the 3D arrangement of LSD1-CoREST1 and the nucleosomal particle within the complex in solution, we performed a series of SAXS measurements. We first characterized each partner separately to assess quality of the samples and to optimize experimental conditions, as described in SI Appendix, Fig. S8. Next, to determine the mutual arrangement of LSD1-CoREST1 and nucleosome, we took advantage of our ability to produce and purify semisynthetic covalent complexes that are inherently homogeneous, as both partners interact in the productive mode by design (SI Appendix, Fig. S5). Covalent complexes between LSD1-CoREST1 and semisynthetic nucleosomes were loaded onto a size exclusion column (Fig. 2C), and fractions corresponding to the 1:1 and 2:1 complexes were concentrated before SAXS investigation. The scattering patterns are shown in Fig. 5 A and B, and values of the derived structural parameters are listed in SI Appendix, Table S4. We generated 3D models by systematically superposing the mutagenesis-validated model of LSD1-CoREST1/DNA complex (Fig. 3) over all nucleotides of the DNA wrapped around the histone core. The best agreement between the calculated scattering pattern and our experimental data was obtained for the model generated as described in the legend to Fig. 5 A and C (χ2 = 1.94). Importantly, the tail of one of the two H3 proteins is located adjacent to the active site cleft of LSD1, in a position fully compatible with binding in the conformation observed in the crystal structure of LSD1-CoREST1/H3 tail complex (19, 22), as shown in Fig. 5C and Movie S1. To illustrate the sensitivity of the SAXS pattern to the mutual arrangement of LSD1/CoREST1 and the nucleosome, we rotated the best fitting model by about 20° with respect to the midplane of the nucleosome. The resulting SAXS pattern exhibits significant differences with the experimental curve (χ2 value = 7.8), as can be observed in SI Appendix, Fig. S8C. Similarly, an increase in χ2 free values from 1.88 to 4.55 was observed when applying the χ2 free analysis presented by Rambo and Tainer (23) to our best model and its rotated version, respectively.
Fig. 5.
Structural models for LSD1-CoREST1/nucleosome 1:1 and 2:1 complexes. SAXS patterns of the 1:1 covalent complex (A) and the 2:1 covalent complex (B). Experimental data are in black, and calculated scattering patterns are shown in red. Data are reported as the intensities I(q) on a logarithmic scale versus the momentum transfer q. The corresponding distributions of reduced residuals are presented in the bottom frames. (C) 3D structure of the 1:1 complex yielding a good fit to the solution scattering curve shown in A. This model was generated as follows. The eight central base pairs of the LSD1-CoREST1/DNA model of Fig. 3 were superimposed to all possible eight base pairs segments of nucleosomal DNA (PDB ID code 1KX5) to generate 138 LSD1-COREST1/nucleosome models. Structures giving steric clashes were not further considered. The remaining models were sorted on the basis of the agreement with the measured scattering curve. The model in which the SANT2 domain is interacting with the (−J7)−(+J1) nucleosomal nucleotides (shown in SI Appendix, Fig. S14) gave the best fit to the SAXS data. The fit was further refined and improved by a small rigid-body search that did not alter the geometry of the SANT2-DNA contact (i.e., a 10° rotation around an axis approximating the axis of the interacting α-helix). The final χ2−value was 1.94 (see SI Appendix for experimental and methodological details). The model of the 2:1 covalent complex is shown in SI Appendix, Fig. S9.
A further validating observation for this model is provided by the properties of the Arg308 and Lys312 mutants. These two residues are not involved in DNA binding (Figs. 3 and 4 and SI Appendix, Table S1) and are located at the point where H3 tail ejects from the nucleosome body in the presented structural model (Fig. 5C). Because of their position, mutations of these residues may result in a diminished ability to capture the entire histone tail, accounting for a reduced efficiency in nucleosome engagement, as observed in our experiments. It is also relevant to notice that the two mutations (P369E, C379D) located at the tip of the tower helices show no effect on nucleosome recognition consistent with their location outside the LSD1-CoREST1/nucleosome contact regions. In essence, the structural model is fully validated by all 12 investigated mutants.
