Using Ezh2-deficient iNKT cells, Dobenecker et al. show that loss of H3K27me3 at the bivalently marked PLZF promoter is an essential link between TCR specificity and iNKT development in mice.
Abstract
The fidelity of T cell immunity depends greatly on coupling T cell receptor signaling with specific T cell effector functions. Here, we describe a chromatin-based mechanism that enables integration of TCR specificity into definite T cell lineage commitment. Using natural killer T cells (iNKT cell) as a model of a T cell subset that differentiates in response to specific TCR signaling, we identified a key role of histone H3 lysine 27 trimethylation (H3K27me3) in coupling iNKT cell TCR specificity with the generation of iNKT cells. We found that the Zbtb16/PLZF gene promoter that drives iNKT cell differentiation possesses a bivalent chromatin state characterized by the simultaneous presence of negative and positive H3K27me3 and H3K4me3 modifications. Depletion of H3K27me3 at the Zbtb16/PLZF promoter leads to uncoupling of iNKT cell development from TCR specificity and is associated with accumulation of iNKT-like CD4+ cells that express a non-iNKT cell specific T cell repertoire. In turn, stabilization of H3K27me3 leads to a drastic reduction of the iNKT cell population. Our data suggest that H3K27me3 levels at the bivalent Zbtb16/PLZF gene define a threshold enabling precise coupling of TCR specificity to lineage commitment.
The development of functionally distinct T lineage cells from early T cell progenitors and the differentiation of peripheral naive T cells into specialized effector cells are governed by differentially composed gene transcription networks (Collins et al., 2009; Koch and Radtke, 2011; Constantinides and Bendelac, 2013; van der Veeken et al., 2013). In turn, the composition and operation mode of these networks are determined greatly by signals derived from the cell surface expressed TCR, as well as by other receptors (Moran et al., 2011; Seiler et al., 2012; Gottschalk et al., 2013; Zarin et al., 2014). The multitude of phenotypes, which could be attained by a developing or activated naive T cell, suggests the existence of gene regulatory mechanisms that enable the highly calibrated yet swift conversion of multiple signaling events into a definitive transcriptional state of genes that serve as master regulators of distinct T cell lineages.
The described mode of gene regulation matches the chromatin mechanism that contributes to the activation of the lineage-specifying genes during pluripotent embryonic stem (ES) cell differentiation (Azuara et al., 2006; Bernstein et al., 2006; Voigt et al., 2012, 2013; Hu et al., 2013). In ES cells, the simultaneous presence of permissive and suppressive histone modifications at gene promoters keeps lineage-specific gene expression at a quasi-stable silent state that could be readily shifted to an active state during ES cell differentiation into various lineages (Azuara et al., 2006; Bernstein et al., 2006). One of the best-studied combinations of permissive and suppressive histone modifications that co-occupy lineage-specific genes in ES cells involves trimethylation of lysine 4 (H3K4me3) and lysine 27 on histone H3 (H3K27me3). The genes associated with these modifications are considered bivalent (Bernstein et al., 2006).
H3K27me3 and H3K4me3 are broadly distributed among different loci in T lineage cells (Chang and Aune, 2007; Wei et al., 2009). The locus-specific changes in relative abundance of H3K27me3 and H3K4me3 pointed to the possible role of chromatin bivalency in the regulation of gene expression during T cell differentiation (Wei et al., 2009). However, the role of bivalency in coupling TCR signal specificity and/or strength to the specific differentiation outcome has not been established.
In this study, we discuss how bivalency at the promoter of the transcription factor PLZF, which drives T cell differentiation into the iNKT lineage, contributes to the coupling of TCR specificity to iNKT cell development.
RESULTS AND DISCUSSION
iNKT cell development is associated with changes in the chromatin state of the PLZF gene
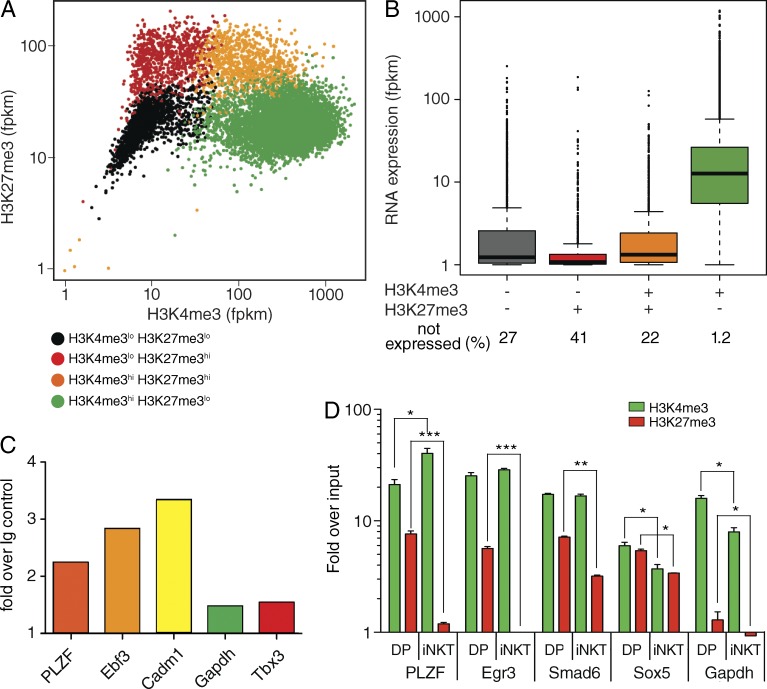
In developing CD4+CD8+ double positive (DP) thymocytes, many of the transcription factor genes that drive T cell differentiation possess bivalent chromatin at their promoters. A genome-wide analysis of H3K4me3 and H3K27me3 distribution in DP thymocytes identified 972 transcriptionally silent genes (Zhang et al., 2012) that display both H3K4me3 and H3K27me3 at their transcriptional start site (TSS; Fig. 1 A). 14% of the silent bivalent genes in DP cells encode various transcription factors, including Bcl11a, Fra-2, and PLZF, that have been implicated in T cell differentiation into specific lineages (Liu et al., 2003; Savage et al., 2008; Lawson et al., 2009; Fig. 1 B; and Table S1). We argue that the bivalent state of the chromatin-encompassing, lineage-specifying genes in T cells operates as a switch that couples the TCR-mediated signaling events to activation of transcription factors that drive T cell differentiation into specific lineages (Wei et al., 2009).
