Abstract
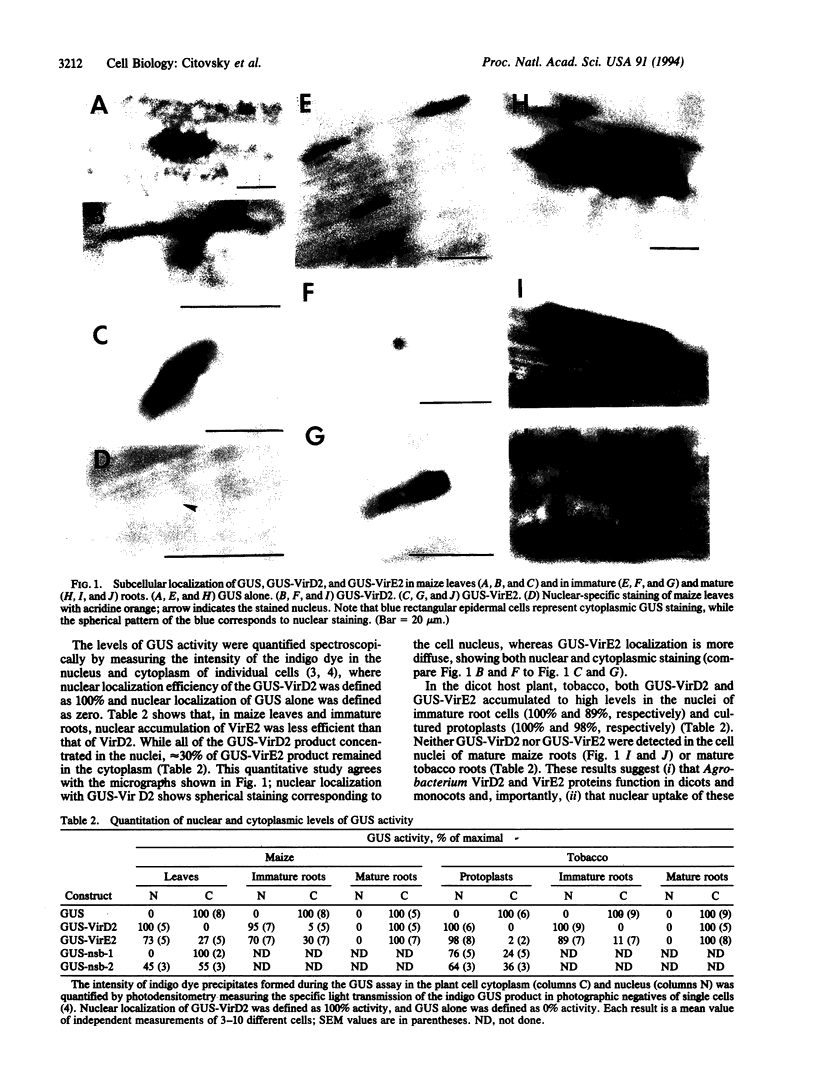
Previously, we have shown that Agrobacterium-plant cell transferred DNA (T-DNA) transport into the host cell nucleus is likely mediated by two specific bacterial proteins, VirD2 and VirE2. Here, we used these proteins to study molecular pathways of nuclear import. First, the role of VirE2 nuclear localization signals (NLSs) in the T-DNA transport pathway was examined by using tobacco plants transgenic for deletion mutants of VirE2. In these plants, the virulence of wild-type Agrobacterium was reduced possibly by competition for the cellular nuclear import machinery. Second, we analyzed the nuclear localization of VirE2 and VirD2 in the nonhost monocot maize. Part of the known recalcitrance of monocots to transformation by Agrobacterium could be due to a potential selectivity in nuclear import pathways in monocotyledonous and dicotyledonous plants. Nuclear transport of VirD2 and VirE2 in maize leaves and roots was compared to that in tobacco protoplasts and roots. Both proteins accumulated in maize leaf and tobacco protoplast nuclei as well as in nuclei of immature root cells. In contrast, VirD2 and VirE2 expressed in mature roots of maize and tobacco remained cytoplasmic. Point mutations of VirE2 nuclear localization signals, NSE 1 and NSE 2, also revealed that, in maize, the NSE 1 signal was mainly responsible for nuclear import; in contrast, both signals functioned independently in tobacco protoplasts.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carrington J. C., Freed D. D., Leinicke A. J. Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell. 1991 Sep;3(9):953–962. doi: 10.1105/tpc.3.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P. J., Ward J. E., Winans S. C., Nester E. W. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J Bacteriol. 1988 Jun;170(6):2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., DE Vos G., Zambryski P. Single-Stranded DNA Binding Protein Encoded by the virE Locus of Agrobacterium tumefaciens. Science. 1988 Apr 22;240(4851):501–504. doi: 10.1126/science.240.4851.501. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Knorr D., Schuster G., Zambryski P. The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell. 1990 Feb 23;60(4):637–647. doi: 10.1016/0092-8674(90)90667-4. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Wong M. L., Zambryski P. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the T-DNA transfer process. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Zupan J., Warnick D., Zambryski P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992 Jun 26;256(5065):1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- Garcia-Bustos J., Heitman J., Hall M. N. Nuclear protein localization. Biochim Biophys Acta. 1991 Mar 7;1071(1):83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Gietl C., Koukolíková-Nicola Z., Hohn B. Mobilization of T-DNA from Agrobacterium to plant cells involves a protein that binds single-stranded DNA. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9006–9010. doi: 10.1073/pnas.84.24.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goelet P., Lomonossoff G. P., Butler P. J., Akam M. E., Gait M. J., Karn J. Nucleotide sequence of tobacco mosaic virus RNA. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5818–5822. doi: 10.1073/pnas.79.19.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Estrella A., Chen Z. M., Van Montagu M., Wang K. VirD proteins of Agrobacterium tumefaciens are required for the formation of a covalent DNA--protein complex at the 5' terminus of T-strand molecules. EMBO J. 1988 Dec 20;7(13):4055–4062. doi: 10.1002/j.1460-2075.1988.tb03299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Estrella A., Van Montagu M., Wang K. A bacterial peptide acting as a plant nuclear targeting signal: the amino-terminal portion of Agrobacterium VirD2 protein directs a beta-galactosidase fusion protein into tobacco nuclei. