Abstract
Drosophila melanogaster plays an important role in molecular, genetic, and genomic studies of heredity, development, metabolism, behavior, and human disease. The initial reference genome sequence reported more than a decade ago had a profound impact on progress in Drosophila research, and improving the accuracy and completeness of this sequence continues to be important to further progress. We previously described improvement of the 117-Mb sequence in the euchromatic portion of the genome and 21 Mb in the heterochromatic portion, using a whole-genome shotgun assembly, BAC physical mapping, and clone-based finishing. Here, we report an improved reference sequence of the single-copy and middle-repetitive regions of the genome, produced using cytogenetic mapping to mitotic and polytene chromosomes, clone-based finishing and BAC fingerprint verification, ordering of scaffolds by alignment to cDNA sequences, incorporation of other map and sequence data, and validation by whole-genome optical restriction mapping. These data substantially improve the accuracy and completeness of the reference sequence and the order and orientation of sequence scaffolds into chromosome arm assemblies. Representation of the Y chromosome and other heterochromatic regions is particularly improved. The new 143.9-Mb reference sequence, designated Release 6, effectively exhausts clone-based technologies for mapping and sequencing. Highly repeat-rich regions, including large satellite blocks and functional elements such as the ribosomal RNA genes and the centromeres, are largely inaccessible to current sequencing and assembly methods and remain poorly represented. Further significant improvements will require sequencing technologies that do not depend on molecular cloning and that produce very long reads.
The genome sequence of the fruit fly Drosophila melanogaster was first reported in 2000 (Adams et al. 2000). This sequence assembly, designated Release 1, represented the single-copy fraction of the genome in 116.2 megabases (Mb) of sequence in 134 large mapped scaffolds containing 1299 sequence gaps and an additional 3.8 Mb in 704 small (<64 kb) unmapped scaffolds. Release 1 was produced by combining a de novo whole-genome shotgun (WGS) sequence assembly, designated WGS1 (Myers et al. 2000), with sequences of mapped BAC and P1 genomic clones, including 29.7 Mb of finished sequences and draft sequences of a tiling path of BAC and P1 clones spanning the euchromatic portion of the genome (Adams et al. 2000).WGS1 and Release 1 were validated by comparison to the available finished genomic sequences and to a BAC-based physical map of the major autosomes (Hoskins et al. 2000).
WGS1 was the first shotgun assembly of a eukaryotic genome and served as a model for sequencing mammalian genomes (Venter et al. 2001; Stark et al. 2007). WGS remains the method of choice in genome sequencing because it is rapid and efficient. However, because eukaryotic genomes typically contain a large fraction of repetitive sequences with complex structures, current WGS sequencing strategies produce fragmented assemblies in which the location, order, and orientation of sequence scaffolds along the chromosomes are poorly determined. Furthermore, tandem and dispersed repetitive sequences including gene families, pseudogenes, transposable elements (TEs), segmental duplications, and simple sequence repeats are poorly represented. This leads to misassembled regions, unmapped regions, and numerous gaps, particularly in heterochromatic regions which often span many megabases of the genome and include vital protein-coding genes and other essential loci. Therefore, physical mapping, cytogenetic mapping, and sequence finishing to improve genome sequence assemblies remain a priority, especially for human (International Human Genome Sequencing Consortium 2004) and model organisms of particular importance in biomedical research.
Because D. melanogaster is a widely used research organism, we have continued to improve the reference genome sequence. Late in 2000, the Release 2 sequence corrected the order and orientation of a few small sequence scaffolds and filled a few hundred small sequence gaps. In 2002, we reported BAC-based finishing of 116.9 Mb of genome sequence in 13 scaffolds spanning the euchromatic portions of the six chromosome arms (Celniker et al. 2002) and an improved WGS assembly (WGS3) including 20.7 Mb of draft-quality sequence in larger scaffolds in the heterochromatic portion of the genome (Celniker et al. 2002; Hoskins et al. 2002). This Release 3 assembly had high sequence accuracy (estimated error rate < 1 in 100,000) and contiguity (37 sequence gaps; seven physical map gaps) in the euchromatic portion of the assembly, and the order and orientation of sequences within the assembly was confirmed by in situ hybridization of 915 BACs to salivary gland polytene chromosomes, representing 96% of the BACs in a tiling path spanning the euchromatic portion of the assembly (Hoskins et al. 2000; Celniker et al. 2002). The euchromatic sequence went through two unpublished revisions in 2004 and 2006 (Releases 4 and 5; http://www.fruitfly.org) to further improve accuracy and completeness. In 2007, we reported on further physical and cytogenetic mapping, and sequence finishing of 15 Mb in the heterochromatic portion of the genome, including essentially all single-copy regions (Hoskins et al. 2007). However, gaps and assembly errors remained due to the difficulties of mapping and finishing in repeat-rich regions. The remaining physical map gaps resulted from the absence of genomic regions from BAC libraries, likely due to incompatibility with molecular cloning or clone instability in E. coli. Sequence gaps within clone-based assemblies resulted from failure of assembly in complex nested repetitive regions. The remaining sequence assembly errors were due to incorrect but self-consistent clone-based sequence assemblies or clone rearrangements. Particularly in heterochromatin, errors in the physical and cytogenetic maps existed due to the presence of repeat-rich sequences.
Despite impressive developments in high-throughput sequencing technology, the production of high-quality finished genome sequences has remained laborious and inefficient. Furthermore, highly repeat-rich genomic regions such as those in centric heterochromatin have remained inaccessible to mapping, sequencing, and assembly. We define the “centric heterochromatin” as the repeat-rich sequences found at the functional centromeres (Sun et al. 2003). “Pericentric heterochromatin” refers to the Mb-scale regions that flank the centromeres and contain large blocks of satellite DNA and other simple-sequence repeats (Supplemental Fig. S1) interspersed with large regions of transposable-element and other middle-repetitive sequences and including essential protein-coding genes. “Telomeric heterochromatin” refers to the subtelomeric regions composed of tandem repeats (Mason and Villasante 2014) and the arrays of telomeric retrotransposons at the most distal chromosome ends (Abad et al. 2004b). By these definitions, the Y chromosome is composed entirely of centric, pericentric, and telomeric heterochromatin.
Here, we report the Release 6 assembly of the D. melanogaster reference genome sequence. Much of the improvement in the sequence is in the mapping, finishing, and assembly of repeat-rich regions in the heterochromatic portions of the genome. Release 6 incorporates (1) additional BAC-based cytogenetic mapping of previously unmapped, unordered, and unoriented sequence scaffolds by fluorescent in situ hybridization (FISH) to mitotic and polytene chromosomes, (2) BAC-based sequence finishing of clones spanning the remainder of the genome physical map guided by comparison to high-resolution BAC restriction fingerprints, and sequence finishing of 10-kb genomic plasmid clones spanning the remainder of the WGS3 assembly, (3) use of cDNA sequences to order and orient scaffolds, (4) incorporation of map and sequence data from other sources, and (5) validation of the sequence assembly by comparison to a whole-genome optical restriction map (Zhou et al. 2007). The resulting genome sequence assembly is a substantially improved reference that spans 143.9 Mb and represents the practical limit of established technologies. Relative to Release 5, Release 6 closes 628 gaps, extends the chromosome arm assemblies into telomeric and pericentric heterochromatin by 5.4 Mb, and increases the Y chromosome assembly 10-fold from ∼242 kb to 3.4 Mb. Further substantial improvement to the reference genome sequence will require new technologies that do not depend on standard molecular cloning. Emerging very-long-read WGS sequencing and assembly technologies will permit efficient production of more complete genome sequences for D. melanogaster and other species.
Results
Cytogenetic mapping of BACs
To improve and extend the anchoring of the genome sequence assembly to the cytogenetic maps of the chromosomes, we performed FISH of labeled BAC probes to Drosophila chromosomes. We selected 68 BACs representing 45 contigs in the Release 5 BAC-based physical map of pericentric heterochromatin (Hoskins et al. 2007). These include the proximal ends of the large contigs spanning the euchromatin and extending into pericentric heterochromatin on six chromosome arms, 14 additional contigs previously mapped to locations in pericentric heterochromatin, and 25 unmapped contigs. To determine the orientations of contigs on the chromosomes, 21 larger contigs were each represented by BACs from both contig ends. The remaining 24 contigs were each represented by one BAC and thus could not be oriented.
We localized BACs on two complementary cytogenetic maps of the heterochromatin. The diploid larval neuroblast mitotic chromosomes provide a complete and unbiased representation of the pericentric, centric, and Y-chromosome heterochromatin (Gatti and Pimpinelli 1992) and have been subdivided into a map of 61 regions with diverse cytological features, designated h1 to h61 (Supplemental Figure S1; Gatti et al. 1994). The map of larval salivary gland polytene chromosomes in the wm4, SuUR Su(var)3-906 strain, in which some domains in pericentric heterochromatin acquire fine banding patterns due to suppression of the normal underreplication of these regions (Andreyeva et al. 2007; Demakova et al. 2007), provides higher spatial resolution but is limited to polytenized regions. BACs were labeled and hybridized to mitotic and polytene chromosomes in parallel experiments using the same BAC DNA preparations. Hybridization of BACs in repeat-rich regions of the genome can result in cross-hybridization to multiple locations. Therefore, in the mitotic and polytene FISH experiments, we measured the relative intensity of each localized signal (Supplemental Table S1) and used the strongest signal to determine the most likely map location of each BAC (Corradini et al. 2003).
We performed mitotic chromosome FISH mapping using the isogenized y1; cn1 bw1sp1 strain (Brizuela et al. 1994) used to produce the BAC libraries and the reference genome sequence (Adams et al. 2000; Hoskins et al. 2000, 2007; Celniker et al. 2002). Of 58 BACs tested, 24 hybridized to unique locations, 29 hybridized to multiple locations but with one unambiguous primary location, and five produced ambiguous results (Fig. 1A–C; Supplemental Figs. S2–S6; Supplemental Table S1). The mitotic FISH data order BAC contigs along chromosomes including the Y chromosome which was not represented by mapped BAC contigs in Release 5 and is inaccessible to polytene FISH due to its strong underreplication in polytene tissues. We also performed several mitotic FISH experiments in which two BACs were hybridized simultaneously to the same chromosome preparation. These experiments provided information on the relative order of contigs (e.g., Supplemental Fig. S2D) but did not have sufficient resolution to determine the orientation of individual contigs. For example, we were unable to orient a contig on chromosome arm 3L using BACs separated by 366 kb (Supplemental Fig. S6). One very large contig on chromosome arm 2R was oriented by single-probe mitotic FISH experiments with probes, BACR27E03 and BACR24N15, separated by 1.6 Mb (Supplemental Table S1); polytene FISH experiments did not orient this contig.