Finally, starting from the 1:1 complex, a model was built for the 2:1 complex in which a second LSD1-CoREST1 was added by applying the nucleosome twofold symmetry to the first copy (SI Appendix, Fig. S9). The calculated scattering pattern of the resulting assembly is in good agreement with the experimental curve (χ2 = 1.6; Fig. 5B), providing further support to the structural model derived from the 1:1 scattering curve.
Discussion
A Stick-and-Catch Model for Nucleosome Recognition.
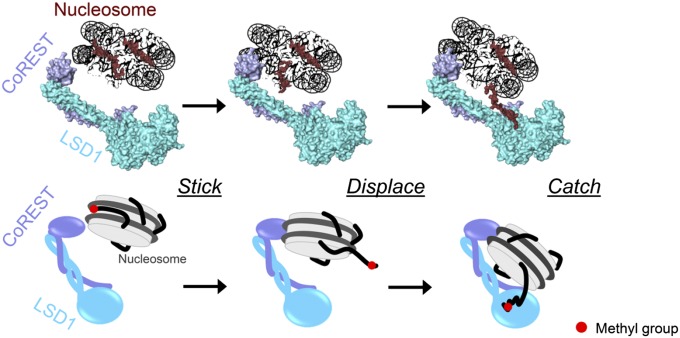
Our experimental work consistently outlines a “stick-and-catch” mode of nucleosome recognition and modification (Fig. 6 and Movie S1). The lack of sequence specificity of DNA association enables the SANT2 domain to search for catalytically competent binding modes. Such an explorative DNA search is a theme common to many nucleosome readers (24, 25). As CoREST scans binding sites along nucleosomal DNA, the histone tails (with specific reference to H3) are displaced from the nucleosome and become available for binding by LSD1 active site. The “displacing” role of DNA binding by CoREST might also apply to tails engaged in internucleosomal interactions or bound to linker DNA. Nonproductive DNA-binding modes are possible that do not trigger H3 tail capture by the active site, and therefore do not form catalytically competent complexes. Consistently, a significant fraction of free nucleosomes was systematically observed in all experiments (Figs. 2C and SI Appendix, Figs. S5 and S6). The covalent linkage implemented by the semisynthetic nucleosomes restricts the binding positions to only a few “productive” interaction modes, resulting in more homogeneous complexes that favored the 3D structural analysis of the SAXS data. This mechanism can be extended to acetylated nucleosome recognition by the multienzymatic LSD1-CoREST-HDAC complex. The hyperacetylated histone N-termini are naturally detached from (or more weakly retained by) the nucleosome core (SI Appendix, Table S3 and Fig. S10) (26, 27). In this way, on binding of the LSD1-SANT2 domain to DNA, the histone tail is proximate to the HDAC active site, which removes acetyl groups from lysine residues. As the tail becomes free from acetylations and is already in the core of the enzyme complex, it is poised to visit the LSD1 active site, which functions only on deacetylated substrate (8, 12). Indeed, binding of the histone tail to nucleosomal DNA would be disfavored by the steric hindrance posed by the protruding tower domain and associated CoREST.
Fig. 6.
A stick-and-catch model for nucleosome recognition by LSD1/CoREST. The SANT2 domain of CoREST associates to nucleosomal DNA. The Lys4-methylated H3 N-terminal tail is thereby displaced from the nucleosomal body and can be captured by the LSD1 active site.