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9534–9537. doi: 10.1073/pnas.87.24.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987 Sep 17;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Howard E. A., Winsor B. A., De Vos G., Zambryski P. Activation of the T-DNA transfer process in Agrobacterium results in the generation of a T-strand-protein complex: Tight association of VirD2 with the 5' ends of T-strands. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4017–4021. doi: 10.1073/pnas.86.11.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E. A., Zupan J. R., Citovsky V., Zambryski P. C. The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell. 1992 Jan 10;68(1):109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987 Dec 20;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T. M., Fromm M., Weissinger A., Tomes D., Schaaf S., Sletten M., Sanford J. C. Transfer of foreign genes into intact maize cells with high-velocity microprojectiles. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4305–4309. doi: 10.1073/pnas.85.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Lassner M. W., Jones A., Daubert S., Comai L. Targeting of T7 RNA polymerase to tobacco nuclei mediated by an SV40 nuclear location signal. Plant Mol Biol. 1991 Aug;17(2):229–234. doi: 10.1007/BF00039497. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., Overman L. B., Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J Mol Biol. 1986 Feb 20;187(4):603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- McClary J. A., Witney F., Geisselsoder J. Efficient site-directed in vitro mutagenesis using phagemid vectors. Biotechniques. 1989 Mar;7(3):282–289. [PubMed] [Google Scholar]
- Nigg E. A., Baeuerle P. A., Lührmann R. Nuclear import-export: in search of signals and mechanisms. Cell. 1991 Jul 12;66(1):15–22. doi: 10.1016/0092-8674(91)90135-l. [DOI] [PubMed] [Google Scholar]
- Raikhel N. Nuclear targeting in plants. Plant Physiol. 1992 Dec;100(4):1627–1632. doi: 10.1104/pp.100.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo M. A., Freed D. D., Carrington J. C. Nuclear transport of plant potyviral proteins. Plant Cell. 1990 Oct;2(10):987–998. doi: 10.1105/tpc.2.10.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J., Dilworth S. M., Laskey R. A., Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991 Feb 8;64(3):615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Shurvinton C. E., Hodges L., Ream W. A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11837–11841. doi: 10.1073/pnas.89.24.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Nester E. W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tinland B., Koukolíková-Nicola Z., Hall M. N., Hohn B. The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7442–7446. doi: 10.1073/pnas.89.16.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona M. J., Schmidt R. J., Raikhel N. V. Monocot regulatory protein Opaque-2 is localized in the nucleus of maize endosperm and transformed tobacco plants. Plant Cell. 1991 Feb;3(2):105–113. doi: 10.1105/tpc.3.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona M. J., Schmidt R. J., Raikhel N. V. Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell. 1992 Oct;4(10):1213–1227. doi: 10.1105/tpc.4.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C., Nester E. W. Association of the virD2 protein with the 5' end of T strands in Agrobacterium tumefaciens. J Bacteriol. 1988 Aug;170(8):3367–3374. doi: 10.1128/jb.170.8.3367-3374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]