Significance
Voltage-gated sodium channels (NaV) are known to form clusters at the membranes of excitable cells; however, what governs their transport is largely unknown. We found that the endoplasmic reticulum (ER) and cis-Golgi associated ubiquitin ligase really interesting new gene (RING) finger protein 121 (RNF121) mediates the degradation and membrane localization of NaV. This apparent quality control of NaV ensures the transport of properly folded channels to the membranes of excitable cells. To our knowledge, this is the first pathologically relevant identification of a voltage-gated ion channel as a substrate for ER-associated protein degradation, whose degradation is governed by an ER- and Golgi-associated E3-ubiquitin ligase.
Keywords: zebrafish, touch response, voltage-gated sodium channel, ubiquitin, escape
Abstract
Following their synthesis in the endoplasmic reticulum (ER), voltage-gated sodium channels (NaV) are transported to the membranes of excitable cells, where they often cluster, such as at the axon initial segment of neurons. Although the mechanisms by which NaV channels form and maintain clusters have been extensively examined, the processes that govern their transport and degradation have received less attention. Our entry into the study of these processes began with the isolation of a new allele of the zebrafish mutant alligator, which we found to be caused by mutations in the gene encoding really interesting new gene (RING) finger protein 121 (RNF121), an E3-ubiquitin ligase present in the ER and cis-Golgi compartments. Here we demonstrate that RNF121 facilitates two opposing fates of NaV channels: (i) ubiquitin-mediated proteasome degradation and (ii) membrane localization when coexpressed with auxiliary NaVβ subunits. Collectively, these results indicate that RNF121 participates in the quality control of NaV channels during their synthesis and subsequent transport to the membrane.
Voltage-gated sodium channels (NaV) are large (∼230 kDa) multipass transmembrane proteins (1). The NaV channel family is comprised of nine members (NaV1.1–NaV1.9), whose activity typically underlies the rising phase of action potentials in excitable cells. In excitable cells, NaV channels form complexes with auxiliary β subunits (NaVβ1–4) in the Golgi apparatus (2), a process that enhances the kinetics and membrane localization of NaV channels (3, 4). In addition to these roles, several NaVβ subunits also function as cell adhesion molecules independent of NaV channels (5). At the axon initial segment (AIS) and nodes of Ranvier of neurons, NaV channels form clusters that facilitate the generation and propagation of action potentials. Although the molecular basis of NaV clustering at these sites has been extensively studied (6), the transport of NaV channels to these sites has been less explored. For instance, to date, only the annexin II light chain (p11) has been shown to associate with and facilitate the transport of NaV1.8 to the plasma membrane (7). Furthermore, subsequent efforts revealed that p11 acts only on NaV1.8 (8). Thus, the transport of other NaV channels remains unclear.
In zebrafish, several studies have explored the contribution of NaV channels and their auxiliary NaVβ subunits through the use of forward and reverse genetics. In brief, impairments in NaV1.1, NaV1.6a, and NaVβ1b have been shown to diminish touch-evoked escape responses and NaV channel activity in Rohon–Beard (RB) sensory neurons (9–11). In addition, two other mutants identified in forward genetic screens have been shown to affect NaV channel activity indirectly. The first, pigu, arises from a mutation in a GPI-transamidase necessary for the proper localization of NaV channels (12). Although the genetic locus of the second mutation, macho (13, 14), has yet to be identified, rough mapping indicates that it lies within a region lacking both NaV channels and auxiliary NaVβ subunits. Collectively, these results indicate that the characterization of touch-unresponsive zebrafish mutants is an efficient strategy to gain insight into the trafficking and function of NaV channels.
In this study, we identified a touch-unresponsive zebrafish mutant (mi500), which was found to be a new allele of the molecularly unidentified motor mutant alligator (13). Electrophysiological analysis revealed that NaV channel activity was severely diminished throughout the sensorimotor circuit in mutants. Further characterization uncovered that NaV channels were not localized at the AIS in mutant RBs, but instead seem to be accumulated within the endoplasmic reticulum (ER) and cis-Golgi compartments. Meiotic mapping and sequence analysis showed that the alligator locus encodes really interesting new gene (RING) finger protein 121 (RNF121), an ER- and cis-Golgi–resident E3-ubiquitin ligase that mediates the ubiquitination of NaV1.6. We found that RNF121 promotes the degradation and membrane transport of NaV1.6. Furthermore, overexpression of NaV1.6 worsened the touch response in rnf121-knockdown larvae, suggesting that an excess amount of NaV exerts proteotoxicity. These findings suggest that the proper transport of NaV channels is attributable to RNF121-mediated quality control of NaV channels within the ER and Golgi apparatus.
Results
The Mutant Phenotype Arises from a Defect in Sensorimotor Coupling.
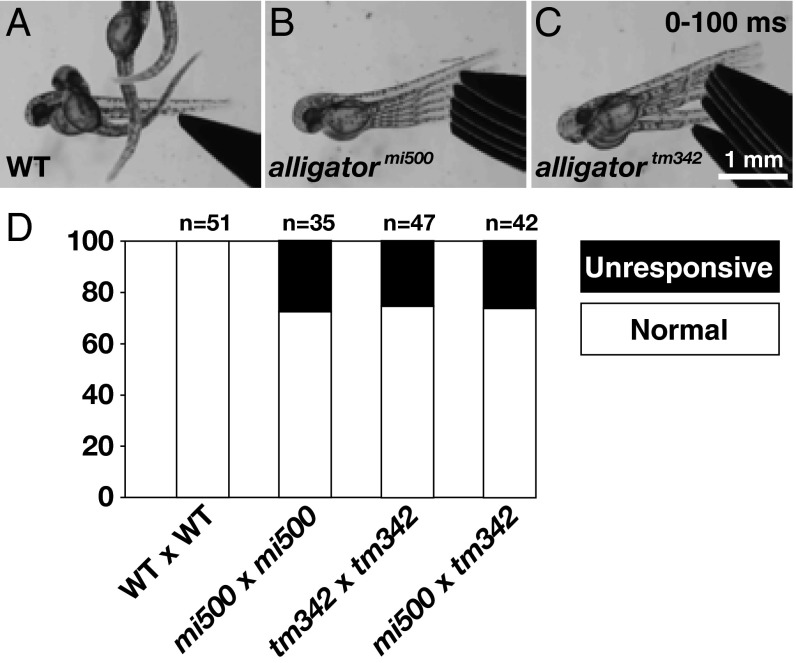
We identified a recessive zebrafish mutant (mi500) in a forward genetic screen for larvae that displayed abnormal touch-evoked motor behaviors. In short, a tactile stimulus delivered to the tail of a WT larva at 48 hours postfertilization (hpf) evoked a brief contraction, followed by a sustained bout of swimming (Fig. 1A). This response, herein referred to as a “normal” response (Materials and Methods), is typically 100% penetrant in WT larvae (Fig. 1D). In contrast to WT progeny, approximately 25% of larvae obtained from incrosses of mi500 heterozygous carriers were completely unresponsive to touch (Fig. 1B). A complementation test with the previously identified alligatortm342 mutant (13) revealed that mi500 was a new allele of this unresolved mutant (Fig. 1C). Because we found that alligatormi500 and alligatortm342 arise from missense and nonsense mutations, respectively (as detailed later), alligatortm342 was chosen for further analysis.
