Abstract
Molecular imaging is a fast growing biomedical research that allows the visual representation, characterization and quantification of biological processes at the cellular and subcellular levels within intact living organisms. In vivo tracking of cells is an indispensable technology for development and optimization of cell therapy for replacement or renewal of damaged or diseased tissue using transplanted cells, often autologous cells. With outstanding advantages of bioluminescence imaging, the imaging approach is most commonly applied for in vivo monitoring of transplanted stem cells or immune cells in order to assess viability of administered cells with therapeutic efficacy in preclinical small animal models. In this review, a general overview of bioluminescence is provided and recent updates of in vivo cell tracking using the bioluminescence signal are discussed.
Keywords: Optical imaging, Bioluminescence, Cell tracking, In vivo imaging, Cell therapy
Introduction
The increasing density of imaging technologies, coupled with the development of cell therapies changes a revolution in cell tracking in vivo. Tracking of transplanted cells via imaging tools by visualizing the fate, function, migration, and homing of the cells using in vivo models would be highly valuable for research on cell-based therapies [1, 2]. Implanted cells can be visualized using anatomical imaging modalities, such as magnetic resonance imaging and computed tomography, which have excellent anatomical resolution and no depth limitation [3–6]; however, these techniques have limitations of low sensitivity, low throughput, and high cost of instrumentation. Due to the highest sensitivity and low cost, optical imaging has been used most commonly for in vivo cell tracking [7, 8].
Direct cell labeling can be a useful tool for evaluation of initial distribution, localization, and migration of administered cells; however, due to signal dilution by mitotic division of labeled cells and persistent signals from the labeled cells even after death, it cannot be used for long-term monitoring of cells [9]. Despite its weaknesses, the direct labeling strategy has been widely used for cell tracking due to the advantages of simplicity and feasibility. Fluorophores, nanoparticles, or radionuclides are commonly used for direct labeling. Radionuclides are very sensitive agents for direct labeling; however, there are radiation hazards and inability of long-term monitoring due to decay of the radionuclides. Among optical imaging tools, bioluminescence imaging (BLI) is an indirect cell labeling technique with reporter genes which is a promising method for cell tracking in small animal models. Bioluminescence is generated by conversion of chemical energy into visible light by the action of luciferase enzymes and their substrates in living animals. This applications in molecular imaging is referred to as BLI [10]. In vivo, BLI has been most commonly used by various researchers for the visualization of a variety of biological events because of its high sensitivity, relative ease of use, and low cost of instrumentation [11]. BLI is based on the detection of light emitted from cells that express light–generating enzymes, such as firefly luciferase (FLuc), Renilla luciferase (RLuc), Gaussia luciferase (GLuc), Metridia luciferase, Vargula luciferase, or bacterial luciferase [12–15]. Results from comparison of BLI reporter proteins are shown in Table 1.
Table 1.
Comparison of different luciferases
| Content | Firefly | Renilla | Gaussia | Bacterial |
|---|---|---|---|---|
| Origin of the species and name | Photoni firefly/Photinus pyralis and Vargula hilgendorfii | Sea pansy/Renilla reniformis | Copepod/Gaussia princeps, Metridia longa and Metridia pacifica | Photobacterium/Vibrio fischeri, haweyi and harveyi |
| Molecular weight | 62 kDa | 36 kDa | ~20 kDa | ~76 kDa |
| Emission max. | 562 nm (550-570 nm) |
482 nm (RLuc8, 547 nm) |
480–600 nm | ~490 nm |
| Emission type | Glow | Flash | Flash | Glow |
| Substrate | D-Luciferin | Coelenterazine | Coelenterazine | Not necessary |
| Cofactor | ATP, Mg2+ | No | No | ATP, FMNH2 |
| Secretion | No | No | Yes | No |
| Reference | [16, 17] | [18, 19] | [20, 21] | [22, 23] |
Weaknesses of BLI are poor spatial resolution, limited penetration depth, and low quantification accuracy due to loss and scatter of light in the body; and, so far, the weaknesses make visualization of inner organs of animal difficult and preclude clinical translation [6]. However, cell tracking using BLI can provide the highest sensitivity in small animal studies due to the absence of endogenous luciferase expression in mammalian cells [24]. This review provides a general overview of bioluminescence and recent updates on cell tracking using bioluminescence imaging modalities for in vivo animal studies.
