Abstract
Dentin matrix protein 1 (DMP1) is a key regulator for osteoblast differentiation and mineralization of the bone extracellular matrix (ECM). Osteocalcin (OCN) is one of the most abundantly expressed non-collagenous protein by osteoblasts in bone. In the present study, we generated a mouse model (OCN-DMP1) that overexpresses full-length DMP1 utilizing the OCN promoter. Expression of genes encoding osteogenic transcription factors and ECM proteins during early post-natal development in male OC-DMP1 and wild type (WT) mice was evaluated in femurs and calvaria. Bones were dissected from n=4 animals at 15, 30, 60 and 90-days of age. Total RNA was isolated, reverse transcribed, and real-time PCR analysis was performed. Results confirmed a difference (p<0.05) in osteogenic gene expression between OC-DMP1 and WT mice at the specified time points. Additionally, a unique osteogenic gene expression profile for calvaria and femur, representing intramembranous and endochondral bone formation was identified. These data suggest that bone-specific DMP1 overexpression has a profound influence on bone development by impacting osteogenic gene expression. This animal model presented here provides new opportunities for analysis of in vivo roles of DMP1 in bone.
Keywords: Osteogenesis, Biomineralization, Craniofacial, Development, Osteoblast, Differentiation
Introduction
Bone is a hierarchical biocomposite tissue composed of a type I collagen framework impregnated with non-collagenous proteins and carbonated hydroxyapatite [1]. Dentin matrix protein 1 (DMP1) is critical for osteoblast differentiation and mineralization of the bone extracellular matrix (ECM) [2–5]. Abnormalities in the expression of bone-matrix proteins results in pathological bone quality, such as in the case of osteogenesis imperfecta, rickets and osteomalacia [6–8]. Mammalian bone development occurs primarily by endochondral ossification in which bone replaces a cartilage scaffold, while the flat bones of the skull are formed by intramembranous ossification. Osteoblast expression of osteogenic genes is a carefully orchestrated process resulting in the formation of healthy bone. Osteocalcin (OCN) is the most abundantly expressed non-collagenous gene by osteoblasts [8] and therefore its promoter would be ideal to drive bone-specific expression of DMP1 during bone development [9].
In the present study, we generated a mouse model that overexpresses full-length DMP1 using the OCN promoter and analyzed for changes in the gene expression profile of key osteogenic transcription factors and ECM proteins during early post-natal development in male mice. Femurs and calvaria tissues were used to investigate changes in osteogenic genes during endochondral and intramembranous ossification, respectively.
Methods
Mouse model
Transgenic mice were generated to overexpress full-length DMP1 using the 1.3kDa promoter region of the BGLAP (OCN) gene (a kind gift from Dr. Gerard Karsenty). The OCN-driven DMP1 overexpression (OC-DMP1) mice were generated in a C57B/L6 genetic background. Genotyping of transgenic mice was performed using genomic DNA extracted from tail biopsies. Primers specific to the incorporated transgene were generated and used for PCR amplification using genomic DNA as the template. The amplicon was identified using an agarose gel. OC-DMP1 mice from one founder were mated until three OC-DMP1+/+ transgenic mouse line was generated. Line with the highest expression of DMP1 was used for further analysis. Age-matched wild type (WT) C57B/L6 male mice were used as a reference to characterize the genetic phenotype of the OC-DMP1 mice. All experiments were performed in accordance to the animal use protocol reviewed and approved by the Institutional Animal Care and Use Committee of the University of Illinois at Chicago, Chicago, IL, USA.
Real-time quantitative PCR
At the time points of 15, 30, 60, and 90-days post-natal, age and weight matched OC-DMP1 and WT mice were euthanized by CO2 asphyxiation followed by cervical dislocation. Femurs and calvaria were immediately dissected and transferred to TRIzol® Reagent (Life Technologies™). Femurs were mechanically cleaned of soft tissue, homogenized in nitrogen using a mortar and pestle, and suspended in TRIzol® Reagent. Calvaria were mechanically cleaned of soft tissue and the sutures removed with surgical scissors prior to homogenization and suspension in TRIzol® Reagent. Total RNA was extracted by following the provided manufacturer’s instructions for TRIzol® Reagent. RNA was subsequently purified using an RNeasy® Mini Kit (Qiagen™) according to the manufacturer’s protocol. SuperScript® III (Life Technologies™) was used for reverse transcription to generate cDNA from the purified femur and calvaria RNAs. Quantitative real-time PCR was performed with FastStart Universal SYBR Green Master (ROX) (Roche) in a StepOnePlus® (Applied Biosystems®) real-time PCR system. RT-PCR primer sequences are provided in Table 1.