Figure 1.
Development of iNKT cells is associated with changes in the chromatin state of the PLZF gene. (A). Genome-wide distribution of H3K4me3 and H3K27me3 in developing thymocytes. The levels of H3K4me3 and H3K27me3 at the TSS (±3 kb; units of reads per million mapped reads, fpkm) were measured by ChIP sequencing of the chromatin derived from ex vivo purified mouse preselection (CD4+, CD8+, TCRβINT, CD69lo) double-positive (DP) thymocytes. Genes were clustered in subgroups according to the levels of H3K4me3 and H3K27me3. The orange color indicates the bivalent H3K4me4hi H3K27me3hi genes. (B) Box plots show the mRNA expression levels of bivalent as compared with H3K27me3+ or H3K4me3+ monovalent, or genes not modified. The numbers indicate the frequency of nonexpressed genes. (C) H3K4me3+ H3K27me3+ bivalency at the indicated gene loci was assessed by sequential ChIP with anti-H3K27me3, followed by anti-H3K4me3 antibody for the indicated promoters. The reChIP data are given as fold over IgG control in the second IP. (D) The transition from bivalent to monovalent H3K4me3hi state was assessed by measuring levels of H3K27me3 (red) and H3K4me3 (green) at the TSS of PLZF and control genes in preselection DP (DP) thymocytes and thymic iNKT cells. The levels of histone modifications were quantified by ChIP-qPCR from three independent experiments performed in triplicate. Error bars are SEM of independent experiments. Significance was determined by unpaired Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To address the role of bivalency in T cell lineage decision, we focused on iNKT cells. The development of iNKT cells can serve as a bona fide example of TCR-driven activation of a transcriptional master regulator of T lineage cell differentiation. The development of iNKT cells depends on signaling mediated by invariant αβTCR that binds lipids presented by MHC class I–like CD1d molecules (Godfrey and Berzins, 2007; Godfrey et al., 2010; Alonzo and Sant’Angelo, 2011; Constantinides and Bendelac, 2013). The signal from the CD1d-restricted αβTCR drives the expression of the transcription factor PLZF that is required for initiation, maintenance, and function of iNKT lineage cells (Kovalovsky et al., 2008; Savage et al., 2008). The PLZF locus appears as bivalent in DP thymocytes (Fig. 1 A). In support of genuine PLZF bivalency, as opposed to a mixture of PLZF alleles that are marked either with H3K4me3 or H3K27me3, we found both H3K4me3 and H3K27me3 to be present on the same nucleosome at the transcriptional start site (TSS) of the PLZF gene by sequential chromatin immunoprecipitation (ChIP; Fig. 1 C). Unlike DP cells, the PLZF gene is H3K4me3 monovalent in developed iNKT cells. The shift of PLZF from bivalency to monovalency during iNKT cells differentiation is gene specific and highly significant as revealed by quantitative PCR-based analysis of H3K27me3 and H3K4me3 levels at the PLZF promoter in preselection DP and thymic iNKT cells (Fig. 1 D). Other T cell lineage–determining transcription factors, like Smad6 and Sox5, are bivalent both in DP cells and in iNKT cells (Fig. 1 D). These results suggested the transition of the PLZF locus from the H3K27me3+ K3K4me3+ bivalent to the K3K4me3+ monovalent state as a prerequisite for iNKT cell differentiation.
Ezh2 deficiency results in increased iNKT cell numbers and altered iNKT cell development
The role of H3K27me3/H3K4me3 bivalency in iNKT cell development has been initially addressed by an artificial and genome-wide shift of the chromatin state from H3K27me3+ to H3K27me3−. This has been achieved by conditional inactivation of the key H3K27me3 methyltransferase Ezh2 selectively in T cells with CD4-cre (Su et al., 2005). The loss of Ezh2 during T cell differentiation has no obvious impact on the overall H3K27me3 levels in the thymus, but was associated with diminished H3K27me3 levels in iNKT cells (Fig. 2 B). The reduction in Ezh2 levels and ensuing loss of H3K27me3 in Ezh2-deficient iNKT cells as compared with their wild-type counterparts was apparent as early as stage 0 (CD1d-Tetramer+, TCRβ+, and CD24+) of iNKT cell development and becomes more pronounced at later stages of iNKT cell differentiation in the thymus (Fig. 2 B, bottom). It is plausible that selective loss of H3K27me3 in Ezh2-deficient iNKT cells reflects a higher proliferation rate of iNKT cells as compared with the rest of DP thymocytes. Active division of the Ezh2-deficient cells facilitates replacement of the histone H3 that has been modified by K27me3 before Ezh2 ablation by de novo synthesized nonmodified histone H3 in the Ezh2-deficient cells (Mosammaparast and Shi, 2010; Pedersen and Helin, 2010).
Figure 2.