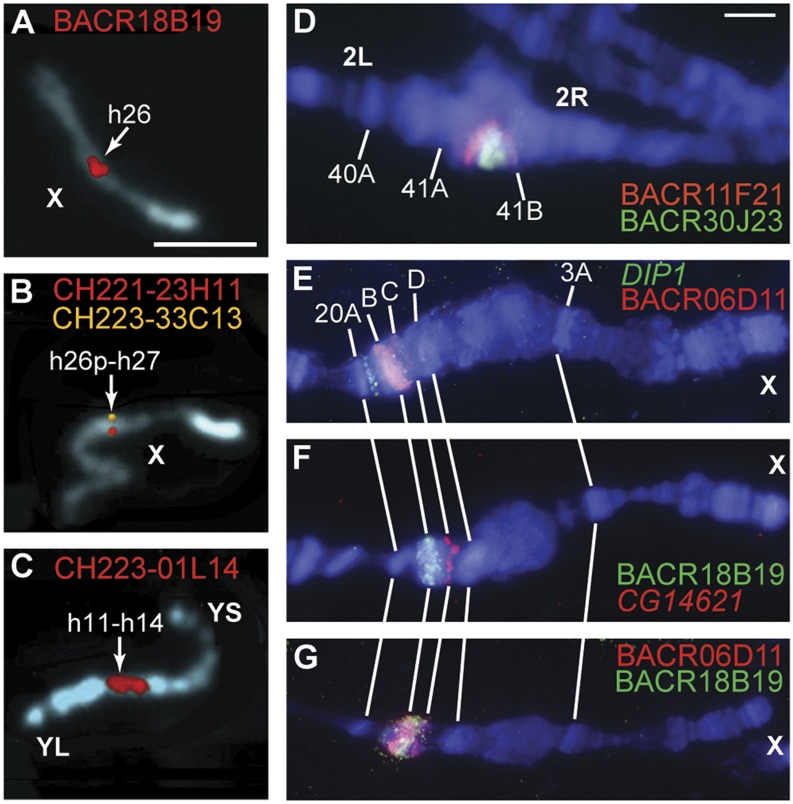
Figure 1.
FISH mapping of BACs on mitotic and polytene chromosomes. (A–C) BAC fluorescent hybridization signals (red, yellow) on mitotic chromosomes stained with DAPI (blue) are shown in pseudocolored images. Arrows indicate numbered divisions in the cytogenetic map of the pericentric and centric heterochromatin of mitotic chromosomes. Scale bar (A) indicates 3 µm. (A) BACR18B19 represents the previously unmapped Release 5 scaffold AABU01001089 and maps to division h26 on the X chromosome. (B) CH221-23H11 (red) represents the proximal end of the Release 5 arm X sequence, and CH223-33C13 (yellow) represents the distal end of the Release 5 XHet scaffold CP000208. Their signals overlap in h26p-h27; “p” indicates a proximal location within cytogenetic division h26. (C) CH223-01L14 represents the previously unmapped Release 5 scaffolds AABU01002700, AABU01002715, and AABU01001895 and produces a strong signal in h11-h14 on the Y chromosome. The long arm (YL) and short arm (YS) of the Y chromosome are indicated. (D–G) BAC and gene fluorescent hybridization signals (red, green) on polytene chromosomes of the wm4, SuUR Su(var)3-906 strain stained with DAPI (blue). Scale bar (D) indicates 3 µm. (D) BACR11F21 (red) and BACR30J23 (green) represent opposite ends of the Release 5 scaffold CP000188 and map to the proximal part of region 41A in the pericentric heterochromatin of chromosome arm 2R. BACR30J23 localizes distal to BACR11F21, orienting the scaffold. (E) BACR06D11 (red) represents the previously unmapped Release 5 scaffold CP000194 and localizes in the 20BC region of the X chromosome, proximal to DIP1 (green) in 20A. (F) BACR18B19 (green) represents the previously unmapped Release 5 scaffold AABU01001089 and also localizes in the 20BC region, distal to CG14621 (red) in 20C. (G) BACR06D11 and BACR18B19 signals overlap, but the strongest BACR06D11 signal is distal to the strongest BACR18B19 signal, suggesting their relative order. Sequence finishing shows that these BACs overlap each other by 40 kb.
We performed polytene chromosome FISH mapping using the wm4, SuUR Su(var)3-906 strain (Andreyeva et al. 2007; Demakova et al. 2007). Of 68 BACs tested, 24 hybridized to unique locations, 26 hybridized to multiple locations but with one unambiguous primary location, three produced ambiguous results, and 15 failed to localize specifically (Supplemental Table S1). The latter set represent two regions that are underreplicated and unbanded even in the wm4, SuUR Su(var)3-906 strain: a region of chromosome arm 3R and the entire Y chromosome. For most BACs derived from the pericentric heterochromatin of the X chromosome and the autosomes, polytene FISH provided sufficient specificity and resolution to determine the order of BACs along the chromosome arms. To improve the quality of the order and orientation information, all BACs in polytenized regions were mapped in additional FISH experiments in which pairs of probes, including probes for genes with known cytogenetic locations, were hybridized simultaneously to the same polytene chromosome preparation (Fig. 1D–G; Supplemental Figs. S7–S13). For 11 of 21 contigs analyzed, the ordering of paired BACs determined the orientation of the contig on the chromosome (Supplemental Table S1). These 11 oriented contigs include the extensions of three large contigs from euchromatin into pericentric heterochromatin (2Lh, 2Rh, 3Lh), another previously oriented contig on the X chromosome (XHet), and seven newly oriented contigs on chromosome arms 2R (one contig), 3L (four contigs), and 3R (two contigs). The success of these experiments depended on chromosomal location rather than contig size (i.e., distance between paired BAC probes). The 10 contigs for which orientation experiments were not successful map to chromosomal locations that are poorly banded or unbanded in the polytene chromosomes used: chromosome arm 2R (three contigs), proximal arm 3L (one contig), proximal arm 3R (four contigs), and the Y chromosome (two contigs).
We integrated the mitotic and polytene FISH data to produce a cytogenetic map (Supplemental Table S1). For BACs that produced ambiguous or conflicting data, or no data by one of the FISH methods, the map location was determined using the data from the other method or the data from a paired BAC in the same contig. The integrated map confirms the Release 5 assignments of 13 of the 14 previously mapped Release 5 heterochromatic BAC contigs to chromosome arms. In Release 5, scaffold CP000212 was mismapped to chromosome arm 3L based on analysis of a P-element transposon inserted in repeat-rich genomic sequence (Hoskins et al. 2007); the new BAC FISH results show that CP000212 maps to the Y chromosome (Supplemental Fig. S5B). In addition, the integrated map provides cytogenetic locations for 22 of 25 Release 5 unmapped BAC contigs. Three of these newly localized contigs map to the X chromosome, three map to chromosome arm 2R, and four map to chromosome arm 3R. The most substantial impact of the mitotic FISH data is on the representation of the Y chromosome: 12 previously unmapped BAC contigs and the one mismapped contig have a unique or primary localization on the Y chromosome, whereas no BACs were mapped to the Y in Release 5.
Two BAC contigs have ambiguous localizations in the cytogenetic map. BACN15B04 hybridized to locations on 3R and the Y chromosome with equal intensities in mitotic FISH and produced no specific labeling in polytene FISH. This ambiguity was resolved during sequence assembly: The corresponding scaffold was incorporated into 3R (see below). CH223-02O07 hybridized to primary locations on the Y chromosome in mitotic FISH and on 3L in polytene FISH. It represents a minimal contig comprised of a single STS and a single small-insert BAC and remains unmapped in Release 6. Finally, BACR19D10 hybridized primarily to the Y chromosome in mitotic FISH and produced no specific labeling in polytene FISH. However, sequence assembly places the corresponding scaffold unambiguously on 3R (see below).
Sequence finishing
To further improve the accuracy and completeness of the reference genome sequence, we used our previously described clone-based sequence finishing strategy (Celniker et al. 2002; Hoskins et al. 2007) to sequence BACs from the genome physical map and 10-kb genomic plasmid clones from the WGS3 assembly. Tiling path BACs for sequence finishing were identified using the genome physical map and alignments of BAC end sequences. Finishing of BAC sequences in regions that were poorly assembled in WGS3 required de novo BAC sequencing, for which we generated plasmid subclone libraries with average insert sizes of 3 kb (Releases 4 and 5) or 9 kb (Release 6) (Methods). The large-insert clone libraries facilitated assembly of repeat-rich sequences. Regions of the WGS3 assembly that were not represented within identified BACs were finished using WGS 10-kb plasmids.
Sequence finishing in highly repetitive regions of the genome is challenging, and the resulting assemblies require independent experimental verification. We used BAC restriction fingerprinting (Marra et al. 1997, 1999) to verify the assemblies of previously finished BAC sequences, guide new sequence assemblies, and resolve discrepancies between overlapping sequences in the genome tiling path. In Releases 5 and 6, we used the verified BAC-based sequences to represent the genome, even when they differed from the WGS3 sequences. Differences between the BAC-based and WGS3 sequences were usually polymorphisms due to transposable element insertion or variation in tandem repeat copy number.
We fingerprinted 1510 BACs including previously sequenced BACs and additional BACs in a redundant tiling path selected from the genome physical map and BAC end-sequence alignments. For each BAC, fingerprints were produced using five restriction enzymes (ApaLI, BamHI, EcoRI, HindIII, XhoI). For 583 BACs, which had regions of sequence for which none of these enzymes produced a fragment within our accurate sizing range (500 bp to 20 kb), fingerprints were collected using five additional restriction enzymes (BglII, NcoI, PstI, EcoRV, PvuII). These enzymes were selected based on desirable distributions of restriction sites within the sequence and for compatibility with our experimental digest conditions. The BAC fingerprint data are provided in Supplemental Data File S1. We used an automated analysis and data tracking system to compare BAC-based sequence assemblies to BAC fingerprints (M Krzywinski, unpubl.). For each BAC, the restriction fingerprints were compared to the in silico restriction digests of the sequence by pairing experimental and in silico fragments that matched within the experimental error of the fingerprint process. For each base position within the BAC-based sequence, the number of digests whose in silico fragment had a matching experimental fragment was determined. Any in silico or experimental fragments that could not be paired were identified. The total size of regions in which m/n in silico fragments were paired was calculated (n = number of digests, m = 0,1,…,n). BACs containing no experimental support (m = 0) or weak support (m = 1,2) were selected for sequence assembly review within these regions, and additional finishing and editing were performed as required. BACs whose fingerprints contained experimental fragments that could not be matched to any in silico fragments were also reviewed for possible missing sequence.
We summarize the improvements in the Release 4 and Release 5 sequence assemblies of the chromosome arms as follows. The Release 4 sequence of the chromosome arms spanned 118.4 Mb. To produce the assembly, the sequences of 216 BACs were finished or improved (Supplemental Table S2), 21 sequence gaps were closed, inversions in the assemblies of chromosome arm 3L and the fourth chromosome were corrected, and the BAC-based sequences were verified using BAC fingerprint analysis. The Release 5 sequence of the chromosome arms represented 120.4 Mb, spanning the euchromatin and extending into pericentric heterochromatin by a total of 4.7 Mb (Hoskins et al. 2007). To produce the assembly, 26 tiling path BACs were finished or improved (Supplemental Table S2), the sequence was extended into telomeric and pericentric heterochromatin, and most remaining euchromatic sequence gaps were closed. The Release 5 chromosome arm sequences contained six physical map gaps and two sequence gaps; five gaps mapped in euchromatin and three mapped in the extensions into pericentric heterochromatin. The six physical map gaps remain in Release 6; five are due to persistent gaps in the physical map that are not represented in the WGS 10-kb genomic plasmid library or in available BAC libraries (Hoskins et al. 2007). The exception is the gap corresponding to the histone gene cluster on chromosome arm 2L at 39D, a tandem gene array spanning >500 kb. This region is represented in BAC and WGS 10-kb plasmid libraries, but the structure and length of the tandem array have prevented assembly of its complete sequence.