Bioluminescent Reporter Genes
Firefly Luciferase
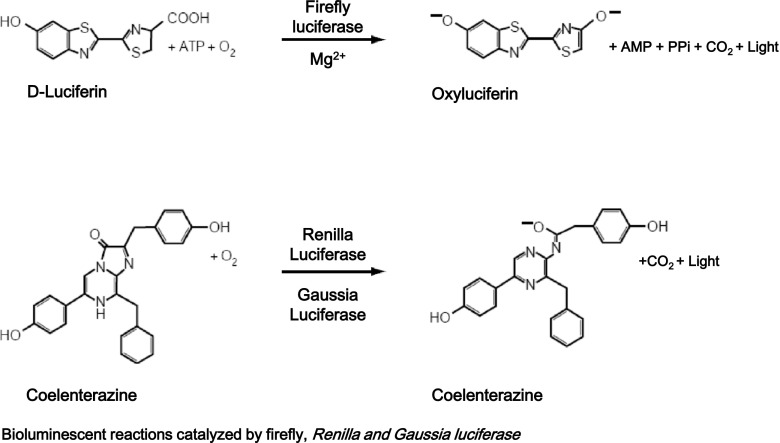
The FLuc-encoding gene cloned from the North American firefly, Photinus pyralis, is the most studied and well characterized bioluminescent reporter gene [16]. BLI using the FLuc-luciferin system has quickly become the standard procedure for in vivo cell tracking. D-Luciferin, a substrate for FLuc, is a small molecule that freely diffuses across the cell membrane. FLuc emits light in the presence of D-luciferin, ATP, Mg2+, and oxygen [17]; the reaction of FLuc and D-luciferin is illustrated in Fig. 1 [25]. Due to the glow kinetics of FLuc, the photon emission from the reaction persists for a relatively long period of time. The higher photon output of the modified FLuc, which is codon-optimized for mammalian cells, has been used prevalently for in vivo cell tracking. FLuc, with a red-shifted spectrum, has recently been investigated for achievement of better tissue penetration and resolution [26]. Light emitted from FLuc is in the yellow-green region, with a peak at 562 nm in basic media (range, 550–570 nm) [27]. However, it is a pH-sensitive enzyme, the peak can shift the emission to red (maximum at 620 nm) in acid media (pH 5–6), as well as by high temperature and heavy metal cations [28]. In addition, differential sensitivity of FLuc to oxidative stress in apoptotic cells has also been reported. Czupryna et al. [29] suggested that reactive oxygen species (ROS) were responsible for the reduction of FLuc activity and could be substantially altered in studies where pH levels and ROS levels become elevated. Also, Kitamaya et al. [30] reported on the thermal instability and pH-sensitive spectral property of firefly luciferase which can affect its use as a sensitive multicolor luminescence label or bioluminescence resonance energy transfer donor. In addition, the isolation of bright, red-shifted variants of Photinus pyralis luciferase in order to minimize light absorption and scattering by tissue for in vivo animal studies has also been reported [31–33].
Fig. 1.
Bioluminescence reactions catalyzed by firefly, Renilla, and Gaussia luciferase
Renilla Luciferase
RLuc purified from the marine organism Renilla reniformis (sea pansy) displays blue-green bioluminescence by catalysis of coelenterate luciferin (coelenterazine) oxidation [18]. RLuc, a 36-kDa monomeric enzyme, can catalyze the non-ATP-dependent oxidation of coelenterazine in the presence of oxygen for generation of a luminescence with a wavelength centered at 482 nm [18]. In contrast to photon emission by FLuc, due to its flash kinetics, photons produced by interaction of RLuc and coelenterazine last a relatively short period of time. For the first time, Bhaumik et al. [34] demonstrated the ability of BLI using RLuc-coelenterazine as a cell tracking strategy in living mice and also validated the possibility of dual imaging with expression of RLuc and FLuc in the same animal subjects. By these data, this strategy has been used in various studies where the tracking of two molecular events is needed, including the cell tracking of two cell populations through use of two different reporter genes. However, RLuc and its substrate also have some limitations, including instability, low permeability of the substrate, and high background signals caused by auto-oxidation [35]. The number of available substrate analogs for RLuc is increasing, and new ones having increased signal emission in vivo has been reported [36]. In this study, coelenterazine-f, −h, and -e analogs showed fourfold to eightfold greater RLuc activity, relative than coelenterazine-native, in RLuc-expressing cells. Light emitted by RLuc is easily absorbed and scattered by tissues due to blue light emitted by RLuc. Accordingly, the imaging performance suffers from poor sensitivity and spatial resolution. Several studies given an effort to solve this disadvantage, red-shifted variant RLuc, so-called RLuc8, was created and described as having an emission light with a maximum wavelength of 660 nm (547 nm peak), which also had greater stability and higher light emission than native RLuc [19, 37, 38].