Fig. 1.
mi500 is a new allele of the touch-unresponsive mutant alligator. Tactile stimuli delivered to the tail of WT (A), alligator allele mi500 (B), or alligator allele tm342 (C). Of note, the images are superimposed video stills of the first 100 ms following a stimulus. (D) Histogram representing the percentage of larvae that were touch-unresponsive or exhibited a normal touch response (Materials and Methods) from incrosses and complementation crosses of alligator alleles mi500 and tm342.
To obtain a more detailed picture of the mutant phenotype, we examined whether mutants retained other motor behaviors, including spontaneous coiling, touch-evoked contractions, and “beat-and-glide” swimming. Spontaneous coiling begins at ∼17 hpf and consists of alternating contractions of the trunk and tail (15). Touch-evoked contractions begins at ∼21 hpf and is characterized by one to four rapid alternating contractions of the trunk and tail in response to tactile stimuli. Finally, beat-and-glide swimming characteristics of adult swimming begins at ∼72 hpf when larvae also orientate dorsoventrally. An assessment of these motor behaviors revealed that mutants exhibit a similar spontaneous coiling frequency and distribution of touch-evoked contractions compared with WT siblings (Tables S1 and S2). However, mutants failed to orientate dorsoventrally and never exhibited beat-and-glide swimming. Of note, mutants did not survive beyond 10 d.
We chose to focus our investigation at 48 hpf, corresponding to the onset of the mutant phenotype. We first determined whether mutants were unresponsive to other sensory stimuli by exposing unrestrained larvae to the noxious agent mustard oil. A puff of mustard oil to the trunk and tail region triggered swimming in WT larvae, whereas a control puff (1% DMSO) had no effect (Fig. S1 A and B). These findings are in agreement with previous reports regarding the effect of mustard oil on zebrafish larvae (16, 17). In contrast to WT larvae, mutants failed to move in response to mustard oil (Fig. S1 C–F). Responsiveness to mustard oil application was further explored through the use of a transgenic line that expresses the Ca2+ indicator GCaMP7a in RB sensory neurons. In response to the application of mustard oil, we observed Ca2+ transients in WT RBs, but not in mutant RBs (Fig. S1 G–J). Lastly, we examined whether mutant skeletal muscle was able to contract by applying caffeine, a ryanodine receptor agonist, to the trunk musculature (Fig. S1K). We found that WT and mutant larvae exhibited muscle contractions following caffeine application (Fig. S1 L–O). Thus, the mutant phenotype arises from a progressive loss of sensorimotor coupling.
NaV Channels Fail to Traffic Properly in Mutants.
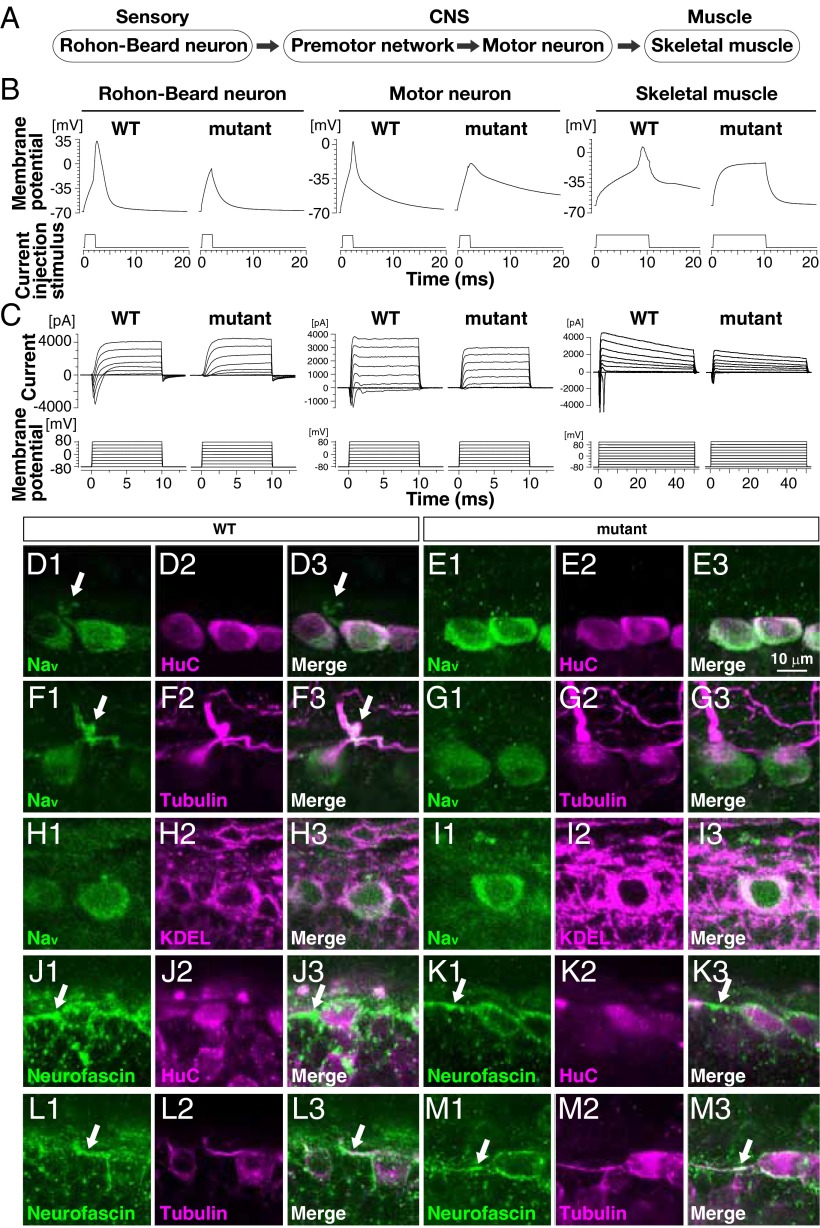
Findings thus far prompted us to assess the electrogenic properties of cells within the zebrafish sensorimotor circuit (Fig. 2A). Whole-cell current-clamp recordings made from RB sensory neurons, motor neurons, and fast-twitch skeletal muscle revealed that the resting membrane potentials of these cells did not differ between WT and mutants (Table S3). Injections of depolarizing current elicited action potentials in WT RBs (n = 10 of 10), motor neurons (n = 11 of 11), and skeletal muscle (n = 5 of 5; Fig. 2B). However, current injections failed to evoke action potentials in all mutant RBs (n = 0 of 10) and in most mutant motor neurons (n = 2 of 7) and skeletal muscle (n = 3 of 8). Subsequent whole-cell voltage-clamp recordings from these cells revealed normal potassium currents, but severely diminished voltage-gated sodium currents (Fig. 2C and Table S3), the loss of which accounts for the lack of sensory-evoked responses in mutants.