The Release 6 chromosome arm sequences are assembled from a tiling path of 1113 BACs and 136 WGS3 scaffolds that subsume the genome physical map (Supplemental Table S2). To produce Release 6, we assembled and finished 139 additional BAC sequences (Supplemental Table S2). In 54 cases, tiling path BACs mapped to Release 5 sequence scaffolds by end-sequence alignment were finished using WGS 10-kb plasmids as templates, and then the BAC-based sequences were verified by comparison to BAC fingerprints. In 80 cases, tiling path BACs were used to construct plasmid subclone libraries and finished by sequencing a combination of BAC subclones and WGS plasmids. Nonredundant portions of 136 WGS3 scaffolds that were not represented in mapped BACs were finished as previously described (Hoskins et al. 2007). Finally, sequences of five BACs identified, sequenced, and assembled in independent work were incorporated (see below).
Assembly of Release 6
To assemble the Release 6 genome sequence, BAC-based and WGS3-based sequences that overlapped one another in BAC contigs or in newly extended sequences were merged into sequence scaffolds. Scaffolds were assigned to chromosome arms and ordered and oriented into sequence assemblies using BAC FISH and other available map information. The Release 5 assembly was divided into chromosome arm sequences spanning the euchromatin and extending into telomeric and pericentric heterochromatin and a separate set of mapped sequence scaffolds in pericentric heterochromatin (e.g., XHet). In Release 6, the chromosome arm assemblies include both the euchromatin and the mapped heterochromatin.
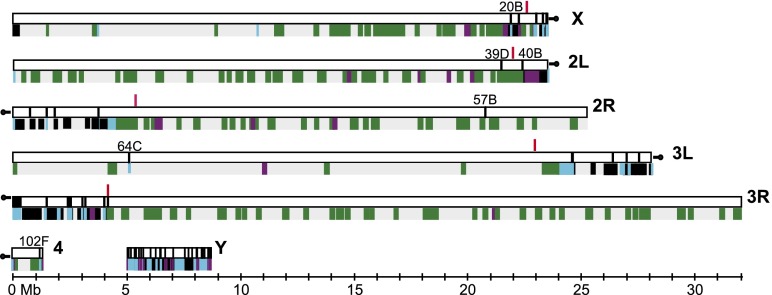
The Release 6 assembly spans 143.9 Mb (142.6 Mb, excluding sequence gaps represented by N’s) (Table 1) and increases the genomic coverage of the reference sequence assembly by 5.7 Mb compared to Release 5. The Release 6 chromosome arm sequences (X, 2L, 2R, 3L, 3R, 4, Y) comprise 137.5 Mb (137.0 Mb without N’s) in 145 mapped sequence scaffolds (summarized in Fig. 2). The sequences of the X chromosome and the autosomes extend from telomeric heterochromatin to deep within pericentric heterochromatin, based on the boundaries between euchromatin and pericentric heterochromatin defined by epigenetic analysis in the modENCODE Project (Fig. 2; Supplemental Table S2; Riddle et al. 2011). The impact on representation of the Y chromosome is particularly significant, increasing from 242 kb in Release 5 to 3.4 Mb in 105 scaffolds in Release 6. The remaining, small WGS3-based scaffolds that are not incorporated into the chromosome arm assemblies are retained in Release 6. They include 49 scaffolds comprising 1.7 Mb that were improved by sequence finishing and 1816 unimproved WGS3 scaffolds comprising 3.8 Mb (Table 2). Many of these small scaffolds were mapped to chromosomal regions in He et al. (2012); we represent those mapping results in Release 6, as described below. Here, we describe details of the assembly of the Release 6 chromosome arms, moving from the telomere to the centromere (X, 2L, 3L) or centromere to telomere (2R, 3R, 4) (lowest to highest numbered divisions along the polytene chromosome map). Further details of the assembly are presented in Supplemental Table S2 and Supplemental Figure S14.
Table 1.
Summary of the Release 6 sequence assembly
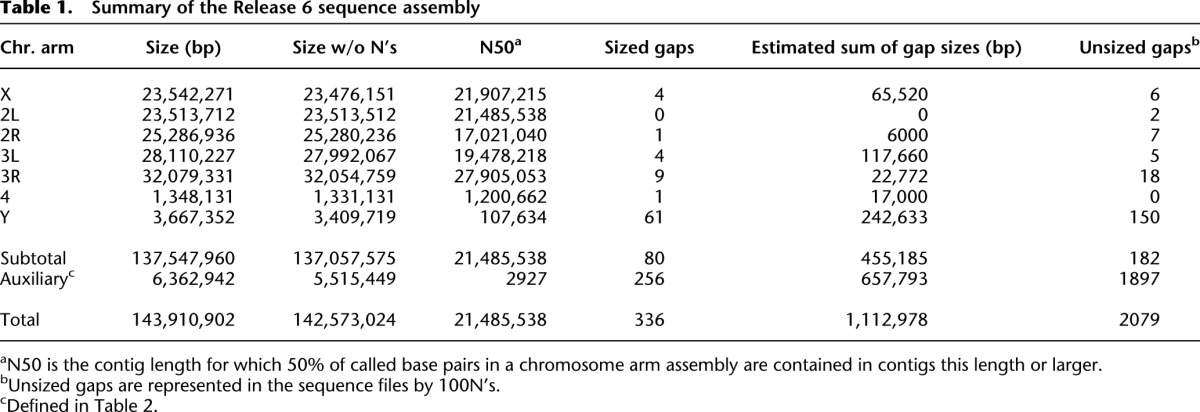
Figure 2.
The Release 6 chromosome arm sequences. Schematic representation of the Release 6 chromosome arm sequences (horizontal white bars). The boundary between euchromatin and pericentric heterochromatin on each arm (Riddle et al. 2011) is indicated (red vertical lines). Clone gaps between sequence scaffolds (vertical black lines) are labeled with their cytogenetic locations on the polytene chromosome map. (Below) Color-coding indicates the sequence release at which each BAC-based sequence was finalized: Release 3 (gray), Release 4 (green), Release 5 (purple), Release 6 (black). Sequences finished in WGS3-based scaffolds are indicated (blue).
Table 2.
Summary of the Release 6 auxiliary sequence scaffolds
The X chromosome
The X chromosome assembly spans 23.5 Mb in six scaffolds with two clone gaps in euchromatin and three in heterochromatin (Fig. 2); there are no mapped scaffolds associated with the short, heterochromatic XR arm. For Release 6, two BACs were finished and assembled near the XL telomere: BACR40C07 extends the sequence by 111,715 bp to the end of the physical map of the telomere (Abad et al. 2004b), and CH221-48I20 closes a subtelomeric 1.688 satellite repeat-rich sequence gap.
The two euchromatic clone gaps (X: 21,907,215; X: 22,260,554) map in polytene division 20B, where there was a single clone gap in Release 5. FISH identified two Release 5 unmapped BAC contigs, represented by BACR06D11 and BACR18B19, that localize within the clone gap and are ordered relative to one another and to flanking gene probes (Fig. 1E–G; Supplemental Table S1). Sequence finishing showed that these BACs overlap and produced a Release 6 sequence scaffold that is oriented by the polytene FISH data. In addition, the sequence distal to the Release 5 gap was extended by finishing of BACR27N03 (subsuming the unmapped scaffold CP000347), and the sequence proximal to the gap was improved by partial finishing of BACR46H24. Thus, the Release 5 clone gap was partially filled by extending the flanking sequences and inserting a new scaffold, leaving two clone gaps. Although it is very rich in transposable element sequences associated with the flamenco (flam) locus (Pélisson et al. 1994), this region maps in euchromatin, distal to the boundary between euchromatin and pericentric heterochromatin (X: 22,628,490) defined in Riddle et al. (2011).
The first clone gap, in pericentric heterochromatin (X: 23,020,991), maps to polytene region 20DE and mitotic region h26p-h27 (Supplemental Table S1). It is flanked on either side by a tandem array of the simple sequence repeat TAGA (Celniker et al. 2002). The gap appears to represent a satellite-like array of ∼50 kb and coincides with the major focus of accumulation of an SNF2-type chromatin remodeler, the X-linked nuclear protein (XNP); the site appears to be a genome-wide regulator of gene silencing (Schneiderman et al. 2009).
The first of the three sequence scaffolds in X pericentric heterochromatin corresponds to Release 5 XHet (CP000208), and its orientation is confirmed by FISH (Supplemental Fig. S7). The second scaffold corresponds to a Release 5 unmapped BAC contig and is ordered but not oriented by FISH (Supplemental Fig. S7). The sequence of this scaffold includes a tandem array of eight degenerate R1 elements, two of which are interrupted by a Circe element (Losada et al. 1999). Together, these two scaffolds extend the Release 6 sequence close to the proximal end of the polytene chromosome map in 20F (Supplemental Table S1). The third scaffold merges three small Release 5 unmapped scaffolds. It is not ordered or oriented with respect to the other scaffolds and is placed in its proximal location based on its sequence content. Similar to the second scaffold, the sequence of the third scaffold includes a tandem array of five degenerate R1 elements, three of which have been disrupted by insertion of a transposable element. This sequence composition suggests that the scaffold derives from an edge of the rDNA locus, which maps more proximally in the X heterochromatin (Losada et al. 1999).
Chromosome arm 2L
The assembly of chromosome arm 2L spans 23.5 Mb in three scaffolds with one clone gap in euchromatin and one in heterochromatin (Fig. 2). The sequence of the first two scaffolds, extending from the telomeric end to the second clone gap, is unchanged from Release 5. As in previous releases, the first clone gap (2L: 21,485,538) represents the tandem array of 100–200 copies of the core histone gene cluster at 39D and is estimated to span >500 kb (Celniker et al. 2002). Proximal to the second clone gap (2L: 22,420,241) at 40B, the sequences corresponding to the proximal end of Release 5 arm 2L (i.e., 2Lh) and 2LHet (CP000215) are merged and extended to produce the third scaffold. This sequence was extended distally into the clone gap at 40B by finishing of BACR39N16. The 40B gap is bordered by five copies of an HMS-Beagle/Invader TE repeat on the distal side and a number of TEs of different classes on the proximal side. The 2L assembly ends with a 26-kb array of the 260-bp subtype of the 1.688 satellite DNA family (Supplemental Fig. S1; Abad et al. 2000). The Release 6 sequence extends 1.5 Mb beyond the euchromatin-heterochromatin boundary (2L: 22,000,975) defined in Riddle et al. (2011) into 2L pericentric heterochromatin and to the proximal end of the polytene map at 40F (Supplemental Table S1; Supplemental Fig. S8).