Gaussia Luciferase
Gaussia luciferase (GLuc) is a small bright luminescent protein which involves chemical reaction higher than the luciferases from Renilla sp. and firefly. GLuc, uses the same substrate coelenterazine which is used for RLuc. GLuc does not require ATP for production of light in cells and tissues; therefore, it is a useful reporter [20]. GLuc has the nature of extracellular secretion; therefore, its concentration in blood shows correlation with expression level in a given biological system [21], and it can be used as a biomarker for longitudinal monitoring of therapy response of systemic metastases [39]. Limitations of GLuc include loss of signal (quenching) by nearby pigmented molecules such as hemoglobin and a flash-type bioluminescence reaction in vivo. To overcome the limitation of the quenching, an optimized microtiter well-based binding assay involving capture of GLuc from blood using a specific antibody has been developed, and use of the assay leads to approximately tenfold increased sensitivity [40]. In an effort to overcome the rapid light decay, Maguire et al. [20] isolated and characterized a GLuc mutant (GLucM43I), which catalyzes enhanced light stability in the presence of Triton-X 100 detergent, suitable for high-throughput applications. The secretory nature of GLuc affords evaluation of luciferase activity without lysis of cells or organisms and is a valuable characteristic for noninvasive detection and quantification of biological processes in animal models [41–43]. However, the secreting nature of the native GLuc enzyme significantly attenuates the in vivo bioluminescent signal from the cell expressing GLuc [44]. Elmer et al. [45] reported that this limitation could be overcome by modified Gluc by addition of a CD8 transmembrane domain, and also they produced the membrane-anchored form of GLuc-positive cells from both mouse and human primary T cells and demonstrated that the cells have a markedly superior in vivo BLI signal when compared with native GLuc positive T cells.
Bacterial Luciferase
Bacterial luciferase (Lux) is distinct in function from FLuc and RLuc. Lux is isolated from Photorhabdus luminescens; its luminescence reaction involves the oxidation of a long-chain aldehyde (RCHO) and reduced flavin mononucleotide, resulting in the production of oxidized flavin mononucleotide and a long-chain fatty acid (RCOOH), along with the emission of blue-green light at 490 nm [22, 23] which has been used for BLI study of bacterial infections [46]. It is a unique bioreporter system with no addition of exogenous substrate due to generation of its own substrate [47]. Contag et al. [48] first demonstrated the feasibility of in vivo BLI for the study of microbial pathogenesis These researchers achieved stable transformation of three strains of Salmonella typhimurium with a plasmid bearing the lux operon from Potorhabdus luminescens. They reported that bacterial pathogenesis appeared unaffected by the lux labeling, and the intensity of the light emitted by the bacteria enabled efficient in vivo monitoring. Less light will be transmitted from Lux than FLuc, which will affect quantification of imaging data because attenuation of light intensity in tissue is greater for short wavelengths than for long wavelengths. Francis et al. [49] constructed a novel Potorhabdus luminescence lux operon by cloning of a Gram-positive ribosome binding site and then generated bioluminescent bacteria labeled with the novel operon, which could be detected noninvasively in vivo. Min et al. [50, 51] developed a quantitative, noninvasive imaging technique that enables monitoring of bioluminescent bacterial migration in living subjects. Using this technique, studies involving the imaging protocol with luciferase-expressing E. coli were a useful approach for quantitative visualization of labeled bacteria in mouse models with tumor xenograft or metastases.