The in vivo relevance of this mechanistic model is supported by several studies. Above all, it is well established that H3 tails interact with DNA in native high-order chromatin, being engaged in intra- and internucleosomal interactions (17, 18). Moreover, it has been demonstrated that CoREST1 is essential to promoting in vivo histone demethylation by favoring association to the nucleosomal substrate (10). It is also notable that CoREST3 exhibits a lower repressive capacity compared with that of CoREST1, which correlates with a weaker binding to DNA (Fig. 4 and SI Appendix, Table S1) (28). Of special relevance, a splicing variant of murine CoREST3 lacking the SANT2 domain shows no repressive activity. Its overexpression leads to increased (rather than decreased, as for CoREST1 and full-length CoREST3) Lys4 methylation levels on target genes, supporting the idea that SANT2 is essential for histone demethylation (29). The weaker binding of LSD1-CoREST to DNA at higher (more physiological) salt concentrations, far from being a shortcoming, is actually an advantageous feature. It prevents a too sticky and unspecific association to chromatin and enables external factors to finely modulate the strength of the interaction. Indeed, DNA binding must be locally enhanced on the specific chromatin domains that have to be targeted by the histone-modifying complexes. Increased binding to DNA can be afforded by local variations in the chromatin environment (e.g., nucleosome packing, local dielectric constant) and by transcription factors that recruit LSD1 complexes to target genes favoring association to DNA. It is also fascinating to observe that just 20 nucleotides of wrapped DNA separate the SANT2–DNA contact point from the site where the H3 tail projects out from the nucleosome body to reach into the LSD1 active site (SI Appendix, Fig. S11). A similar DNA length is known to be covered by the histone H3 tails lying along the nucleosomal DNA helix (18, 27, 30). These features highlight the ergonomics and functional significance of the LSD1-CoREST architecture. The length and slight curvature of the tower domain is such that the clamping grip of the enzyme on the nucleosome disk has the proper design to inherently induce the detachment of the histone tail from the nucleosomal DNA.
Our studies reveal the competitive nature of the molecular mechanisms underlying nucleosome recognition and histone tail binding and modification. The central role is played by the noncatalytic domains, and in this regard, the helical tower and associated CoREST are unique features of LSD1, as they are absent in its LSD2 homolog as well as in the histone demethylases of the jumonji family (31–33). The variety of the noncatalytic domains and subunits dictates the differential biological functions specifically exerted by each histone-modifying enzyme. Such diversity, which remains mostly unexplored, underlies different architectures and modes of nucleosome recognition, providing opportunities for innovative “noncatalytic” inhibitors of chromatin-modifying processes.
Materials and Methods
Protein expression, purification, and mutant preparation were performed using standard methods and established protocols, as described in SI Appendix (22, 34–36). For the preparation of semisynthetic histones, lyophilized H3 Lys4Cys-Cys110Ala histone was dissolved in 1 M Hepes/NaOH at pH 7.8, 4 M guanidinium chloride, 10 mM l-Met, 10 mM DTT. Alkylation reaction for the installation of dimethyl-Lys analog in position 4 was performed in the same buffer, using a final 50-mM concentration of the 1-methyl-1-(prop-2-ynyl)aziridinium chloride alkylating agent (synthesized as described in SI Appendix), following previously published protocols (20). The complete installation of the propargylamine analog was confirmed by ESI-ITMS (LCQFleet Thermo Scientific Ion Trap) mass spectrometry (SI Appendix, Fig. S12), indicating that 100% H3 molecules were modified. Fluorescence polarization, chromatography-based assays, and SAXS experiments were carried out using established methodologies as detailed in SI Appendix. Fluorescence polarization experiments were performed on PHERAstar FS and CLARIOstar plate readers (BMG Labtech). SAXS data were collected on the Nanostar laboratory instrument (IBBMC), on SWING beamline at the SOLEIL synchrotron, and on BM29 beamline at ESRF synchrotron.
Supplementary Material
Acknowledgments
We thank Andrea V. Gómez for support in the CoREST3 experiments. We thank Federica Corana (Centro Grandi Strumenti, Pavia) for the excellent support in mass spectrometry analysis. V.S. was a recipient of a EMBO short-term fellowship. F.F. is supported by a career development award from the Armenise-Harvard foundation and by the Programma Giovani Ricercatori Rita Levi-Montalcini from MIUR. We acknowledge SOLEIL and ESRF synchrotrons for provision of radiation facilities and their staff during data collection. This work was supported by the Fondazione Cariplo (2010.0778), Associazione Italiana Ricerca sul Cancro (IG-11342 and IG-15208), and Italian Ministry of Education, University and Research (Progetto Epigen).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.A.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419468112/-/DCSupplemental.