Chromosome arm 2R
The assembly of chromosome arm 2R spans 25.3 Mb in six scaffolds with one clone gap in euchromatin and four in heterochromatin (Fig. 2). The first scaffold merges three Release 5 unmapped BAC contigs and localizes within pericentric heterochromatin at the proximal end of the polytene map in 41A; it is weakly ordered by mitotic FISH but not oriented (Supplemental Table S1; Supplemental Figs. S3, S8). It contains the gene CG45781 (Ozkan et al. 2013), and the gene annotation is improved by our cDNA IP04839, which merges three gene models (CG42644, CG40378, CG43676) (Supplemental Fig. S15A). The second scaffold corresponds to the Release 5 2RHet scaffold CP000188 and is oriented by polytene FISH (Fig. 1D). The third scaffold corresponds to the Release 5 2RHet scaffold CP000218, and the fourth scaffold corresponds to a group of Release 5 2RHet scaffolds that are linked within a BAC contig. These two scaffolds are weakly ordered, and the fourth scaffold is weakly oriented, by mitotic FISH; polytene FISH provides only the localization of these scaffolds (Supplemental Table S1). The fifth scaffold merges the Release 5 sequences of the most distal 2RHet scaffold (CP000219) and the proximal end of the arm 2R sequence (i.e., 2Rh), and it extends from pericentric heterochromatin at 41B to the euchromatic clone gap at 57B (2R: 20,780,707), which persists from Release 1 (Hoskins et al. 2000) and is not represented in available BAC libraries. The sixth scaffold extends from the 57B gap to the distal end of the assembly, which was extended in Release 6 by 27,751 bp toward the tip of the arm by sequencing of BACR11J01 and overlaps the physical map of the 2R telomere (Abad et al. 2004b).
Chromosome arm 3L
The assembly of chromosome arm 3L spans 28.1 Mb in seven scaffolds with one clone gap in euchromatin and five in heterochromatin (Fig. 2). The first scaffold is identical to Release 5 and extends from the telomeric end to the euchromatic clone gap at 64C (3L: 5,107,766). This gap was reduced in size by 171 kb in Release 5 by sequence finishing of BACR15L14 and two flanking WGS3-based sequences (Supplemental Table S2). It is flanked on either side by fragments of the transposable element BEL. The gap is not represented in available BAC libraries. The second scaffold extends from the 64C gap, past the boundary between euchromatin and pericentric heterochromatin (3L: 22,962,476) defined in Riddle et al. (2011), and merges the proximal end of the Release 5 arm 3L sequence (i.e., 3Lh) to the end of the BAC contig, subsuming the most distal Release 5 3LHet scaffold (CP000343).
The third sequence scaffold corresponds to a WGS3 scaffold (211000080752) that includes copies of the 353-bp subtype of the 1.688 satellite DNA family. This satellite is concentrated at h48p in the mitotic map (Supplemental Fig. S1; Abad et al. 2000) and at PAA-PAB in the Plato Atlantis (PA) division in the polytene map (Andreyeva et al. 2007). PA is polytenized in SuUR Su(var)3-906 strains but is not polytenized in wild-type strains. In Release 6, the finished sequence of the scaffold is assembled at the cytogenetic location of the major satellite block.
The fourth scaffold corresponds to two Release 5 3LHet scaffolds, CP000225 and CP000224, which are merged by sequence finishing of BACR11L06. The scaffold is oriented by mitotic and polytene FISH (Supplemental Table S1; Supplemental Figs. S4, S10). The fifth and sixth scaffolds finish and extend the Release 5 3LHet scaffolds CP000210 and CP000192, respectively, and the two scaffolds are ordered and oriented by polytene FISH (Supplemental Table S1). Additionally, the sequencing of BACR22B20 extends the fifth scaffold into the 361-bp repeat (another variant of the 1.688 satellite DNA family) located in h52d (Supplemental Fig. S1; A Villasante, unpubl.). Finally, the seventh scaffold extends the Release 5 scaffold CP000217, which was mistakenly placed in 2RHet in the Release 5 sequence assembly. This proximal scaffold is ordered but not oriented by FISH (Supplemental Table S1).
Chromosome arm 3R
The assembly of chromosome arm 3R spans 32.1 Mb in 18 scaffolds with no gaps in euchromatin and 17 clone gaps in pericentric heterochromatin (Fig. 2). The first eight scaffolds are small (20 kb to 87 kb), and only the fourth is represented in the Release 5 physical map. The fourth scaffold was finished using BACN36O04, the seventh was finished using CH221-27P10, and the remainder are WGS3-based. The BACs CH221-29J09 and CH221-27P10 were identified and sequenced in an independent project (A Villasante, unpubl.). These two BACs and the eight WGS3 scaffolds were identified because they contain dodeca satellite sequences found in heterochromatin region h53 (Supplemental Fig. S1; Abad et al. 1992; Losada et al. 2000; Andreyeva et al. 2007). The order and orientation of these eight scaffolds have been determined using a physical map of the region (A Villasante, unpubl.), and the assignments of four scaffolds (211000022280666, 211000022280600, 211000022279535, 211000022279847) to Chromosome 3 are verified by independent mapping results (He et al. 2012).
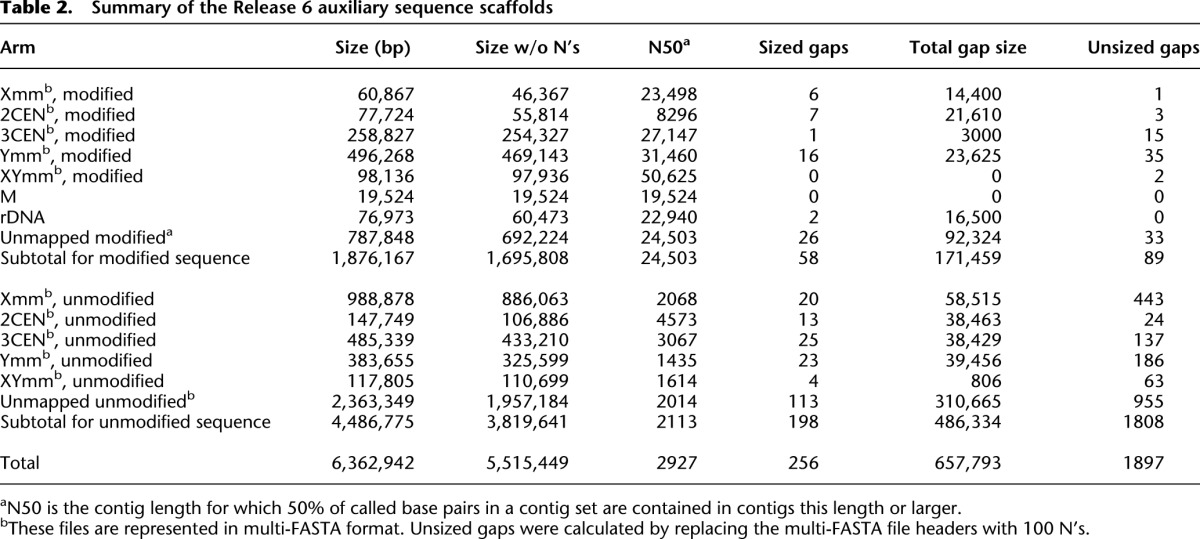
The ninth through the thirteenth scaffolds are ordered and oriented into a “super scaffold” by a cDNA (MIP31562) isolated in the modENCODE Project (Graveley et al. 2011) that encodes a new gene Myosin 81F (Myo81F; CG45784) at a very proximal location in 3R pericentric heterochromatin (Fig. 3A). Alignment of the 6604-bp cDNA sequence to the Release 6 genome defines 22 exons spanning >2.5 Mb with four clone gaps in unsized introns. To validate the Myo81F gene structure and the assembly of the super scaffold, we mapped RNA-seq reads from the modENCODE developmental time course (Graveley et al. 2011) to the cDNA sequence. Myo81F is expressed in the larval and pupal stages, most abundantly in white prepupae aged 12 h and 24 h with RPKM expression levels of 2.9 and 2.7, respectively. All exon junctions in the cDNA are verified by alignments of junction-spanning reads (range = 5 to 55 reads). Annotation of the cDNA sequence indicates a polyadenylated mRNA with a 195-nt 5′ UTR, a 75-nt 3′ UTR, and a long open reading frame encoding a 2082-aa protein. The protein has a myosin motor domain, three IQ motifs, and two sets of MyTH4 (Myosin Tail Homology 4), Ubiquitin, and FERM (4.1 protein, Ezrin, Radixin, and Moesin) domains (Fig. 3B). This structure is similar to class VII myosins, but the protein also contains two PH-like (Pleckstrin Homology-like) domains which are found only in class X myosins (Sellers 2000). The closest homolog is the unmapped Drosophila pseudoobscura gene GA22220 (20 exons spanning 33 kb), and together the two proteins may represent a new myosin class. Within the super scaffold assembly, the ninth scaffold merges three Release 5 BAC contigs, two unmapped and CP000221 in 3RHet, and is extended proximally by the dodeca-containing BACR19P07, which was identified and sequenced in an independent project (A Villasante, unpubl.). The tenth scaffold merges two Release 5 unmapped BAC contigs, and the eleventh corresponds to another unmapped BAC contig. The twelfth and thirteenth scaffolds are WGS3 scaffolds that are mapped only by sequence alignment to the Myo81F cDNA. Despite spanning >2.5 Mb, FISH experiments on nine BACs within the super scaffold (Supplemental Table S1; Supplemental Figs. S4, S12) had insufficient resolution to provide strong evidence for the orientation of the assembly on the chromosome arm.
Figure 3.
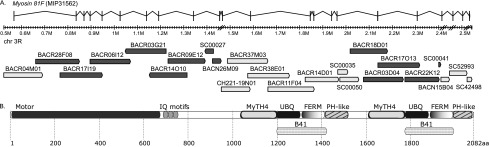
Myosin 81F links five Release 6 sequence scaffolds in pericentric heterochromatin. (A) The cDNA MIP31562 defines a new gene Myosin 81F that spans >2.5 Mb in the pericentric heterochromatin of chromosome arm 3R. The cDNA sequence was used in assembling five Release 5 BAC contigs, three unmapped (white boxes) and two on 3RHet (shaded boxes), into a series of five ordered and oriented Release 6 sequence scaffolds (Supplemental Fig. S14, 3RHet.9–13). Four unsized clone gaps between the scaffolds are indicated by double diagonal lines and map within introns of the gene. MIP31562 is 6604 bp in length, and its genomic alignment defines 22 exons. (B) The long ORF of MIP31562 encodes a 2082-aa protein with a myosin motor domain, three IQ motifs, and two sets of MyTH4 (Myosin Tail Homology 4), Ubiquitin (UBQ), FERM (4.1 protein, Ezrin, Radixin, and Moesin), and PH-like (Pleckstrin Homology-like) domains. The UBQ and FERM domains together are known as the multidomain Band 4.1 (B41) (Sellers 2000).