Imaging of Bioluminescence
BLI has grown to be the method for optical tracking of cells in small laboratory animals. BLI can enable simultaneous visualization of monitoring for the expression of two divergent luciferase proteins by use of their specific substrates, i.e., the dissimilarity in the substrates for each luciferase makes it possible to selectively distinguish between the luminescent reactions for each enzyme. The imaging dual luciferase gene (red codon optimized firefly luciferase and a green click beetle luciferase) activities in the same animals could reduce variations from individual differences of the experimental animals [52]. This type of experimental design is particularly useful when trying to acquire accurate, quantitative data on gene expression by simultaneously monitoring two events in real time. Wang et al. [53] performed the imaging of stem cell differentiation for prediction of the treatment response to stem cell therapy using the dual reporter system consisting of the RLuc gene driven by the mouse stem cell virus constitutive promoter and the FLuc gene driven by the endothelial cell-specific tie-2 promoter. Luciferases from different species, depending on different substrates and emitting at distinct wavelengths have been used to analyze in vivo bioluminescence imaging of transplanted neural stem cells in the brain [54]. Bioluminescence imaging of Fluc and Rluc provided the real-time monitor of tumor cells and hUC-MSCs administered simultaneously in breast cancer therapy [55].
BLI has been further applied to the in vivo study for simultaneous monitoring of multiple cellular events. Nakajima et al. [56] developed a novel reporter assay system that emits three different colors (green, orange, and red light) using a single substrate. They demonstrated that this system enabled direct comparison of two or more transcriptional activities and/or interactions with transcription factors in the same cell population in a one-step reaction with a single luciferin. Kitayama et al. [57] who first reported on a multicolor luciferase assay system, successfully demonstrated two independent promoter activities simultaneously within the same cells and precisely compared their characteristics in vivo using green- and red-emitting luciferases. The multicolor luciferase system can be easily applied in various studies, including high-throughput monitoring of gene expression, signal transduction cascade, and protein-protein interaction in both in vitro and in vivo studies.
Application of Optical Imaging for Cell Tracking
Tracking of Stem Cells
Stem cell therapy is an emerging therapeutic strategy for the introduction of stem cells into damaged or diseased tissues for the recovery of abnormal tissues, and has become a promising tissue regeneration modality for the treatment of irreversible cardiac and brain injury [58–62]. The therapeutic effect of stem cell therapy may be related to the generation of target tissue by differentiation capacity of the cell and also to the secretion and delivery of cytokines by the cells [63]. The study of the in vivo behavior of transplanted stem cells should be conducted in animals for the development of clinical stem cell therapy. Once injected systemically via the vascular route, the injected cells migrate away from the initial injection site toward target sites and can be observed in gastrointestinal tissues, kidney, lung, liver, thymus, and skin [64]. Migration routes differ significantly according to the characteristics of the administered cells, and tracking the administered cells in vivo was almost impossible using classic biological techniques, except for labor-intensive postmortem histology. BLI has been a valuable tool for use in longitudinal assessment of transplanted stem cell fate both in vitro and in vivo, by labeling cells with a constitutively expressing bioluminescent reporter gene. Several groups have studied stem cell migration, survival, and morphological differentiation using BLI. Studies using BLI imaging for the treatment of myocardial ischemia with various adult stem cells have been undertaken and assessed the fates of the implanted stem cells [65, 66]. They also reported that bone marrow mononuclear cells exhibited a higher survival rate than mesenchymal stem cells (MSCs), adipose stromal cells, and skeletal myoblasts, leading to more robust preservation of heart function. Kim et al. [67] used the FLuc reporter gene in order to visualize survival of grafted hair stem cells in animal models (Fig. 2). In that study, the signal intensity from hair stem cells with BLI reporter gene was drastically reduced within the first 4 days after stem cell transplantation, probably due to apoptosis. Several studies using BLI reporter imaging have demonstrated successful in vivo visualization of administered cardiomyocytes purified from embryonic stem cells and the beneficial effect of cardiac function by cell administration following myocardial ischemia [68–70]. However, lack of long-term survival potential of engrafted therapeutic cells in vivo is one of the limitations in stem cell therapy. One in vivo study investigated the methods by improving cardiomyoblast survival and biodistribution, including overexpression of BCL2 genes [71].
