Abstract
Electrophysiological recording of action potentials from sensory neurons of mosquitoes provides investigators a glimpse into the chemical perception of these disease vectors. We have recently identified a bitter sensing neuron in the labellum of female Aedes aegypti that responds to DEET and other repellents, as well as bitter quinine, through direct electrophysiological investigation. These gustatory receptor neuron responses prompted our sequencing of total mRNA from both male and female labella and tarsi samples to elucidate the putative chemoreception genes expressed in these contact chemoreception tissues. Samples of tarsi were divided into pro-, meso- and metathoracic subtypes for both sexes. We then validated our dataset by conducting qRT-PCR on the same tissue samples and used statistical methods to compare results between the two methods. Studies addressing molecular function may now target specific genes to determine those involved in repellent perception by mosquitoes. These receptor pathways may be used to screen novel repellents towards disruption of host-seeking behavior to curb the spread of harmful viruses.
Keywords: Molecular Biology, Issue 94, Gustation, insect, Aedes aegypti, electrophysiology, mosquito, RNA-seq, qRT-PCR, taste, chemosensory
Introduction
Compounds like DEET, Picaridin, Citronellal and IR3535 have been shown to effectively repel mosquitoes, including the important disease vector Aedes aegypti1,2. We record action potentials from sensory neurons associated with specific gustatory sensilla to determine the cells involved with mosquito repellency. Coupled with downstream sequencing of expressed genes in these tissues, we may identify the genes most likely mediating the responses of these cells in order to screen new compounds for improved repellency capabilities.
RNA-seq is a powerful tool, quickly becoming standard for tracking temporal and spatial changes in gene expression. RNA-seq analyses of insect chemosensory appendages and organs have been used to uncover molecular receptors in several insect species3-5, greatly improving on conventional PCR-based searches gene by gene6. Insects represent the most diverse animal class, presenting many opportunities to study the connection between genes and unique phenotypes. RNA-seq technology can be employed on any living insect tissue. Likewise, electrophysiological recording from sensory cells within uniporous gustatory sensilla can be achieved in many different insect species. The pairing of these two techniques allows researchers to narrow the set genes involved in an observed chemosensory phenotype. Different species will present specific challenges, but may inform the connection between chemosensory receptor genes and a chemosensory adaptation. The size and morphology of chemosensory sensilla is variable and may require extensive troubleshooting when recording action potentials to reduce noise and identify repeatable signals. Dissections of chemosensory organs may be trivial or delicate and time consuming, depending on morphology and size of the insect. Recovery of high-quality RNA may require some troubleshooting as well, such as avoiding certain pigments during tissue collection.
While demonstrating the effects of repellent compounds through behavioral trials is direct and informative, this approach is time intensive and broad with respect to mechanism of action. Electrophysiology coupled with RNA-seq allows for more specific analyses of what drives avoidance behaviors in insects. Once the “toolkit” of chemical discrimination has been identified in an insect species, more specific attempts to improve on known repellents are possible. Receptors and associated proteins in sensory cells responsible for these behaviors may be expressed heterologously for direct chemical screening. Furthermore, molecular modeling can predict which chemicals will elicit strong responses from these receptors7.
The snapshot of all active genes in a narrow set of chemosensory tissues may also be useful in identifying similar genes in other species. Using sequence homology and expression similarities, researchers may form sets of molecular receptors most likely mediating responses to repellents that are broadly effective on insects. We present the following protocol to aid researchers in deconstructing insect chemosensory pathways and to persuade more to delve into the neuroethology of non-model and economically important insects.
Protocol
1. Rearing Ae. aegypti adults
Hatch eggs in approximately ¾ inch water in shallow tray. Overcrowding will reduce the size of adults.
Rear larvae at 25 °C (12-hL:12-hD) and feed with ground fish food. NOTE: Overfeeding may reduce survival rates.
Remove pupae individually by Pasteur pipette daily and transfer to plastic dishes (9 cm × 5.5 cm) inside small containment buckets with fine mesh lids, thus establishing 24 hr age groupings.
Feed adult mosquitoes 10% sucrose solution by cotton ball placed on the screen walls of a cage housing the mosquitoes in an environmental chamber at 27 °C and 70% relative humidity under the same photoperiod as larvae.
