Abstract
The Escherichia coli respiratory complex II paralogs succinate dehydrogenase (SdhCDAB) and fumarate reductase (FrdABCD) catalyze interconversion of succinate and fumarate coupled to quinone reduction or oxidation, respectively. Based on structural comparison of the two enzymes, equivalent residues at the interface between the highly homologous soluble domains and the divergent membrane anchor domains were targeted for study. This included the residue pair SdhB-R205 and FrdB-S203, as well as the conserved SdhB-K230 and FrdB-K228 pair. The close proximity of these residues to the [3Fe-4S] cluster and the quinone binding pocket provided an excellent opportunity to investigate factors controlling the reduction potential of the [3Fe-4S] cluster, the directionality of electron transfer and catalysis, and the architecture and chemistry of the quinone binding sites. Our results indicate that both SdhB-R205 and SdhB-K230 play important roles in fine tuning the reduction potential of both the [3Fe-4S] cluster and the heme. In FrdABCD, mutation of FrdB-S203 did not alter the reduction potential of the [3Fe-4S] cluster, but removal of the basic residue at FrdB-K228 caused a significant downward shift (>100mV) in potential. The latter residue is also indispensable for quinone binding and enzyme activity. The differences observed for the FrdB-K228 and Sdh-K230 variants can be attributed to the different locations of the quinone binding site in the two paralogs. Although this residue is absolutely conserved, they have diverged to achieve different functions in Frd and Sdh.
Keywords: Complex II, succinate dehydrogenase, iron-sulfur cluster, quinone binding site
1. Introduction
Escherichia coli is a versatile facultative anaerobe. During aerobic growth, the respiratory enzyme succinate dehydrogenase (SdhCDAB) mediates electron transfer (ET) from succinate to ubiquinone (UQ) and thus plays a pivotal role in the ET chain and oxidative phosphorylation. Under anaerobic conditions, the paralog fumarate reductase (FrdABCD) acts as an electron sink to regenerate menaquinone (MQ) by utilizing fumarate as the terminal electron acceptor in lieu of oxygen. Three dimensional crystal structures of these two paralogs show they share an overall architecture wherein the soluble SdhAB/FrdAB dimers are anchored to the inner surface of the cytoplasmic membrane by their respective hydrophobic anchors, SdhCD or FrdCD [1–3]. Coupling of the succinate-fumarate and quinone-quinol reactions is achieved via the ET relay, which consists of a flavin adenine dinucleotide (FAD) in SdhA/FrdA and three [Fe-S] clusters in SdhB/FrdB [4,5]. In the case of SdhCDAB, a heme moiety in the membrane intrinsic domain may also partake in electron transfer, but its incorporation into the holoenzyme is not essential for function [6].
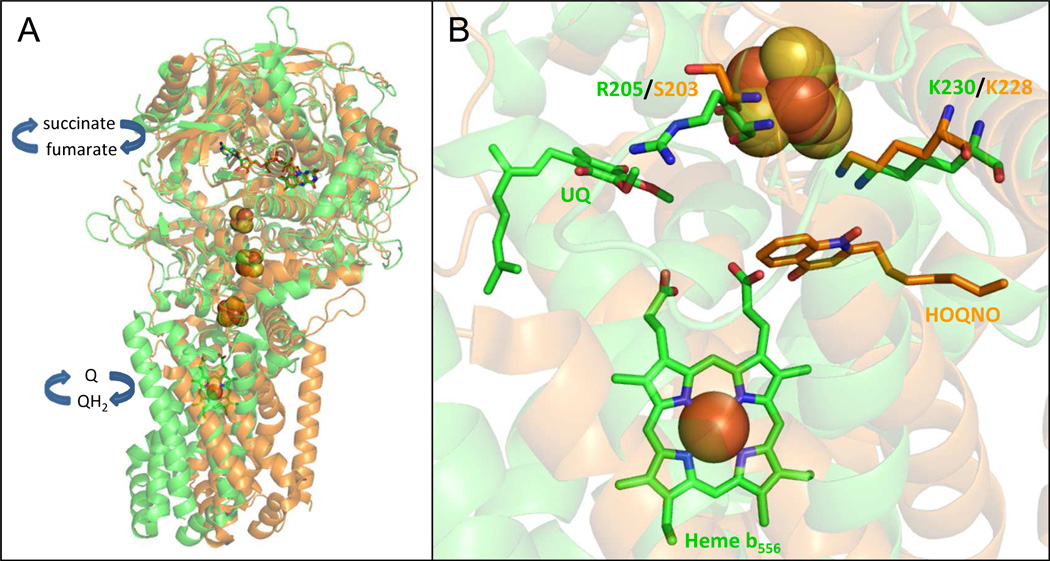
Superimposition of the three-dimensional structures of SdhCDAB (PDB:1NEK) and FrdABCD (PDB:1KF6) shows that the dicarboxylate binding site, the FAD cofactor, and the [Fe-S] clusters have essentially identical spatial arrangements (RMSD = 1.69 Å for subunit A, 1.75 Å for subunit B) (Fig. 1A). Despite the high level of similarity observed between SdhAB and FrdAB, there are subtle differences in sequence and biophysical properties of the cofactors that determine the directionality of enzyme function (i.e., succinate oxidation coupled to quinone reduction versus quinol oxidation coupled to fumarate reduction). For instance, the equivalent, but non-conserved, SdhA-Q50 and FrdAE49 residues dictate dicarboxylate oxidation or reduction by exerting a coulombic effect on the electronic state of the FAD cofactor [7]. In SdhB, the native reduction potentials (Em values) of the [2Fe-2S], [4Fe-4S] and [3Fe-4S] clusters are +10mV, −175mV and +65mV respectively, and thus favor electron flow towards the quinone-binding site (Q-site) [8,9]. In contrast, the Em values of the [Fe-S] clusters in FrdB are poised ([2Fe-2S], −20mV; [4Fe-4S], −320mV; [3Fe-4S], −70mV) [10,11] such that ET from the Q-site to the FAD is favored. The residue pair SdhB-H207/FrdB-T205 was recently demonstrated to contribute to the high and low reduction potentials of the [3Fe-4S] cluster in SdhCDAB and FrdABCD, respectively [12]. An additional level of catalytic control is exerted by the type of quinone preferentially utilized by the two paralogs. The Em of the succinate/fumarate couple is +30mV. The higher Em value of +110mV for the UQ/UQH2 transition ensures electrons flow away from the FAD in Sdh whereas the lower Em value of −74 mV for the MQ/MQH2 transition directs electrons towards the FAD in Frd [4]. Finally, catalysis may also be controlled by the type of subunits assembled. In Ascaris suum, the mitochondrial Complex II may act as either a Sdh or Frd depending on the stage of parasitic life cycle and which subunits are expressed [13,14]. Recently, the direction of electron transfer was also shown to be crucial for mitochondrial energy production in tumor microenvironments wherein Complex II behaves like a fumarate reductase [15].
Figure 1.