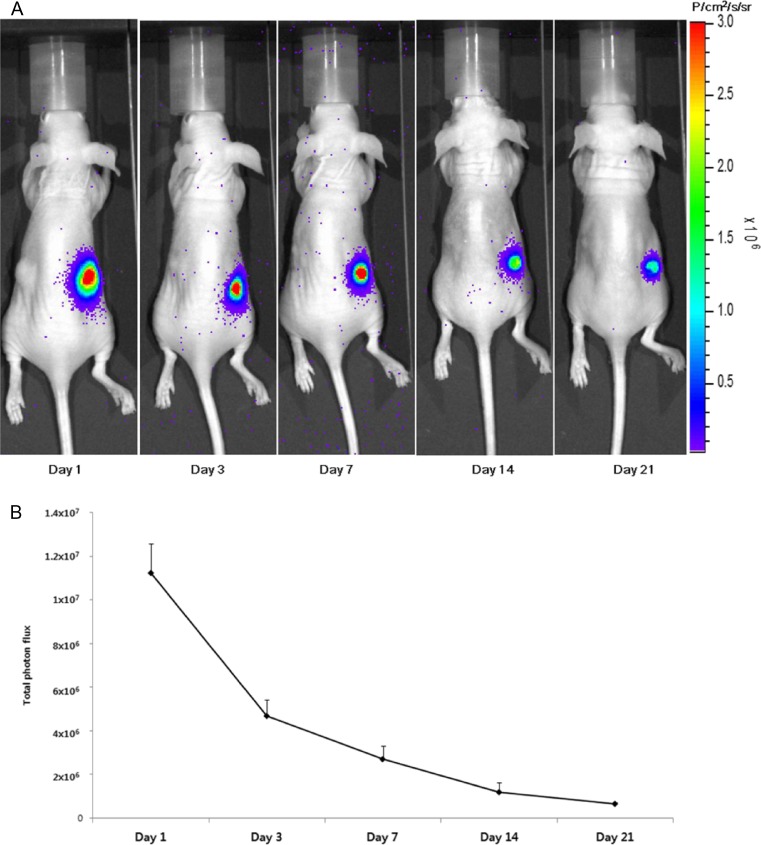
Fig. 2.
a For the bioluminescence imaging study, cell mixtures (1 × 106 newborn mouse fibroblasts [NFs] expressing enhanced firefly luciferase [NF-effluc], 1 × 106 primary epithelial cells in a total volume of 100 μl) were injected into the hypodermis (on the right flank). Parent NFs and epithelial cells of the same number and volume were injected into the left flank. Bioluminescence imaging was acquired at 1, 3, 7, 14, and 21 days after inoculation. NF-effluc-inoculated sites were well visualized following an injection of D-luciferin but not at parental NF inoculation sites. b A decrease of total photon flux of approximately 60 % was observed on day 3, 70 % on day 7, 80 % on day 14, and 90 % on day 21 at the NF-effluc inoculation sites. (Reprinted with permission of Kim et al. [67])
Long-term in vivo monitoring of implanted stem cells can be feasibly performed using BLI reporter gene techniques. Many studies have been conducted in order to enhance the homing of systemically delivered MSCs toward target sites. The chemokine stromal-derived factor-1 and its ligand, CXCR4, play an important role in regulating the homing of engrafted stem cells to infracted areas [72]. Multiple studies have demonstrated that overexpression of CXCR4 on MSCs was effective in accelerating mobilization of these cells toward ischemic areas [72–74]. The mobility of MSCs to the target tissue can be easily verified using in vivo BLI reporter gene strategies as well. BLI imaging could provide a significant amount of data, such as the real-time and long-term visualization of transplanted stem cells; therefore, the in vivo dynamics of stem cells, including migration, proliferation, death, and even differentiation can be monitored noninvasively using BLI. It allows the high-throughput screening of various factors affecting therapeutic results of transplanted stem cells in various in vivo animal models [75, 76].