Loss of Ezh2 results in increased iNKT cell numbers and development into NKT2 cell. (A) iNKT cells in thymus, spleen, and peripheral lymph nodes of Ezh2fl/fl; CD4-cre (Ezh2−/−) and littermate controls were detected as CD1d-Tetramer+ and TCRβ+ cells. (bottom) Percentage (left) or absolute number (right) of CD1d-Tetramer+ and TCRβ+ cells in the thymus. Representative plots of >5 independent experiments with >15 mice in total are shown. Each dot represents a single mouse. (B) Flow cytometry histograms show levels of H3K27me3 in DP, CD4+ single positive (CD4+ SP), and iNKT cells (top) and Ezh2, H3K27me3, and PLZF in subpopulations of developing iNKT cell stages defined as: stage 0 (CD1d-Tetramer+, TCRβ+, and CD24+); stage 1 (CD1d-Tetramer+, TCRβ+, CD24−, CD44lo, and NK1.1−); stage 2 (CD1d-Tetramer+, TCRβ+, CD24−, CD44+, and NK1.1−); and stage 3 (CD1d-Tetramer+, TCRβ+, CD24−, CD44+, and NK1.1+). Representative plots of three independent experiments with more than five mice in total are shown. Gray shaded area, WT; red line, Ezh2-deficient cells. (C) Deficiency in Ezh2 leads to altered iNKT cell development and increased number of IL-4–producing cells. The subpopulations of developing iNKT cells were defined by expression of CD44 and NK1.1 on CD1d-Tetramer+ TCRβ+ CD24− cells. (middle) Absolute number of iNKT cells. (right) Absolute number of IL-4–producing iNKT cells. Representative plots of more than three independent experiments with three or more mice in total are shown. Each dot represents a single mouse. Significance for all data was determined by unpaired Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01. ≤ 0.05.
Loss of Ezh2 and H3K27me3 in iNKT cells was associated with a major change in iNKT cell differentiation. A detailed analysis of iNKT cell development revealed a >3-fold increase in frequency and absolute numbers of Ezh2-deficient thymic iNKT cells that express the invariant CD1d-restricted TCR (Fig. 2 A). This increase is attributed to a rise in the subpopulation of developing iNKT cells that matches the stage 2 (CD44hi NK1.1−) of the iNKT cell development (Fig. 2 C, middle). Notably, the CD44hi NK1.1− cells may not only represent a transient stage in iNKT cell development, but rather a sub-population of terminally differentiated iNKT cells defined as NKT2 subset. Normally, NKT2 cells express PLZF at levels higher than NKT1 or NKT17 iNKT cells (Lee et al., 2013) and produce preferentially Th2 cytokines, in response to activation. Consistent with the NKT2 phenotype, the Ezh2-deficient iNKT cells were greatly enriched for IL-4–expressing cells and preferentially produce IL-4 in response to polyclonal in vitro activation with phorbole-12-myristate-13-acetate (PMA) in combination with ionomycin (Fig. 2 C, right). The observed phenotype of the Ezh2-deficient thymic iNKT cells is similar to the phenotype of iNKT cells that develop in the absence of the lysine demethylase-like protein Jarid2 (Pereira et al., 2014). Both Ezh2 and Jarid2 are components of the PRC2 complex that catalyzes methylation of lysine 27 of histone 3. Collectively, these data point to the importance of PRC2 in iNKT cell development.
Ezh2 is required for coupling of TCR specificity to iNKT cell generation
The increase in iNKT cell numbers in Ezh2fl/fl; CD4-cre mice was dwarfed by the appearance of thymic and splenic T cells that express PLZF protein in the absence of the iNKT-specific, CD1d-restricted TCR (Fig. 3 A). These Ezh2-deficient TCR+ iNKT-like cells accumulate at large numbers in peripheral lymphoid organs (Fig. 3 A). Similar to iNKT cells that are selected and triggered by MHC class 1–like (CD1d) bound lipid antigens, the Ezh2-deficient iNKT-like cells express high levels of CD5, which is indicative of antigen-driven T cell selection (Fig. 3 A).
Figure 3.
Ezh2 is required for coupling TCR specificity to iNKT cell generation. (A) The size of the iNKT-like CD1d-Tetramer− TCRβ+ PLZF+ cell population was analyzed in thymi and spleens of Ezh2fl/fl; CD4-cre (Ezh2−/−) and Ezh2fl/fl control (WT) littermates. Flow cytometry histograms show the expression of CD5 in iNKT cells (CD1d-Tetramer+ TCRβ+ PLZF+) and iNKT-like cells (CD1d-Tetramer− TCRβ+ PLZF+). Gray shaded area, Ezh2fl/fl control [WT], iNKT; blue shaded area, Ezh2fl/fl; CD4-cre [Ezh2−/−], iNKT; and red shaded area, Ezh2fl/fl; CD4-cre [Ezh2−/−], iNKT-like. (bottom) Absolute number and percentage of iNKT-like (PLZF+ CD1d-Tet−) cells in thymus and spleen of Ezh2fl/fl control (WT, black diamonds) and Ezh2fl/fl; CD4-cre (red circles) mice. Representative plots of >5 independent experiments with 10 or more mice in total are shown. Each dot represents a single mouse. (B) Vβ-usage of CD4+ and CD8+ αβT cells, as well as iNKT and iNKT-like cells. (top) Vβ-usage of TCRβ+ CD4+ T cells (white bars) or TCRβ+ CD8+ T cells (gray bars) from Ezh2fl/fl control (WT) or Ezh2fl/fl; CD4-cre (Ezh2−/−) mice. (middle) Vβ-usage of iNKT cells from Ezh2fl/fl control (WT, white bars) or Ezh2fl/fl; CD4-cre (Ezh2−/−, gray bars) mice. (bottom), Vβ-usage of iNKT-like cells from Ezh2fl/fl; CD4-cre (Ezh2−/−) mice. Nine mice per genotype were analyzed in three independent experiments, and the error bars represent SD. (C) Development of iNKT and iNKT-like cells in the presence of ovalbumin-specific transgenic (OT-II) TCR. (left) Representative frequencies of CD1d-Tetramer− PLZF+ iNKT-like cells in thymus and spleen of Ezh2fl/fl; CD4-cre (Ezh2−/−); OT-II and Ezh2fl/fl (WT); OT-II littermate controls are shown out of three independent experiments with five or more mice in total. (middle) Production of IL-4 and IFN-γ measured by intracellular staining of iNKT-like cells after stimulation with PMA/ionomycin. Representative dot-plots of two independent experiments with more than three mice shown. (right) Representative dot-plots of three independent experiments with more than five mice show the expression of Vβ5.1/5.2 and Vα2 in TCRβ+ CD4+ SP cells from Ezh2fl/fl (WT); OT-II control mice (left), TCRβ+ CD4+ SP cells from Ezh2fl/fl; CD4-cre (Ezh2−/−); OT-II (middle), and iNKT-like cells from Ezh2fl/fl; CD4-cre mice (Ezh2−/−); OT-II (right). Numbers indicate the percentage of cells in the corresponding quadrants. Significance was determined by unpaired Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
The presence of the Ezh2-deficient iNKT-like cells suggested that loss of Ezh2 and H3K27me3 causes an uncoupling of TCR specificity and PLZF expression. Consistent with this model, the TCR Vβ repertoire expressed by the iNKT-like cells (Fig. 3 B, bottom) was more similar to the diverse Vβ repertoire of conventional αβTCR-bearing T cells (Fig. 3 B, top) than to the limited Vβ (Vβ2, 7, and 8) repertoire of wild-type iNKT cells (Fig. 3 B, middle). Notably, Ezh2 deficiency has no impact on the Vβ repertoire displayed by αβTCR bearing T cells (Fig. 3 B, top).
