Abstract
Chitosan (CS) and dextran sulfate (DS) are charged polysaccharides (glycans), which form polyelectrolyte complex-based nanoparticles when mixed under appropriate conditions. The glycan nanoparticles are useful carriers for protein factors, which facilitate the in vivo delivery of the proteins and sustain their retention in the targeted tissue. The glycan polyelectrolyte complexes are also ideal for protein delivery, as the incorporation is carried out in aqueous solution, which reduces the likelihood of inactivation of the proteins. Proteins with a heparin-binding site adhere to dextran sulfate readily, and are, in turn, stabilized by the binding. These particles are also less inflammatory and toxic when delivered in vivo. In the protocol described below, SDF-1α (Stromal cell-derived factor-1α), a stem cell homing factor, is first mixed and incubated with dextran sulfate. Chitosan is added to the mixture to form polyelectrolyte complexes, followed by zinc sulfate to stabilize the complexes with zinc bridges. The resultant SDF-1α-DS-CS particles are measured for size (diameter) and surface charge (zeta potential). The amount of the incorporated SDF-1α is determined, followed by measurements of its in vitro release rate and its chemotactic activity in a particle-bound form.
Keywords: Chemistry, Issue 95, Dextran sulfate, chitosan, glycan, SDF-1α, nanoparticle, polyelectrolyte complex
Introduction
Dextran sulfate (DS) and chitosan (CS) are polysaccharides with multiple substituted negatively charged sulfate groups (in DS), or positively charged amine groups (deacetylated CS). When mixed in an aqueous solution, the two polysaccharides form polyelectrolyte complexes through electrostatic interactions. The resulting complexes may form large aggregates that will be phase-separated from the aqueous solution (precipitates), or small particles that are water dispersible (colloids). The specific conditions that contribute to these outcomes have been extensively studied, and have been summarized and illustrated in detail in a recent review 1. Among these conditions, two basic requirements for producing water dispersible particles are the oppositely charged polymers must 1) have significantly different molar mass; and 2) be mixed in a non-stoichiometric ratio. These conditions will allow the charge-neutral complexed polymeric segments generated by charge neutralization to segregate and form the core of the particle, and the excess polymer to form the outer shell 1. The glycan particles described in this protocol are intended for pulmonary delivery, and are designed to be net negatively charged, and of nanometer dimensions. The negative surface charge reduces the likelihood of cellular uptake of the particles 2,3. Particles of nanometer dimension facilitate the passage through the distal airways. To achieve this goal, the amount of DS used in this preparation is in excess of CS (weight ratio 3:1); and high-molecular-weight DS (weight-average MW 500,000) and low-molecular-weight CS (MW range 50–190 kDa, 75–85% deacetylated) are used.
SDF-1α is a stem cell homing factor, which exerts the homing function through its chemotactic activity. SDF-1α plays an important role in homing and maintenance of hematopoietic stem cells in the bone marrow, and in recruitment of progenitor cells to the peripheral tissue for injury repair 4,5. SDF-1α has a heparin-binding site in its protein sequence, which allows the protein to bind to heparin/heparan sulfate, form dimers, be protected from protease (CD26/DPPIV) inactivation, and interact with target cells via the cell surface receptors 6-8. DS has similar structural properties as heparin/heparan sulfate; thus, the binding of SDF-1α to DS would be similar to that of its natural polymeric ligands.
In the following protocol, we describe the preparation of SDF-1α-DS-CS nanoparticles. The procedures represent one of the formulations that have been previously studied 9. The protocol is originally adapted from an investigation of VEGF-DS-CS nanoparticles 10. A small scale preparation is described, which can be easily scaled up with the same stock solutions and preparation conditions. After preparation, the particles are characterized by examining their size, zeta potential, extent of SDF-1α incorporation, in vitro release time, and activity of the incorporated SDF-1α.
Protocol
1. Preparation of SDF-1α Glycan Nanoparticles
Owing to the purpose of in vivo delivery, sterilize all containers, pipettes, and tips used in the preparation.
Prepare the following stock solutions in UltraPure Water: 1% dextran sulfate; 1 M NaOH (sterile filtered with a PES membrane); 0.1% chitosan in 0.2% glacial acetic acid (filter through 0.8 and 0.22 μm filters consecutively and adjust pH to 5.5 with NaOH afterward); 0.1 M ZnSO4; 15% mannitol; and 0.92 mg/ml SDF-1α (stored in aliquots at 80 °C, and kept at 4 °C once thawed).
Sterilize stock solutions through 0.22 μm filter membranes. Assess endotoxin levels in the prepared solution with limulus amebocyte lysate (LAL) gel clot assay. Ensure that the levels are <0.06 EU/ml.
Add 0.18 ml UltraPure water to a 1.5 ml glass vial containing a magnetic stir bar. Set the stir speed at 800 rpm. Add 0.1 ml 1% dextran sulfate and stir for 2 min. Add 0.08 mg SDF-1α (0.087 ml of 0.92 mg/ml SDF-1α solution) and stir for 20 min.