(A) Superimposition of E. coli SdhCDAB (green) and FrdABCD (orange). The paralogs share similar architectures of the electron transfer relay and the soluble domains. The FAD molecule is represented by sticks and the iron-sulfur clusters (from top to bottom: [2Fe-2S], [4Fe-4S] and [3Fe-4S]) are drawn in spheres. (B) The residues examined in this study are in close proximity to the quinone binding sites and the [3Fe-4S] cluster in Sdh and Frd. In both panels, the two views were generated after a three-dimensional alignment of SdhCDAB (PDB:1NEK, co-crystallized with native UQ) with FrdABCD (PDB:1KF6, co-crystallized with exogenous HOQNO, a menasemiquinone analog that acts as a competitive Q-site inhibitor) using PyMOL v.1.2r1 (DeLano Scientific LLC.).
A number of factors have been suggested to govern [Fe-S] cluster Em values, including (i) the local H-bonding network; (ii) the local electrostatic field, (iii) solvent accessibility, (iv) backbone amides and (v) the type of amino acid residues coordinating the cluster [9,16–22]. As pointed out above, the structures of SdhCDAB and FrdABCD are essentially identical, especially in the regions encompassing the ET relay connecting the dicarboxylate substrate and their respective Q-sites. Differences in reduction potentials observed for the [Fe-S] clusters in SdhB and FrdB must arise due to the differences in the environments surrounding the individual clusters. The overall similarity between the [Fe-S] subunits and enzymes makes them ideal model systems to study the effects of cluster environments on observed Em values.
In this study, we examined residues that are located at the interface between the soluble and membrane anchor domains, and also in close proximity to the [3Fe-4S] cluster and the Q-site: SdhB-R205 and SdhB-K230, as well as the equivalent residues FrdB-S203 and FrdB-K228 (Fig. 1B). Using a combination of site-directed mutagenesis, redox potentiometry, electron paramagnetic resonance (EPR) spectroscopy and kinetic analysis, we probed how protein modulation of cofactor Em value is achieved and whether this affected enzyme catalysis. Our results extend upon a previous study that showed FrdB-K228 is a critical residue required for Q-binding and oxidation/reduction [23], and we elaborate here on its influence on the Em value of the [3Fe-4S] cluster.
2. Materials and Methods
Strains, Plasmids and Mutagenesis
E. coli laboratory strain TG1 (supE hsdΔ5 thi Δ (lac-proAB) F´[traD36 proAB+ lacIq lacZΔM15]; GE Healthcare) was used for mutagenesis. Strain DW35 (ΔfrdABCD, sdhC::kan) [24] was used for all enzyme expression and growth studies. Expression of SdhCDAB and FrdABCD variants were anaerobically induced by using the plasmids pFAS and pH3, respectively [25,26]. Mutations were constructed using primers from Qiagen and the QuikChange protocol from Stratagene. All recombinant plasmids were verified by DNA sequencing.
Enzyme Expression and Preparation
E. coli strain DW35 harboring the different sdhCDAB or frdABCD constructs was grown for 16 hours at 37 °C in Terrific broth, using a 1 % starting inoculum, with appropriate antibiotics. Isolated membranes enriched in either enzyme were prepared by repeated passage through an Avestin Emulsiflex for cell lysis followed by differential ultracentrifugation to isolate the cytoplasmic membrane [6]. All final membrane preparations containing activated enzymes were suspended in 100 mM MOPS / 5 mM EDTA / 1 mM malonate at pH 7.
Growth Studies
Aerobic growth on succinate and anaerobic growth on glycerol-fumarate (G-F) were carried out as previously described [27,28]. A Klett-Summerson colorimeter equipped with a no. 66 filter was used to monitor bacterial growth.
SDS-PAGE and Flavin Quantification
Protein concentrations were estimated by the Lowry method [29] with the inclusion of 1 % (w/v) sodium dodecyl sulfate in the mixture [30]. 30 µg of protein was resolved on a 12 % SDS-PAGE gel and visualized by Coomassie blue staining. Fluorometric quantification of the covalent flavin of Sdh was carried out in triplicate as described [31], using 5 mg of protein as starting material.
Enzyme Assays
Succinate dependent reduction of MTT (2-(4,5-dimethyl-2-thiazolyl)-3,5-diphenyl-2H-tetrazolium bromide; Δ = 17 mM−1 cm−1) was measured spectrophotometrically at 570 nm in the presence of 750 µM PMS (phenazine methosulfate) and 0.1 % Triton X-100 [32]. Succinate-dependent reduction of Q0 (2,3-dimethoxy-5-methyl-1,4-benzoquinone; Δ = 0.73 mM−1 cm−1) and fumarate-dependent oxidation of lapachol (LPCH; 2-hydroxy-3-(3-methyl-2-butenyl)-1,4-naphthoquinone; Δ = 2.66 mM−1 cm−1), were monitored at 410 nm and 481 nm, respectively. Turnover numbers were calculated based on covalent flavin concentrations. All assays were carried out using isolated membranes enriched in SdhCDAB or FrdABCD.
HOQNO Fluorescence Quench (FQ) Titrations
2-n-heptyl-4-hyroxyquinoline-N-oxide (HOQNO) binding was monitored by fluorescence according to van Ark and Berden [33]. 2 µL aliquots of 0.1 mM HOQNO were added to the sample, and fluorescence was measured at an excitation wavelength of 341 nm and an emission wavelength of 479 nm. Titrations were performed at protein concentrations of 0.50, 0.75, and 1.00 mg mL−1. The concentration of HOQNO binding sites and the dissociation constant of HOQNO were estimated by fitting the data according to Okun et al. [34,35]. The number of dissociable inhibitor binding sites for FrdABCD was assumed to be 1 [34,36]. The dissociation constants for HOQNO binding were not determined for SdhCDAB as HOQNO does not bind with high affinity to SdhCDAB.
Redox Titrations and EPR Spectroscopy
Redox titrations were carried out anaerobically under argon at 25 °C on SdhCDAB or FrdABCD-enriched membranes at a total protein concentration of approximately 30 mg mL−1 in 100 mM MOPS / 5 mM EDTA (pH 7.0). The following redox mediators were used at a concentration of 25 µM: quinhydrone, 2,6-dichloroindophenol, 1,2-naphthoquinone, toluylene blue, phenazine methosulfate, thionine, duroquinone, methylene blue, resorufin, indigotrisulfonate, indigodisulfonate, anthraquinone-2-sulfonic acid, phenosafranine, benzyl viologen, and methyl viologen. EPR spectra were recorded using either a Bruker Elexsys E500 or a Bruker ESP300 EPR spectrometer each equipped with an Oxford Instruments ESR900 flowing helium cryostat. Spectra of the [3Fe-4S] cluster were determined at 12 K while that of heme b were determined at 9 K, both at a microwave power of 20 mW, a frequency of 9.38 GHz, and a modulation amplitude of 10 Gpp at 100 kHz. Presented data were gathered from two independent potentiometric titrations for each variant enzyme.