For electrophysiology, use 5-10 day old animals. In general, larger animals will produce better results. For RNA isolation, use animals that are within a single 24 hr age grouping in the 5-10 day old range.
2. Preparation of Chemicals
Determine a suitable concentration of electrolyte (e.g. 1-10 mM) that elicits minimal activity from selected sensory neurons. 10 mM NaCl or 1mM KCl are reasonable electrolytes to test baseline activity when beginning an experiment.
Dissolve each experimental chemical in appropriate solvent (if not water, use ethanol or DMSO) that has little or no effect on spike activity when compared to electrolyte alone. Final concentrations of 10% ethanol or 1% DMSO are reasonable starting concentrations.
Select appropriate (e.g. quinine for bitter or sucrose for sweet) control chemicals that are well-described feeding deterrents or stimulants in insects.
3. Electrophysiology (tip recording8; Figure 1)
Form glass electrodes by mechanically pulling glass capillaries. Optimize heat and pull settings to pull glass to form appropriately shaped recording electrodes. Carefully break off end of glass electrode using forceps under a dissecting microscope to create electrode tip opening that is wide enough to envelope target sensillum, but small enough to avoid contact with neighboring structures.
Under a microscope, visually identify by morphology and position the target sensilla/sensillum to ensure repeatability of tip recording. Prep animals to allow free access to sensilla from angle of electrode entry.
Cold anesthetize adult insects in freezer at -20 °C. Avoid damage to peripheral neurons by overexposure to cold temperatures. Cut narrow strips of cellophane tape with razor blade on glass slide. Immobilize whole insect on a glass slide using narrow strips.
Insert a chemically sharpened tungsten wire into the dorsal thorax or eye to serve as the indifferent (ground) electrode.
Fill glass electrode made in step 3.1 with a stimulating solution of interest. Using an inserted syringe, fill capillary from pulled end to blunt end. Flick out all air bubbles, holding pulled end down. Insert a silver wire (chloriding optional) into the glass electrode made in step 3.1 to serve as recording and stimulating electrode.
Connect electrodes to a preamplifier that is designed for recordings from contact chemoreceptive sensilla in insects. This preamplifier (300 Hz-3 kHz bandpass filter) will reduce the settling time to capture neuronal activity as close to stimulus introduction as possible. Collect, store and analyze electrical signals using a microcomputer equipped with spike recording software. NOTE: Grounding the recording setup properly may drastically improve signal-to-noise ratio.
Stimulate sensilla with a control solution (electrolyte only) to ensure functionality. Count spikes starting 200 msec following the stimulus artifact for all preparations.
Randomize the order of test chemicals for all experiments, except dose–response studies, to eliminate bias. For dose–response studies, expose sensilla to increasing concentrations of the experimental chemical. Allow a minimum of 3 min between stimulations to ensure adequate recovery. NOTE: This requirement may vary depending on insect and sensilla. Threshold is defined as that concentration for which standard error does not overlap with the standard error for the lowest concentration tested.
4. RNA Isolation and Sequencing (Figure 2)
Prepare the tightly fitting RNAse-free pestles by cooling them on dry ice before tissue disruption.Prepare accompanying RNase-free pestles that fit snap cap tubes tightly. Keep collection tubes closed unless actively dissecting to avoid excess condensation.
Using filtered aspirator, place 30 to 40 cold-anesthetized mosquitoes (15 sec in -20 °C freezer) on cold stage and sort by sex. Place animals dorsal side down, grasping wings to not damage taste appendages.
Using two pairs of fine forceps, dissect paired labella or tarsi from males or females under a dissecting microscope and limit inclusion of adjacent tissues. NOTE: 500 paired labella yields approximately 800-1,000 nanograms of total mRNA. 400 individual tarsi (5 segments each) yield 1,200-1,800 nanograms of total mRNA.
With one pair of gasping-forceps, lightly grasp mosquito proboscis just proximal to labella exposing inner stylets. Using other pair of collection-forceps, remove labella and add tissue to side of cold collection tube.