The uncoupling between the TCR specificity and iNKT cell development in the absence of Ezh2 was underscored further by the appearance of iNKT-like cells in mice that express transgenic MHC class II restricted ovalbumin-specific αβTCR OT-II (OT-II-TCR). Due to the allelic inclusion within the TCRα gene locus (Borgulya et al., 1992), the OTII-TCR mice can express not only the transgenic Vα2 but also other TCR Vα chains that could be paired with the transgenic Vβ5. Regardless of the described relative diversity of the TCR repertoire in the OT-II-TCR mice, the Vβ5 TCR does normally not support iNKT cell development. In fact, the OTII-TCR; Ezh2fl/fl mice have no iNKT cells. As opposed to the control mice, deficiency in Ezh2 resulted in the generation of large numbers of CD1d-Tetramer− and PLZF+ iNKT-like cells that express the transgenic Vβ5 (Fig. 3 C, left). Nearly all of the iNKT-like cells in the OT-II-TCR; Ezh2fl/fl; CD4-Cre mice express Vβ5 and ∼40% of the cells co-express Vβ5 and Vα2, albeit at levels lower than in control CD4+ SP cells (Fig. 3 C, right; and not depicted). Similar to the wild-type iNKT cells, the Ezh2-deficient iNKT-like cells derived from the OT-II-TCR; Ezh2fl/fl; CD4-Cre mice were able to produce large amounts of IL-4 and IFN-γ in response to TCR stimulation (Fig. 3 C, middle) whereas CD4+ SP cells from either OT-II-TCR; Ezh2fl/fl control mice or OT-II-TCR; Ezh2fl/fl; CD4-Cre mice produced only negligible amounts (not depicted).
Accumulation of Ezh2-deficient iNKT and iNKT-like cells promotes development of bystander CD8+ T cells
Alteration in iNKT cell development in the absence of Ezh2 led to secondary changes in αβTCR-bearing T cell subpopulations. The increase in the total number of IL-4–producing iNKT and iNKT-like cells results in elevated IL-4 levels in the serum of Ezh2fl/fl; CD4-cre mice (unpublished data). Accordingly, Ezh2fl/fl; CD4-cre mice display a selective increase in the number of memory-like (bystander) CD8 cells, which are known to accumulate proportionally to the IL-4–producing iNKT cell numbers (Weinreich et al., 2009; Fig. 4 A). Unlike memory CD8+ T cells, which express both T-box transcription factors Eomes and T-bet (Intlekofer et al., 2005, 2008), bystander CD8+ T cells express only Eomes but produce IFN-γ after in vitro stimulation like memory cells (Weinreich et al., 2009). The CD8+ T cells in Ezh2fl/fl; CD4-cre mice fulfill both criteria (Fig. 4 C). The abundance of thymic T cell subpopulations, such as CD4+ SP and DP, were not affected by Ezh2 deficiency, confirming the accumulation of bystander CD8+ T cells and excluding a generally altered αβT cell development in the absence of Ezh2 (Fig. 4 B).
Figure 4.
Accumulation of Ezh2-deficient iNKT and iNKT-like cells results in the development of bystander CD8+ T cells. (A) Generation of memory-like (bystander) CD8+ T cells was quantified by flow cytometry. Numbers indicate percentage of cells of defined surface phenotype (left). Histograms (right) show surface expression of the memory T cell markers (CD24lo, CD44hi, and CD122hi) on thymic and splenic CD8+ T cells derived from Ezh2fl/fl; CD4-cre (red line) and Ezh2fl/fl (WT) littermate controls (shaded area). Data are representative of 5–7 experiments with 2–3 mice per genotype each. (B) Absolute number of CD4+ CD8+ double positive (DP), CD4+ CD8− single positive (CD4+ SP), and CD4− CD8+ single positive (CD8+ SP) thymocytes from Ezh2fl/fl control (WT, black symbols) or Ezh2fl/fl; CD4-cre (red symbols) mice are shown. Data represents three independent experiments with eight mice per genotype analyzed. Each dot represents a single mouse. (C) Expression of Eomes and T-bet in ex vivo isolated CD8+ splenic T cells (top) and IFN-γ production after PMA/ionomycin stimulation (middle), were measured by intracellular FACS. (bottom) Quantification of percentage of IFN-γ positive CD8+ splenocytes isolated from Ezh2fl/fl control (WT, black diamonds) or Ezh2fl/fl; CD4-cre (red circles) mice. Data represents two independent experiments with four or more mice analyzed. Each dot represents a single mouse. Significance was determined for all data by unpaired Student’s t test: **, P ≤ 0.01; ***, P ≤ 0.001.