Fig. 2.
NaV channel activity and membrane localization are diminished in mutants. (A) Schematic of the sensorimotor circuit in zebrafish. (B) Whole-cell current-clamp recordings showed that action potentials are elicited in WT RBs, motor neurons, and fast-twitch skeletal muscle. Current injection failed to initiate an action potential in mutant RB cells and most motor neurons and fast-twitch skeletal muscle. (C) Whole-cell voltage-clamp recordings made from indicated cells showing that voltage-dependent inward currents are missing in mutant RBs and motor neurons and significantly diminished in skeletal muscle (Table S3). (D–M) Immunohistochemical labeling of the following proteins in WT and mutant RBs: pan-NaV, the neuronal RNA-binding protein HuC, acetylated α-tubulin common to axons, ER and cis-Golgi proteins containing KDEL tetrapeptides at the C terminus, and neurofascin common to the AIS. Arrows indicate NaV and neurofascin found in the proximal tubulin-positive processes. Note that NaV proteins in the proximal tubulin-positive processes is observed in WT RB cells but not in mutants, whereas neurofascin accumulates at the AIS in WT and mutant RBs.
We next performed whole-mount immunohistochemistry to determine the expression profile of NaV channels in mutants. In WT larvae, NaV protein was detected in the cell bodies of large, dorsal spinal-cord neurons at 48 hpf (Fig. 2D). The size, location, and coexpression of HuC protein identified these cells as RB sensory neurons. A closer examination of NaV’s subcellular distribution within RBs revealed that NaV protein colocalized with proteins containing the KDEL motif, a common marker of proteins within the ER and cis-Golgi compartments (Fig. 2H). NaV protein was also observed in proximal tubulin-positive neurites (Fig. 2F), which were also positive for the AIS marker neurofascin (Fig. 2 J and L). Taken together, these results are consistent with the transport of NaV protein from their origin of synthesis and place of maturation in the ER and cis-Golgi compartments to one of their functional destinations at the AIS in WT RBs. In comparison, NaV protein was detected in the ER and cis-Golgi compartments of mutants (Fig. 2 E and I), but was noticeably absent from the AIS of mutant RBs (Fig. 2 G, K, and M). Furthermore, NaV protein appeared to be accumulated within the ER and cis-Golgi compartments of mutant RBs (Fig. 2I). These results suggest that a failure of NaV channels to traffic to the membranes of excitable cells might underlie the mutant phenotype.
The alligator Locus Encodes for RNF121, an E3-Ubiquitin Ligase.
The mutant locus was meiotically mapped onto chromosome 21 near rnf121 (Fig. S2A), a 331-aa ER-associated E3-ubiquitin ligase (18). RNF121 is a six-transmembrane domain protein whose amino and carboxyl termini are located within the ER (19). The cytosolic RING-finger motif, which mediates the ubiquitination of target proteins, is located between transmembrane domains five and six (Fig. S2B). Sequence analysis of rnf121 from alligatortm342 uncovered a nonsense mutation at leucine 39 (L39X), which is before the first transmembrane domain of RNF121. Likewise, analysis of rnf121 from alligatormi500 revealed a missense mutation at valine 232 (V232A), an absolutely conserved amino acid within the RING-finger motif of RNF121.
To confirm that rnf121 is the causative gene in alligator mutants, we sought to restore touch responsiveness in mutants through the injection of WT RNF121 RNA, and recapitulate the mutant phenotype in WT larvae through knockdown of RNF121. To this end, one-cell stage embryos obtained from incrosses of mutant carriers were injected with RNA encoding WT zebrafish RNF121 (RNF121WT) or the V232A mutant version (RNF121V232A). Embryos were then raised until 48 hpf and examined for behavioral responses to touch. We observed a significant increase in the percentage of touch-responsive larvae in mutant clutches injected with RNF121WT RNA compared with uninjected mutant clutches (P < 0.001, χ2 test; Fig. S2C), whereas no difference was observed in mutant clutches injected with RNF121V232A RNA. Similarly, human RNF121WT, but not its mutant version (RNF121V228A), significantly restored touch response.
We next sought to induce the mutant phenotype through interfering with the production of RNF121 protein in WT larvae via injection of an antisense morpholino oligonucleotide (MO) designed to block the translation of RNA encoding RNF121. WT larvae injected with 1 or 5 ng of the RNF121 antisense MO showed a dose-dependent increase in the number of touch-unresponsive larvae (Fig. S2D), whereas all larvae injected with a control MO responded to tactile stimuli. The RNA rescue and MO phenocopy, together with the two different mutations in rnf121, demonstrate that rnf121 is the causative gene in alligator mutants.