With one pair of forceps, lightly grasp mosquito leg at junction of tibia and first tarsal segment. Using other pair of forceps, remove tarsi and add tissue to side of cold collection tube. NOTE: Take into account the cell types at the interface of wanted and unwanted tissue. While it is important to limit the inclusion of neighboring tissue, it is equally important to not omit portions of desired tissue. The final sample must accurately represent the entire organ of interest. Create a precise boundary with the grasping forceps such that collection forceps can precisely liberate by scraping or grabbing only the desired tissue.
Periodically, spin down collection tube briefly in 0 °C centrifuge at 9,000 x g to keep tissue at bottom of tube. If moisture accumulates in collection tubes during dissection, add 50uL of cold Trizol reagent to cover collected tissue.
Using anti-smudge lab marker, clearly label one RNase-free 1.5 ml snap cap tube for each tissue type for collection. Using 1.5 ml tube rack and liquid nitrogen, freeze then grind tissue into fine powder (fine pieces if in Trizol). Repeat once. Bring total volume of Trizol to 1 ml and resuspend ground tissue by vortexing.
Add 200 ul of chloroform and mix thoroughly. After 5 min at room temperature, spin down in centrifuge at 4 °C and 11,000 x g for 15 min. Carefully add aqueous layer directly to a genomic DNA filter column. Spin down at 11,000 x g for 5 min. Add equal volume of RNase-free 70% ethanol to flow-through and mix by pipet. Transfer liquid to an RNA collection column and continue RNA extraction per the instructions provided.
Quantify and assess the purity of total RNA on a precision spectrophotometer and proceed with any standard RNA sequencing method.
5. Quantitative RT-PCR Validation of RNA Sequencing (Figure 3)
Select as many genes as possible to compare relative gene expression levels over a dynamic range, both in predicted sequence abundance and presumed chemosensory gene function.
Design primer pairs for each target gene to amplify a specific 100-180 basepair PCR product. Exclude non-specific gDNA amplification by ensuring at least one primer per set spans an intron boundary. Alternatively, use DNase treatment to reduce genomic contamination.
Collect insect tissue as described in previous section. Calculate how much tissue is required to provide enough RNA to complete all qRT-PCR reactions. NOTE: Biological triplicate and technical triplicate reactions will provide maximum confidence in results.
Calculate relative gene quantification as Xtarget-(Ct[target]-Ct[reference]) for each target gene and average for each replicate, both biological and technical. Use housekeeping gene(s) to normalize Ct values between biological replicates and to root the Y-axis scale in comparisons of relative expression.
Assess the similarity of RNA-seq and qRT-PCR datasets using the regression-based, bootstrap equivalence procedure9, iteratively applied, to the data from each tissue. NOTE: This procedure identifies the smallest “region of similarity” that can be constructed around the observed RNA-seq and qRT-PCR data, and allow the relative gene expression as measured by qRT-PCR to be considered statistically equivalent to the measurement of relative gene expression by RNA-seq.
Representative Results
The trace recordings of action potentials from Ae. aegypti gustatory sensilla (Figure 1) demonstrate the effectiveness of direct stimulation with a range of chemicals. This technique can be used to quantify responses to any stimulating chemical by counting spikes of a given amplitude and duration over a reasonable time range (generally less than 500 ms). Trace recordings must be readily reproducible under a given set of experimental conditions. Otherwise, the observed physiological responses may represent uncommon phenotypes among individuals of the test species.
Raw RNA-seq data requires filtration and accurate mapping before being presented as rounded RPKM (the number of mapped reads per kilobase length of transcript per million reads mapped) or FPKM (the number of read fragments per kilobase length of transcript per million reads mapped) numerical values (Figure 2). The distinction between these two RNA-seq normalization methods has been described in two seminal works10,11. One advantage of total mRNA sequencing is the ability to detect the expression of large gene families simultaneously. The visual presentation of the expression of many genes at once allows for a unique understanding of the molecular toolset for a given tissue. Comparisons between sexes, developmental stages or tissue types can be made rapidly.
Biological replicates for RNA-seq datasets may be limited by cost or difficulty of tissue collection. qRT-PCR offers the ability to validate small subsets of total gene expression at less cost and requires less source RNA. Side by side data comparisons (Figure 3A) allow for greater confidence in RNA-seq results, as the two methods rely on different chemistries for gene abundance estimation. Statistical equivalence functions (Figure 3B) demonstrate the limits of certainty in each case.