The relative order and orientation of the fourteenth, fifteenth, and sixteenth scaffolds is based on a gene model defined by homology that links the three scaffolds into a super scaffold (Supplemental Fig. S15B). A new gene model was identified within a set of Release 5 3RHet and unmapped scaffolds by sequence similarity to the ion channel gene Piezo (MA Crosby, pers. comm.). The new gene model, named Piezo-like (Pzl; CG45783), is represented in FlyBase R5.47 by five models that are fragments of the nearly complete new gene model. In Release 6, Pzl spans >709 kb (3R: 2,554,124…3,263,573) in 19 exons and two clone gaps within unsized introns. To validate the Pzl gene structure and the assembly of the super scaffold, we mapped RNA-seq reads from the modENCODE developmental time course (Graveley et al. 2011) to the gene model. Pzl is expressed in the pupal stages, most abundantly in white prepupae aged 24 h and 2 d with RPKM expression levels of 0.4 and 0.5, respectively. The RNA-seq data produce an improved gene model with three additional exons encoding a protein of 2173 aa (Supplemental Data File S2). All exon junctions in the improved gene model are verified by alignments of junction-spanning reads (range = 5 to 33 reads) including all exon junctions that span clone gaps between sequence scaffolds. Within the super scaffold, the fourteenth scaffold corresponds to the Release 5 3RHet scaffold CP000207; mitotic FISH confirms its location (Supplemental Table S1; Supplemental Fig. S4). The fifteenth scaffold corresponds to a Release 5 unmapped BAC contig; FISH of BACR19D10 indicates a location on the Y chromosome (Supplemental Table S1), but the scaffold is placed on 3R based on the Pzl gene model and its validation by RNA-seq analysis, and this chromosome assignment is consistent with independent mapping experiments (He et al. 2012). The sixteenth scaffold corresponds to the Release 5 3RHet scaffold CP000220 and is oriented by polytene FISH (Supplemental Table S1; Supplemental Fig. S13), thus orienting the super scaffold.
The seventeenth and most distal scaffold in 3R pericentric heterochromatin corresponds to the Release 5 3RHet scaffold CP000190 and is ordered and oriented by polytene FISH (Supplemental Table S1). Arm 3R is distinguished from the other arms because the boundary between the pericentric heterochromatin and the euchromatin falls within a clone gap (3R: 4,174,178) (Riddle et al. 2011), likely due to the presence of unclonable repeats. Finally, the eighteenth scaffold spans the euchromatin in a single gap-free contig that is identical to the Release 5 arm 3R sequence.
The fourth chromosome
The assembly of Chromosome 4 spans 1.35 Mb in two scaffolds with one clone gap (Fig. 2); there are no mapped scaffolds associated with the short, heterochromatic 4L arm. At the centromere-proximal end, the first clone in the Release 5 tiling path (BAC05L22) was determined to be chimeric. The first 24,043 bp of the BAC (and thus the Release 5 arm 4 sequence) instead represent a segment of chromosome arm 3R. In Release 6, this segment is replaced with a WGS3-based sequence (211000022278700) that extends proximally from the retained portion of the BAC-based sequence by 3427 bp (Supplemental Table S2). The Release 6 sequence begins with a TTATATTA tandem repeat that extends for 374 bp; this repeat may derive from the TTATA satellite located at h59 in the pericentric heterochromatin of Chromosome 4 (Supplemental Fig. S1). The remainder of the sequence assembly of the chromosome is identical to Release 5. The clone gap at 102F (4: 1,200,662) is flanked on either side by the simple repeat ATAAATT and is not represented in available BAC libraries.
The Y chromosome
The assembly of the heterochromatic Y chromosome spans 3.67 Mb in 105 scaffolds ranging from 630 bp to 491 kb (Fig. 2) and is 10-fold larger than the Release 5 assembly (347 kb). The greatly improved coverage of the Y in Release 6 is due to analysis of Y-linked genes (Fig. 4), identification of Y-localized BAC contigs by mitotic FISH (Supplemental Table S1; Fig. 1C; Supplemental Fig. S5), and BAC-based sequence finishing (Supplemental Fig. S14). The locations of scaffolds in the assembly are based in large part on the known cytogenetic locations of genes and other Y-specific sequences
Figure 4.
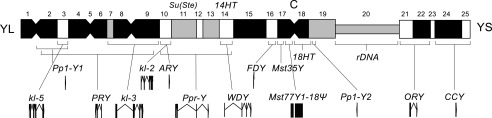
Genes in the Release 6 assembly of the Y chromosome. The locations of genes on the mitotic cytogenetic map of the long (YL) and short (YS) arms of the Y chromosome are indicated. The chromosome is divided into 25 heterochromatic regions, h1 through h25, and the location of the centromere (C) is indicated. The locations of highly repetitive sequence blocks at the Su(Ste) locus, the 14HT satellite, the 18HT satellite, and the rDNA locus are indicated. Genes newly represented in the Release 6 assembly are Pp1-Y1, polycystine-related-Y (PRY), Aldehyde reductase Y (ARY), WD40 Y (WDY), flagrante delicto Y (FDY), Mst35Y, Mst77FY1-18, and Coiled-Coils Y (CCY), and those partially represented in Release 5 and completely represented in Release 6 are male fertility factor kl-5 (kl-5), male fertility factor kl-3 (kl-3), male fertility factor kl-2 (kl-2), Ppr-Y, Pp1-Y2, and Occludin-Related Y (ORY). The FDY gene has been tentatively placed in region h16. Cytogenetic map locations of the genes with citations are indicated in the text.
The first three scaffolds in the Release 6 assembly contain the Y-linked genes Pp1-Y1 at h1-h3 in the mitotic map (Carvalho et al. 2001), kl-5 in h3 (Gepner and Hays 1993; R Kurek, S Bonaccorsi, H Buenemann, and M Gatti, unpubl.) and PRY in h2-h9 (Carvalho et al. 2000; Koerich et al. 2008), respectively. The map location of the third scaffold is supported by FISH of BACN19P02 (Supplemental Table S1). The fourth through the seventh scaffolds are ordered and oriented by the kl-3 gene in h7-h9 (Carvalho et al. 2000; Koerich et al. 2008), and the map locations of the fourth and sixth are supported by FISH of CH223-16D09 and CH223-01L14, respectively (Supplemental Table S1). The eighth and ninth scaffolds contain kl-2 (Carvalho et al. 2000; Koerich et al. 2008) and ARY (Vibranovski et al. 2008), respectively, both of which map in h10. The map location of the ninth scaffold is supported by FISH of BACR02I01 (Supplemental Table S1); it corresponds to the Release 5 scaffold CP000212, which was previously mismapped to 3RHet based on a P-element insertion (Hoskins et al. 2007). The tenth through the 87th scaffolds correspond to fragments of the Suppressor of Stellate tandem repeats (Su[Ste]) (Balakireva et al. 1992) identified in small WGS3 scaffolds. The 88th scaffold also contains Su(Ste) repeats. FISH experiments localized this complex cluster of repeats in h11 (Palumbo et al. 1994).
The 89th through the 92nd scaffolds correspond to Release 5 unmapped BAC contigs mapped by FISH of BACR05E05, BACR16A16, BACN37M16, and BACR09J05, respectively (Supplemental Table S1). The 93rd scaffold contains the Y-linked gene Ppr-Y in h10-h14 (Carvalho et al. 2001) and merges two BAC contigs; the map location is supported by FISH of BACR04I06 and BACR46N08 (Supplemental Table S1). The 94th scaffold contains the WDY gene in h14 (Vibranovski et al. 2008) and merges two BAC contigs; the map location is supported by FISH of BACR35K24, BACR12H05, and CH223-46N17 (Supplemental Table S1). The 95th and 96th scaffolds correspond to contigs mapped by FISH of BACR04L22 and BACR26A08, respectively (Supplemental Table S1). The 97th scaffold corresponds to BACR28N05 and contains the 14HT satellite block located in h14 (Abad et al. 2004a). The 98th scaffold contains the FDY gene (AB Carvalho and A Clark, unpubl.). Since the sequence of this WGS3-based scaffold includes TART-A elements, and region h16 contains primarily degenerate TART-A elements (Abad et al. 2004a), we tentatively place the scaffold in h16. However, experiments using a rearranged Y chromosome (Kennison 1981; Carvalho et al. 2000) place FDY at h1-h3 (AB Carvalho, unpubl.).
The 99th scaffold corresponds to the previously sequenced BACR07N15 and contains the Mst35Y pseudogenes within a large palindrome located in h17 (Mendez-Lago et al. 2011); the 100th scaffold contains additional sequence of this palindrome. The 101st through the 105th scaffolds contain the Mst77Y genes and pseudogenes in h17-h18 (Russell and Kaiser 1993; Krsticevic et al. 2010), the 18HT satellite block within the previously sequenced BACR26J21 in h18 (Agudo et al. 1999; Mendez-Lago et al. 2009), Pp1-Y2 (Carvalho et al. 2001) in proximal h19 (Abad et al. 2004a), ORY in h21-h22 (Carvalho et al. 2001), and CCY in h24-h25 (Carvalho et al. 2001; Koerich et al. 2008), respectively. The genome sequence of Pp1-Y2 was improved by alignment of a cDNA (MIP26157) identified in the modENCODE Project (Graveley et al. 2011) that defines 5′ and 3′ UTRs and the translation start site; the first AUG codon is four codons upstream of the previous FlyBase annotation (FlyBase 5.42).
Chromosome assignments of small scaffolds
There remain 1863 scaffolds representing 6.4 Mb (5.5 Mb without N’s) of genomic sequences that have not been incorporated into the chromosome arm assemblies. These scaffolds range in size from 544 bp to 88,768 bp. The sequences of 49 of these scaffolds representing 1.9 Mb (1.7 Mb without N’s) have been improved by sequence finishing; the remainder are unimproved scaffolds from the WGS3 assembly (Table 2).
He et al. (2012) mapped many of the Release 5 unmapped scaffolds to chromosomal regions using a series of mutant Drosophila strains bearing large chromosomal deletions and rearrangements. They performed comparative genome hybridization using DNA isolated from embryos of specific mutant genotypes and oligonucleotide microarrays representing the entire Release 5 sequence. Oligonucleotide sequences that did not hybridize to DNA of a particular deletion genotype were inferred to map within the deleted region. These data were consistent with the Release 5 cytogenetic map, validating the method. In Release 6, the scaffolds that were not incorporated into chromosome arm sequences were assigned to “auxiliary sequence files” based on the He et al. (2012) data: Xmm, 2CEN, 3CEN, Ymm, XYmm, and U (unmapped) (Methods). For each set, the corresponding Release 6 scaffolds are represented as a series of individual scaffold sequences in FASTA format.
Two additional sequence files are included in Release 6. To represent the large tandem ribosomal RNA gene arrays present in the pericentric heterochromatin of the X and Y chromosomes (bobbed loci), we used the published rDNA sequence (GenBank: X01475) (Roiha and Glover 1981) to identify WGS3 unassembled reads containing rDNA sequences. Four WGS 10-kb plasmids were sequenced and assembled to represent a portion of the rDNA repeats (auxiliary sequence file “rDNA”). To represent the mitochondrial genome of the reference strain, we used the published mtDNA sequence (GenBank: U37541) (Clary et al. 1982), which is a composite of sequences from the Canton S and Oregon R strains, to identify the corresponding WGS3 scaffold (GenBank: AABU01002389), and finished the sequence. The nucleotide sequence of the gene-containing region is 99% identical to the previously published sequence, and the translated protein sequences are 100% identical. The nucleotide sequences of the mitochondrial tRNAs are identical. Most of the sequence differences are within the 3.8-kb A + T repeat region, which is 95% identical.
Validation of the Release 6 assembly by whole-genome restriction mapping
To validate the Release 6 sequence assembly, we produced a whole-genome NheI restriction map of the reference strain using a commercial optical mapping platform (OpGen) and compared it to the in silico restriction map of the sequence assembly (Methods). The whole-genome restriction map assembly comprises 58 contigs with an average coverage depth of 55× and a combined length of 213 Mb (Supplemental Data File S3).