Loss of RNF121 Does Not Induce the ER Stress Response.
Members of the ER-associated RNF proteins have been shown to be involved in the regulation of protein levels in the ER through ubiquitin-mediated proteasome degradation (19). This function is of particular importance when the accumulation of misfolded proteins in the ER triggers the activation of the unfolded protein response (UPR), a cellular process designed to remove unwanted proteins via ER-associated degradation. To address whether the UPR was activated in mutants, we examined the expression of BiP and CHOP and the alternative splicing of XBP1, which are typically induced during the UPR. RT-PCR analysis of untreated WT and mutant larvae, and larvae treated with a low dose (0.5 μM) or high dose (2 μM) of the ER-stress inducer tunicamycin revealed the following (Fig. S3A). In the absence of tunicamycin, the levels of BiP and CHOP were equivalent in WT and mutants, whereas the alternative splicing of XBP1 was not detected in either. In WT larvae, up-regulation of BiP and CHOP as well as the alternative splicing of XBP1 was observed only when larvae were treated with a high dose of tunicamycin (Fig. S3B). In mutans, however, the expression of BiP and CHOP and the alternative splicing of XBP1 were induced following application of tunicamycin at both doses. Taken together, these results show that the UPR is not active in mutants despite their elevated sensitivity to ER-stress inducers.
RNF121 Increases Membrane Localization of NaV Channels in the Presence of β-Subunits.
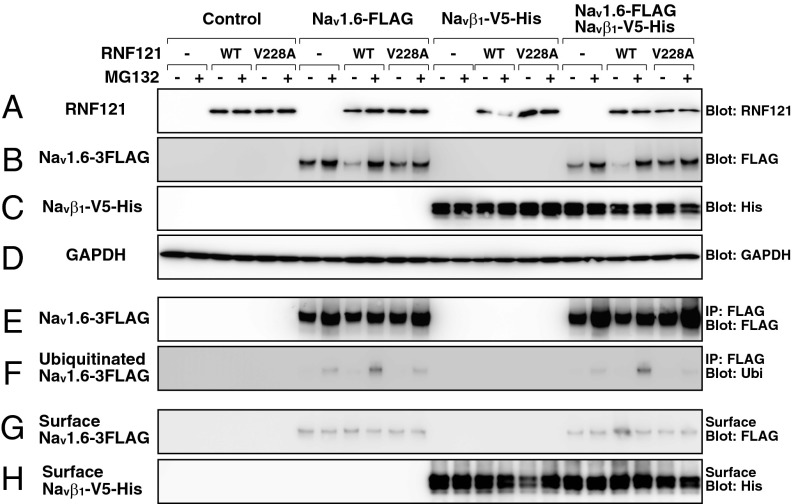
We next examined how RNF121 affects NaV channels, whose activity was diminished in mutants. To this end, we assayed recombinant expression of NaV1.6, NaVβ1, RNF121WT, and RNF121V228A in HEK293T cells. Western blotting of whole-cell extracts revealed that untransfected HEK293T cells lack endogenous expression of RNF121, whereas cells transfected with human RNF121WT or RNF121V228A expressed RNF121 protein at levels unaffected by the proteasome inhibitor MG132 (P > 0.13, t test, n = 4; Fig. 3A). Furthermore, immunofluorescence revealed RNF121 to be colabeled with protein disulfide isomerase, a marker of the ER and cis-Golgi compartments (Fig. S2 E–L). When NaV1.6 was coexpressed with RNF121WT, we observed a reduction of NaV1.6 protein from whole-cell extracts in the absence of MG132 (P < 0.05, n = 4; Fig. 3B). To determine whether the reduction of NaV1.6 was the consequence of ubiquitin-mediated degradation of NaV1.6 by RNF121WT, we treated cells with the proteasome inhibitor MG132. Treatment with MG132 restored NaV1.6 protein levels comparable to NaV1.6 expressed alone (P > 0.5, n = 4) and increased the amount of NaV1.6 that was ubiquitinated (P < 0.05, n = 4; Fig. 3 E and F). A similar phenomenon was not observed in cells cotransfected with NaV1.6 and mutant RNF121V228A. These results indicate that RNF121 regulates the quantity of NaV1.6 protein through the constitutive activity of the ubiquitin-dependent proteasome pathway.
Fig. 3.
RNF121 facilitates ubiquitination and membrane localization of Nav1.6 in HEK293T cells. Protein extracts from cells transfected with RNF121WT or RNF121V228A, NaV1.6-FLAG, and/or NaVβ1-V5-His expression vectors. Proteasome activity was inhibited by MG132. Whole-cell extracts probed with anti-RNF121 (A), anti-FLAG (B), anti-His (C), or anti-GAPDH (D). Assessing the ubiquitination of NaV1.6 from whole-cell extracts was achieved by immunoprecipitation with anti-FLAG, followed by probing with anti-FLAG (E) or anti-ubiquitin (F), which represents total and ubiquitinated NaV1.6-FLAG, respectively. Membrane localization of NaV1.6-FLAG and NaVβ1-V5-His assayed through incubation of cells in biotin, followed by purification of biotinylated proteins and probing with anti-FLAG (G) or anti-His (H), respectively. Note that ubiquitination of NaV1.6-FLAG was enhanced when coexpressed with RNF121WT, and that membrane localization of NaV1.6-FLAG increased when RNF121WT and NaVβ1-V5-His were coexpressed.