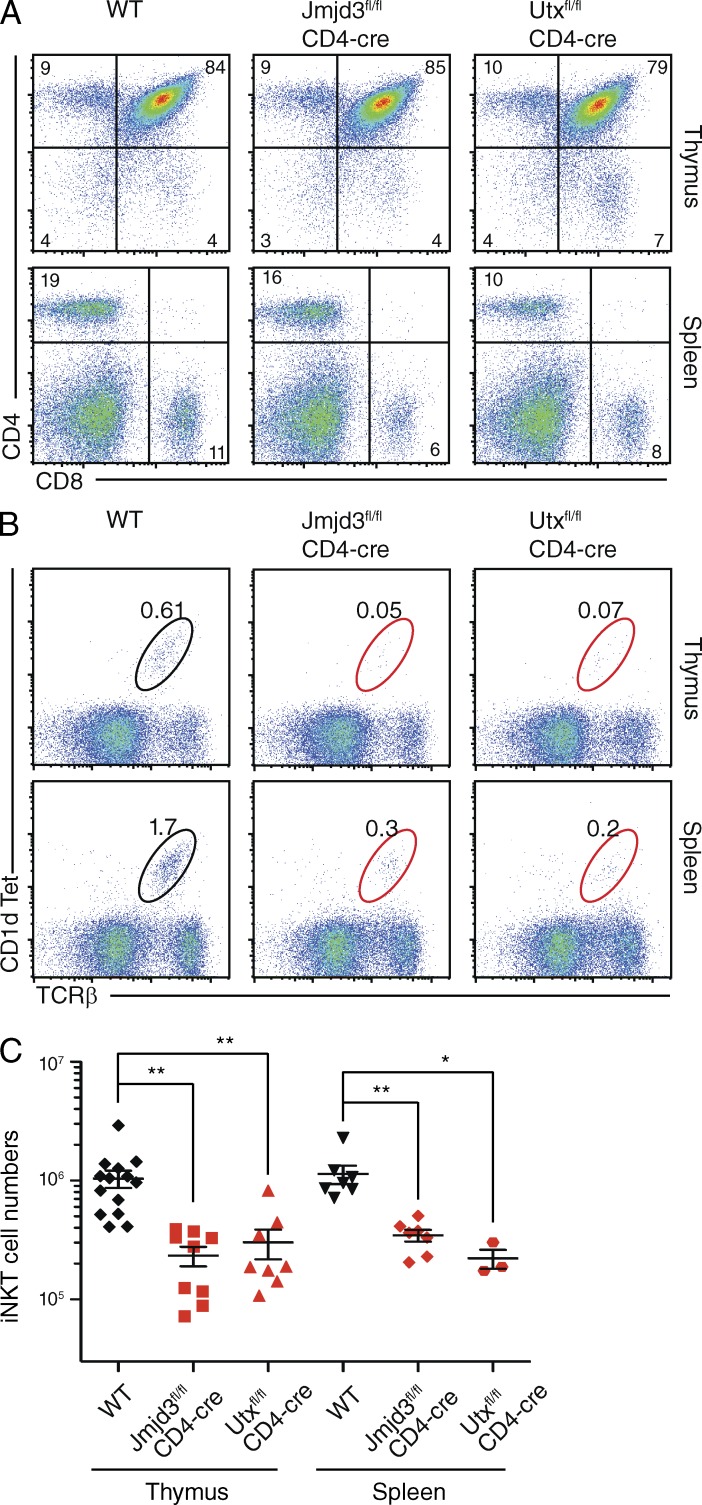
The H3K27me3 demethylases Jmjd3 and Utx are essential for iNKT cell generation
The critical role of H3K27me3 in the regulation of PLZF gene expression and iNKT cell differentiation has been further underscored by the profound impairment in iNKT cell differentiation caused by the stabilization of H3K27me3 levels. To prevent the enzymatic removal of H3K27me3 in T cells, we used conditional inactivation of genes encoding the H3K27me3-specific demethylases Utx and Jmjd3 (Agger et al., 2007; Lan et al., 2007; Mosammaparast and Shi, 2010; Pedersen and Helin, 2010) in developing thymocytes. Deficiency in either of these demethylases had no major effect on T cell development in the thymus or on the generation of peripheral αβTCR-expressing CD4+ or CD8+ T cells (Fig. 5 A). However, lack of Utx or Jmjd3 resulted in reduced expression of PLZF mRNA (not depicted) and an associated major reduction in frequencies (Fig. 5 B and not depicted) and total numbers (Fig. 5 C) of thymic and peripheral iNKT cells that express a CD1d-specific TCR.
Figure 5.
The histone H3K27me3 demethylases Jmjd3 and Utx are required for iNKT cell generation. Thymi and spleens of mice with T cell–specific deficiency of Jmjd3 (Jmjd3fl/fl; CD4-cre) or Utx (Utxfl/fl; CD4-cre) were examined for the surface expression of CD4 and CD8 (A) and CD1d-Tet and TCRβ (B) by flow cytometry. Numbers indicate the percentage of cells in the corresponding gates. Representative dotplots from two independent experiments with more than three mice are shown. (C) Quantification of iNKT cell numbers in thymus and spleen of Jmjd3fl/fl; CD4-cre, Utxfl/fl; CD4-cre, and WT controls, from more than three independent experiments with three or more mice. Each dot represents a single mouse. Significance was determined by unpaired Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01.
These findings reinforced the model where coupling between the TCR specificity/signaling and iNKT lineage decision is determined by a balance between H3K27me3 and H3K4me3 at the PLZF gene promoter. In turn, this model predicts that TCR-driven transition of developing DP cells into iNKT cells requires the selective recruitment of H3K27me3-specific demethylases to the PLZF or other gene loci, e.g., Samd7, which transition from a bivalent to a monovalent state during iNKT generation from DP cells. Indeed, in iNKT cells, Utx was more abundant at the PLZF or Smad7 gene promoters as compared with the same loci in DP cells (unpublished data). Contrary to the bivalent genes, the levels of Utx at the promoters of H3K4me3 monovalent and constitutively expressed genes, such as β-actin, are similar in DP and iNKT cells (unpublished data). Although deficiency in Jmjd3 also results in hampered PLZF expression, no specific recruitment of Jmjd3 to the PLZF promoter in iNKT cells could be detected.