Add 0.33 ml 0.1% chitosan dropwise and stir for 5 min. Change stir speed to the maximum and add 0.1 ml 0.1 M ZnSO4 slowly with a 0.1 ml syringe (over 1 min).
Return stir speed to 800 rpm and stir for 30 min. Add 0.4 ml 15% mannitol and stir for 5 min. Transfer the reaction mixture to a 1.5 ml microfuge tube. Centrifuge at 16,000 x g at 4 °C for 10 min. Note: The presence of mannitol facilitates the resuspension of the particles after centrifugation. Depending on the compactness of the pellet, mannitol concentration can be varied from 0% to 5%. Complete resuspension of the particles after each centrifugation is critical to avoid aggregates in the final suspension.
Aspirate supernatant and use a pipette to remove the last drop of liquid slowly. Add 0.2 ml 5% mannitol. Suspend the pellet with a 0.5 ml, 29 G needle syringe. Add 1 ml 5% mannitol. Centrifuge at 16,000 x g for 15 min.
Repeat step 1.6.
Aspirate the supernatant. Resuspend the pellet in 0.2 ml 5% mannitol. Store the particle suspension at 4 °C. Note: The particle suspension can also be stored frozen at -80 °C or lyophilized. Including 5% mannitol in the suspension is essential to prevent aggregation of the particles after freezing and thawing or after lyophilization and rehydration. Mannitol can be removed by centrifugation of the suspension after the storage.
2. Measurement of Particle Size and Zeta Potential
The particle size and zeta potential are analyzed by dynamic light scattering and electrophoretic light scattering, respectively, with a particle analyzer indicated in Material List.
Set up the following parameters for particle size measurement: Accumulation times: 70; Repeat times: 4; Temperature: 25 °C; Diluent: water; Intensity adjustment: auto.
Dilute the SDF-1α-DS-CS sample with water (10-fold dilution). Load 100 μl of the sample to a disposable UV cuvette such as an Eppendorf cuvette. Insert the cuvette into the cell holder. Wait for the intensity adjustment to reach “Optimum” (~10,000 cps). Start data acquisition.
After the measurement is completed, record the cumulants results of diameter (nm) and polydispersity index. Average the results obtained from each of the 4 repeated readings and calculate the standard deviation.
Load 500 μl of the 10-fold diluted particle sample to a flow cell for zeta potential measurement. Set up the following parameters for the measurement: Accumulation times: 10; Repeat times: 5; Temperature: 25 °C; Diluent: water; Intensity adjustment: auto. Record the results of the zeta potential (mV). Average the result obtained from each of the 5 repeated readings, and calculate the standard deviation.
3. Quantification of SDF-1α in the Particles
Dilute free form SDF-1α to concentrations of 0.01, 0.02, 0.03, 0.04, and 0.05 mg/ml in 1.3x Laemmli sample buffer. Dilute 6 µl SDF-1α-DS-CS samples with 40 µl 1.3x Laemmli sample buffer. (Prepare 4x Laemmli stock buffer containing 0.25 M Tris-HCl, pH 7.5, 8% SDS, 40% glycerol, 0.05 mg/ml bromophenol blue, and 8% 2-mercaptoethanol. Divide the stock solution into small aliquots and keep at -20 °C for single use.)
Heat the samples at 100 °C for 10 min. Vortex the samples twice (each for 10 sec at maximum speed) during the 10 min heating time in order to dissociate the particles completely. Cool down the samples to RT for 2 min. Centrifuge at 10,000 x g for 0.5–1 min to bring down the water condensate. Vortex again to mix well. Note: At this point, the sample solution should be clear with no visible precipitate present.
Load 10 µl sample/well to a 4–20% SDS gel. Run electrophoresis at 200 V for 20 min. Stain the gel with Coomassie blue protein stain.
Examine the protein band density of SDF-1α using a molecular imager and densitometry analysis software. Calculate the quantity of SDF-1α in the particles against a standard curve constructed with free SDF-1α standards.
4. In Vitro Release Assay
Mix SDF-1α glycan particles with Dulbecco’s phosphate buffered saline without calcium and magnesium (D-PBS) at a 1:1 ratio (v/v).
Divide the particle suspension into 0.05 ml aliquots in 1.5 ml tubes. Load the samples to the bottom of the tubes. Avoid introducing air bubbles or disturbing the surface of the liquid. Seal the top of the tubes with Parafilm. NOTE: In doing so, the liquid will remain at the bottom during the subsequent top-to-bottom rotation on the tube mixer.
Rotate the tubes at 37 °C on a Rotating Micro-Tube Mixer. Remove aliquots from the tubes at the designated times and immediately centrifuge the samples at 16,000 x g for 10 min at 4 °C.