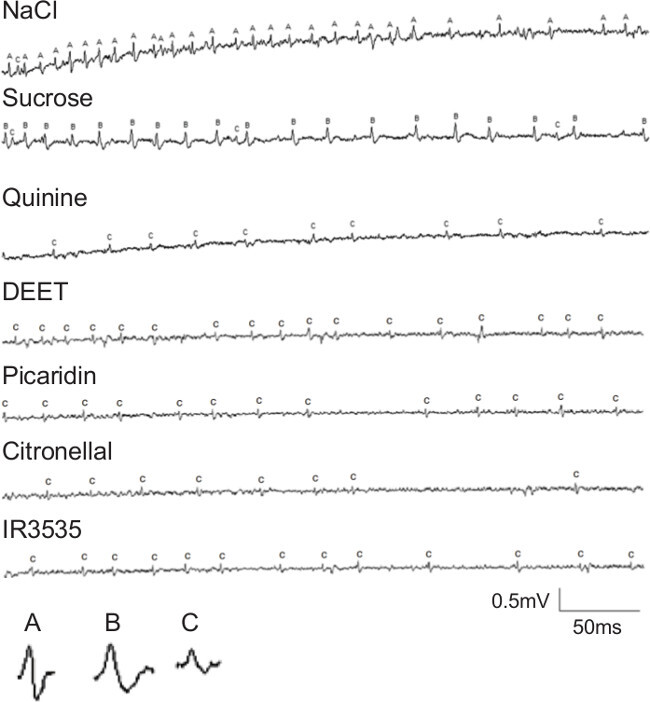 Figure 1: Electrophysiological recordings from a medium-sized sensillum on the labellum of Ae. aegypti females, modified from cited work12. Traces show responses to 100 mM NaCl, 100 mM sucrose, 1 mM quinine, 0.8% DEET, 0.8% Picaridin, 0.8% Citronellal and 0.8% IR3535. The three spikes with differing amplitudes or shapes are labeled a, b or c. These three spikes correspond with three sensory neuron subtypes with differing sensitivities: salt, sugar, and bitter, respectively.
Figure 1: Electrophysiological recordings from a medium-sized sensillum on the labellum of Ae. aegypti females, modified from cited work12. Traces show responses to 100 mM NaCl, 100 mM sucrose, 1 mM quinine, 0.8% DEET, 0.8% Picaridin, 0.8% Citronellal and 0.8% IR3535. The three spikes with differing amplitudes or shapes are labeled a, b or c. These three spikes correspond with three sensory neuron subtypes with differing sensitivities: salt, sugar, and bitter, respectively.
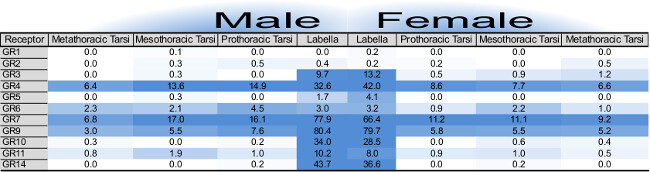 Figure 2: Table of gustatory receptor gene expression as determined by RNA-seq for 8 tissue samples, modified from cited work13. Individual cells report RPKM values for each gustatory gene annotation, male tissues (left) and female tissues (right). Numerical values are rounded to the nearest tenth. Heat-map color intensity is capped at highest RPKM and the midpoint is scaled for each gene family individually. There is only one gene family shown in this case. Please click here to view a larger version of this figure.
Figure 2: Table of gustatory receptor gene expression as determined by RNA-seq for 8 tissue samples, modified from cited work13. Individual cells report RPKM values for each gustatory gene annotation, male tissues (left) and female tissues (right). Numerical values are rounded to the nearest tenth. Heat-map color intensity is capped at highest RPKM and the midpoint is scaled for each gene family individually. There is only one gene family shown in this case. Please click here to view a larger version of this figure.