3. Results
3.1 Variant enzyme design and expression
Comparison of the E. coli SdhCDAB X-ray crystallographic structure with endogenous UQ bound (PDB:1NEK) [3] with that of FrdABCD with the menasemiquinone HOQNO bound (PDB:1KF6) [2] reveals distinct differences between the two paralogs at the interface between the electron transfer subunit and the membrane intrinsic dimer. Figure 1B shows the difference in Q-site orientation relative to the [3Fe-4S] clusters between the two enzymes. The guanidinium moiety of SdhB-R205 is oriented towards the SdhCDAB Q-site, while the Nε-amide of FrdB-K228 points towards both the Frd quinone-binding site and the backbone carbonyl oxygen of FrdB-C204. Table 1 lists the residues examined in this study, along with their potential H-bonding partners. The Lys residue is conserved between both enzymes, while FrdB-S203 is equivalent to SdhB-R205. Reciprocal mutations were made at the SdhB-R205/FrdB-S203 position to determine whether residues at this position play a role in defining the [3Fe-4S] cluster reduction potential and/or bias for quinone reduction or quinol oxidation in the two enzymes. The conserved Lys residue was also examined using the substitutions FrdB-K228R, FrdB-K228E, FrdB-K228L and SdhB-K230L. SDS-PAGE and covalent flavin analyses on SdhCDAB and FrdABCD-enriched membranes indicated that all variant enzymes were overexpressed, assembled, and correctly targeted to the cytoplasmic membrane (Supplemental Fig. 1).
Table 1.
Hydrogen bonding partners for residues examined in present study. H-bonds listed are between the side chains of the residues in question and the indicated partners. PDB files 1NEK and 1KF6 were used to anaylze Sdh and Frd structures, respectively [2,3].
| Residue | H-bond partner | Distance (Å) |
|---|---|---|
| SdhB-R205 | Oδ1, SdhD-D82 | 3.1 |
| HOH310 | 3.5 | |
| HOH328 | 2.7 | |
| HOH319 | 2.8 | |
| SdhB-K230 | Backbone C=O, SdhB-C206 | 3.1 |
| HOH351 | 3.0 | |
| HOH313 | 2.8 | |
| FrdB-S203 | Nδ2, FrdB-N164 | 2.8 |
| FrdB-K228 | Backbone C=O, FrdB-C204 | 3.1 |
| Oε1, FrdB-Q225 | 2.7 | |
| O4, HOQNO | 3.1 | |
3.2 Growth experiments
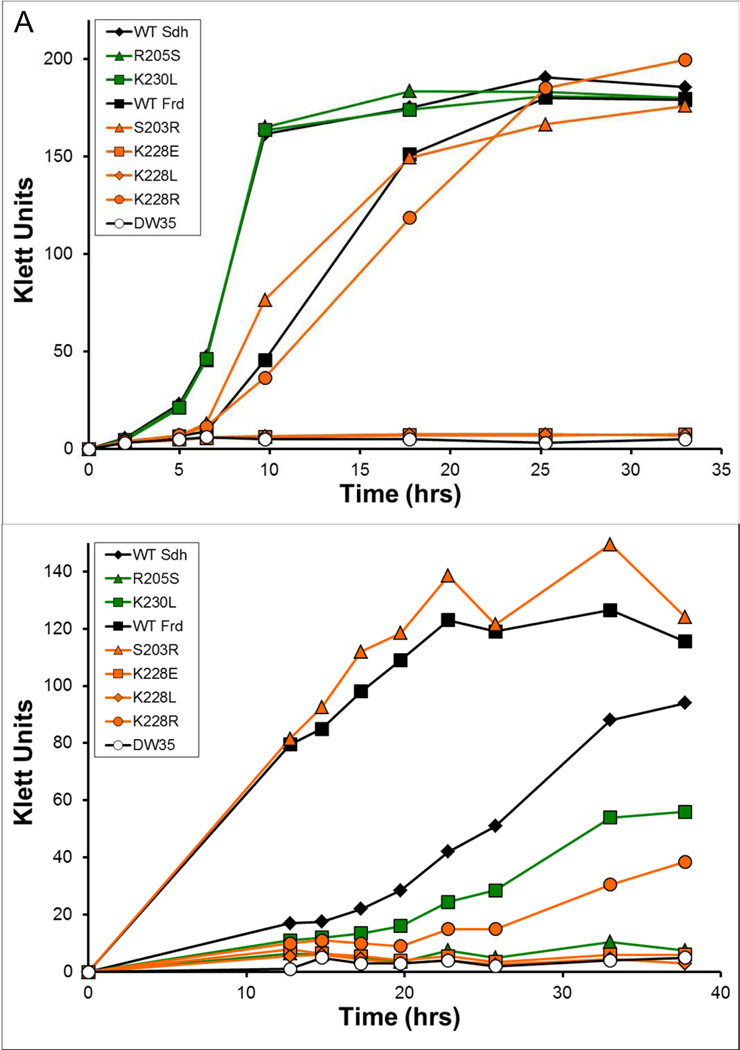
The in vivo activities of the variant enzymes were examined by monitoring their abilities to support growth in minimal medium (Fig. 2). The parent strain, DW35, lacks functional SdhCDAB and FrdABCD and therefore did not grow aerobically with succinate as electron donor nor anaerobically with G-F as the donor-acceptor pair [24]. Overexpression of SdhCDAB in DW35 resulted in better growth on succinate whereas overexpression of FrdABCD led to better growth on G-F. This was expected based on previous findings [26]. Figure 2A shows that Sdh enzymes containing the point mutations SdhB-R205S and SdhB-K230L were able to support aerobic respiratory growth on succinate at a rate similar to the WT enzyme. The Frd constructs carrying the FrdB-S203R and FrdB-K228R mutations were also able to support growth on succinate. However, mutation of FrdB-K228 to Glu or Leu abolished the ability to complement growth. Differences in growth were more apparent when the cells were forced to respire anaerobically on G-F (Fig. 2B). FrdB-S203R was the only variant that supported comparable growth on G-F relative to the WT Frd enzyme. The SdhB-R205S, FrdB-K228E and FrdB-K228L variants did not support growth on G-F, but limited growth was observed when DW35 cells expressed the SdhB-K230L or FrdB-K228R variant enzymes.
Figure 2.
Growth of E. coli DW35 complemented with plasmids encoding wild-type and variant enzymes. Aerobic growth on succinate minimal medium (A) and anaerobic growth on glycerol-fumarate minimal medium (B) were examined. Growth curves were collected from two separate and independent bacterial cultures at the times indicated.
Enzyme activity
The non-physiological succinate:PMS/MTT assay, which measures enzymatic succinate oxidation activity and only requires the functional SdhAB or FrdAB domain, is an excellent reporter assay for correct enzyme assembly. Table 2 shows that all membrane preparations containing the Sdh and Frd variants had significant succinate:PMS/MTT activities, indicating that the mutations in SdhB/FrdB did not disrupt enzyme targeting and assembly to the cytoplasmic membrane. Interestingly, the SdhB-K230L variant had a succinate:PMS/MTT turnover rate that was approximately 80% higher than the WT Sdh enzyme.
Table 2.