Separate the supernatants and pellets, and reconstitute the pellet with 0.05 ml D-PBS. Store the samples at −20 °C. After all the samples are collected, examine the supernatants and pellets on a SDS gel as described above.
5. Migration Assay
This assay measures the chemotactic activity of SDF-1α. Interaction of SDF-1α with its receptor (CXCR4) on the cell surface causes migration of the cell towards the SDF-1α gradient. In this assay, the cells are loaded into an upper well (separated by a semipermeable membrane from a lower well) and SDF-1α solution into a lower well.
Dilute SDF-1α (free or particle-bound form) with migration buffer (RPMI-1640 medium containing 0.5% bovine serum albumin) in a 3-fold serial dilution to final concentrations of 100, 33, 11, 3.7, 1.2, 0.41, 0.14, and 0.05 ng/ml.
Add 0.6 ml of the diluted SDF-1α solution or migration buffer only (negative control) to a well in the 24-well plate. Add 0.57 ml migration buffer to a well (input cell control). Incubate at 37 °C for 30 min. Place a permeable cell culture insert such as Transwell (pore size 5 μm, diameter 6.5 mm) on top of the lower well. Load 0.1 ml Jurkat cells (5 × 105) into the Transwell insert. Load 0.03 ml cells directly to the input cell control well. Incubate the plate at 37 °C for 2 hr in a 5% CO2 incubator.
Remove the Transwell inserts. Transfer the cells that have migrated to the lower wells to a 4 ml polystyrene tube. Count the migrated cells with a flow cytometer.
Calculate migration as a percentage of the input cell number (cell number in the input cell control well x 100/30) after subtraction of the numbers in negative controls (cells migrated to the wells containing only migration buffer).
Representative Results
The size and zeta potential of the prepared SDF-1α-DS-CS particles are determined with a particle analyzer. Figure 1 shows the analysis of the size measurement. From the cumulants results obtained from four repeated measurements, the average hydrodynamic diameter of the SDF-1α-DS-CS particles is 661 ± 8.2 (nm) and the polydispersity is 0.23 ± 0.02. The result of the zeta potential measurement is shown in Figure 2. From the five repeated measurements, the zeta potential of the SDF-1α-DS-CS particles is -24.8 ± 0.5 mV. As mentioned above, the presence of 5% mannitol in the SDF-1α-DS-CS particle suspension is essential for prevention of particle aggregation during a freezing and thawing process. Figure 3 shows the size measurement of the particles suspended in water and frozen at -80 °C. A distinct aggregation peak is visible after thawing.
The amount of SDF-1α in the SDF-1α-DS-CS particles is estimated by SDS gel electrophoresis. Figure 4A shows a Coomassie blue-stained SDS gel with SDF-1α standards, DS-CS nanoparticle (control particles without SDF-1α incorporation, Ctrl NP) and SDF-1α-DS-CS nanoparticle (SDF-1α NP) samples loaded. A standard curve is constructed from the density analysis of the SDF-1α standard bands (Figure 4B). Calculation from the standard curve indicates that the SDF-1α concentration in the SDF-1α NP sample is 0.03 mg/ml (average of duplicate). Since the dilution of the sample with the sample buffer is (6+40)/6, the original sample concentration is 0.23 mg/ml. As the final volume of the particle preparation is 0.2 ml, total SDF-1α in the final suspension is 0.046 mg. Considering the input mass of SDF-1α in the reaction mixture is 0.08 mg, the entrapment efficiency of SDF-1α in the DS-CS particles is 57%. Chitosan has been previously reported to stain with Coomassie blue. The staining, however, is not significant in the gel shown in Figure 4A. It is, perhaps, related to the quantity of chitosan (particle matrix) loaded on the gel. In any case, a complete dissociation and separation of SDF-1α from the particle matrix is necessary for accurately assessing the amount of the incorporated protein. This gel also shows that SDF-1α is not detectably degraded by heating at 100 °C for 10 min or by the vortex process, as no bands are found between the SDF-1α protein band and the dye front.
The in vitro release rate of SDF-1α from the nanoparticles is determined by incubation of the particles in 50% D-PBS at 37 °C for 7 days. At 0 hr, 3 hr, 8 hr, 24 hr, 48 hr, and 7 days, aliquots of the samples are removed and immediately centrifuged. The released SDF-1α (in the supernatant) is separated from the particle-bound SDF-1α (pellet). The samples are run on a SDS gel, and the amount of SDF-1α in the supernatants and pellets are determined by Coomassie staining and densitometry analysis. A typical SDS gel of the analysis is shown in Figure 5. As demonstrated, the particle bound SDF-1α is minimally released after a 7-day incubation.
The activity of the particle-bound SDF-1α is measured by a migration assay (chemotactic assay), and compared to that of free SDF-1α. In this assay, SDF-1α in the DS-CS nanoparticles is serially diluted with a migration buffer, and incubated with Jurkat cells for 2 hr. The migrated cells are counted with a flow cytometer. The data (with SDF-1α concentrations of 0.05–11 ng/ml) are plotted with non-linear regression fitting, and the EC50 is calculated. As shown in Figure 6, the particle-bound SDF-1α induced the same extent of migration as that of free SDF-1α at all concentrations measured. The EC50 values of the free and particle-bound forms of SDF-1α are 0.55 and 0.45 ng/ml, respectively.