Table 1.
Real-time PCR primer sequences
| Gene | Sense (5′→3′) | Anti-sense (5′→3′) |
|---|---|---|
| OCN | CTCCTGAGAGTCTGACAAAGCCTT | GCTGTGACATCCATTACTTGC |
| DMP1 | CCCAAAGGAACACAAGGAGA | TTCGCTGAGGTTTTGACCTT |
| RUNX2 | CCTGAACTCTGCACCAAGTC | GAGGTGGCAGTGTCATCATC |
| OSX | AGCGACCACTTGAGCAAACATC | CGGCTGATTGGCTTCTTCTTCC |
| ALP | GTGCCAGAGAAAGAGAGAGA | TTTCAGGGCATTTTTCAAGGT |
| COL1A1 | CCCCAACCCTGGAAACAGAC | GGTCAGGTTCAGTTGGTCAAAGG |
| COL1A2 | GCTCAGCTTTGTGGATACGCG | GTCAGAATACTGAGCAGCAAAG |
Statistics
Data are expressed as mean ± standard deviation with n=4 unique, independent samples being investigated for each gene. Two-way ANOVA were performed followed by two-tailed Student’s t-test post-hoc analysis using SPSS (IBM®) with p < 0.05 considered statistically significant.
Results
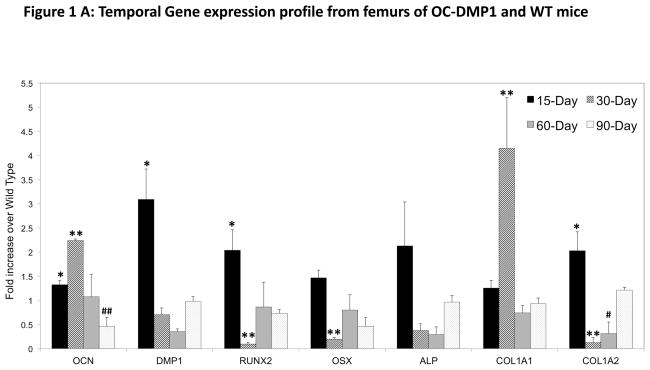
Temporal changes in the osteogenic gene profile was observed in the cDNA obtained from femurs and calvaria at the specified time points (Fig 1A & B). The OCN gene expression pattern in the femur significantly increased at 15-days and peaks with a greater than 2-fold increase in expression at 30-days with a subsequent decrease until OCN expression is significantly less than WT at 90-days. Interestingly, DMP1 expression peaks at 15-days with a 3-fold increase over age-matched WT, then stays within the range of WT expression for the duration of the study. Additionally, osteogenic transcription factors RUNX2 and OSX share a similar trend, although RUNX2 displays a significant increase at 15-days, while OSX does not. However, both transcription factors are significantly downregulated at 30-days. Femur collagen expression is unique for the COL1A1 and COL1A2 genes. COL1A1 is expressed significantly more in OC-DMP1 femurs only at 30-days, while COL1A2 expression is significantly increased at 15-days, but decreased at 30 and 60-day time points. Expression of ALP, an early gene marker for mineralized matrix formation did not significantly differ at any time point between mice.
Figure 1. Changes in the temporal gene expression profile in OC-DMP1 and WT mice.
(A) RNA was extracted from the femurs at defined time points as stated in the “Materials and Methods” section. Q-PCR was performed and the gene expression profiles display fold changes of OC-DMP1 mouse data normalized to wild type mouse gene expression for each gene, at each time point (n=4 unique femurs). Statistical significance indicates a difference (p<0.05) between OC-DMP1 and wild type gene expression for each gene at 15-days (*), 30-days (**), 60-days (#), 90-days (##) as determined by one-way ANOVA.