Steady-state kinetics and HOQNO binding of variant enzymes. In the succinate:PMS/MTT assay, succinate was used as electron donor and reduction of MTT was measured in the presence of 750 µM PMS. In the succinate:Q0 and lapachol:fumarate assays, rates of Q0 reduction and lapachol oxidation were monitored at varying concentrations of the quinone analogue. kcat and Km values were obtained by plotting v against [S] and fitted to the Michaelis–Menten equation using Solver and at least 8 activity measurements at different Q0 and lapachol concentrations. In HOQNO titrations, Frd enzymes were titrated with increasing amounts of HOQNO until increases in fluorescence were observed. This was repeated at protein concentrations of 0.50 mg mL−1, 0.75 mg mL−1 and 1.00 mg mL−1 such that the concentration of binding sites and dissociation constants (Kd) can be determined. Error: <10 % of reported value. ND: not determined.
| Succinate:PMS/MTT | Succinate:Q0 | Lapachol:Fumarate | Kd, HOQNO | |||
|---|---|---|---|---|---|---|
| kcat (s−1) | kcat (s−1) | Km (mM) | kcat (s−1) | Km (mM) | (nM) | |
| WT Sdh | 13.4 | 33.9 | 0.15 | 11.9 | 0.10 | ND |
| SdhB-R205S | 17.4 | 15.0 | 0.22 | 5.1 | 0.10 | ND |
| SdhB-K230L | 24.5 | 35.6 | 0.17 | 9.2 | 0.07 | ND |
| WT Frd | 6.6 | 7.1 | 0.06 | 126.1 | 0.22 | 5 |
| FrdB-S203R | 8.6 | 6.3 | 0.09 | 53.5 | 0.14 | 5 |
| FrdB-K228E | 4.4 | 0.7 | ND | 0.1 | ND | 110 |
| FrdB-K228L | 5.4 | 0.6 | ND | 0.1 | ND | 110 |
| FrdB-K228R | 7.3 | 5.5 | 0.06 | 0.6 | ND | 80 |
Measurements of succinate-dependent reduction of Q0 (succinate:Q0 oxidoreductase activity) and fumarate-dependent oxidation of lapachol (LPCH:fumarate oxidoreductase assay) address the ability of the variant enzymes to perform catalysis with quinones in opposing directions (Table 2). In both assays, the SdhB-K230L variant showed catalytic values that were comparable to the WT Sdh enzyme. The reciprocal variants SdhB-R205S and FrdB-S203R both retained significant catalytic activities (>40% of respective WT enzymes). The most significant decreases in catalytic function were observed in the FrdB-K228E and FrdB-K228L variants, which had approximately 10% of WT succinate:Q0 activity but negligible LPCH:fumarate activity. The related variant, FrdB-K228R, retained ~80% of WT enzyme activity in the succinate:Q0 assay but had negligible LPCH:fumarate activity. If a quinone analog with a 2-isoprenoid side chain (Q2) was used, the membrane-bound FrdB-K228R enzyme retained 43% of its succinate-Q2 oxidoreductase activity, in reasonable agreement with the Q0-analogue. However, if the membranes were treated with even a small amount (0.006%) of Triton X-100, the FrdB-K228R enzyme lost measurable activity within 10 minutes, suggesting the stability of the enzyme had been compromised in this variant.
3.3 HOQNO binding
We examined the quinone binding affinities of the FrdB variants by carrying out FQ titrations of the enzyme using the menaquinol analogue HOQNO. This compound fluoresces in the unbound state and when bound to the protein, its fluorescence is quenched. Membrane preparations from DW35 cells devoid of Sdh and Frd expression, as well as DW35 carrying high-copy number plasmids encoding the functional sdh operon, do not bind HOQNO and thus were not examined. Table 2 shows the WT Frd enzyme bound HOQNO with an apparent dissociation constant (Kd) of 5 nM, consistent with previous studies [36]. No change in the affinity for HOQNO was observed in the FrdB-S203R variant. All three FrdB-K228 variants, including FrdB-K228R, caused the Kd value to increase more than 10-fold (80–110 nM range) compared to the WT Frd enzyme, in agreement with the role of FrdB-K228 in quinone binding [23].
3.4 EPR spectroscopy of the [3Fe-4S] cluster and heme b
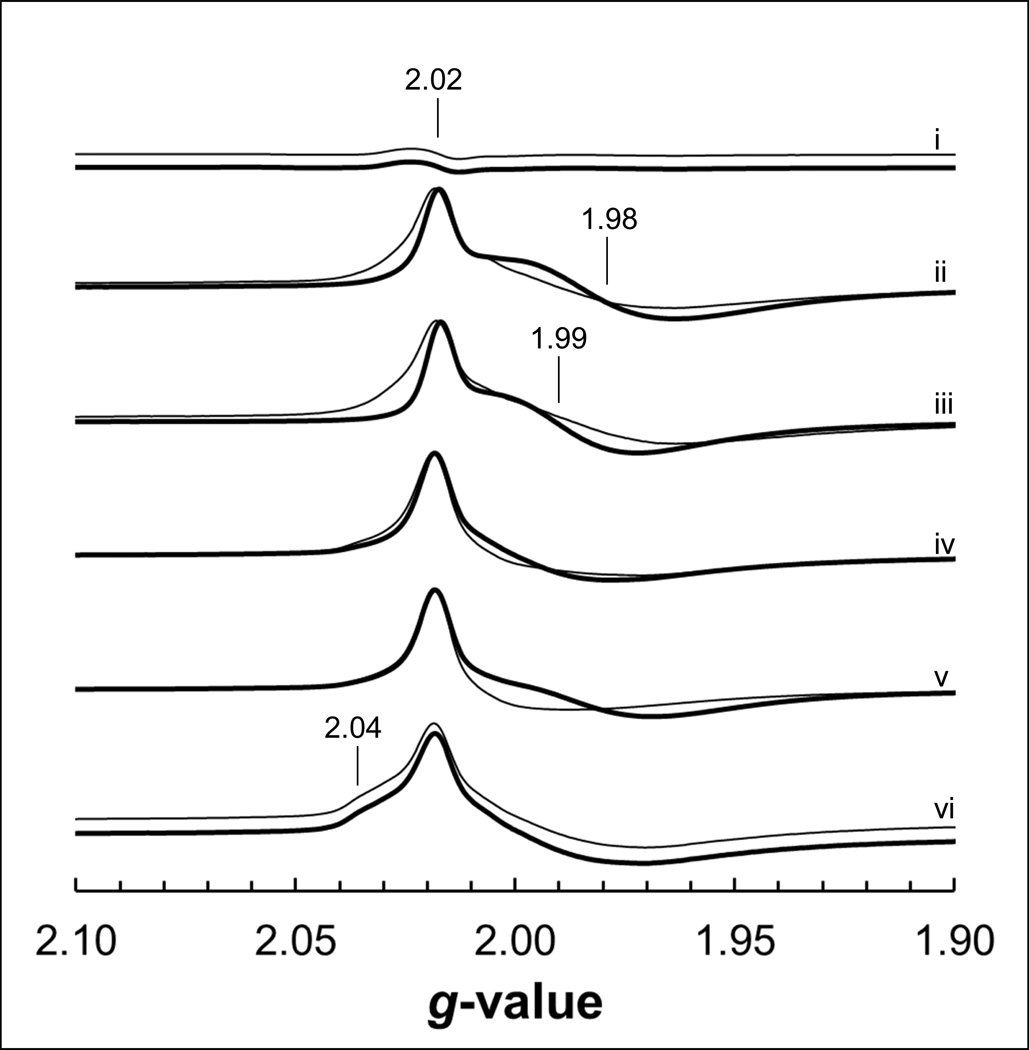
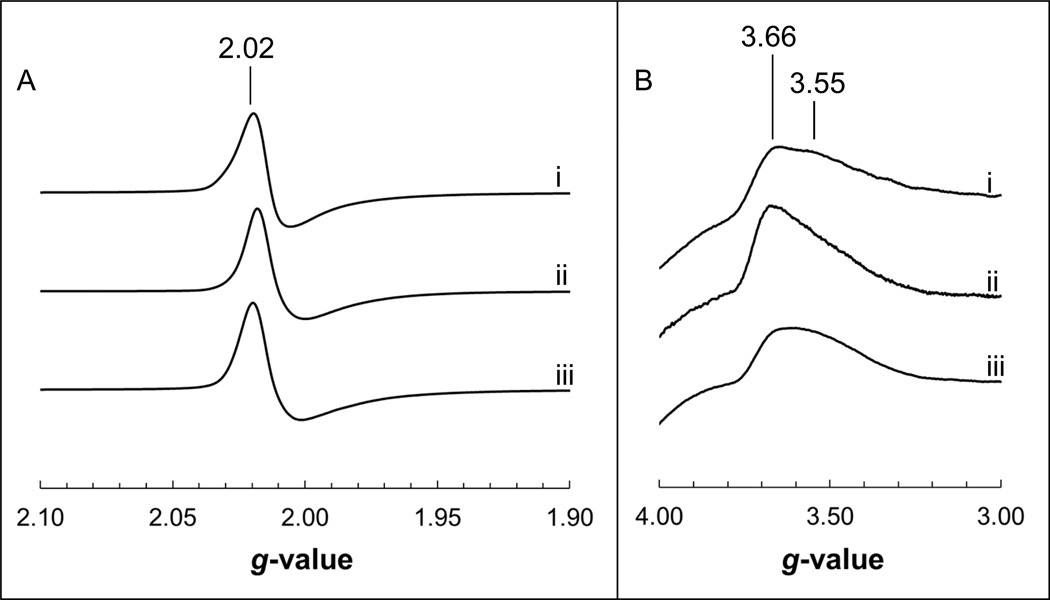
The cofactors which are in closest proximity to the variants studied herein are the [3Fe-4S] clusters of Frd and Sdh, as well as the heme b moiety of Sdh. The biophysical properties of these cofactors were examined by EPR spectroscopy to probe structure-function relationships in the SdhB/FrdB variants. The wild-type enzymes exhibit broadly similar oxidized spectra corresponding to the [3Fe-4S] cluster, comprising a peak at g = 2.02 and a trough immediately upfield which is significantly broader in the case of FrdABCD [37]. In the FrdB-K228E/ L/R variants, the EPR line shapes of the [3Fe-4S] cluster were narrower than that of the WT Frd enzyme, and is most noticeable in the FrdB-K228L variant (note the position of the trough in the absence of HOQNO in Fig. 3). In addition, the FrdB-K228R variant also showed a distinct shoulder at g = 2.04 that is absent in the WT signal. No major changes were observed in the oxidized spectra of the SdhB-R205S, SdhB-K230L and FrdB-S203R variants (Figs. 3 and 4).