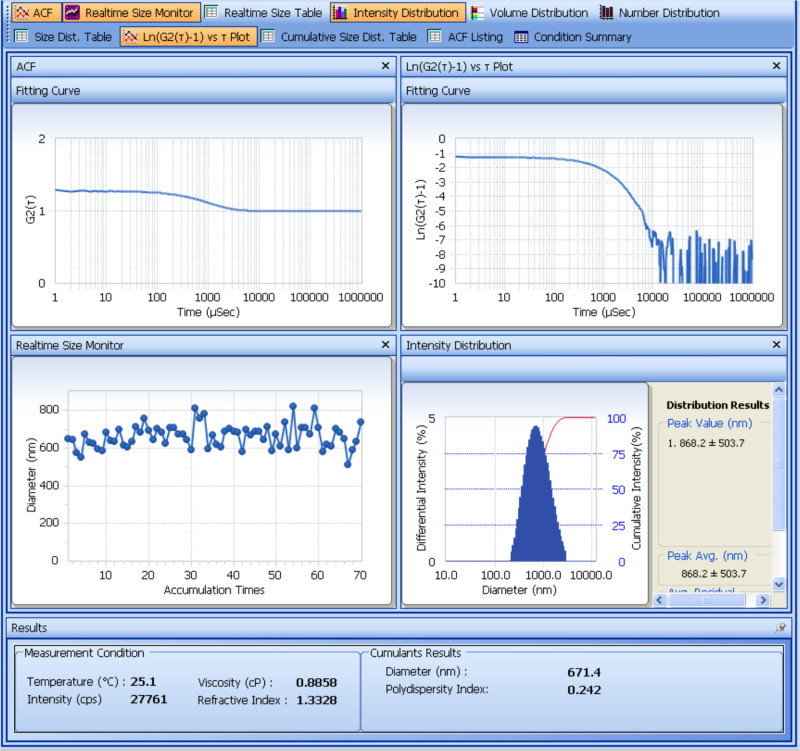 Figure 1. Analysis screen shot showing dynamic light scattering measurement of particle size. The top two panels show the plots of the autocorrelation function, G2(τ), and logarithm of [G2(τ)-1] over time. The instrument's digital signal processor (correlator) expresses the detected light intensity as a function of delay time (τ). The plot shows that SDF-1α-DS-CS particles caused an exponential decay of the intensity between 0.2-2 msec. To obtain the average decay constant <Γ>, a program analyzes the Ln[G2(τ)-1] plot with cumulants fitting. The first order coefficient of the derived polynomial equation is assigned to <Γ>, which is proportional to the slope of the curve. The second order coefficient divided by the square of <Γ> is the polydispersity index, which describes the intensity distribution width. From <Γ> the average diffusion coefficient (<D>) and particle diameter (d) are calculated as: Γ=D·q and D = kBT/3πη0d (Stoke-Einstein equation). The middle two panels show the fluctuation of the size reading during the 70 accumulation measurements, and the size distribution calculated with respect to intensity in the measurement. The cumulants diameter and polydispersity index are shown at the bottom of the screen.
Figure 1. Analysis screen shot showing dynamic light scattering measurement of particle size. The top two panels show the plots of the autocorrelation function, G2(τ), and logarithm of [G2(τ)-1] over time. The instrument's digital signal processor (correlator) expresses the detected light intensity as a function of delay time (τ). The plot shows that SDF-1α-DS-CS particles caused an exponential decay of the intensity between 0.2-2 msec. To obtain the average decay constant <Γ>, a program analyzes the Ln[G2(τ)-1] plot with cumulants fitting. The first order coefficient of the derived polynomial equation is assigned to <Γ>, which is proportional to the slope of the curve. The second order coefficient divided by the square of <Γ> is the polydispersity index, which describes the intensity distribution width. From <Γ> the average diffusion coefficient (<D>) and particle diameter (d) are calculated as: Γ=D·q and D = kBT/3πη0d (Stoke-Einstein equation). The middle two panels show the fluctuation of the size reading during the 70 accumulation measurements, and the size distribution calculated with respect to intensity in the measurement. The cumulants diameter and polydispersity index are shown at the bottom of the screen.