In conclusion, we propose that the bivalent chromatin at the PLZF promoter operates as a switch that converts signaling cues into an irreversible lineage decision. In ES cells, Utx is present in multiprotein complexes with the H3K4 methyltransferase MLL4 (Cho et al., 2007; Issaeva et al., 2007; Smith et al., 2011) and the C terminus of MLL4 is responsible for MLL4’s interaction with the core subunits of the MLL4 complex (Kim et al., 2014). We propose that accumulation of MLL4, as well as the associated Utx, can function as a proxy of the TCR signal strength. The definitive shift to PLZF gene expression will happen as soon as MLL4 and, associated with it, Utx, reach a level that alters the equilibrium between H3K4me3 and H3K27me3. Once expressed, the PLZF proteins will drive further iNKT cell differentiation by binding to its gene targets, which are responsible for iNKT cell differentiation and maintenance. The suggested mechanism is likely to operate at other transcription factor genes, where distinct levels of bivalency may contribute to differential TF expression in response to TCR signals of varying strength (Leishman et al., 2002; Baldwin et al., 2004; Zhou et al., 2004; Kronenberg and Rudensky, 2005).
MATERIALS AND METHODS
Antibodies.
The following antibodies were purchased from either BD or eBioscience: CD3e (145-2C11), CD4 (RM4-5), CD8α (53–6.7), TCRβ (H57-597), IL4, CD24 (M1/69), CD44 (IM7), CD122 (TM-β1), IFN-γ, Eomes (Dan11mag), and T-bet (4B10). APC-conjugated CD1d-PBS-57 (αGalCer analogue) loaded and unloaded tetramers were provided by the Tetramer Facility of the US National Institutes of Health. The monoclonal antibody against PLZF (D-9) was purchased from Santa Cruz Biotechnology, Inc. The Jmjd3 antibody was provided by Y. Shi (Boston Children’s Hospital, Boston, MA) and the Utx antibody (A302-374A) was purchased from Bethyl Laboratories. Anti-H3K4me3 (17–614) and anti-H3K27me3 (07–449) antibodies for ChIP-seq were purchased from Millipore.
Mice.
Ezh2fl/fl; CD4-cre mice were previously described (Su et al., 2005) and are on a mixed background of C57BL/6 and 129/Sv. Jmjd3fl/fl; CD4-cre mice. Utxfl/fl; CD4-cre mice are on the C57BL/6 background. Animals were maintained under specific pathogen–free conditions. All animal experiments were conducted in accordance with local and institutional guidelines.
Standard ChIP assays and ChIP-seq.
ChIP was performed as previously described (Lee et al., 2006); Goldberg et al., 2010). In brief, 107 cells were used for ChIP-seq experiments and 106 cell for ChIP-qPCR experiments. The cells were cross-linked with 0.5% formaldehyde at room temperature for 10 min. Chromatin was sonicated to 300–500 bp in RIPA buffer with 0.3 M NaCl. 1–10 µg antibodies were preincubated with Dynabead Protein A/G (Invitrogen) for at least 8 h before incubating with sonicated chromatin overnight. After overnight incubation, beads were washed in modified RIPA wash buffer (100 mM LiCl [for H3K27me3] or 300 mM LiCl [for H3K4me3]) and one time in Tris EDTA buffer (10 mM Tris, pH 8.0, and 1 mM EDTA). After overnight cross-link reversal at 65°C, RNase digestion, and Proteinase K digestion, ChIP DNA and input DNA were purified using the QIAquick PCR purification kit (QIAGEN). For regular ChIP and for validation of ChIP-Seq, ChIP DNA was analyzed via qPCR using SYBR Green PCR Master Mix and the LightCycler 480 (both from Roche). Primer sequences are available upon request.
For ChIP-Seq, 30 µl of ChIP DNA were used to generate blunt-ended DNA using reagents supplied with the Epicenter DNA EndRepair kit (Epicenter Biotechnologies) according to the manufacturer’s instructions. The end-repaired DNA was purified using the QIAquick PCR purification kit. Using Klenow Fragment (3′ to 5′ exo-, NEB) “A” bases were added to the DNA. The DNA was purified using the MinElute kit. T4 DNA ligase (NEB) was used for ligation of Illumina/Solexa adapters to the DNA fragments. The adaptor-ligated DNA was purified with the MinElute kit. The DNA fragments were subjected to 18 cycles of PCR using the Illumina/Solexa primers 1.0 and 2.0 to generate the ChIP-Seq libraries. The ChIP-Seq libraries were purified with the MinElute kit.
Samples were sequenced on the Illumina Hiseq2000 platform for 50 cycles, and raw sequencing data were processed using the CASAVA_v1.8.2 software to generating fastq files. Sequencing reads were aligned to the mouse genome (mm9) using Bowtie v0.12.7 (Langmead et al., 2009). Reads were kept if they aligned with two errors or less and did not align to more than one location in the genome. A 25-bp density coverage map was created by extending each read for 100 bp to account for average library fragment length and mapping the number of reads per 25-bp bin using igvtools (Thorvaldsdóttir et al., 2013). Values in each sample were normalized to fpkm values by calculating the fraction of mapped reads per bin in 1,000,000 total reads.
For comparative analysis of promoter regions, the number of aligned reads in the area surrounding the transcriptional start site (±3 kb) of each gene was used.
Quantitative PCR.
Chromatin from 106 FACS-sorted iNKT (TCRβ+ CD1d-Tetramer+) and psDP (CD4+CD8+, TCRint, and CD69lo) cells was prepared as described above. Quantitative real-time PCR was performed using SYBR Green (Roche) on a Lightcycler 480 (Roche). Primer sequences are available upon request. Data are presented as mean ± SD.
Sequential ChIP.