Figure 3.
EPR spectra of the oxidized [3Fe-4S] cluster in Frd. Membranes from DW35 (i) and those overexpressing WT Frd (ii), FrdB-S203R (iii), FrdB-K228E (iv), FrdB-K228L (v) and FrdB-K228R (vi) were incubated for 5 minutes after addition of ethanol (thin lines) or 0.5 mM HOQNO/ethanol (thick lines), followed by addition of 0.2 mM ferricyanide and incubating for 2 an additional minutes. Spectral amplitudes were adjusted to the oxidized WT Frd signals for comparison.
Figure 4.
EPR spectra of the oxidized [3Fe-4S] (A) and heme b (B) in Sdh. Spectra of WT Sdh (i), SdhB-R205S (ii) and SdhB-K230L (iii) were obtained during redox titrations wherein Sdh was fully oxidized (ambient potential poised at least 100 mV higher than the experimentally determined midpoint potential). Spectral amplitudes were adjusted to the oxidized WT Sdh signals for comparison.
As reported previously [34,36,38] and observed in Fig. 3, the addition of HOQNO elicits an additional peak trough at g = 1.98 in the [3Fe-4S] cluster signal that is specific to the Frd enzyme. As expected, the addition of HOQNO to the FrdB-S203R mutant resulted in appearance of a peak-trough signal; however, it is centered at g = 1.99. Surprisingly, the peak-trough signal did not appear when we added HOQNO to the FrdB-K228R and FrdB-K228E variants, but appeared when HOQNO was added to the FrdB-K228L variant.
We also examined the oxidized heme b signal in the mutant Sdh variants (Fig. 4B). The WT Sdh enzyme exhibits a broad signal that comprises two overlapping peaks located at g = 3.66 and g = 3.55 [6,39,40]. In the SdhB-R205S variant, the two components observed in the wild-type enzyme collapse into a single archetypal highly anisotropic low spin (HALS) heme spectrum with a single ramp-type feature corresponding to gz [41,42]. Such an effect has previously been observed in Sdh variants that affect the proton channel [39] and the planarity of the heme [40]. No major changes were observed in the oxidized heme spectrum of the SdhB-K230L variant.
Finally, the reduction potentials (Em values) of the [3Fe-4S] cluster and heme b were determined and are shown in Table 3. The SdhB-R205S variant caused the Em values of the [3Fe-4S] cluster and the heme b to decrease by 39 mV and 34 mV, respectively. The reciprocal variant, FrdB-S203R, did not cause a change in the Em value of the [3Fe-4S] cluster. The SdhB-K230L variant only caused modest changes in the Em values of the [3Fe-4S] cluster and heme, which were decreased by 15mV and increased by 15mV, respectively. In Frd, mutation of the equivalent Lys residue resulted in much larger variations in the Em value of the [3Fe-4S] cluster. The FrdB-K228R variant, which likely retains a positive charge, elicited a modest 18 mV decrease in its Em value. The FrdB-K228E and FrdB-K228L variants, both of which eliminate a basic residue whose side chain interacts with MQ and the backbone amide oxygen of FrdB-Cys104, caused the Em value of the [3Fe-4S] cluster to decrease to −172mV and −177mV, respectively.
Table 3.
Midpoint potentials of the [3Fe-4S] cluster and heme b. Redox titrations on membrane preparations containing each construct were carried out at pH 7 and fitted to the Nernst equation. 200 µL samples poised at varying reduction potentials were frozen with liquid nitrogen-chilled ethanol and analyzed by EPR. The signal at g = 2.02 was used to determine the midpoint potential of the [3Fe-4S] cluster while the g = 3.66 signal was used for the heme b. Since Frd does not assemble a heme b cofactor, no values were reported. The error in Em values is approximately ± 10 mV.
| [3Fe-4S] | Heme b | |
|---|---|---|
| WT Sdh | +67 | +10 |
| SdhB-R205S | +28 | −24 |
| SdhB-K230L | +52 | +25 |
| WT Frd | −67 | -- |
| FrdB-S203R | −67 | -- |
| FrdB-K228E | −172 | -- |
| FrdB-K228L | −177 | -- |
| FrdB-K228R | −85 | -- |
4. Discussion
In this study, we examined the equivalent residues SdhB-R205 and FrdB-S203, as well as the conserved pair SdhB-K230 and FrdB-K228. These residues are located at the interface between the soluble and membrane anchor domains and allow us to study: i) protein control of cofactor midpoint potentials because of their close proximity to the [3Fe-4S] cluster (and the heme b in Sdh), ii) quinone binding and oxidation/reduction, and iii) differences in Sdh and Frd that lead to directionality of ET and catalysis.