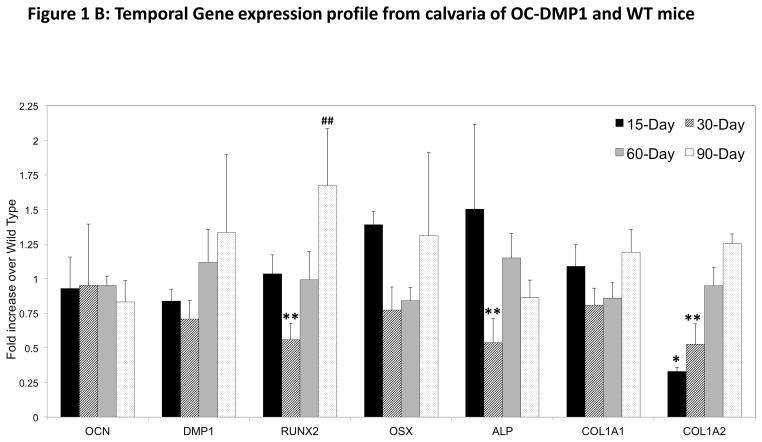
(B): RNA was extracted from the calvaria at defined time points. Q-PCR was performed and the gene expression profiles display fold changes of OC-DMP1 mouse data normalized to wild type mouse gene expression for each gene, at each time point (n=4 unique calvaria). Statistical significance indicates a difference (p<0.05) between OC-DMP1 and wild type gene expression for each gene at 15-days (*), 30-days (**), 60-days (#), 90-days (##) as determined by one-way ANOVA.
Osteogenic gene expression pattern in the calvaria is overall unique when compared to the femurs. At no point investigated for this study did OCN or DMP1 expression in OC-DMP1 calvaria change significantly in comparison to WT mice. Transcription factor RUNX2 did however display a significant decrease in expression at 30-days, followed by an upward trend leading to an increase at 90-days. Expression of OSX followed a similar temporal trend in the calvaria of OC-DMP1 mice, but was at no point different than WT mice. Similarly, COL1A1 did not display any significant changes at any time point. However, expression of COL1A2 increased consistently from 15 to 90-days of age, being downregulated in OC-DMP1 calvaria at 15 and 30-days of age. ALP was also downregulated in transgenic calvaira at the 30-day time point.
Discussion
Our objective was to i) investigate the implications of bone-specific DMP1 overexpression on the temporal expression of osteogenic genes and ii) determine if the expression pattern differed between bones formed by endochondral and intramembranous ossification in vivo. Our data indicate a difference in the temporal expression of osteogenic genes in OC-DMP1 during early post-natal development when compared to WT mice. Comparing the expression profiles of genes in the femurs formed by endochondral ossification and calvaria formed by intramembranous ossification further indicated that the difference seen in OC-DMP1 mice is unique to each bone system and the pronounced expression in femurs. Overall, the biggest differences in gene expression were seen at 15 and 30-days of age. These gene expression data are unique to the OC-DMP1 mice, however, they temporally support the findings of our group’s previous CMV-promoter driven ubiquitous overexpression of DMP1 mouse model [10].
In femurs, upregulation of genes encoding the ECM proteins OCN, DMP1 and COL1A2 at 15-days is most likely dictated by the increased RUNX2 expression at this time point. Interestingly, both transcription factors and COL1A2 were down regulated at 30-days, despite a significant increase in OCN expression. These findings may be explained through further investigation into possible feedback signaling loops or compensatory mechanisms within osteoblasts that may be triggered due to upregulation of the prior mentioned genes at the 15-day time point. Analogously, increased OCN expression at 30-days may be the result of delayed osteogenic signaling from the many upregulated genes at the 15-day time point. Furthermore, as OCN expression falls back to WT levels and then below that of WT levels at 60 and 90-days, respectively, it is reasonable to understand why most osteogeneic genes in OC-DMP1 femurs settle back to WT levels by 90-days of age.