As NaV channels are typically coupled to auxiliary NaVβ subunits at the plasma membrane (2), we examined whether the coexpression of NaVβ1 influenced the degradation of NaV1.6 by RNF121. The coexpression of RNF121WT and RNF121V228A had little effect on the levels of NaVβ1 in whole cells and at the cell surface (P > 0.3, n = 4; Fig. 3 C and H). In addition, the coexpression of NaVβ1 did not affect the ubiquitination and degradation of NaV1.6 by RNF121WT (P > 0.5, n = 4; Fig. 3 B, C, and F). However, a closer examination of the surface fraction revealed that coexpression of NaVβ1 and RNF121WT lead to an increase in surface localized NaV1.6 (P < 0.05, n = 4; Fig. 3G). Furthermore, the application of MG132 eliminated the increase in surface-localized NaV1.6. Thus, RNF121 facilitates the ubiquitination and proteasome-mediated degradation of NaV1.6, but is also capable of promoting membrane localization of NaV1.6 when coexpressed with NaVβ1.
Overexpression of NaVβ1 Can Compensate for a Reduction in RNF121.
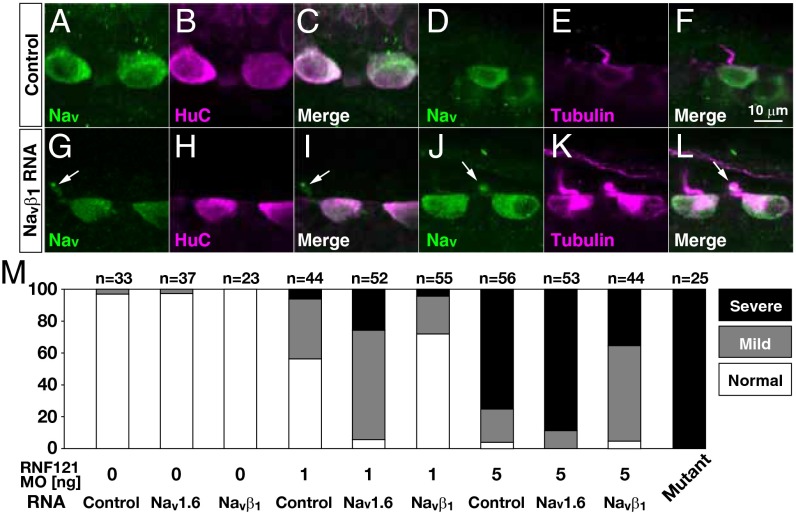
We further explored the in vivo ability of NaVβ1 to affect membrane localization of NaV channels, and the touch responsiveness of larvae in the absence of RNF121. To this end, RNA encoding NaVβ1 or luciferase (control) was injected into one-cell embryos obtained from incrosses of heterozygous alligator carriers. Although we did not observe an increase in the percentage of touch-responsive larvae (NaVβ1, P > 0.8; control, P > 0.8, χ2 test), we did observe labeling of NaV protein within the proximal tubulin-positive processes of some mutant RBs injected with NaVβ1 RNA (n = 4 of 10; Fig. 4 G–L), but not in any mutants injected with control RNA (n = 10; Fig. 4 A–F). Thus, overexpression of NaVβ1 can partially restore membrane localization of NaV channels in the absence of RNF121, but is insufficient to restore touch responsiveness.
Fig. 4.
Overexpression of NaVβ1 partially compensates for the loss of RNF121. (A–L) Overexpression of NaVβ1 restores NaV localization at the proximal tubulin-positive processes in some RBs. Immunohistochemical labeling of RBs for the following proteins in mutants injected with control RNA (luciferase; A–F) or RNA encoding NaVβ1 (G–L): pan-NaV, HuC enriched in RB cell bodies, and acetylated α-tubulin common to axons. Arrows highlight NaV localization in the proximal tubulin-positive processes. (M) Control, NaV1.6, or NaVβ1 RNA was injected into WT embryos with or without RNF121 antisense MO (1 or 5 ng). Histograms represents the percentage of larvae displaying a touch response. Note that overexpression of NaV1.6 diminished touch responsiveness, whereas overexpression of NaVβ1 partially restored the touch responsiveness in morpholino injected larvae. Touch responses were classified as described in Materials and Methods.
We next used the RNF121 antisense MO to investigate the ability of NaVβ1 to restore NaV channel function when RNF121 protein levels are reduced, rather than completely eliminated. Larvae coinjected with varying doses of RNF121 antisense MO (1 ng or 5 ng) and a fixed amount of NaVβ1 RNA (200 pg) exhibited an increase in touch responsiveness compared with larvae injected with MO alone (Fig. 4M). This result is consistent with residual RNF121 interacting with NaVβ1 to increase surface expression of NaV1.6. Conversely, the overexpression of NaV1.6 was found to further decrease touch responsiveness in larvae injected with the varying doses of RNF121 morpholino. Thus, the ability of NaVβ1 to promote the transport of NaV channels appears to vary with the amount of NaV protein present, which is regulated by RNF121.
Discussion
The work reported here began with the isolation of a new allele of the recessive zebrafish mutant alligator (13) that produces progeny unresponsive to sensory stimuli beginning on the second day of development. Here we reveal that alligator arises from mutations in the ER- and cis-Golgi–associated E3-ubiquitin ligase RNF121, which is required for functional NaV channels to reach the membrane of excitable cells. Collectively, our results indicate that RNF121 plays an essential role in the quality control of NaV channel synthesis.
Both Alleles of alligator Appear to Be Null Alleles.
We found that alligatortm342 and alligatormi500 arise from a nonsense and missense mutation in RNF121, respectively. As the alligatortm342 mutation truncates RNF121 before the first membrane-spanning domain, this allele likely represents a null allele. By comparison, the consequence of the valine-to-alanine substitution (RNF121V232A) in alligatormi500 was not immediately clear. Several findings suggests that this mutation eliminates the enzymatic activity of RNF121. First, the missense mutation was found in a completely conserved valine residue of the enzymatic RING-finger domain. Second, recombinant expression of RNF121V228A in HEK293T cells indicates that the substitution does not affect protein expression. Third, the coexpression of RNF121V228A and NaV1.6 in HEK293T cells failed to increase the amount of ubiquitinated NaV1.6. Finally, the behavioral phenotype of the two alleles were indistinguishable. Thus, both alleles of alligator appear to be null alleles.
Transport of NaV Channels.