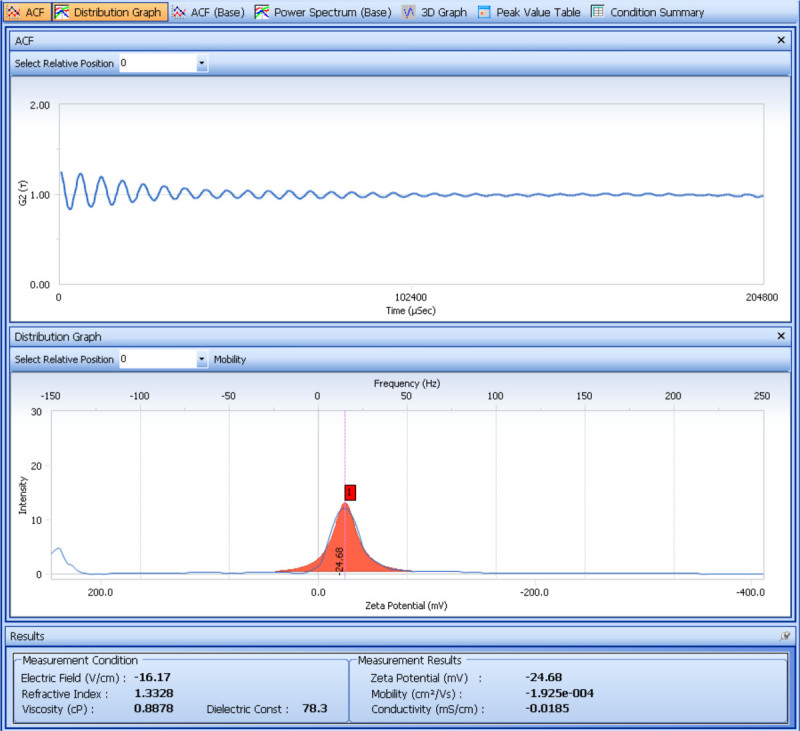 Figure 2. Analysis screen shot showing electrophoretic light scattering measurement of particle zeta potential. The top panel shows a G2(τ) over time plot. The gradually decaying oscillating function is typically observed in charged particles that are subjected to electrophoresis and light scattering. The function reflects the frequency shift (Doppler shift) caused by the moving particles in the electrical field. A Fourier transformation of G2(τ) gives the power spectrum (intensity vs. frequency plot) shown in the middle panel. The center of the frequency shift peak defines the mobility of the particles, from which the zeta potential is calculated. The calculated zeta potential is shown at the bottom of the screen.
Figure 2. Analysis screen shot showing electrophoretic light scattering measurement of particle zeta potential. The top panel shows a G2(τ) over time plot. The gradually decaying oscillating function is typically observed in charged particles that are subjected to electrophoresis and light scattering. The function reflects the frequency shift (Doppler shift) caused by the moving particles in the electrical field. A Fourier transformation of G2(τ) gives the power spectrum (intensity vs. frequency plot) shown in the middle panel. The center of the frequency shift peak defines the mobility of the particles, from which the zeta potential is calculated. The calculated zeta potential is shown at the bottom of the screen.
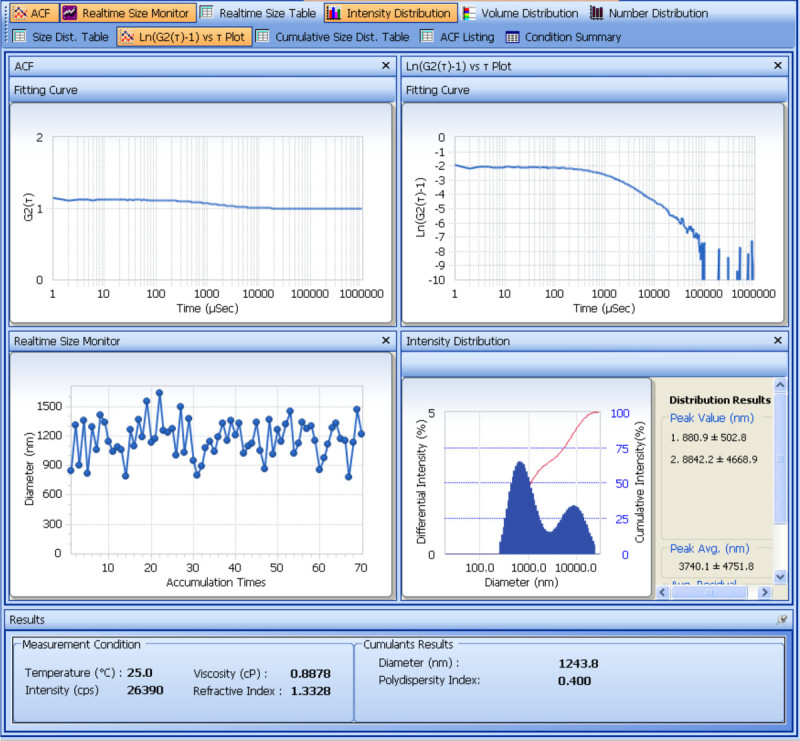 Figure 3. Particle size measurement of SDF-1α-DS-CS particles after freezing and thawing in water. In the absence of mannitol, the particles form aggregates after freezing and thawing. Note the formation of an additional intensity/size peak and the increase in cumulants diameter and polydispersity compared to that of Figure 1.