4.1 FrdB-K228 and SdhB-K230
Based on the structural alignment depicted in Figure 1 and the H-bonding partners listed in Table 1, we hypothesized that quinone binding and oxidation/reduction would be impaired in the FrdB-K228 variants. A previous study had shown the FrdB-K228R and FrdB-K228L variants were unable to reduce or oxidize exogenous quinone species [23]. In agreement, the FrdB-K228L variant examined herein also had negligible succinate:Q0 and LPCH:fumarate activities. The FrdB-K228R variant showed significant loss of LPCH:fumarate activity, but surprisingly retained significant succinate:Q0 activity when assayed in membranes (Table 2). Upon detergent treatment, the enzyme rapidly lost activity, consistent with the negligible activity (<1%) observed with the purified enzyme [23]. In addition to the lack of LPCH:fumarate activity shown here, the FrdB-K228R variant is also unable to utilize MQ1H2 and UQ1 as substrates [23]. The decreased activity of the FrdB-K228R membrane-bound enzyme may reflect an altered pKa for the reaction since an Arg residue may have a higher pKa than the Lys residue that is normally present. Overall, the loss of activity in the FrdB-K228 variant enzymes suggests the importance of an amino acid side chain with a basic pKa value at this position. The extended H-bonding network involving FrdB-C204, FrdB-Q225, FrdD-W14, and MQ (Supplemental Fig. 2A) most likely helps to decrease the pKa of FrdB-K228 to allow proton exchange at physiological pH. We also examined whether the FrdB-K228 variants, similar to the FrdC-E29L variant, were trapped in an intermediary state during catalysis where the menasemiquinone is stabilized [36,38]. However, in Q-pool coupling assays using dithionite-fumarate as the donor-acceptor pair, we did not observe significant increases in the semiquinone EPR spectra of the variants (data not shown). The presence of the semiquinone EPR signal itself is in stark contrast to the experiments done on the E. coli respiratory nitrate reductase, wherein mutation of a Lys residue that hydrogen bonds to MQ/UQ abolishes the semiquinone signal in redox titrations [43,44].
Mutation of FrdB-K228 also demonstrated that this residue plays a pivotal role in defining the biophysical properties of the [3Fe-4S] cluster. The Glu/Leu/Arg variants all resulted in changes in the EPR line shape of the oxidized [3Fe-4S] spectrum, which suggests subtle changes in the H-bonding network and microenvironment surrounding FrdB-K228, the [3Fe-4S] cluster and the MQ molecule. In addition, the expected appearance of an EPR peak-trough signal at g = 1.98 upon HOQNO addition is either attenuated or absent, consistent with the observed increases in the Kd for HOQNO [34,36]. The FrdB-K228L and FrdB-K228E variants also elicited large decreases in the Em values of the [3Fe-4S] cluster (ΔEm = −110 mV and −105 mV respectively) while only a moderate change was observed in the FrdB-K228R variant (ΔEm = −18 mV). The magnitude of change in the Em value of the [3Fe-4S] cluster in the FrdB-K228E/L variants is consistent with disruption of a H-bonding network connected to the iron-sulfur cluster and the MQ molecule [17,45,46].
In Sdh, the conserved residue SdhB-K230 does not interact with the Q-site. The residue was once thought to be a component of a proposed H+ channel leading to the Q-site [47], but site-directed mutagenesis and crystallography data suggest otherwise [39,48]. Not surprisingly, we did not observe changes in catalytic activities in the SdhB-K230L variant. It had been suggested the redox properties of the [3Fe-4S] cluster might be altered in the SdhB-K230L variant [48]. Indeed, given the FrdB-K228L variant removed an H-bond donor to FrdB-C204 (a ligand to the [3Fe-4S] cluster) and resulted in a 110 mV decrease in the Em value of the [3Fe-4S], we would also expect the parallel mutation in Sdh to yield a similar shift in Em value. However, we were surprised that this variant only elicited a 15 mV decrease in the Em value of the [3Fe-4S] cluster compared to the WT Sdh enzyme. Since the Lys residue points into a hydrophilic milieu, the likely explanation is that additional water molecules can fill the void in the SdhB-K230L variant when the amino group is removed.
4.2 FrdB-S203 and SdhB-R205
We constructed the reciprocal mutations FrdB-S203R and SdhB-R205S in hopes of replicating a recent study on the SdhB-H207/FrdB-T205 pair that elicited an opposing effect on the [3Fe-4S] cluster Em value [12]. Although the SdhB-R205S variant decreased the Em values of both the [3Fe-4S] cluster and heme b, no change in the Em value of the former was observed in the FrdB-S203R variant. The small changes in Em values are consistent with minor perturbations within the protein milieu without disrupting the H-bonding network surrounding the [3Fe-4S] cluster or the heme [12,39,40]. The SdhB-R205S variant also had a noticeable effect on the EPR lineshape of the heme. The shift towards the “sharp” species at g = 3.66 may be attributed to the heme adopting a more planar, rather than a saddled or mixed, conformation [40]. We have also shown that variants that perturb the H-bonding network involving the water channel caused the low-spin oxidized heme b spectrum to shift towards the g = 3.66 species [39], as was observed here.
The Sdh structure (PDB:1NEK) shows a distinct water channel leading from the cytoplasm, through the enzyme, to the Q-site and presumably acts as a proton shuttle for UQ reduction [3,47,48]. The guanidinium side chain of SdhB-R203 forms H-bonds to the three terminal water molecules (HOH310, HOH328, HOH319) closest to UQ in the native structure, as well as to the side chain of SdhD-D82. The latter residue is involved in a complex H-bonding network that also includes SdhCR31, SdhD-Y83 and UQ (Supplemental Fig. 2B), and its replacement by Leu abolished 98% and 85% of succinate:Q0 and plumbagin:fumarate activities, respectively [28]. If the role of SdhB-R205 is to stabilize the water molecules closest to UQ for H+ exchange, then this explains why the SdhB-R205S variant caused a larger decrease in succinate:Q0 activity than the FrdB-S203R variant. Given that both reciprocal variants can catalyze in vitro reactions and complement the DW35 strain in vivo, it is obvious that this pair of equivalent residues is non-essential for enzyme function. Furthermore, there does not appear to be any correlation between enzyme turnover and the midpoint potentials of any of the redox cofactors in Sdh and Frd.
4.3 Conclusions
We have examined equivalent residues in Sdh and Frd which have evolutionarily diverged (SdhB-R205/FrdB-S203) or remained conserved (SdhB-K230/FrdB-K228). These two pairs of residues provide an interesting insight into the bidirectional, but biased, enzymatic functionalities of these two homologous enzymes because they are positioned between the highly conserved electron transfer subunit and the highly variable membrane anchor domain. Although SdhB-K230 and FrdB-K228 have remained conserved throughout evolution, their roles have clearly diverged, reflecting the different locations of the Q-sites in Sdh and Frd. The former residue is a component of a water network that may be involved in shuttling H+ towards the Q-site in Sdh. In Frd, the Lys residue is involved in quinone binding; in addition, the hydrogen bonding functionality of this residue to the backbone amide bond oxygen of FrdB-C204 appears to be an important contributor in establishing the midpoint potential of the [3Fe-4S] cluster.