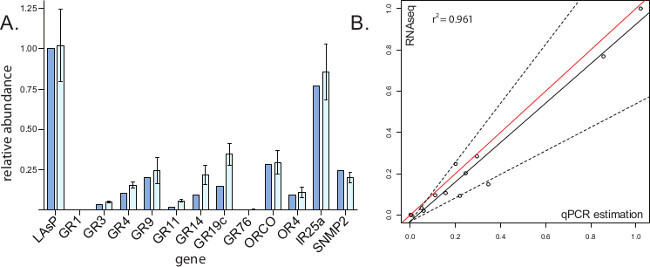 Figure 3: Comparison of qRT-PCR data and RNA-seq data, modified from cited work13.(A) Each chemoreception gene of the horizontal axis is represented by two data columns. The left column represents relative expression as determined by RNA-seq; RPKM values of chemoreception genes were divided by that of housekeeping gene AaegLAsP to generate numerical proportions in this tissue. The right column represents relative expression as determined by qRT-PCR. First, biological replicate Ct values for labella genes were normalized using AaegLAsP as a standard. Second, replicate Ct values for each gene were used to calculate relative expression values (Etarget-(Ct[target]-Ct[reference])). The error bars indicate standard deviation of individually calculated relative expression values. (B) RNA-seq and qRT-PCR datasets were compared in a separate analysis. The vertical axes represent relative gene abundance as demonstrated by RNA-seq data, with a value of 1 being attributed to housekeeping gene AaegLAsP and all other gene expression being reported relative to this value. The horizontal axes represent relative gene abundance as predicted by qRT-PCR, with a value of 1 being attributed to housekeeping gene AaegLAsP and all other gene expression being reported relative to this value. Red lines represent exact equivalence. Dashed lines represent boundaries of the 'region of similarity' for slope, calculated by the statistical equivalence algorithm9. Solid black lines represent the least-squares regression of qRT-PCR equivalence to RNA-seq quantification for the 12 target genes and AaegLAsP housekeeping gene, each represented by circle data point. Please click here to view a larger version of this figure.
Figure 3: Comparison of qRT-PCR data and RNA-seq data, modified from cited work13.(A) Each chemoreception gene of the horizontal axis is represented by two data columns. The left column represents relative expression as determined by RNA-seq; RPKM values of chemoreception genes were divided by that of housekeeping gene AaegLAsP to generate numerical proportions in this tissue. The right column represents relative expression as determined by qRT-PCR. First, biological replicate Ct values for labella genes were normalized using AaegLAsP as a standard. Second, replicate Ct values for each gene were used to calculate relative expression values (Etarget-(Ct[target]-Ct[reference])). The error bars indicate standard deviation of individually calculated relative expression values. (B) RNA-seq and qRT-PCR datasets were compared in a separate analysis. The vertical axes represent relative gene abundance as demonstrated by RNA-seq data, with a value of 1 being attributed to housekeeping gene AaegLAsP and all other gene expression being reported relative to this value. The horizontal axes represent relative gene abundance as predicted by qRT-PCR, with a value of 1 being attributed to housekeeping gene AaegLAsP and all other gene expression being reported relative to this value. Red lines represent exact equivalence. Dashed lines represent boundaries of the 'region of similarity' for slope, calculated by the statistical equivalence algorithm9. Solid black lines represent the least-squares regression of qRT-PCR equivalence to RNA-seq quantification for the 12 target genes and AaegLAsP housekeeping gene, each represented by circle data point. Please click here to view a larger version of this figure.
Discussion
The most challenging aspect of recording action potentials from gustatory sensilla is deciding which responses are “normal.” When employing single gustatory sensillum tip recording the first time for a given insect species, the total number and sensitivities of the gustatory receptor neurons (GRNs) are likely unknown. Many preliminary recordings are first required to decide the range and concentrations of chemicals to test. In this instance, we began with the observation that a single GRN responded to low concentrations of DEET (Figure 1). Subsequently, we examined responses within the same sensillum to a range of concentrations of chemicals known to be bitter or aversive to other insects and observed a similar spike shape and amplitude (Figure 1). These observations suggested that a single GRN subtype responds to these aversive compounds. For comparison, we tested sugary compounds known to stimulate feeding in mosquitoes to see if a similar spike occurred. We observed a different, larger spike in this case. Similarly, we observed a third, intermediate-sized spike in response to salt stimulation, bringing the total GRN subtype count to three.