Figure 3. Particle size measurement of SDF-1α-DS-CS particles after freezing and thawing in water. In the absence of mannitol, the particles form aggregates after freezing and thawing. Note the formation of an additional intensity/size peak and the increase in cumulants diameter and polydispersity compared to that of Figure 1.
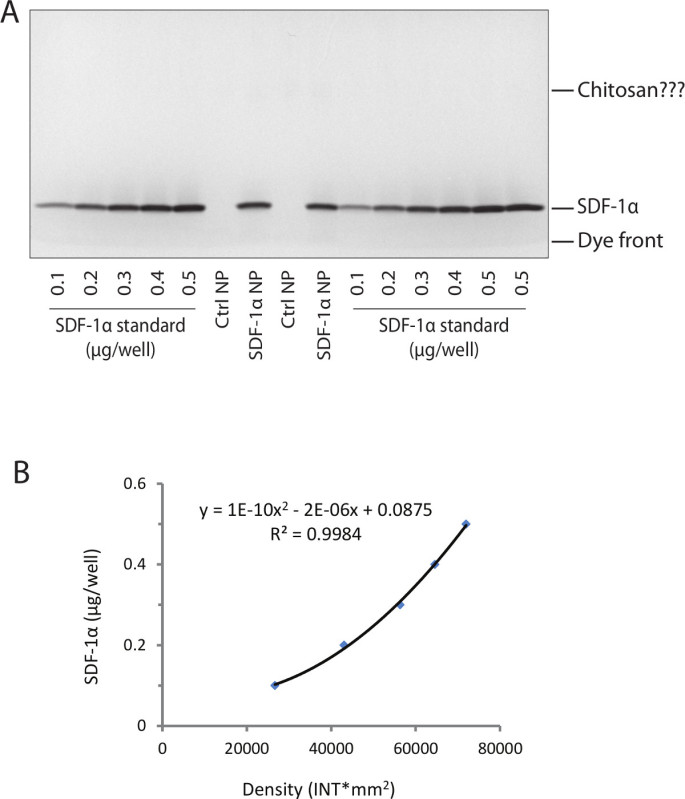 Figure 4. Quantification of SDF-1α incorporation in DS-CS nanoparticles. (A) Photograph of Coomassie-stained SDS gel loaded with free SDF-1α standards, DS-CS nanoparticle (Ctrl NP), and SDF-1α-DS-CS nanoparticle (SDF-1α NP) samples as indicated. The SDF-1α protein bands and the gel running dye front are marked. The Ctrl NP samples are not stained with Coomassie blue and the chitosan bands (wide) are vaguely visible. (B) The bands are analyzed with a densitometer, from which the standard curve is constructed with the free SDF-1α standards (average of duplicate). The amount of SDF-1α in the nanoparticles is calculated against the standard curve, which gives an average value of 0.3 µg/well (10 µl).
Figure 4. Quantification of SDF-1α incorporation in DS-CS nanoparticles. (A) Photograph of Coomassie-stained SDS gel loaded with free SDF-1α standards, DS-CS nanoparticle (Ctrl NP), and SDF-1α-DS-CS nanoparticle (SDF-1α NP) samples as indicated. The SDF-1α protein bands and the gel running dye front are marked. The Ctrl NP samples are not stained with Coomassie blue and the chitosan bands (wide) are vaguely visible. (B) The bands are analyzed with a densitometer, from which the standard curve is constructed with the free SDF-1α standards (average of duplicate). The amount of SDF-1α in the nanoparticles is calculated against the standard curve, which gives an average value of 0.3 µg/well (10 µl).
 Figure 5. SDS gel showing SDF-1α in vitro release time course. Coomassie-stained SDS gel shows the amount of released SDF-1α (in supernatants) and bound SDF-1α (in pellets) after incubation of SDF-1α-DS-CS nanoparticles at 37 °C for various periods of time. Re-printed with permission from9.
Figure 5. SDS gel showing SDF-1α in vitro release time course. Coomassie-stained SDS gel shows the amount of released SDF-1α (in supernatants) and bound SDF-1α (in pellets) after incubation of SDF-1α-DS-CS nanoparticles at 37 °C for various periods of time. Re-printed with permission from9.