Tracking of Immune Cells
In addition to stem cells, immune cells are also attractive targets for tracking research using BLI techniques. Immune cells such as T cells, natural killer cells, and dendritic cells play important roles in cancer immunotherapy [77, 78]. Immune cell migration is a key aspect of development of the immune system and in mediation of an immune response. Therefore, the ability to monitor the migration and fate of immune cells noninvasively under in vivo conditions is helpful to understanding the role of immune cells in various disease conditions and to devising rational therapeutic strategies and is also critical to optimizing the strategies. BLI could track migration to sites of inflammation and measure the life span of adoptively transferred T cells for tumor immunotherapy [79]. Rabinovich et al. [80] developed an enhanced version of FLuc that can be transduced into primary mouse T cells and could provide sufficient sensitivity in tracking <10,000 T cells within living mice. Edinger et al. [81] demonstrated homing of cytokine induced killer cells derived from splenocytes, which were retrovirally transduced with both green fluorescent protein and FLuc, to sites of tumor growth followed by tumor eradication. BLI has also been proven useful in noninvasive monitoring of engrafted T cells in mouse models of graft versus host disease [82]. Nguyen et al. [83] performed noninvasive and longitudinal monitoring of in vivo dynamics of regulatory T cells (Tregs) using luciferase expressing mouse Tregs in a mouse model of allogenic bone marrow transplantation. They monitored robust proliferation of Tregs in secondary lymphoid organs, with the early migration and infiltration to peripheral tissues. Shin et al. [84] demonstrated the recruitment of macrophages to the atherosclerotic lesion in an in vivo animal model using BLI. In addition, they reported that macrophage chemotaxis is higher in mechanical atherosclerosis than in lipogenic atherosclerosis. Using BLI, Lee et al. [85] monitored migration of macrophages to chemically induced inflammation in a mouse model over 21 days.
Tracking of Bacteria
A number of recent reports have demonstrated that bacteria are capable of targeting both primary and metastatic tumors. Bioluminescent bacteria were generated by transforming MG1655 with an expression plasmid (pLux) that contains the luxCDABE operon from Photobacterium leognathi. This molecular imaging protocol is a powerful approach for the quantitative visualization of the distribution of bacteria in mouse tumor models [50].
Prospective and Conclusions
BLI is one of the most popular and sensitive techniques for the in vivo monitoring of cell-based therapy. Application of the BLI approach for monitoring of transplanted stem cells or immune cells associates viability of administered cells with therapeutic efficacy in preclinical animal models of various diseases. However, BLI still has weaknesses of poor tissue penetration of light photons and poor quantification by absorption and scatter in tissues in vivo. A promising potential lies in the development of new bioluminescence proteins capable of producing stronger and longer wavelength light; a large number of researchers are working on these issues. Development of BLI technologies that deliver more sensitive, quantitative, and three-dimensional information is on the way as well. In vivo BLI technology plays a pivotal role in the investigation of preclinical cell tracking studies, and its contribution to the studies will become more critical and indispensable by the advancement of both BLI reporter proteins and BLI technologies.
Acknowledgments
Funding
This research was supported by a grant from the Medical Cluster R&D Support Project of Daegu Gyeongbuk Medical Innovation Foundation, Republic of Korea (2013), grants from the National Nuclear R&D Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (No. 2012M2A2A7014020, 2009–0078234) and grants from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea. (A111345).
Conflicts of Interest
The authors Jung Eun Kim, Kalimuthu Senthilkumar, Byeong-Cheol Ahn declare that they have no conflict of interest.
Disclosure
The manuscript does not contain clinical studies or patient data.
Animal Studies
The in vivo mouse data are reprinted with permission from Kim et al. [67].