Supplementary Material
Acknowledgments
This work was funded by the Canadian Institutes of Health Research (grant MDP89735 to J.H.W.), the National Institutes of Health (grant GM61606 to G.C.) and the Department of Veterans Affairs, Office of Research and Development, Biomedical Laboratory Research Division (Merit award BX001077 to G.C.). Infrastructure funding was provided by the Canada Foundation for Innovation. Q.T. was supported by an Alberta Heritage Foundation for Medical Research and Canadian Institutes of Health Research Graduate Studentships.
Abbreviations
- Em
midpoint potential
- EPR
electron paramagnetic resonance
- ET
electron transfer
- FAD
flavin adenine dinucleotide
- FQ
fluorescence quench
- FrdABCD
fumarate reductase
- G-F
glycerol-fumarate
- HOQNO
2-n-heptyl-4-hyroxyquinoline-N-oxide
- LPCH
lapachol
- MQ/MQH2
oxidized/reduced menaquinone
- Q-site
quinone binding site
- SdhCDAB
succinate dehydrogenase
- UQ/UQH2
oxidized/reduced ubiquinone
References
- 1.Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. Structure of the Escherichia coli fumarate reductase respiratory complex. Science. 1999;284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- 2.Iverson TM, Luna-Chavez C, Croal LR, Cecchini G, Rees DC. Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J. Biol. Chem. 2002;277:16124–16130. doi: 10.1074/jbc.M200815200. [DOI] [PubMed] [Google Scholar]
- 3.Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 4.Cecchini G, Schroder I, Gunsalus RP, Maklashina E. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta. 2002;1553:140–157. doi: 10.1016/s0005-2728(01)00238-9. [DOI] [PubMed] [Google Scholar]
- 5.Maklashina E, Cecchini G. The quinone-binding and catalytic site of complex II. Biochim. Biophys. Acta. 2010;1797:1877–1882. doi: 10.1016/j.bbabio.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. Escherichia coli succinate dehydrogenase variant lacking the heme b. Proc. Natl. Acad. Sci. 2007;104:18007–18012. doi: 10.1073/pnas.0707732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maklashina E, Iverson TM, Sher Y, Kotlyar V, Andrell J, Mirza O, Hudson JM, Armstrong FA, Rothery RA, Weiner JH, Cecchini G. Fumarate reductase and succinate oxidase activity of Escherichia coli complex II homologs are perturbed differently by mutation of the flavin binding domain. J. Biol. Chem. 2006;281:11357–11365. doi: 10.1074/jbc.M512544200. [DOI] [PubMed] [Google Scholar]
- 8.Condon C, Cammack R, Patil DS, Owen P. The succinate dehydrogenase of Escherichia coli. Immunochemical resolution and biophysical characterization of a 4-subunit enzyme complex. J. Biol. Chem. 1985;260:9427–9434. [PubMed] [Google Scholar]
- 9.Cheng VW, Ma E, Zhao Z, Rothery RA, Weiner JH. The iron-sulfur clusters in Escherichia coli succinate dehydrogenase direct electron flow. J. Biol. Chem. 2006;281:27662–27668. doi: 10.1074/jbc.M604900200. [DOI] [PubMed] [Google Scholar]
- 10.Morningstar JE, Johnson MK, Cecchini G, Ackrell BA, Kearney EB. The high potential iron-sulfur center in Escherichia coli fumarate reductase is a three-iron cluster. J. Biol. Chem. 1985;260:13631–13638. [PubMed] [Google Scholar]
- 11.Cammack R, Patil DS, Weiner JH. Evidence that centre 2 in Escherichia coli fumarate reductase is a [4Fe-4S]cluster. Biochim. Biophys. Acta. 1986;870:545–551. doi: 10.1016/0167-4838(86)90264-5. [DOI] [PubMed] [Google Scholar]
- 12.Ruprecht J, Iwata S, Rothery RA, Weiner JH, Maklashina E, Cecchini G. Perturbation of the quinone-binding site of complex II alters the electronic properties of the proximal [3Fe-4S] iron-sulfur cluster. J. Biol. Chem. 2011;286:12756–12765. doi: 10.1074/jbc.M110.209874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amino H, Osanai A, Miyadera H, Shinjyo N, Tomitsuka E, Taka H, Mineki R, Murayama K, Takamiya S, Aoki T, Miyoshi H, Sakamoto K, Kojima S, Kita K. Isolation and characterization of the stage-specific cytochrome b small subunit (CybS) of Ascaris suum complex II from the aerobic respiratory chain of larval mitochondria. Mol. Biochem. Parasitol. 2003;128:175–186. doi: 10.1016/s0166-6851(03)00074-4. [DOI] [PubMed] [Google Scholar]
- 14.Iwata F, Shinjyo N, Amino H, Sakamoto K, Islam MK, Tsuji N, Kita K. Change of subunit composition of mitochondrial complex II (succinate-ubiquinone reductase/quinol-fumarate reductase) in Ascaris suum during the migration in the experimental host. Parasitol. Int. 2008;57:54–61. doi: 10.1016/j.parint.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Tomitsuka E, Kita K, Esumi H. An anticancer agent, pyrvinium pamoate inhibits the NADH-fumarate reductase system--a unique mitochondrial energy metabolism in tumour microenvironments. J. Biochem. 2012;152:171–183. doi: 10.1093/jb/mvs041. [DOI] [PubMed] [Google Scholar]
- 16.Rothery RA, Weiner JH. Alteration of the iron-sulfur cluster composition of Escherichia coli dimethyl sulfoxide reductase by site-directed mutagenesis. Biochemistry. 1991;30:8296–8305. doi: 10.1021/bi00098a003. [DOI] [PubMed] [Google Scholar]
- 17.Langen R, Jensen GM, Jacob U, Stephens PJ, Warshel A. Protein control of iron-sulfur cluster redox potentials. J. Biol. Chem. 1992;267:25625–25627. [PubMed] [Google Scholar]
- 18.Jensen GM, Warshel A, Stephens PJ. Calculation of the redox potentials of iron-sulfur proteins: the 2-/3-couple of [Fe4S*4Cys4] clusters in Peptococcus aerogenes ferredoxin Azotobacter vinelandii ferredoxin I, and Chromatium vinosum high-potential iron protein. Biochemistry. 1994;33:10911–10924. doi: 10.1021/bi00202a010. [DOI] [PubMed] [Google Scholar]
- 19.Kowal AT, Werth MT, Manodori A, Cecchini G, Schroder I, Gunsalus RP, Johnson MK. Effect of cysteine to serine mutations on the properties of the [4Fe-4S] center in Escherichia coli fumarate reductase. Biochemistry. 1995;34:12284–12293. doi: 10.1021/bi00038a024. [DOI] [PubMed] [Google Scholar]
- 20.Stephens PJ, Jollie DR, Warshel A. Protein Control of Redox Potentials of Iron-Sulfur Proteins. Chem. Rev. 1996;96:2491–2514. doi: 10.1021/cr950045w. [DOI] [PubMed] [Google Scholar]
- 21.Chen K, Bonagura CA, Tilley GJ, McEvoy JP, Jung YS, Armstrong FA, Stout CD, Burgess BK. Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential. Nat. Struct. Biol. 2002;9:188–192. doi: 10.1038/nsb751. [DOI] [PubMed] [Google Scholar]