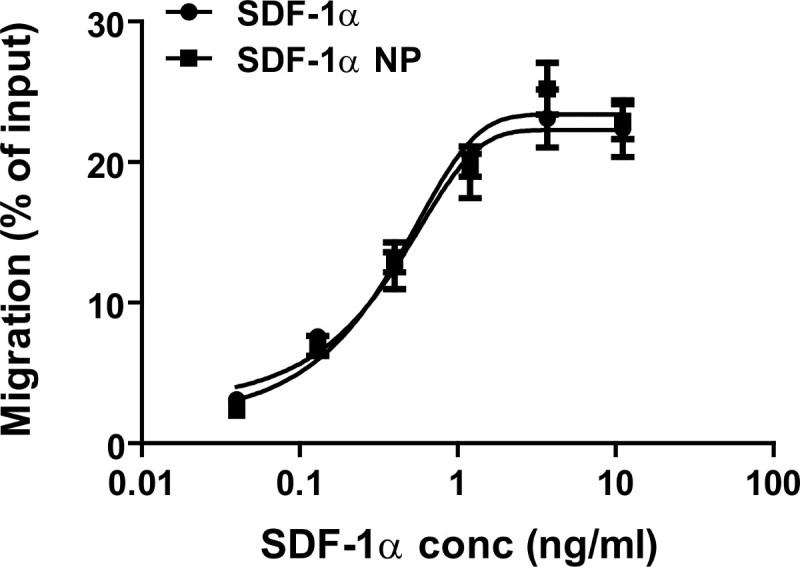 Figure 6. Dose−response curves of SDF-1α (circles) and SDF-1α nanoparticle (NP) (squares) in a Jurkat cell migration assay. SDF-1α in both free and nanoparticle bound forms exhibited the same activity with EC50 values of 0.55 and 0.45 ng/ml, respectively. Re-printed with permission from9.
Figure 6. Dose−response curves of SDF-1α (circles) and SDF-1α nanoparticle (NP) (squares) in a Jurkat cell migration assay. SDF-1α in both free and nanoparticle bound forms exhibited the same activity with EC50 values of 0.55 and 0.45 ng/ml, respectively. Re-printed with permission from9.
Discussion
As mentioned above, the DS-CS nanoparticles are formed through charge neutralization between polyanion (DS) and polycation (CS) molecules. Since the charge interaction occurs readily during the molecular collision, the concentration of the polymer solutions and the stirring speed during mixing is critical for the size of the resultant particles. A general trend is that more diluted DS and CS solutions 15 and higher stirring speed result in smaller particles.
The formulation of the SDF-1α glycan nanoparticles can be varied. For example, the amounts and ratios of SDF-1α/DS/CS used in this protocol are 0.08/0.33/1 (mg/mg/mg), respectively. Depending on the intended use of SDF-1α nanoparticles, these ratios can change. If the amount of SDF-1α added to the mixture is 0.04 mg (0.04/0.33/1, mg/mg/mg) instead of 0.08 mg, the particles will be smaller, ~650 nm, and the SDF-1α entrapment efficiency slightly greater, 70–80% 9. However, when delivered in vivo, a greater amount of the glycan matrix will be present per SDF-1α. If a smaller particle size is desired, the formulation of SDF-1α glycan nanoparticles can be further modified. One alternative is to use a smaller molecular weight chitosan, as demonstrated in Ref 15. It should be noted, however, that chitosan has a high affinity for endotoxin (negatively charged lipopolysaccharides); thus, a test of endotoxin level or potentially a purification is necessary for the use of any chitosan products prior to in vivo delivery. The second alternative is to load SDF-1α onto pre-formed small DS-CS nanoparticles. Since the latter can be manipulated more readily in particle size and SDF-1α has a high affinity for DS, loading a small amount of SDF-1α to the outer shell of the DS-CS particles can result in smaller particles.
Compared with other types of nanoparticles (e.g., PLGA11, polyanhydride12, or gelatin13 nanoparticles), the DS-CS nanoparticles have particular advantages and limitations. The major advantages of the DS-CS nanoparticles are: 1) the particle preparation does not involve organic solvents or vigorous sonication. It is, therefore, suitable for protein factor incorporation, as gentle preparation reduces the likelihood of protein inactivation; 2) the matrix of the particles has a similar structural property as extracellular matrix, which makes the particles compatible with tissue environment and is less toxic and inflammatory; and 3) glycan particle-bound SDF-1α mimics a natural binding relationship between the protein and the extracellular matrix, which explains the full chemotactic activity of SDF-1α after being incorporated into the glycan particles. The fully active SDF-1α in the particle-bound form allows it to serve as a stationary homing factor in tissue environment. The limitations of the DS-CS nanoparticles are: 1) the diameters of the particles are generally greater than the above mentioned particles; and 2) easily collapse in salt solutions, which may not be suitable for direct injection into blood circulation.
The particle-bound SDF-1α is found to be minimally released from the nanoparticles after a seven-day incubation. This phenomenon is related to the high affinity of SDF-1α for heparin, which also contributes to its function in vivo. Since proteins with heparin binding domains will generally have different affinities for heparin, their release rates will likely differ. For example, in a VEGF165-DS-CS nanoparticle prepared in a similar formulation, 44% of the incorporated VEGF165 was released in the first hour and 29% was released in the next 47 hr during incubation at 37 °C14. Therefore, the in vitro release pattern for a specific protein needs to be individually tested.