References
- 1.Cao F, Lin S, Xie X, Ray P, Patel M, Zhang X, et al. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113(7):1005–14. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeiser R, Nguyen VH, Beilhack A, Buess M, Schulz S, Baker J, et al. Inhibition of CD4+ CD25+ regulatory T-cell function by calcineurin-dependent interleukin-2 production. Blood. 2006;108(1):390–9. doi: 10.1182/blood-2006-01-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman SC, Galpoththawela C, Gilad AA, Bulte JW, Walczak P. Long-term MR cell tracking of neural stem cells grafted in immunocompetent versus immunodeficient mice reveals distinct differences in contrast between live and dead cells. Magn Reson Med. 2011;65(2):564–74. doi: 10.1002/mrm.22613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acton P, Zhou R. Imaging reporter genes for cell tracking with PET and SPECT. Q J Nucl Med Mol Imaging. 2005;49(4):349–60. [PubMed] [Google Scholar]
- 5.Zhang Y, Ruel M, Beanlands RS, Dekemp RA, Suuronen EJ, DaSilva JN. Tracking stem cell therapy in the myocardium: applications of positron emission tomography. Curr Pharm Design. 2008;14(36):3835–53. doi: 10.2174/138161208786898662. [DOI] [PubMed] [Google Scholar]
- 6.Ahn BC. Applications of molecular imaging in drug discovery and development process. Curr Pharm Biotechnol. 2011;12(4):459–68. doi: 10.2174/138920111795163904. [DOI] [PubMed] [Google Scholar]
- 7.Chao F, Shen Y, Zhang H, Tian M. Multimodality molecular imaging of stem cells therapy for stroke. BioMed Res Int. 2013;2013:849819. [DOI] [PMC free article] [PubMed]
- 8.Yang CY, Hsiao JK, Tai M-F, Chen ST, Cheng HY, Wang JL, et al. Direct labeling of hMSC with SPIO: the long-term influence on toxicity, chondrogenic differentiation capacity, and intracellular distribution. Mol Imag Biol. 2011;13(3):443–51. doi: 10.1007/s11307-010-0360-7. [DOI] [PubMed] [Google Scholar]
- 9.Kraitchman DL, Bulte JW. In vivo imaging of stem cells and beta cells using direct cell labeling and reporter gene methods. Arterioscler Thromb Vasc Biol. 2009;29(7):1025–30. doi: 10.1161/ATVBAHA.108.165571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keyaerts M, Caveliers V, Lahoutte T. Bioluminescence imaging: looking beyond the light. Trends Mol Med. 2012;18(3):164–72. doi: 10.1016/j.molmed.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Baker M. Whole-animal imaging: the whole picture. Nature. 2010;463(7283):977–80. doi: 10.1038/463977a. [DOI] [PubMed] [Google Scholar]
- 12.Verhaegen M, Christopoulos TK. Recombinant Gaussia luciferase. Overexpression, purification, and analytical application of a bioluminescent reporter for DNA hybridization. Anal Chem. 2002;74(17):4378–85. doi: 10.1021/ac025742k. [DOI] [PubMed] [Google Scholar]
- 13.Thompson EM, Nagata S, Tsuji FI. Cloning and expression of cDNA for the luciferase from the marine ostracod Vargula hilgendorfii. Proc Natl Acad Sci U S A. 1989;86(17):6567–71. doi: 10.1073/pnas.86.17.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenz W, Cormier M, O’Kane D, Hua D, Escher A, Szalay A. Expression of the Renilla reniformis luciferase gene in mammalian cells. J Biolumin Chemilumin. 1996;11(1):31–7. doi: 10.1002/(SICI)1099-1271(199601)11:1<31::AID-BIO398>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 15.Nealson K, Hastings JW. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979;43(4):496. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraga H. Firefly luminescence: a historical perspective and recent developments. Photochem Photobiol Sci. 2008;7(2):146–58. doi: 10.1039/b719181b. [DOI] [PubMed] [Google Scholar]
- 17.Close DM, Xu T, Sayler GS, Ripp S. In vivo bioluminescent imaging (BLI): noninvasive visualization and interrogation of biological processes in living animals. Sensors. 2010;11(1):180–206. doi: 10.3390/s110100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz WW, McCann RO, Longiaru M, Cormier MJ. Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc Natl Acad Sci U S A. 1991;88(10):4438–42. doi: 10.1073/pnas.88.10.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel. 2006;19(9):391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 20.Maguire CA, Deliolanis NC, Pike L, Niers JM, Tjon-Kon-Fat L-A, Sena-Esteves M, et al. Gaussia luciferase variant for high-throughput functional screening applications. Anal Chem. 2009;81(16):7102–6. doi: 10.1021/ac901234r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tannous BA. Gaussia luciferase reporter assay for monitoring biological processes in culture and in vivo. Nat Protoc. 2009;4(4):582–91. doi: 10.1038/nprot.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziegler M, Baldwin TO. Biochemistry of bacterial bioluminescence. Curr Top Bioenerg. 1981;12:65–113. doi: 10.1016/B978-0-12-152512-5.50008-7. [DOI] [Google Scholar]