We aligned the whole-genome map to the Release 6 sequence (Methods; Supplemental Data File S4), resulting in alignment of 129 Mb of the Release 6 sequence to 50 map contigs with a combined length of 198 Mb. Thus, the map assembly is partially redundant; alignment to the sequence assembly identifies 68.7 Mb in overlapping, redundant maps. The alignment shows a high level of matching of ordered restriction fragments across the sequence assemblies of the X chromosome and the autosomes, including essentially complete coverage of the euchromatin and substantial coverage of telomeric and pericentric heterochromatin (Supplemental Table S3). The aligned regions represent 129 Mb (96.5%) of the 134 Mb in the sequence assemblies of these chromosome arms. Due to the reduced representation of the X (75%) and Y (25%) chromosomes relative to the autosomes in the mixed population of XX females and XY males used to construct the whole-genome map, we collected lower coverage of the X and Y chromosomes in single-molecule maps. This contributed to a less complete alignment to the X chromosome sequence assembly and a failure to assemble reliable map contigs representing the Y chromosome. We also did not identify alignments to partially mapped or unmapped sequence scaffolds. The comparison of the whole-genome map to the Release 6 sequence does not identify any order or orientation errors within the aligned regions of the sequence assembly.
At the six telomeric regions represented in the Release 6 chromosome arm assemblies, the whole-genome map is consistent with both the physical maps of these telomeres (Abad et al. 2004b) and the sequence assemblies. The whole-genome map assemblies do not define discrete chromosome ends. Instead, near each telomere, there is an abrupt reduction in the depth of coverage in the single-molecule optical maps that suggests the position of the most common chromosome end in a population (Supplemental Fig. S16). At each of the six telomeres represented in the alignment, we trimmed the whole-genome map at the restriction site nearest this position (Methods). At the XL, 2L, 2R, and 3R telomeres, the whole-genome map extends beyond the sequence assembly by 30 kb, 39 kb, 46 kb, and 19 kb, respectively (Supplemental Table S3; Supplemental Fig. S17). In contrast, at the 3L and 4R telomeres, the sequence assembly extends beyond the whole-genome map by 5.0 kb and 5.1 kb, respectively (Supplemental Table S3; Supplemental Fig. S17). The Release 6 sequence includes the telomere-associated sequences (TAS repeats in the subtelomeric regions) at four of the six assembled telomeric regions (2L, 2R, 3L, 3R), and it extends into the telomeric transposable element sequences (TAHRE, TART, and HeT-A) at four telomeres (XL, 2L, 2R, 4R).
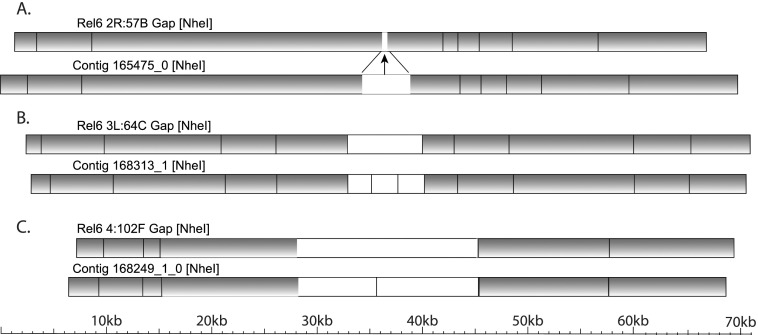
The whole-genome map spans seven clone gaps between sequence scaffolds and measures the gap sizes (Supplemental Table S3). In euchromatin, the gap at 2R: 57B is estimated to be 2.6 kb, the gap at 3L: 64C is estimated to be 7.2 kb, and the gap at 4: 102F is estimated to be 17 kb (Fig. 5). In pericentric heterochromatin, the gap at 2L: 40B (2L: 22,420,241), which was previously estimated to span ∼100 kb in Release 3 (Yasuhara et al. 2003), is estimated to be 92 kb. Also, in pericentric heterochromatin, the gap at 3L: PACp (3L: 26,400,914) is estimated to be 72 kb, and the gap at 3R: PAF (3R: 4,012,024) spans 39 kb (Supplemental Fig. S18). The gap at 3L: PAEd (3L: 27,549,160) falls within a deletion in the whole-genome map with breakpoints in the sequence assembly 55 kb to the left and 18 kb to the right of the gap. In the Release 6 sequence, the 3L: 64C and 4: 102F gaps are represented by 7 kb and 17 kb of N’s, respectively; all other clone gaps between sequence scaffolds are represented by 100 N’s (Supplemental Table S2).
Figure 5.
Measurement of three euchromatic clone gaps by whole-genome optical restriction mapping. Alignments of the Release 6 genomic sequence (Rel6) to whole-genome optical restriction map contigs (Contig) at clone gaps between sequence scaffolds (arrows) are diagrammed. Aligned NheI restriction fragments (shaded boxes), unaligned fragments (white boxes), and alignment points (lines connecting NheI restriction sites in the sequence and the map) are indicated. The euchromatic clone gaps at (A) 2R: 57B, (B) 3L: 64C, and (C) 4: 102F are spanned by whole-genome map contigs, providing estimates of the gap sizes (Supplemental Table S3).
We identified no discrepancies in the relative order of scaffolds or restriction fragments in the alignment of the whole-genome map to the Release 6 chromosome arm sequences. However, we did identify rare discrepancies that are consistent with insertion-deletion polymorphisms segregating within the reference strain or that may identify local assembly errors in repeat-rich regions. We identified 41 discrepancies larger than 4 kb (Supplemental Table S4). Ten are deletions with respect to the genome sequence and could therefore be evaluated. Five of these correspond to previously documented differences between the WGS3 and BAC-based sequence assemblies and were attributed to polymorphisms within the reference strain: four transposable element insertions and one large tandem duplication (Celniker et al. 2002). The other five correspond to two transposable element insertions, two tandem duplications, and one tandem repeat copy number variant. The whole-genome optical restriction map validates the Release 6 chromosome arm sequences and identifies rare polymorphisms and local regions of incomplete sequence finishing.
Discussion
The accuracy, contiguity, and coverage of the Release 6 reference sequence of the D. melanogaster genome represent the limits of what is practically achievable with established clone-based methods for mapping, sequencing, and assembling a complex eukaryotic genome. We used FISH to both mitotic and polytene chromosomes to produce an exhaustive BAC-based cytogenetic map. We used the WGS3 assembly and WGS sequence reads representing as much of the genome as can be stably cloned in Escherichia coli. We assembled and finished sequences of the regions represented in the BAC map and the WGS reads to near the limit of the information available in mate-pair sequence reads and BAC restriction fingerprints. These regions include repeat-rich telomeric and pericentric heterochromatin and a substantial fraction of the Y chromosome. We integrated the map and sequence data, and available data from others, to produce a comprehensive clone-based genome sequence assembly. Finally, we validated the order and orientation of sequences in the assemblies of six chromosome arms (X, 2L, 2R, 3L, 3R, 4) by comparison to a whole-genome optical restriction map. Our approach, integrating BAC-based and WGS data, improved accuracy and completeness compared to what would have resulted from either strategy alone. New technologies will be required to substantially improve on this reference genome sequence. Specifically, long sequence reads that do not require molecular cloning will be required to span the remaining gaps, particularly in the highly repeat-rich heterochromatin.
We reached the practical limits of BAC FISH mapping to chromosomes. In terms of the mapping resolution available in the polytene chromosome FISH experiments, the set of scaffolds that were ordered and oriented by alignment to the Myo81F cDNA sequence and that span >2.5 Mb in 3R pericentric heterochromatin were represented by nine BACs in polytene FISH experiments, but these experiments did not determine the orientation of this large set of scaffolds due to the poor banding of polytene chromosomes in this very centromere-proximal region. In terms of available probes, six small Release 5 unmapped BAC contigs were not included in the FISH mapping experiments. Four of these were integrated into mapped locations in chromosome arm assemblies during sequence finishing, and two (representing WGS3 scaffolds smaller than 25 kb) are represented in 3CEN (3CEN.1 and 3CEN.31). Based on our experience, FISH would be an inefficient method for mapping the remaining sequence scaffolds.
There remain a few regions in the Release 6 sequence that could be improved with further directed mapping and finishing. For example, there are several identified, unsequenced BACs that contain dodeca satellite repeats and are therefore likely to derive from the h54-h56 region of arm 3R (A Villasante, unpubl.). The 4.1-kb WGS3 scaffold AABU01002623 also contains dodeca repeats and is represented in the Release 6 sequence file 3CEN. These unfinished sequences may derive from a clone gap in the 3R assembly. Similarly, the X: 20B region, which includes two clone gaps of unknown size, might be improved with focused effort. This region is a complex nest of fragmented TE sequences and is associated with the flam locus and production of piRNAs. Zanni et al. (2013) reported that the Release 5 unmapped scaffold CP000194 (Release 5 U: 964336–1041768) containing piRNA cluster 17, includes a 30-kb segmental duplication of a portion of the flam locus. This scaffold is incorporated in Release 6 in the newly mapped scaffold between the two gaps in the X: 20B region. We determined that further work in such regions would not provide significant improvements. In general, it will be more efficient to apply emerging new technologies to further improve the reference genome sequence, rather than to rely on conventional clone-based methods.
In producing Release 6, we corrected the map locations and orientations of Release 5 sequence scaffolds and filled sequence gaps within them. However, with the exception of the chimeric BAC identified at the left end of the Chromosome 4 assembly, we did not break Release 5 scaffolds. The FlyBase Release 5 gene annotation did not include models that spanned clone gaps between sequence scaffolds, so all of the Release 5 gene models could be migrated to Release 6. FlyBase has moved the Release 5 gene annotation onto the Release 6 sequence (Dos Santos et al. 2014). Here, we report the discovery of two new genes that link Release 6 scaffolds into super scaffolds. The Release 6 sequence, particularly the pericentric heterochromatin and the Y chromosome, may lead to the discovery of additional new genes and other functional elements.
We defined two new, very large protein-coding genes in the pericentric heterochromatin of chromosome arm 3R. Myo81F spans >2.5 Mb, and Pzl spans >700 kb. These genes permitted a series of sequence scaffolds to be ordered and oriented, and the improved gene structures will facilitate genetic analyses of their functions. Myo81F is the largest gene identified to date in the D. melanogaster genome, though the incompletely assembled Y-chromosome fertility factors kl-5, kl-3 and ks-1 may ultimately prove to be larger (for review, see Gatti and Pimpinelli 1992). Heterochromatic genes are not a unique feature of the Drosophila genome. For example, the human potassium channel gene KCNJ18 maps within a gap in the reference human genome sequence in pericentric heterochromatin and was not represented in sequence databases prior to its discovery and association with thyrotoxic hypokalemic periodic paralysis (Ryan et al. 2010).