References
- 1.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467(7315):562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ, Sixma TK. The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat Commun. 2014;5:3291. doi: 10.1038/ncomms4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada K, et al. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472(7344):448–453. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- 4.Canzio D, et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature. 2013;496(7445):377–381. doi: 10.1038/nature12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith E, Shilatifard A. The chromatin signaling pathway: Diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40(5):689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron R, Binda C, Tortorici M, McCammon JA, Mattevi A. Molecular mimicry and ligand recognition in binding and catalysis by the histone demethylase LSD1-CoREST complex. Structure. 2011;19(2):212–220. doi: 10.1016/j.str.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forneris F, Binda C, Battaglioli E, Mattevi A. LSD1: Oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem Sci. 2008;33(4):181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Forneris F, et al. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J Biol Chem. 2006;281(46):35289–35295. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- 9.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J Biol Chem. 2005;280(50):41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 10.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Shi YJ, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19(6):857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, et al. Crystal structure of human histone lysine-specific demethylase 1 (LSD1) Proc Natl Acad Sci USA. 2006;103(38):13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, et al. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell. 2006;23(3):377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006;13(7):626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- 16.Aasland R, Stewart AF, Gibson T. The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21(3):87–88. [PubMed] [Google Scholar]
- 17.Kulaeva OI, et al. Internucleosomal interactions mediated by histone tails allow distant communication in chromatin. J Biol Chem. 2012;287(24):20248–20257. doi: 10.1074/jbc.M111.333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pepenella S, Murphy KJ, Hayes JJ. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma. 2014;123(1-2):3–13. doi: 10.1007/s00412-013-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M, et al. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol. 2007;14(6):535–539. doi: 10.1038/nsmb1255. [DOI] [PubMed] [Google Scholar]
- 20.Simon MD, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128(5):1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tahirov TH, et al. Mechanism of c-Myb-C/EBP beta cooperation from separated sites on a promoter. Cell. 2002;108(1):57–70. doi: 10.1016/s0092-8674(01)00636-5. [DOI] [PubMed] [Google Scholar]
- 22.Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J Biol Chem. 2007;282(28):20070–20074. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 23.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496(7446):477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Ani G, Malik SS, Eastlund A, Briggs K, Fischer CJ. ISWI remodels nucleosomes through a random walk. Biochemistry. 2014;53(27):4346–4357. doi: 10.1021/bi500226b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol Cell Biol. 2011;31(23):4746–4759. doi: 10.1128/MCB.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brower-Toland B, et al. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346(1):135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol. 2008;28(1):227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrios AP, et al. Differential properties of transcriptional complexes formed by the CoREST family. Mol Cell Biol. 2014;34(14):2760–2770. doi: 10.1128/MCB.00083-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Upadhyay G, Chowdhury AH, Vaidyanathan B, Kim D, Saleque S. Antagonistic actions of Rcor proteins regulate LSD1 activity and cellular differentiation. Proc Natl Acad Sci USA. 2014;111(22):8071–8076. doi: 10.1073/pnas.1404292111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ettig R, Kepper N, Stehr R, Wedemann G, Rippe K. Dissecting DNA-histone interactions in the nucleosome by molecular dynamics simulations of DNA unwrapping. Biophys J. 2011;101(8):1999–2008. doi: 10.1016/j.bpj.2011.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang R, et al. LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol Cell. 2013;49(3):558–570. doi: 10.1016/j.molcel.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karytinos A, et al. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284(26):17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruidenier L, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276(1):19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 35.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272(3):301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 36.Villa F, et al. Crystal structure of the catalytic domain of Haspin, an atypical kinase implicated in chromatin organization. Proc Natl Acad Sci USA. 2009;106(48):20204–20209. doi: 10.1073/pnas.0908485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.