The purpose of preparation of the SDF-1α-DS-CS nanoparticles is to deliver SDF-1α to the lung and establish a stem cell homing signal in the tissue. The particles can also be delivered to other tissues for the same purpose, although the specific nanoparticle format is not required for solid tissue injection. The fact that SDF-1α is stabilized by DS in the nanoparticle matrix and is minimally released from the particles supports stem cell recruitment principles, and the particle is expected to be beneficial for tissue regeneration.
Since the protein-glycan nanoparticles have a similar matrix to that of cell surface glycosaminoglycans and extracellular matrix, it is possible that the particle-incorporated proteins can be exchanged into these matrixes (The negatively charged extracellular matrix molecules can compete with the binding of the positively charged protein) in vivo. This exchange can alter the protein release kinetics, and, thus, needs to be further investigated after the initial in vitro characterization.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was supported by NIH grants: HL671795, HL048743, and HL108630.
References
- Delair T. Colloidal polyelectrolyte complexes of chitosan and dextran sulfate towards versatile nanocarriers of bioactive molecules. Eur J Pharm Biopharm. 2011;78(1):10–18. doi: 10.1016/j.ejpb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Morachis JM, Mahmoud EA, Almutairi A. Physical and chemical strategies for therapeutic delivery by using polymeric nanoparticles. Pharmacol Rev. 2012;64(3):505–519. doi: 10.1124/pr.111.005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue ZG, et al. Surface charge affects cellular uptake and intracellular trafficking of chitosan-based nanoparticles. Biomacromolecules. 2011;12(7):2440–2446. doi: 10.1021/bm101482r. [DOI] [PubMed] [Google Scholar]
- Ghadge SK, Muhlstedt S, Ozcelik C, Bader M. SDF-1alpha as a therapeutic stem cell homing factor in myocardial infarction. Pharmacol Ther. 2011;129(1):97–108. doi: 10.1016/j.pharmthera.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Sharma M, Afrin F, Satija N, Tripathi RP, Gangenahalli GU. Stromal-derived factor-1/CXCR4 signaling: indispensable role in homing and engraftment of hematopoietic stem cells in bone marrow. Stem Cells Dev. 2011;20(6):933–946. doi: 10.1089/scd.2010.0263. [DOI] [PubMed] [Google Scholar]
- Sadir R, Baleux F, Grosdidier A, Imberty A, Lortat-Jacob H. Characterization of the stromal cell-derived factor-1alpha-heparin complex. J Biol Chem. 2001;276(11):8288–8296. doi: 10.1074/jbc.M008110200. [DOI] [PubMed] [Google Scholar]
- Amara A, et al. Stromal cell-derived factor-1alpha associates with heparan sulfates through the first beta-strand of the chemokine. J Biol Chem. 1999;274(34):23916–23925. doi: 10.1074/jbc.274.34.23916. [DOI] [PubMed] [Google Scholar]
- Sadir R, Imberty A, Baleux F, Lortat-Jacob H. Heparan sulfate/heparin oligosaccharides protect stromal cell-derived factor-1 (SDF-1)/CXCL12 against proteolysis induced by CD26/dipeptidyl peptidase IV. J Biol Chem. 1074;279(42):43854–43860. doi: 10.1074/jbc.M405392200. [DOI] [PubMed] [Google Scholar]
- Yin T, et al. SDF-1alpha in glycan nanoparticles exhibits full activity and reduces pulmonary hypertension in rats. Biomacromolecules. 2013;14(11):4009–4020. doi: 10.1021/bm401122q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Vitharana SN, Peek LJ, Coop T, Berkland C. Polyelectrolyte complexes stabilize and controllably release vascular endothelial growth factor. Biomacromolecules. 2007;8(5):1607–1614. doi: 10.1021/bm061211k. [DOI] [PubMed] [Google Scholar]
- McCall RL, Sirianni RW. PLGA nanoparticles formed by single- or double-emulsion with vitamin E-TPGS. J Vis Exp. 2013. p. 51015. [DOI] [PMC free article] [PubMed]
- Carrillo-Conde BR, Roychoudhury R, Chavez-Santoscoy AV, Narasimhan B, Pohl NL. High-throughput synthesis of carbohydrates and functionalization of polyanhydride nanoparticles. J Vis Exp. 2012. p. 3967. [DOI] [PMC free article] [PubMed]
- Xu J, Amiji M. Therapeutic gene delivery and transfection in human pancreatic cancer cells using epidermal growth factor receptor-targeted gelatin nanoparticles. J Vis Exp. 2012. p. e3612. [DOI] [PMC free article] [PubMed]
- Lauten EH, et al. Nanoglycan complex formulation extends VEGF retention time in the lung. Biomacromolecules. 2010;11(7):1863–1872. doi: 10.1021/bm100384z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz C, Domard A, Viton C, Pichot C, Delair T. Versatile and efficient formation of colloids of biopolymer-based polyelectrolyte complexes. Biomacromolecules. 2004;5(5):1882–1892. doi: 10.1021/bm049786+. [DOI] [PubMed] [Google Scholar]