The agreement between the Release 6 sequence and the whole-genome optical restriction map validates the sequence assembly in the aligned regions of the chromosome arm sequences. The unaligned regions of the sequence assembly are repeat-rich, located near gaps between scaffolds, or have an insufficient number of restriction fragments to identify significant alignments to map contigs. The alignment also measures the sizes of seven clone gaps between sequence scaffolds. Three physical map gaps in the euchromatin at 2R: 57B, 3L: 64C, and 4: 102F are spanned by aligned map contigs, and the largest of these gaps measures just 17 kb. These persistent gaps in the genome physical map are much smaller than the cloned insert sizes of available BAC libraries (Hoskins et al. 2007; Venken et al. 2009), but none is spanned by identified BACs. The remaining unsized clone gaps in euchromatin are associated with two large repeat-rich regions: The pair of gaps at X: 20B is associated with the complex nest of diverse TE sequences near the flam locus, and the gap at 2L: 39D corresponds to the tandem repeat of core histone genes spanning at least 500 kb. Four clone gaps in heterochromatin are also spanned by aligned map contigs. The largest of these spans 92 kb, and the smallest is contained within a deletion of at least 73 kb in the whole-genome map. In addition to measuring the sizes of these gaps, the whole-genome map confirms the relative order and orientation of the aligned sequence scaffolds. The use of longer DNA molecules to produce additional optical map data might permit estimation of the sizes of additional gaps (Zhou et al. 2007), and the use of additional restriction enzymes might provide sufficient information to identify alignments to additional sequence scaffolds.
The goal of genome sequencing projects is to determine the complete and continuous sequences of the chromosomes. A number of known genetic elements in Drosophila heterochromatin are incompletely represented in Release 6. These include the most distal portions of the telomeres, the rDNA tandem gene arrays on the X and Y chromosomes, and the parts of the centric and pericentric heterochromatin enriched for highly repetitive satellite sequences that represent the functional centromeres and flanking regions. Thus, further improvements in technologies to produce more complete genome sequences, ultimately true “end-to-end” chromosome assemblies, remain a high priority.
Methods
Cytogenetic mapping of BACs
BACs selected for cytogenetic mapping were colony-purified and end-sequenced to verify clone identities (data not shown). BAC DNA was prepared by the standard alkaline lysis method. To limit bias in the interpretation of FISH results, BAC preparations were assigned a code number, and identical coded samples were delivered to the Dimitri laboratory for mitotic FISH and the Zhimulev laboratory for polytene FISH. The BAC FISH results were recorded without knowledge of the clone identities or expected map locations.
FISH to mitotic chromosomes from the isogenized y1; cn1 bw1 sp1 reference strain (Brizuela et al. 1994) was performed as described in Accardo and Dimitri (2010).
FISH to polytene chromosomes was performed as described in Saunders (2004) with modifications described here. wm4, SuUR Su(var)3-906 larvae were grown at 25°C in uncrowded vials on standard fly food. Salivary glands were dissected in Ephrussi-Beadle saline (Ephrussi and Beadle 1936) and fixed in a 3:1 mixture of ethanol and acetic acid for 30 min at −20°C, squashed in 45% acetic acid, snap-frozen in liquid nitrogen, and stored in 70% ethanol at −20°C. Squashes of polytene chromosomes were incubated in 2× SSC for 1 h at 65°C, washed three times for 5 min in 2× SSC at room temperature, denatured in 2× SSC, 0.07 N NaOH for 0.5 min, dehydrated in increasing concentrations of cold ethanol (70%, 80%, 100%) for 3–5 min each, and air dried. DNA probes were labeled with biotin-16-dUTP or digoxigenin-11-dUTP (Roche) in random-primed reactions with the Klenow fragment of DNA polymerase I. Labeled probes were added to hybridization solution (50% formamide, 2× SSC, 10% dextran sulphate, 1.0% sonicated salmon sperm DNA) to a final amount of 0.1–0.2 µg per slide. Hybridization was performed overnight at 37°C in a humid chamber. Unbound probes were removed with three 15-min washes in 0.2× SSC at 42°C. Slides were stained with avidin-FITC and rhodamine anti-DIG conjugate in blocking solution (0.1% BSA, 1× DIG-blocking reagent [Roche]) for 30 min at 37°C in a humid chamber and washed three times for 5 min with 4× SSC, 0.1% Tween-20. Finally, 10 µl of antifade solution (2.5 mg/mL of 1,4-diazobicyclo-[2.2.2]-octane in 2× SSC [Sigma]) with DAPI were added before examination by fluorescence microscopy. In addition to BAC DNAs, marker gene DNA probes (Supplemental Table S5) were used to correlate polytene regions with mitotic regions.
Sequence finishing
BAC-based sequencing was performed using plasmid subclone libraries. For Releases 4 and 5, BACs were used to construct plasmid libraries with inserts of ∼3 kb (Celniker et al. 2002). For Release 6, BACs were used to construct plasmid libraries with inserts of ∼9 kb using the previously described approach with the following modifications. BAC DNA was fragmented by shearing (HydroShear, GeneMachines). Fragments were size-selected and cloned in pBR194b, a derivative of the pBR194c vector used to construct the 10-kb WGS plasmid libraries (Adams et al. 2000). The medium-copy origin of replication in these vectors is compatible with larger inserts. To construct pBR194b, a polylinker was inserted between the paired BstXI cloning sites in pBR194c. Plasmid libraries constructed in pBR194b were transformed into E. coli DH10B by electroporation to reduce selection against clones with larger inserts. A sample of subclones that were sequenced to completion during BAC-based finishing have insert sizes of 9.5 (±1.8) kb (data not shown).
Sequence finishing of BACs and WGS 10-kb plasmids was conducted as previously described (Celniker et al. 2002; Hoskins et al. 2007). BAC-based sequence assembly was verified by comparison to BAC restriction fingerprints (Supplemental Data File S1). BAC fingerprints were particularly valuable in regions of complex repetitive sequences and for estimating copy number in tandem repeats, as previously described (Celniker et al. 2002). Sequence quality was estimated as described (Celniker et al. 2002); the single-copy portions of all finished sequences have an estimated error rate of less than 1/100,000.
Approximately 1% of BACs used as templates in sequence finishing for Releases 3 to 6 had acquired an insertion of an E. coli transposon. We observed insertions of Tn10, IS3 and IS5 elements. These insertions were identified by comparison of the finished BAC sequences to a database of E. coli transposon sequences. Following the validation of finished BAC sequences by comparison to BAC fingerprints, the inserted transposon sequences were removed from the finished BAC-based sequences. Researchers using BAC clones in their experiments should be aware of this phenomenon, since E. coli transposon insertions might impair the functions of cloned genes in transgenic constructs.
BAC restriction fingerprinting
BAC fingerprints were generated using an agarose gel-based restriction enzyme methodology as previously described (Mathewson et al. 2007), with the following restriction enzyme digestion conditions optimized for each enzyme: 2 units of enzyme were used for EcoRV; 5 units of enzyme were used for ApaLI, BglII, EcoRI, HindIII, NcoI, PstI, PvuII, and XhoI; 20 units of enzyme were used for BamHI. Digestion time was 1 h for BgII, EcoRI, EcoRV, HindIII, NcoI, PstI, and PvuII; 2 h for ApLI, BamHI, and XhoI. All digests were performed at 37°C. Gel images were acquired using a Molecular Dynamics Fluorimager 595. Lane tracking of the digitized gel images was manually adjusted using Image software (www.sanger.ac.uk/resources/software/image/), and restriction fragments were identified and sized automatically using BandLeader software (Fuhrmann et al. 2003).
Sequence assembly
Sequence assemblies of chromosome arms were produced as previously described (Celniker et al. 2002). Alignments to previous genome sequence releases and BAC projects were generated using MUMmer v3 (Kurtz et al. 2004). Gaps in the sequence assembly are represented by sets of N’s. Clone gaps are represented by 100 N’s unless otherwise noted. Sequence gaps are represented by sets of N’s corresponding to the estimated gap size. For details of the genome sequence assembly, see Supplemental Table S2 and Supplemental Figure S14.
Assignment of scaffolds to auxiliary sequence files
We used WUBLASTv2.0.0605 (W Gish, unpubl.; http//blast.advbiocomp.com) to identify Release 5 scaffolds not represented in the tracked finishing work for Release 6. Scaffolds with fewer than 30 mismatches were considered to be redundant and were subsumed; those with more than 30 mismatches (defined as the scaffold length minus blast match_length) were considered to be nonredundant and were included as scaffolds in Release 6. We used the assignments of these small Release 5 sequence scaffolds reported in He et al. (2012) to generate five separate Release 6 multi-FASTA files: sequences mapping to the X chromosome (Xmm), sequences mapping to the centric region of the second chromosome (2CEN), sequences mapping to the centric region of the third chromosome (3CEN), sequences mapping to the Y chromosome (Ymm), sequences mapping to the X or Y chromosome (XYmm), and remaining unmapped sequences (ArmU).
Whole-genome optical restriction mapping
High molecular weight genomic DNA in agarose plugs was prepared from a mixed-sex population of adults of the isogenized y1; cn1 bw1 sp1 reference strain (Brizuela et al. 1994) as previously described (Hoskins et al. 2000). At OpGen, Inc., DNA molecules larger than 150 kb were processed on the ARGUS Whole Genome Mapping System to yield single-molecule NheI optical restriction maps. These maps were assembled using commercial software to produce a de novo whole-genome map consensus (Supplemental Data File S3). First, 146,884 single-molecule maps longer than 350 kb were assembled into seed contigs. Next, the seed contigs were reassembled and extended using 451,089 single-molecule maps longer than 250 kb.
The resulting whole-genome map was aligned to the in silico NheI restriction map of the Release 6 sequence assembly using MapSolver v10.0 (OpGen) and default parameters (threshold = 4.0). The alignment was reviewed and edited in MapSolver. We deleted artifactual interchromosomal alignments between the set of redundant map contigs aligned to the regions flanking the gap at 2L: 39D (associated with the core histone gene cluster) and sequences flanking the gap at X: 20D (associated with a tandem array of the satellite-like repeat TAGA). We deleted an artifactual alignment between a map contig and a 27.6-kb segment of the Y chromosome (Y: 2,539,554…2,567,134). We trimmed the map assembly at the six telomeric regions represented in chromosome arm sequence assemblies to remove suspicious restriction fragments extending beyond discrete locations in the consensus map assembly where coverage in single-molecule maps abruptly dropped, suggesting the true locations of chromosome ends (Supplemental Fig. S16). These suspicious fragments were trimmed using the Hide feature, but they are retained in the alignment file (Supplemental Data File S4). We hid four restriction fragments with a combined length of 19.6 kb at the XL telomere, ten fragments (78.1 kb) at the 2L telomere, three fragments (7.3 kb) at the 2R telomere, four fragments (38.4 kb) at the 3L telomere, 18 fragments (138.4 kb) at the 3R telomere, and four fragments (54.3 kb) at the 4R telomere.
Data access
The Release 6 genome assembly has been submitted to the NCBI Assembly database (http://www.ncbi.nlm.nih.gov/assembly/) under accession number GCA_0000012154. BAC and WGS-based sequence data have been submitted to NCBI GenBank (http://www.ncbi.nlm.nih.gov/genbank) under the accession numbers listed in Supplemental Table S2. The sequences of the cDNAs IP04839 and MIP31562 and the cloning vector pBR194b have been submitted to GenBank under accession numbers BT128705, BT150454, and KM891592, respectively.