Our recombinant expression assay demonstrated that RNF121 contributes to NaV1.6 protein levels. Recordings from several types of excitable cells also established that RNF121 is required for the transport of functional NaV channels to the membrane of these cells. Taken together with the reported spatial expression of NaV orthologs in zebrafish [RBs, NaV1.1 and NaV1.6 (11); skeletal muscle, NaV1.4; motor neurons, NaV1.5 and NaV1.6 (20)], RNF121 is at least required for NaV channel complexes composed of these four α-subunits. However, given its ubiquitous expression in larvae (21), RNF121 might contribute to the quality control of all NaV channels.
We noted two paradoxical effects of RNF121 on NaV1.6 channels in our study. The first was the observation that, although the coexpression of RNF121WT and NaV1.6 caused an overall decrease in the total amount of NaV1.6 protein in HEK293T cells, at the same time, it also caused an increase in the amount of NaV1.6 protein at the cell surface. These findings led us to conclude that RNF121 potentiates the process of transporting NaV1.6 to the membrane. The second paradoxical observation was that the inhibition of protein degradation by MG132 negated the ability of NaVβ1 to potentiate the transport of NaV1.6 to the membrane, a finding that suggests that the constitutive clearance of NaV channels (properly folded or otherwise) is necessary for the transport of NaV channels to the membrane. Taken together, these results indicate that the quality control of NaV channels by RNF121 is an essential process for their transport to the membrane.
We found that the touch responsiveness of larvae decreased concomitantly with RNF121 protein levels (i.e., WT > 1 ng MO > 5 ng MO > null mutant). Unexpectedly, we also uncovered an apparent interplay between NaV1.6 and NaVβ1 protein levels. In larvae lacking RNF121 activity (alligator mutants), overexpression of NaVβ1 restored some transport of NaV channels to the membrane in RBs, but failed to restore touch responsiveness in mutant larvae. The most likely explanations for this observation is that the amount of NaV channel transported is insufficient to restore activity throughout sensorimotor circuit. Indeed, touch responsiveness of RNF121 morphants was partially restored by overexpression of NaVβ1 and deteriorated by overexpression of NaV1.6.
Cellular Model for the Loss of RNF121.
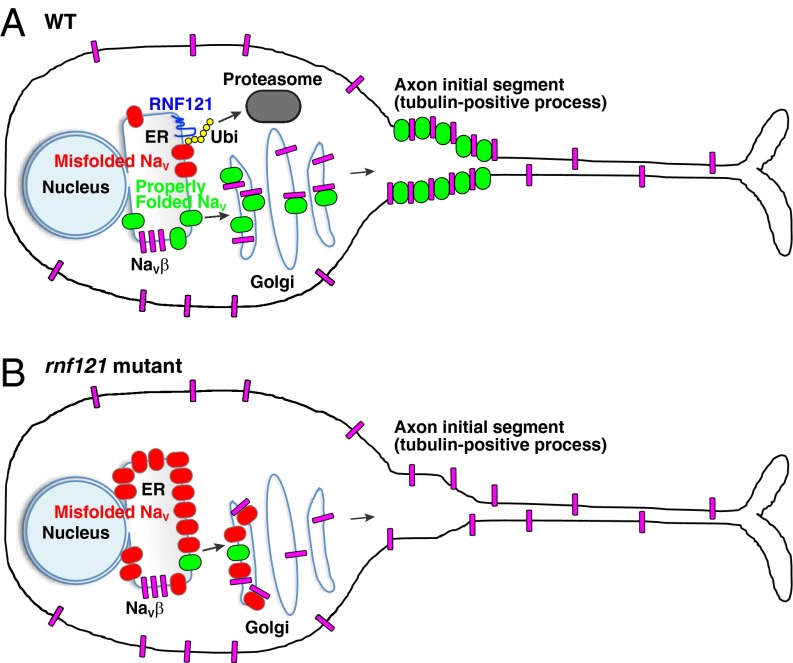
Collectively, our findings suggest the following model for RNF121 (Fig. 5). NaV channels, being composed of 24 transmembrane-spanning segments, are intrinsically susceptible to misfolding during synthesis in the ER. In WT cells, RNF121 facilitates the ubiquitination of misfolded NaV proteins, which marks them for proteasome-mediated degradation, thereby serving as a quality-control step. In the absence of RNF121, misfolded NaV proteins accumulate in the ER and cis-Golgi compartments, where it sequesters available NaVβ subunits. The ensuing shortage of NaVβ subunits in the Golgi impedes the transport of any properly folded NaV proteins.
Fig. 5.
A model of RNF121-mediated quality control of Nav channels. (A) A WT neuron wherein RNF121 mediates ubiquitination of misfolded NaV channels marking them for proteasome-mediated degradation. Properly folded NaV channels (green) associate with NaVβ subunits (magenta) in the Golgi apparatus and are transported to the AIS. Of note, some NaVβ subunits are transported to the membrane independent of NaV channels. (B) An rnf121 mutant neuron wherein misfolded NaV channels (red) accumulate in the ER and cis-Golgi compartments, which, over time, depletes NaVβ subunits, preventing them from forming complexes with properly folded NaV channels, causing an impairment of NaV transport.
Our model also suggests that a reduction in NaVβ protein levels alone could impair the transport of properly folded NaV channels, an effect that would be expected to diminish NaV channel activity in excitable cells. Consistent with this notion is the finding that knocking down NaVβ1b in zebrafish reduces NaV channel activity in RBs and the touch responsiveness of larvae (9). Although a reduction in the touch responsiveness of mice lacking NaVβ1 has not been reported (22), mice may functionally compensate through the expression of additional β-subunits.
Materials and Methods
Animals.
Zebrafish were bred and raised according to guidelines set forth by the National Institute of Genetics of Japan. The alligator allele mi500 (alligatormi500) was isolated in an N-ethyl-N-nitrosourea mutagenesis. The alligator allele tm342 (alligatortm342) was provided by the European Zebrafish Resource Center. The zebrafish transgenic line Tg(SAIGFF213A) expresses a modified GAL4 in RB sensory neurons (23), whereas the Tg(UAS:GCaMP7a) and Tg(UAS:RFP) transgenic line drives the calcium indicator GCaMP7a and RFP, respectively, under the control of UAS promoter (24).
Behavioral Analysis.