Sequential ChIP was performed as previously described (Voigt et al., 2012). In brief, preselection DP (psDP) thymocytes were MACS sorted by depletion with anti-CD3, anti-CD25, anti-CD44, and anti-Ter119. Chromatin from psDP cells was first IPed with an anti-H3K27me3 antibody and the bound material from the first immunoprecipitation was eluted with 30 µl of 0.1% SDS and 30 mM DTT in TE for 1 h at 37°C under constant agitation. Beads were washed once and volume adjusted to 1.5 ml for the second IP with anti-H3K4me3 antibody and eluted as described above. To control for antibody carryover from the first immunoprecipitation, IgG was used as control in the second immunoprecipitation and the ChIP signals from the second immunoprecipitation were normalized to this IgG sample. Sequential-ChIPs were analyzed by qPCR with the following primers: Cadm1-F, 5′-CTCCTGCTGTTGCTCCTTTC-3′; Cadm1-R, 5′-TTCCATAGCTACGGCTCCTG-3′; Ebf3-F, 5′-GGCCAGCTTGGCTACATAGA-3′; Ebf3-R, 5′-TCTTCCTTCCACCCTCACAC-3′; Gapdh-F, 5′-TCCCCTCCCCCTATCAGTTC-3′; Gapdh-R, 5′-GACCCGCCTCATTTTTGAAA-3′; Tbx3-F, 5′-TGGAACCCGAAGAAGACGTA-3′; Tbx3-R, 5′-ATCTCTGTACCCCGCTTGTG-3′; PLZF-F, 5′-AGCCCTTGCCTGTACAAAGA-3′; PLZF-R, 5′-TGCCTCACCAACCTTTCTTC-3′.
Bivalent genes.
Bivalent genes were defined as genes in which the sum of all fpkm values within the transcriptional start site region (±3 kb) of H3K4me3 and H3K27me3 ChIPs exceeded those of the IgG control ChIP by at least 1.5-fold (Bernstein et al., 2006).
Preparation of reads from RNA sequencing.
Fastq reads from the Rothenberg laboratory (Zhang et al., 2012) were realigned to the mouse reference genome mm9 using Tophat (Kim et al., 2013) to account for splicing, alternative promoter usage, and insertions and deletions. Subsequently, the cufflinks RNA-seq analysis tool cuffdiff (Trapnell et al., 2013) was used to assess differential gene expression, alternative promoter usage, and splicing variation between experimental datasets. The resulting fpkm values were used for further data analysis and visualization.
Flow cytometry.
Single-cell suspensions from tissues were prepared. APC-conjugated CD1d-PBS-57 loaded and unloaded. Tetramers were obtained from the tetramer facility of the US National Institutes of Health. PLZF staining was done as previously described (Savage et al., 2008). Cells were stained for surface markers first, washed, and then fixed with the permeabilization and fixation buffer (Foxp3 Staining Buffer Set; eBioscience). Intracellular PLZF was detected with 2 µg/ml mouse monoclonal antibody D-9 (Santa Cruz Biotechnology, Inc.) and FITC-conjugated rat anti–mouse IgG1 (BD). Cells were stained for 1 h with Eomes (Dan11mag; eBioscience), T-bet (4B10; eBioscience) followed by secondary antibody staining in Foxp3 Staining Buffer Set (eBioscience). Data were analyzed by FlowJo (Tree Star).
In vitro IL-4 production.
Thymocytes were isolated and plated at 106 cells/ml. The cells were stimulated with 50 ng/ml PMA and 1.5 µg ionomycin (Sigma-Aldrich) for 5 h with Brefeldin A (Sigma-Aldrich) added for the final 3 h. The cells were surface stained and then stained with anti–IL-4 (BD) via a fix/perm kit (BD) and analyzed by flow cytometry.
Statistical analysis.
Statistical analysis was performed in Prism (GraphPad Software) with the unpaired Student’s t test for total cell numbers and frequencies. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Online supplemental material.
Table S1 lists four categories of genes separated by the epigenetic marks at their TSS. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20141499/DC1.
Supplementary Material
Acknowledgments
We thank J. Shi for the Jmjd3 antibody, K. Velinzon for cell sorting, J. Guevarra for high-throughput sequencing, P. Voigt for help with the sequential-ChIP, and A. Rudensky for reading the manuscript.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- DP
- double positive
- iNKT cell
- invariant NK T cell
- H3K27me3
- histone H3 lysine 27 trimethylation
- TSS
- transcriptional start site
References
- Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., and Helin K.. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 449:731–734 10.1038/nature06145 [DOI] [PubMed] [Google Scholar]
- Alonzo E.S., and Sant’Angelo D.B.. 2011. Development of PLZF-expressing innate T cells. Curr. Opin. Immunol. 23:220–227 10.1016/j.coi.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., and Fisher A.G.. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8:532–538 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- Baldwin T.A., Hogquist K.A., and Jameson S.C.. 2004. The fourth way? Harnessing aggressive tendencies in the thymus. J. Immunol. 173:6515–6520 10.4049/jimmunol.173.11.6515 [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 125:315–326 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Borgulya P., Kishi H., Uematsu Y., and von Boehmer H.. 1992. Exclusion and inclusion of alpha and beta T cell receptor alleles. Cell. 69:529–537 10.1016/0092-8674(92)90453-J [DOI] [PubMed] [Google Scholar]
- Chang S., and Aune T.M.. 2007. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat. Immunol. 8:723–731 10.1038/ni1473 [DOI] [PubMed] [Google Scholar]
- Cho Y.W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G.R., Copeland T.D., Kalkum M., and Ge K.. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282:20395–20406 10.1074/jbc.M701574200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A., Littman D.R., and Taniuchi I.. 2009. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat. Rev. Immunol. 9:106–115 10.1038/nri2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides M.G., and Bendelac A.. 2013. Transcriptional regulation of the NKT cell lineage. Curr. Opin. Immunol. 25:161–167 10.1016/j.coi.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey D.I., and Berzins S.P.. 2007. Control points in NKT-cell development. Nat. Rev. Immunol. 7:505–518 10.1038/nri2116 [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Stankovic S., and Baxter A.G.. 2010. Raising the NKT cell family. Nat. Immunol. 11:197–206 10.1038/ni.1841 [DOI] [PubMed] [Google Scholar]