Supplementary Material
Acknowledgments
We thank S. Richards, D. Wheeler, D. Muzny, S. Scherer, and R.A. Gibbs for assistance in the production of the Release 4 sequence of chromosome arm 3L; T. Murphy at NCBI for thorough quality control of the Release 6 sequence submission; M.A. Crosby for sharing her observation that Piezo is homologous to a fragmented set of five Release 5 gene models; V. Gvozdev for 28S rDNA and SCLR plasmids; and E. Frise for computer systems support. This work was supported by NIH grants P50 HG00750 (G.M.R.), R01 HG00747 (G.H.K.), and R01 HG002673 (S.E.C.) and performed under U.S. Department of Energy Contracts DE-AC0376SF00098 and DE-AC02-05CH11231, University of California. I.F.Z. was supported by grant 13-04-40137 from the Russian Federation; E.N.A. was supported by grant 12-04-00874-a from the Russian Federation; P.D. was supported by a grant from the Instituto Pasteur-Fondazione Cenci Bolognetti; A.V. was supported by Ministerio de Economía y Competitividad grant BFU2011-30295-C02-01; and A.B.C. was supported by NIH grant R01 GM064590 and grants from Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). We dedicate this work in memory of our dear colleague and friend, Alfredo Villasante, who died during the final preparation of this manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.185579.114.
Freely available online through the Genome Research Open Access option.
References
- Abad JP, Carmena M, Baars S, Saunders RD, Glover DM, Ludena P, Sentis C, Tyler-Smith C, Villasante A. 1992. Dodeca satellite: a conserved G+C-rich satellite from the centromeric heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci 89: 4663–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abad JP, Agudo M, Molina I, Losada A, Ripoll P, Villasante A. 2000. Pericentromeric regions containing 1.688 satellite DNA sequences show anti-kinetochore antibody staining in prometaphase chromosomes of Drosophila melanogaster. Mol Gen Genet 264: 371–377. [DOI] [PubMed] [Google Scholar]
- Abad JP, de Pablos B, Agudo M, Molina I, Giovinazzo G, Martin-Gallardo A, Villasante A. 2004a. Genomic and cytological analysis of the Y chromosome of Drosophila melanogaster: telomere-derived sequences at internal regions. Chromosoma 113: 295–304. [DOI] [PubMed] [Google Scholar]
- Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. 2004b. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol 21: 1613–1619. [DOI] [PubMed] [Google Scholar]
- Accardo MC, Dimitri P. 2010. Fluorescence in situ hybridization with Bacterial Artificial Chromosomes (BACs) to mitotic heterochromatin of Drosophila. Methods Mol Biol 659: 389–400. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Agudo M, Losada A, Abad JP, Pimpinelli S, Ripoll P, Villasante A. 1999. Centromeres from telomeres? The centromeric region of the Y chromosome of Drosophila melanogaster contains a tandem array of telomeric HeT-A- and TART-related sequences. Nucleic Acids Res 27: 3318–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyeva EN, Kolesnikova TD, Demakova OV, Mendez-Lago M, Pokholkova GV, Belyaeva ES, Rossi F, Dimitri P, Villasante A, Zhimulev IF. 2007. High-resolution analysis of Drosophila heterochromatin organization using SuUR Su(var)3-9 double mutants. Proc Natl Acad Sci 104: 12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakireva MD, Shevelyov Y, Nurminsky DI, Livak KJ, Gvozdev VA. 1992. Structural organization and diversification of Y-linked sequences comprising Su(Ste) genes in Drosophila melanogaster. Nucleic Acids Res 20: 3731–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizuela BJ, Elfring L, Ballard J, Tamkun JW, Kennison JA. 1994. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics 137: 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Lazzaro BP, Clark AG. 2000. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci 97: 13239–13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Dobo BA, Vibranovski MD, Clark AG. 2001. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci 98: 13225–13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol 3: research0079.1–0079.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary DO, Goddard JM, Martin SC, Fauron CM, Wolstenholme DR. 1982. Drosophila mitochondrial DNA: a novel gene order. Nucleic Acids Res 10: 6619–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradini N, Rossi F, Verni F, Dimitri P. 2003. FISH analysis of Drosophila melanogaster heterochromatin using BACs and P elements. Chromosoma 112: 26–37. [DOI] [PubMed] [Google Scholar]
- Demakova OV, Pokholkova GV, Kolesnikova TD, Demakov SA, Andreyeva EN, Belyaeva ES, Zhimulev IF. 2007. The SU(VAR)3-9/HP1 complex differentially regulates the compaction state and degree of underreplication of X chromosome pericentric heterochromatin in Drosophila melanogaster. Genetics 175: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM. 2014. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res 43: D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi B, Beadle GWA. 1936. Technique of transplantation for Drosophila. Am Nat 70: 218–225. [Google Scholar]
- Fuhrmann DR, Krzywinski MI, Chiu R, Saeedi P, Schein JE, Bosdet IE, Chinwalla A, Hillier LW, Waterston RH, McPherson JD, et al. 2003. Software for automated analysis of DNA fingerprinting gels. Genome Res 13: 940–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Pimpinelli S. 1992. Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet 26: 239–275. [DOI] [PubMed] [Google Scholar]
- Gatti M, Bonaccorsi S, Pimpinelli S. 1994. Looking at Drosophila mitotic chromosomes. Methods Cell Biol 44: 371–391. [DOI] [PubMed] [Google Scholar]
- Gepner J, Hays TS. 1993. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc Natl Acad Sci 90: 11132–11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Caudy A, Parsons L, Rosebrock A, Pane A, Raj S, Wieschaus E. 2012. Mapping the pericentric heterochromatin by comparative genomic hybridization analysis and chromosome deletions in Drosophila melanogaster. Genome Res 22: 2507–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Nelson CR, Berman BP, Laverty TR, George RA, Ciesiolka L, Naeemuddin M, Arenson AD, Durbin J, David RG, et al. 2000. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science 287: 2271–2274. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Smith CD, Carlson JW, Carvalho AB, Halpern A, Kaminker JS, Kennedy C, Mungall CJ, Sullivan BA, Sutton GG, et al. 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol 3: research0085–0085.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, Wan KH, Park S, Mendez-Lago M, Rossi F, et al. 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316: 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945. [DOI] [PubMed] [Google Scholar]
- Kennison JA. 1981. The genetic and cytological organization of the Y chromosome of Drosophila melanogaster. Genetics 98: 529–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerich LB, Wang X, Clark AG, Carvalho AB. 2008. Low conservation of gene content in the Drosophila Y chromosome. Nature 456: 949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsticevic FJ, Santos HL, Januario S, Schrago CG, Carvalho AB. 2010. Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster. Genetics 184: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Abad JP, Agudo M, Villasante A. 1999. The analysis of Circe, an LTR retrotransposon of Drosophila melanogaster, suggests that an insertion of non-LTR retrotransposons into LTR elements can create chimeric retroelements. Mol Biol Evol 16: 1341–1346. [DOI] [PubMed] [Google Scholar]
- Losada A, Abad JP, Agudo M, Villasante A. 2000. Long-range analysis of the centromeric region of Drosophila melanogaster chromosome 3. Chromosome Res 8: 651–653. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res 7: 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M, Kucaba T, Sekhon M, Hillier L, Martienssen R, Chinwalla A, Crockett J, Fedele J, Grover H, Gund C, et al. 1999. A map for sequence analysis of the Arabidopsis thaliana genome. Nat Genet 22: 265–270. [DOI] [PubMed] [Google Scholar]
- Mason JM, Villasante A. 2014. Subtelomeres in Drosophila and other Diptera. In Subtelomeres (ed. Louis EJ, Becker MM), pp. 211–225. Springer-Verlag, Berlin/Heidelberg. [Google Scholar]
- Mathewson CA, Schein JE, Marra MA. 2007. Large-scale BAC clone restriction digest fingerprinting. Curr Protoc Hum Genet 53: 5.19.1–5.19.21. [DOI] [PubMed] [Google Scholar]
- Mendez-Lago M, Wild J, Whitehead SL, Tracey A, de Pablos B, Rogers J, Szybalski W, Villasante A. 2009. Novel sequencing strategy for repetitive DNA in a Drosophila BAC clone reveals that the centromeric region of the Y chromosome evolved from a telomere. Nucleic Acids Res 37: 2264–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Lago M, Bergman CM, de Pablos B, Tracey A, Whitehead SL, Villasante A. 2011. A large palindrome with interchromosomal gene duplications in the pericentromeric region of the D. melanogaster Y chromosome. Mol Biol Evol 28: 1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. 2000. A whole-genome assembly of Drosophila. Science 287: 2196–2204. [DOI] [PubMed] [Google Scholar]
- Ozkan E, Carrillo RA, Eastman CL, Weiszmann R, Waghray D, Johnson KG, Zinn K, Celniker SE, Garcia KC. 2013. An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo G, Bonaccorsi S, Robbins LG, Pimpinelli S. 1994. Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics 138: 1181–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélisson A, Song SU, Prud’homme N, Smith PA, Bucheton A, Corces VG. 1994. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J 13: 4401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, Gorchakov AA, Jaffe JD, Kennedy C, Linder-Basso D, et al. 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res 21: 147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiha H, Glover DM. 1981. Duplicated rDNA sequences of variable lengths flanking the short type I insertions in the rDNA of Drosophila melanogaster. Nucleic Acids Res 9: 5521–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SR, Kaiser K. 1993. Drosophila melanogaster male germ line-specific transcripts with autosomal and Y-linked genes. Genetics 134: 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, da Silva MR, Soong TW, Fontaine B, Donaldson MR, Kung AW, Jongjaroenprasert W, Liang MC, Khoo DH, Cheah JS, et al. 2010. Mutations in potassium channel Kir2.6 cause susceptibility to thyrotoxic hypokalemic periodic paralysis. Cell 140: 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RD. 2004. In situ hybridization to polytene chromosomes. Methods Mol Biol 247: 279–287. [DOI] [PubMed] [Google Scholar]
- Schneiderman JI, Sakai A, Goldstein S, Ahmad K. 2009. The XNP remodeler targets dynamic chromatin in Drosophila. Proc Natl Acad Sci 106: 14472–14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. 2000. Myosins: a diverse superfamily. Biochim Biophys Acta 1496: 3–22. [DOI] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Le HD, Wahlstrom JM, Karpen GH. 2003. Sequence analysis of a functional Drosophila centromere. Genome Res 13: 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods 6: 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Vibranovski MD, Koerich LB, Carvalho AB. 2008. Two new Y-linked genes in Drosophila melanogaster. Genetics 179: 2325–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara JC, Marchetti M, Fanti L, Pimpinelli S, Wakimoto BT. 2003. A strategy for mapping the heterochromatin of chromosome 2 of Drosophila melanogaster. Genetica 117: 217–226. [DOI] [PubMed] [Google Scholar]
- Zanni V, Eymery A, Coiffet M, Zytnicki M, Luyten I, Quesneville H, Vaury C, Jensen S. 2013. Distribution, evolution, and diversity of retrotransposons at the flamenco locus reflect the regulatory properties of piRNA clusters. Proc Natl Acad Sci 110: 19842–19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Bechner MC, Place M, Churas CP, Pape L, Leong SA, Runnheim R, Forrest DK, Goldstein S, Livny M, et al. 2007. Validation of rice genome sequence by optical mapping. BMC Genomics 8: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.