Electrophysiological recording preparations in insects are inherently variable. Target gustatory sensilla are typically very small and must be positioned at the appropriate angle to access with the recording electrode without touching glass to the cuticle. To establish a conductive bridge to gustatory neurons, the terminal pore of the sensillum must be open to the contained lymph to establish direct contact with the recording electrode’s electrolyte. We have observed blockage of sensilla openings either by foreign debris or excreted and dried material from within the target sensillum. To avoid these impediments, care must be taken when rearing and handling insects to avoid contact with the target sensilla. In addition to choosing undamaged, healthy test insects, biological factors such as age, feeding state, mating state and zeitgeber time should be considered when establishing controls to reduce variability in recordings.
It may be difficult to obtain a responsive preparation consistently. Many trials may be necessary to improve the signal-to-noise ratio enough to resolve individual spikes. Experimental variations on the aforementioned preparations may lead to better results. It is advisable to establish a robust response on your recording apparatus for your test species using a common insect spike elicitor, like sucrose or quinine. Once a gustatory sensillum is responding, these responses should be reproducible. The minimum length of time between stimulations should be experimentally determined examining spike dynamics under different temporal constraints. Excessive stimulation should be avoided to preserve GRN health and to prevent sensillum lymph degradation. In the case that response sensitivity decreases rapidly after first stimulation, a greater number of preps must be obtained to provide confidence that these responses are physiologically typical for the given species.
The solubility of some test chemicals can create difficulty making stimulating electrodes, as some solvents may affect GRN activity, even at low final concentrations. These considerations can be addressed through trial and error and with appropriate controls. Any solvent used should be added to the control electrolyte and other test solutions to ensure consistency of responses. Immobilizing small insects, thus providing direct access to target sensilla without damaging fragile tissues, may require trial and error.
The main limitation of electrophysiology is the unfortunate separation of observed neuronal responses and potential downstream behavioral responses. Therefore, it is advantageous to test responses to chemicals that have been shown to elicit specific behavioral responses in order to discuss the significance of the results in an ecological context. Tip recordings allow for very accurate detection of cellular activity, making this technique ideal for assessing transgenic animals or those exhibiting phenotypic differences in behavior. These recordings also offer more information about neuronal responses, such as temporal and amplitude dynamics, than systems relying on visual reporters of neuronal activity. In some instances, neural responses can be quantified more easily than behavioral data12.
Tissue collection for RNA-seq and qRT-PCR is straightforward but requires continuous concentration and attention to preserving cellular material through cold storage and avoidance of excess moisture. RNase activity can affect RNA quality; therefore tissue must only contact clean surfaces throughout collection and RNA extraction. Though the troubleshooting required for qRT-PCR is well documented, the most likely pitfalls occur early in the process when designing primers and selecting target and housekeeping genes. Genes that predictably represent evenly spaced points along a line of relative expression level are useful for comparing RNA-seq and qRT-PCR datasets (Figure 3). For example, selecting genes for qPCR that show RPKM values of 0, 10, 20, 30, etc., respectively, offer a predictable order of abundance to be tested.
RNA-seq was successful at highlighting lowly expressing genes in our survey of gustatory tissues (Figure 2). The reliable gene build for Ae. aegypti (AaegL1.3, http://aaegypti.vectorbase.org/GetData/Downloads/) and prior chemoreception gene identification14 made scoring gene expression relatively easy. For these purposes, insects with available gene annotations and identified gene families of interest are valuable model systems. Determining an accurate threshold for gene expression vs. background noise is speculative, but methods of determining reasonable cutoffs are available15-17. Here we determined an RPKM level of complete confidence and a lower RPKM level of decreased confidence based on precedent9 by which we framed our discussion of results.
In the future we hope to dissect other chemosensory tissues in both the mosquito and other economically important insects to extend our knowledge of the genes mediating insect repellency. We have established several candidate gustatory genes that are most likely mediating responses to aversive compounds by comparing gene sequences to species with available functional data13. We are generating transgenic lines of mosquitoes missing these individual chemosensory genes. We will use tip recordings to assess the role of these genes on the animal’s ability to detect known repellents and assess behavioral responses of these mutants to repellents. In the instance that these candidate genes do not significantly affect repellency, more resolution may be necessary to more precisely determine gene expression or protein presence in a single sensillum or cell. Lowly expressed receptors may be critical targets for screening novel repellents, along with ubiquitously expressed co-receptors. Unfortunately, in situ hybridization and immunolocalization has proven incredibly difficult in insect gustatory tissue18.