- 22.Cheng VW, Rothery RA, Bertero MG, Strynadka NC, Weiner JH. Investigation of the environment surrounding iron-sulfur cluster 4 of Escherichia coli dimethylsulfoxide reductase. Biochemistry. 2005;44:8068–8077. doi: 10.1021/bi050362p. [DOI] [PubMed] [Google Scholar]
- 23.Maklashina E, Hellwig P, Rothery RA, Kotlyar V, Sher Y, Weiner JH, Cecchini G. Differences in protonation of ubiquinone and menaquinone in fumarate reductase from Escherichia coli. J. Biol. Chem. 2006;281:26655–26664. doi: 10.1074/jbc.M602938200. [DOI] [PubMed] [Google Scholar]
- 24.Westenberg DJ, Gunsalus RP, Ackrell BA, Sices H, Cecchini G. Escherichia coli fumarate reductase frdC and frdD mutants. Identification of amino acid residues involved in catalytic activity with quinones. J. Biol. Chem. 1993;268:815–822. [PubMed] [Google Scholar]
- 25.Blaut M, Whittaker K, Valdovinos A, Ackrell BA, Gunsalus RP, Cecchini G. Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J. Biol. Chem. 1989;264:13599–13604. [PubMed] [Google Scholar]
- 26.Maklashina E, Berthold DA, Cecchini G. Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J. Bacteriol. 1998;180:5989–5996. doi: 10.1128/jb.180.22.5989-5996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer ME, Guest JR. Isolation and properties of fumarate reductase mutants of Escherichia coli. J. Bacteriol. 1973;114:563–570. doi: 10.1128/jb.114.2.563-570.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J. Biol. Chem. 2006;281:32310–32317. doi: 10.1074/jbc.M607476200. [DOI] [PubMed] [Google Scholar]
- 29.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 30.Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 31.Singer TP, Edmondson DE. Structure, properties, and determination of covalently bound flavins. Methods Enzymol. 1980;66:253–264. doi: 10.1016/0076-6879(80)66466-0. [DOI] [PubMed] [Google Scholar]
- 32.Kita K, Vibat CR, Meinhardt S, Guest JR, Gennis RB. One-step purification from Escherichia coli of complex II (succinate: ubiquinone oxidoreductase) associated with succinate-reducible cytochrome b556. J. Biol. Chem. 1989;264:2672–2677. [PubMed] [Google Scholar]
- 33.Van Ark G, Berden JA. Binding of HQNO to beef-heart sub-mitochondrial particles. Biochim. Biophys. Acta. 1977;459:119–127. doi: 10.1016/0005-2728(77)90014-7. [DOI] [PubMed] [Google Scholar]
- 34.Rothery RA, Weiner JH. Interaction of a menaquinol binding site with the [3Fe-4S] cluster of Escherichia coli fumarate reductase. Eur. J. Biochem. 1998;254:588–595. doi: 10.1046/j.1432-1327.1998.2540588.x. [DOI] [PubMed] [Google Scholar]
- 35.Okun JG, Lummen P, Brandt U. Three classes of inhibitors share a common binding domain in mitochondrial complex I (NADH:ubiquinone oxidoreductase) J. Biol. Chem. 1999;274:2625–2630. doi: 10.1074/jbc.274.5.2625. [DOI] [PubMed] [Google Scholar]
- 36.Rothery RA, Seime AM, Spiers AM, Maklashina E, Schroder I, Gunsalus RP, Cecchini G, Weiner JH. Defining the Q-site of Escherichia coli fumarate reductase by site-directed mutagenesis, fluorescence quench titrations and EPR spectroscopy. Febs. J. 2005;272:313–326. doi: 10.1111/j.1742-4658.2004.04469.x. [DOI] [PubMed] [Google Scholar]
- 37.Manodori A, Cecchini G, Schroder I, Gunsalus RP, Werth MT, Johnson MK. [3Fe-4S] to [4Fe-4S] cluster conversion in Escherichia coli fumarate reductase by site-directed mutagenesis. Biochemistry. 1992;31:2703–2712. doi: 10.1021/bi00125a010. [DOI] [PubMed] [Google Scholar]
- 38.Hagerhall C, Magnitsky S, Sled VD, Schroder I, Gunsalus RP, Cecchini G, Ohnishi T. An Escherichia coli mutant quinol:fumarate reductase contains an EPR-detectable semiquinone stabilized at the proximal quinone-binding site. J. Biol. Chem. 1999;274:26157–26164. doi: 10.1074/jbc.274.37.26157. [DOI] [PubMed] [Google Scholar]
- 39.Cheng VW, Johnson A, Rothery RA, Weiner JH. Alternative sites for proton entry from the cytoplasm to the quinone binding site in Escherichia coli succinate dehydrogenase. Biochemistry. 2008;47:9107–9116. doi: 10.1021/bi801008e. [DOI] [PubMed] [Google Scholar]
- 40.Tran QM, Fong C, Rothery RA, Maklashina E, Cecchini G, Weiner JH. Out of plane distortions of the heme b of Escherichia coli succinate dehydrogenase. PloS. One. 2012;7:e32641. doi: 10.1371/journal.pone.0032641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salerno JC. Cytochrome electron spin resonance line shapes, ligand fields, and components stoichiometry in ubiquinol-cytochrome c oxidoreductase. J. Biol. Chem. 1984;259:2331–2336. [PubMed] [Google Scholar]
- 42.Berry EA, Walker FA. Bis-histidine-coordinated hemes in four-helix bundles: how the geometry of the bundle controls the axial imidazole plane orientations in transmembrane cytochromes of mitochondrial complexes II and III and related proteins. J. Biol. Inorg. Chem. 2008;13:481–498. doi: 10.1007/s00775-008-0372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertero MG, Rothery RA, Boroumand N, Palak M, Blasco F, Ginet N, Weiner JH, Strynadka NC. Structural and biochemical characterization of a quinol binding site of Escherichia coli nitrate reductase A. J. Biol. Chem. 2005;280:14836–14843. doi: 10.1074/jbc.M410457200. [DOI] [PubMed] [Google Scholar]
- 44.Lanciano P, Magalon A, Bertrand P, Guigliarelli B, Grimaldi S. High-stability semiquinone intermediate in nitrate reductase A (NarGHI) from Escherichia coli is located in a quinol oxidation site close to heme bD. Biochemistry. 2007;46:5323–5329. doi: 10.1021/bi700074y. [DOI] [PubMed] [Google Scholar]
- 45.Denke E, Merbitz-Zahradnik T, Hatzfeld OM, Snyder CH, Link TA, Trumpower BL. Alteration of the midpoint potential and catalytic activity of the rieske iron-sulfur protein by changes of amino acids forming hydrogen bonds to the iron-sulfur cluster. J. Biol. Chem. 1998;273:9085–9093. doi: 10.1074/jbc.273.15.9085. [DOI] [PubMed] [Google Scholar]
- 46.Kolling DJ, Brunzelle JS, Lhee S, Crofts AR, Nair SK. Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters. Structure. 2007;15:29–38. doi: 10.1016/j.str.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J. Biol. Chem. 2006;281:7309–7316. doi: 10.1074/jbc.M508173200. [DOI] [PubMed] [Google Scholar]
- 48.Ruprecht J, Yankovskaya V, Maklashina E, Iwata S, Cecchini G. Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site. J. Biol. Chem. 2009;284:29836–29846. doi: 10.1074/jbc.M109.010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.