- Goldberg A.D., Banaszynski L.A., Noh K.M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X., et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 140:678–691 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk R.A., Martins A.J., Sjoelund V.H., Angermann B.R., Lin B., and Germain R.N.. 2013. Recent progress using systems biology approaches to better understand molecular mechanisms of immunity. Semin. Immunol. 25:201–208 10.1016/j.smim.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Garruss A.S., Gao X., Morgan M.A., Cook M., Smith E.R., and Shilatifard A.. 2013. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 20:1093–1097 10.1038/nsmb.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Intlekofer A.M., Banerjee A., Takemoto N., Gordon S.M., Dejong C.S., Shin H., Hunter C.A., Wherry E.J., Lindsten T., and Reiner S.L.. 2008. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 321:408–411 10.1126/science.1159806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I., Zonis Y., Rozovskaia T., Orlovsky K., Croce C.M., Nakamura T., Mazo A., Eisenbach L., and Canaani E.. 2007. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 27:1889–1903 10.1128/MCB.01506-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Sharma A., Dhar S.S., Lee S.H., Gu B., Chan C.H., Lin H.K., and Lee M.G.. 2014. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 74:1705–1717 10.1158/0008-5472.CAN-13-1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U., and Radtke F.. 2011. Mechanisms of T cell development and transformation. Annu. Rev. Cell Dev. Biol. 27:539–562 10.1146/annurev-cellbio-092910-154008 [DOI] [PubMed] [Google Scholar]
- Kovalovsky D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.J., Im J.S., et al. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9:1055–1064 10.1038/ni.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg M., and Rudensky A.. 2005. Regulation of immunity by self-reactive T cells. Nature. 435:598–604 10.1038/nature03725 [DOI] [PubMed] [Google Scholar]
- Lan F., Bayliss P.E., Rinn J.L., Whetstine J.R., Wang J.K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 449:689–694 10.1038/nature06192 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson V.J., Maurice D., Silk J.D., Cerundolo V., and Weston K.. 2009. Aberrant selection and function of invariant NKT cells in the absence of AP-1 transcription factor Fra-2. J. Immunol. 183:2575–2584 10.4049/jimmunol.0803577 [DOI] [PubMed] [Google Scholar]
- Lee T.I., Johnstone S.E., and Young R.A.. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1:729–748 10.1038/nprot.2006.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Holzapfel K.L., Zhu J., Jameson S.C., and Hogquist K.A.. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14:1146–1154 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leishman A.J., Gapin L., Capone M., Palmer E., MacDonald H.R., Kronenberg M., and Cheroutre H.. 2002. Precursors of functional MHC class I- or class II-restricted CD8alphaalpha(+) T cells are positively selected in the thymus by agonist self-peptides. Immunity. 16:355–364 10.1016/S1074-7613(02)00284-4 [DOI] [PubMed] [Google Scholar]
- Liu P., Keller J.R., Ortiz M., Tessarollo L., Rachel R.A., Nakamura T., Jenkins N.A., and Copeland N.G.. 2003. Bcl11a is essential for normal lymphoid development. Nat. Immunol. 4:525–532 10.1038/ni925 [DOI] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N., and Shi Y.. 2010. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79:155–179 10.1146/annurev.biochem.78.070907.103946 [DOI] [PubMed] [Google Scholar]
- Pedersen M.T., and Helin K.. 2010. Histone demethylases in development and disease. Trends Cell Biol. 20:662–671 10.1016/j.tcb.2010.08.011 [DOI] [PubMed] [Google Scholar]
- Pereira R.M., Martinez G.J., Engel I., Cruz-Guilloty F., Barboza B.A., Tsagaratou A., Lio C.W., Berg L.J., Lee Y., Kronenberg M., et al. 2014. Jarid2 is induced by TCR signalling and controls iNKT cell maturation. Nat. Commun. 5:4540 10.1038/ncomms5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M.P., Mathew R., Liszewski M.K., Spooner C.J., Barr K., Meng F., Singh H., and Bendelac A.. 2012. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13:264–271 10.1038/ni.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E., Lin C., and Shilatifard A.. 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 25:661–672 10.1101/gad.2015411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su I.H., Dobenecker M.W., Dickinson E., Oser M., Basavaraj A., Marqueron R., Viale A., Reinberg D., Wülfing C., and Tarakhovsky A.. 2005. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 121:425–436 10.1016/j.cell.2005.02.029 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., and Mesirov J.P.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14:178–192 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., and Pachter L.. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31:46–53 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veeken J., Arvey A., and Rudensky A.. 2013. Transcriptional control of regulatory T-cell differentiation. Cold Spring Harb. Symp. Quant. Biol. 78:215–222 10.1101/sqb.2013.78.020289 [DOI] [PubMed] [Google Scholar]
- Voigt P., LeRoy G., Drury W.J. III, Zee B.M., Son J., Beck D.B., Young N.L., Garcia B.A., and Reinberg D.. 2012. Asymmetrically modified nucleosomes. Cell. 151:181–193 10.1016/j.cell.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P., Tee W.W., and Reinberg D.. 2013. A double take on bivalent promoters. Genes Dev. 27:1318–1338 10.1101/gad.219626.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T.Y., Watford W.T., et al. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 30:155–167 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M.A., Takada K., Skon C., Reiner S.L., Jameson S.C., and Hogquist K.A.. 2009. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 31:122–130 10.1016/j.immuni.2009.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarin P., Wong G.W., Mohtashami M., Wiest D.L., and Zúñiga-Pflücker J.C.. 2014. Enforcement of γδ-lineage commitment by the pre-T-cell receptor in precursors with weak γδ-TCR signals. Proc. Natl. Acad. Sci. USA. 111:5658–5663 10.1073/pnas.1312872111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.A., Mortazavi A., Williams B.A., Wold B.J., and Rothenberg E.V.. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 149:467–482 10.1016/j.cell.2012.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Mattner J., Cantu C. III, Schrantz N., Yin N., Gao Y., Sagiv Y., Hudspeth K., Wu Y.P., Yamashita T., et al. 2004. Lysosomal glycosphingolipid recognition by NKT cells. Science. 306:1786–1789 10.1126/science.1103440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.