Once we have established which genes mediate these physiological responses and behaviors, we may express these genes in cell systems that are amenable to high-throughput screening of candidate repellents. In silico approaches that use structural modeling7 may also be useful in predicting which types of compounds will elicit strong responses from specific receptors.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors thank Bryan T. Vinyard of the USDA, Agricultural Research Service, Henry A. Wallace Beltsville Agricultural Research Center, Biometrical Consulting Service, Beltsville, MD for statistical analyses. This work was supported in part by a grant to J.C.D. from the Deployed War Fighter Protection (DWFP) Research Program funded by the Department of Defense through the Armed Forces Pest Management Board (AFPMB).
References
- Klun JA, Khrimian A, Debboun M. Repellent and deterrent effects of SS220, picaridin, and DEET suppress human blood feeding by Aedes aegypti, Anopheles stephensi, and Phlebotomus papatasi. J. Med. Entomol. 2006;43(1):34–39. doi: 10.1603/0022-2585(2006)043[0034:radeos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Dickens JC, Bohbot JD. Mini review: Mode of action of mosquito repellents.Pestic. Biochem. Physiol. 2013;106(3):149–155. [Google Scholar]
- Pitts RJ, Rinker DC, Jones PL, Rokas A, Zwiebel LJ. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics. 2011;12(271) doi: 10.1186/1471-2164-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Slone JD, Rokas A, Berger SL, Liebig, et al. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLOS Genet. 2012;8(8):e1002930. doi: 10.1371/journal.pgen.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao M, Fan W, Fang S, Lu MJ, Kondo R, Li W. Transcriptional profiling of adult Drosophila antenna by high-throughput sequencing. Zoological Studies. 2013;52(42) [Google Scholar]
- Bohbot J, Pitts RJ, Kwon HW, Rützler M, Robertson HM, Zwiebel LJ. Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol. Biol. 2007;16(5):525–537. doi: 10.1111/j.1365-2583.2007.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kain P, Boyle SM, Tharadra SK, Guda T, Pham C, et al. Odour receptors and neurons for DEET and new insect repellents. Nature. 2013;502(7472):507–512. doi: 10.1038/nature12594. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hodgson ES, Lettvin JY, Roeder KD. The physiology of a primary chemoreceptor unit. Science. 1955;122(3166):417–418. doi: 10.1126/science.122.3166.417-a. [DOI] [PubMed] [Google Scholar]
- Robinson AP, Duursma RA, Marshall JD. A regression-based equivalence test for model validation: shifting the burden of proof. Tree Physiol. 2005;25(7):903–913. doi: 10.1093/treephys/25.7.903. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JL, Shields VDC, Dickens JC. Gustatory receptor neuron responds to DEET and other insect repellents in the yellow-fever mosquito, Aedes aegypti. Naturwiss. 2013;100(3):269–273. doi: 10.1007/s00114-013-1021-x. [DOI] [PubMed] [Google Scholar]
- Sparks JT, Vinyard BT, Dickens JC. Gustatory receptor expression in the labella and tarsi of Aedes aegypti. Insect Biochem. Mol. Biol. 2013;43(12):1161–1171. doi: 10.1016/j.ibmb.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Kent LB, Walden KKO, Robertson HM. The Gr family of candidate gustatory and olfactory receptors in the yellow-fever mosquito Aedes aegypti. Chem. Senses. 2008;33(1):79–93. doi: 10.1093/chemse/bjm067. [DOI] [PubMed] [Google Scholar]
- Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009;5(12):e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. A model based criterion for gene expression calls using RNA-seq data. Theory Biosci. 2013;132(3):159–164. doi: 10.1007/s12064-013-0178-3. [DOI] [PubMed] [Google Scholar]
- Hart T, Komori HK, LaMere S, Podshivalova K, Salomon DD. Finding active genes in deep RNA-seq gene expression studies. BMC Genomics. 2013;14(778) doi: 10.1186/1471-2164-14-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Morita H. Molecular and cellular designs of insect taste receptor system. Front. Cellu. Neurosci. 2010;4(20):1–16. doi: 10.3389/fncel.2010.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]