Larval behaviors were recorded at 48 hpf by using a high-speed camera (HAS-220; Ditect) at 200 frames per second as previously described (12). Tactile stimuli were delivered to the tail by using a pair of forceps. Responses of larvae to five successive tactile stimuli were classified as follows: normal (responses observed in four or five of the trials), mildly reduced (responses observed in two or three of the trials), or severely reduced (responses observed in none or one of the trials). Mustard oil (100 μM allyl isothiocyanate in 1% DMSO) was applied by a puff (20 psi, 10 ms) through a micropipette (diameter, 20 μm).
Calcium Imaging.
Tg(SAIGFF213A;UAS:GCaMP7a;UAS:RFP) triple transgenic larvae were used for Ca2+ imaging in RB sensory neurons. Sample preparation and confocal imaging were performed as described previously (25). Ca2+ transients were evoked in RB neurons by bath application of mustard oil.
Electrophysiology.
Electrophysiological recordings from larval zebrafish (48–60 hpf) were obtained from neurons and muscle by using previously described methods (26, 27). Recordings were made with an Axon MultiClamp 700B amplifier (Molecular Devices), low-pass filtered at 5 kHz, and sampled at 10 kHz. Data were acquired and analyzed by using pClamp10.
Mapping, Cloning, mRNA Rescue, and Antisense Knockdown.
A mutant carrier fish was crossed with a WIK strain for meiotic mapping. The following microsatellite markers were used:
z4074: CAGAGTTTATGGGGATCAGCGG, GGCCGACACAGTTACAGGCC.
kif4a: CACTCAGCAGAAGTAAAATTCAGCC, GAGACTTCAGTTTCAGGTTCTCC.
rnf121: CAGGGACAGTTCTGGCTG, AACATTTGAATATGTGTTTGTGTCTGTGTG.
Cloning, mRNA rescue, and antisense knockdown were carried out by using the following primers, MOs, and methods as described previously (25).
zRNF121: GGATCCGCCGCCACCATGGCAGGGGTGTTTGAGGTG, CTCGAGTTACTCCAAACCCAGGATGTAATTGATGAG.
hRNF121: GGATCCGCCGCCACCATGGCGGCAGTGGTGGAG, CTCGAGCTATTCCAGGCCCAGGATGTAG.
zRNF121 MO: GCCATCTTTAGGCTTACAGCCCTGC.
Control MO: CCTCTTACCTCAGTTACAATTTATA.
Constructs.
Full-length human NaV1.6 was obtained from Promega and subcloned into pFC27K with a C-terminal 3xFLAG tag. Human NaVβ1-V5-His expression construct (28) was provided by L. Isom, University of Michigan, Ann Arbor, MI. Full-length human RNF121 was cloned in pCS2+ expression vector.
Immunohistochemistry.
Immunostaining of zebrafish larvae was performed as described previously (12). The following antibodies were used: anti-NaV (1:500, SP19; Sigma), anti-HuC/D (1:500, 16A11; Thermo Fisher), anti-acetylated α-tubulin (1:2,000, 6-11B-1; Sigma), anti-KDEL (1:500, 10C3; Stressgen), anti-neurofascin (1:500, rabbit anti-FIGQY, gift from M. Rasband, Baylor College of Medicine, Houston, TX), Alexa 488-conjugated anti-rabbit IgG, and Alexa 568-conjugated anti-mouse IgG (1:500; Thermo Fisher). Immunofluorescence in HEK293T cells was performed by using the following antibodies: anti-RNF121 (1:500; Sigma), anti-PDI (1:500, 1D3; Enzo), Alexa 568-conjugated anti-rabbit IgG, and Alexa 488-conjugated anti-mouse IgG (1:500; Thermo Fisher). Fluorescent images were captured by using a confocal microscope (SP5; Leica).
Transfection, Immunoprecipitation, and Western Blotting.
Transfection into HEK293T cells, immunoprecipitation, and Western blots were performed as described previously (29). Anti-FLAG affinity gel (Sigma) and a cell surface protein isolation kit (Pierce) were used for immunoprecipitation and surface protein isolation, respectively. Anti-DDDDK-tag (1:2,000, FLA-1; MBL), anti-RNF121 (1:500; Sigma), anti–His-tag (1:2,000, OGHis; MBL), anti-GAPDH (1:2,000, 6C5; Acris), anti-ubiquitin (1:500, FK2; Enzo), HRP-conjugated anti-mouse IgG, and HRP-conjugated anti-rabbit IgG (1:2,000; Thermo Fisher) were used in immunoreaction enhancer solution (Toyobo). The intensity of bands was quantified using ImageJ (National Institutes of Health) and statistically analyzed by t test.
Supplementary Material
Acknowledgments
We thank Drs. Lori L. Isom (University of Michigan) and Matthew N. Rasband (Baylor College of Medicine) for providing NaVβ1 constructs and anti-FIGQY antibody, respectively; and Drs. Daisuke Morito and Kazuhiro Nagata (Kyoto Sangyo University) for helpful discussion. The transgenic line was distributed through the National BioResource Project, Japan. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.H.), the Takeda Science Foundation (H.H.), the Inamori Foundation (H.H.), a Collaborative Research Grant from the National Institute of Genetics (to S.E.L.), and National Institute of Neurological Disorders and Stroke Grant R01 NS054731 (to J.Y.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB689077).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414002112/-/DCSupplemental.