Remarkably, OC-DMP1 calvaria display an entirely unique expression profile that differs from the femurs of the same animal. OCN expression does not deviate significantly from WT mice at any time point. In this context, it is reasonable to understand why there is far less deviation for all genes investigated in the calvaria when compared to the femurs. However, the 30-day time point does stand out as the minima for most genes investigated, which can be explained by the significantly decreased RUNX2 expression at 30-days. These data in the calvaria agree with femur data and suggests that this time point is of exceptional interest to our study of osteogenesis in the OC-DMP1 mouse model.
Conclusion
These data in our present study suggest that bone-specific DMP1 overexpression has a profound influence on bone development by perturbing osteogenic gene expression. These findings are unique to the ossification method by which the bone system is formed, with the osteogenic gene profile being altered to a greater extend in those formed via endochondral ossification. In a temporal context, it is reasonable to suggest that rapid bone formation occurring during those time points most influenced by DMP1-overexpression may result in altered bone structure or function. Further investigation is required to fully characterize bones formed by endochondral and intramembranous ossification in OC-DMP1 mice.
Footnotes
Declaration of Interest
All of the author’s of this manuscript declare no conflicts of interest with these data or findings.
References
- 1.Leaver AG, Triffitt JT, Holbrook IB. Newer knowledge of non-collagenous protein in dentin and cortical bone matrix. Clin Orthop Relat Res. 1975;(110):169–92. doi: 10.1097/00003086-197507000-00039. [DOI] [PubMed] [Google Scholar]
- 2.Narayanan K, Srinivas R, Ramachandran A, Hao J, Quinn B, George A. Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1. Proc Natl Acad Sci U S A. 2001;98(8):4516–21. doi: 10.1073/pnas.081075198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng JQ, Zhang J, Dallas SL, Lu Y, Chen S, Tan X, Owen M, Harris SE, MacDougall M. Dentin matrix protein 1, a target molecule for Cbfa1 in bone, is a unique bone marker gene. J Bone Miner Res. 2002;17(10):1822–31. doi: 10.1359/jbmr.2002.17.10.1822. [DOI] [PubMed] [Google Scholar]
- 4.Feng JQ, Ward LM, Lie S, Lu Y, Xie Y, Yuan B, Yu X, Rauch F, Davis SI, Zhang S, Rios H, Drezner MK, Quarles LD, Bonewald LF, White KE. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. 2006;38(11):1310–5. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling Y, Rios HF, Myers ER, Lu Y, Feng JQ, Boskey AL. DMP1 depletion decreases bone mineralization in vivo: an FTIR imaging analysis. J Bone Miner Res. 2005;20(12):2169–77. doi: 10.1359/JBMR.050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickson IR, Millar EA, Veis A. Evidence for abnormality of bone-matrix proteins in osteogenesis imperfecta. Lancet. 1975;2(7935):586–7. doi: 10.1016/s0140-6736(75)90173-7. [DOI] [PubMed] [Google Scholar]
- 7.Takagi Y, Veis A, Sauk JJ. Relation of mineralization defects in collagen matrices to noncollagenous protein components. Identification of a molecular defect in dentinogenesis imperfect. Clin Orthop Relat Res. 1983;(176):282–90. [PubMed] [Google Scholar]
- 8.Lian JB, Gundberg CM. Osteocalcin. Biochemical considerations and clinical applications. Clin Orthop Relat Res. 1988;(226):267–91. [PubMed] [Google Scholar]
- 9.Groot CG, Danes JK, Blok J, Hoogendijk A, Hauschka PV. Light and electron microscopic demonstration of osteocalcin antigenicity in embryonic and adult rat bone. Bone. 1986;7(5):379–85. doi: 10.1016/8756-3282(86)90259-0. [DOI] [PubMed] [Google Scholar]
- 10.Bhatia A, Albazzazz M, Espinoza Orias AA, Inoue N, Miller LM, Acerbo A, George A, Sumner DR. Overexpression of DMP1 accelerates mineralization and alters cortical bone biomechanical properties in vivo. J Mech Behav Biomed Mater. 2012;5(1):1–8. doi: 10.1016/j.jmbbm.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]