References
- 1.Cantrell AR, Catterall WA. Neuromodulation of Na+ channels: An unexpected form of cellular plasticity. Nat Rev Neurosci. 2001;2(6):397–407. doi: 10.1038/35077553. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt JW, Catterall WA. Biosynthesis and processing of the alpha subunit of the voltage-sensitive sodium channel in rat brain neurons. Cell. 1986;46(3):437–444. doi: 10.1016/0092-8674(86)90664-1. [DOI] [PubMed] [Google Scholar]
- 3.Isom LL, et al. Primary structure and functional expression of the beta 1 subunit of the rat brain sodium channel. Science. 1992;256(5058):839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- 4.Isom LL, et al. Functional co-expression of the beta 1 and type IIA alpha subunits of sodium channels in a mammalian cell line. J Biol Chem. 1995;270(7):3306–3312. doi: 10.1074/jbc.270.7.3306. [DOI] [PubMed] [Google Scholar]
- 5.Isom LL, Catterall WA. Na+ channel subunits and Ig domains. Nature. 1996;383(6598):307–308. doi: 10.1038/383307b0. [DOI] [PubMed] [Google Scholar]
- 6.Rasband MN. The axon initial segment and the maintenance of neuronal polarity. Nat Rev Neurosci. 2010;11(8):552–562. doi: 10.1038/nrn2852. [DOI] [PubMed] [Google Scholar]
- 7.Okuse K, et al. Annexin II light chain regulates sensory neuron-specific sodium channel expression. Nature. 2002;417(6889):653–656. doi: 10.1038/nature00781. [DOI] [PubMed] [Google Scholar]
- 8.Poon WY, Malik-Hall M, Wood JN, Okuse K. Identification of binding domains in the sodium channel NaV1.8 intracellular N-terminal region and annexin II light chain p11. FEBS Lett. 2004;558(1-3):114–118. doi: 10.1016/S0014-5793(03)01512-6. [DOI] [PubMed] [Google Scholar]
- 9.Fein AJ, Wright MA, Slat EA, Ribera AB, Isom LL. scn1bb, a zebrafish ortholog of SCN1B expressed in excitable and nonexcitable cells, affects motor neuron axon morphology and touch sensitivity. J Neurosci. 2008;28(47):12510–12522. doi: 10.1523/JNEUROSCI.4329-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Low SE, et al. Na(v)1.6a is required for normal activation of motor circuits normally excited by tactile stimulation. Dev Neurobiol. 2010;70(7):508–522. doi: 10.1002/dneu.20791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pineda RH, Heiser RA, Ribera AB. Developmental, molecular, and genetic dissection of INa in vivo in embryonic zebrafish sensory neurons. J Neurophysiol. 2005;93(6):3582–3593. doi: 10.1152/jn.01070.2004. [DOI] [PubMed] [Google Scholar]
- 12.Nakano Y, et al. Biogenesis of GPI-anchored proteins is essential for surface expression of sodium channels in zebrafish Rohon-Beard neurons to respond to mechanosensory stimulation. Development. 2010;137(10):1689–1698. doi: 10.1242/dev.047464. [DOI] [PubMed] [Google Scholar]
- 13.Granato M, et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- 14.Ribera AB, Nüsslein-Volhard C. Zebrafish touch-insensitive mutants reveal an essential role for the developmental regulation of sodium current. J Neurosci. 1998;18(22):9181–9191. doi: 10.1523/JNEUROSCI.18-22-09181.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saint-Amant L, Drapeau P. Time course of the development of motor behaviors in the zebrafish embryo. J Neurobiol. 1998;37(4):622–632. doi: 10.1002/(sici)1097-4695(199812)37:4<622::aid-neu10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 16.Prober DA, et al. Zebrafish TRPA1 channels are required for chemosensation but not for thermosensation or mechanosensory hair cell function. J Neurosci. 2008;28(40):10102–10110. doi: 10.1523/JNEUROSCI.2740-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Low SE, et al. Touch responsiveness in zebrafish requires voltage-gated calcium channel 2.1b. J Neurophysiol. 2012;108(1):148–159. doi: 10.1152/jn.00839.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darom A, Bening-Abu-Shach U, Broday L. RNF-121 is an endoplasmic reticulum-membrane E3 ubiquitin ligase involved in the regulation of beta-integrin. Mol Biol Cell. 2010;21(11):1788–1798. doi: 10.1091/mbc.E09-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araki K, Nagata K. Protein folding and quality control in the ER. Cold Spring Harb Perspect Biol. 2011;3(11):a007526. doi: 10.1101/cshperspect.a007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novak AE, et al. Embryonic and larval expression of zebrafish voltage-gated sodium channel alpha-subunit genes. Dev Dyn. 2006;235(7):1962–1973. doi: 10.1002/dvdy.20811. [DOI] [PubMed] [Google Scholar]
- 21.Thisse B, Thisse C. 2004 Fast Release Clones: A High Throughput Expression Analysis. Available at zfin.org. Accessed February 6, 2015.
- 22.Chen C, et al. Mice lacking sodium channel beta1 subunits display defects in neuronal excitability, sodium channel expression, and nodal architecture. J Neurosci. 2004;24(16):4030–4042. doi: 10.1523/JNEUROSCI.4139-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muto A, et al. Genetic visualization with an improved GCaMP calcium indicator reveals spatiotemporal activation of the spinal motor neurons in zebrafish. Proc Natl Acad Sci USA. 2011;108(13):5425–5430. doi: 10.1073/pnas.1000887108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muto A, Kawakami K. Prey capture in zebrafish larvae serves as a model to study cognitive functions. Front Neural Circuit. 2013;7:110. doi: 10.3389/fncir.2013.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirata H, et al. accordion, a zebrafish behavioral mutant, has a muscle relaxation defect due to a mutation in the ATPase Ca2+ pump SERCA1. Development. 2004;131(21):5457–5468. doi: 10.1242/dev.01410. [DOI] [PubMed] [Google Scholar]
- 26.Drapeau P, Ali DW, Buss RR, Saint-Amant L. In vivo recording from identifiable neurons of the locomotor network in the developing zebrafish. J Neurosci Methods. 1999;88(1):1–13. doi: 10.1016/s0165-0270(99)00008-4. [DOI] [PubMed] [Google Scholar]
- 27.Buss RR, Drapeau P. Physiological properties of zebrafish embryonic red and white muscle fibers during early development. J Neurophysiol. 2000;84(3):1545–1557. doi: 10.1152/jn.2000.84.3.1545. [DOI] [PubMed] [Google Scholar]
- 28.Patino GA, et al. A functional null mutation of SCN1B in a patient with Dravet syndrome. J Neurosci. 2009;29(34):10764–10778. doi: 10.1523/JNEUROSCI.2475-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirata H, Ogino K, Yamada K, Leacock S, Harvey RJ. Defective escape behavior in DEAH-box RNA helicase mutants improved by restoring glycine receptor expression. J Neurosci. 2013;33(37):14638–14644. doi: 10.1523/JNEUROSCI.1157-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.