- 23.Tu S-C, Mager HI. Biochemistry of bacterial bioluminescence. Photochem Photobiol. 1995;62(4):615–24. doi: 10.1111/j.1751-1097.1995.tb08708.x. [DOI] [PubMed] [Google Scholar]
- 24.de Almeida PE, van Rappard JR, Wu JC. In vivo bioluminescence for tracking cell fate and function. Am J Physiol Heart Circ Physiol. 2011;301(3):H663–71. doi: 10.1152/ajpheart.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn BC. Molecular imaging; biological and medical applications. Republic of Korea: Kyungpook National University Press; 2009. [Google Scholar]
- 26.Liang Y, Walczak P, Bulte JW. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. J Biomed Opt. 2012;17(1):0160041–6. doi: 10.1117/1.JBO.17.1.016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seliger H, Buck J, Fastie W, McElroy W. The spectral distribution of firefly light. J Gen Physiol. 1964;48(1):95–104. doi: 10.1085/jgp.48.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seliger H, McElroy W. The colors of firefly bioluminescence: enzyme configuration and species specificity. Proc Natl Acad Sci U S A. 1964;52(1):75. doi: 10.1073/pnas.52.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Czupryna J, Tsourkas A. Firefly luciferase and RLuc8 exhibit differential sensitivity to oxidative stress in apoptotic cells. PLoS One. 2011;6(5):e20073. doi: 10.1371/journal.pone.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitayama A, Yoshizaki H, Ohmiya Y, Ueda H, Nagamune T. Creation of a thermostable firefly luciferase with pH-insensitive luminescent color. Photochem Photobiol. 2003;77(3):333–8. doi: 10.1562/0031-8655(2003)077<0333:COATFL>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 31.Kajiyama N, Nakano E. Isolation and characterization of mutants of firefly luciferase which produce different colors of light. Protein Eng. 1991;4(6):691–3. doi: 10.1093/protein/4.6.691. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro E, Lu C, Baneyx F. A set of multicolored Photinus pyralis luciferase mutants for in vivo bioluminescence applications. Protein Eng Des Sel. 2005;18(12):581–7. doi: 10.1093/protein/gzi066. [DOI] [PubMed] [Google Scholar]
- 33.Branchini BR, Ablamsky DM, Davis AL, Southworth TL, Butler B, Fan F, et al. Red-emitting luciferases for bioluminescence reporter and imaging applications. Anal Biochem. 2010;396(2):290–7. doi: 10.1016/j.ab.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Bhaumik S, Gambhir S. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci U S A. 2002;99(1):377–82. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luker KE, Luker GD. Applications of bioluminescence imaging to antiviral research and therapy: multiple luciferase enzymes and quantitation. Antiviral Res. 2008;78(3):179–87. doi: 10.1016/j.antiviral.2008.01.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H, Doyle TC, Wong RJ, Cao Y, Stevenson DK, Piwnica-Worms D, et al. Characterization of coelenterazine analogs for measurements of Renilla luciferase activity in live cells and living animals. Mol Imag. 2004;3(1):43–54. doi: 10.1162/153535004773861714. [DOI] [PubMed] [Google Scholar]
- 37.Loening AM, Dragulescu-Andrasi A, Gambhir SS. A red-shifted Renilla luciferase for transient reporter-gene expression. Nat Methods. 2010;7(1):5–6. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods. 2007;4(8):641–3. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 39.Griesenbach U, Vicente CC, Roberts MJ, Meng C, Soussi S, Xenariou S, et al. Secreted Gaussia luciferase as a sensitive reporter gene for in vivo and ex vivo studies of airway gene transfer. Biomaterials. 2011;32(10):2614–24. doi: 10.1016/j.biomaterials.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Bovenberg MSS, Degeling MH, Tannous BA. Enhanced Gaussia luciferase blood assay for monitoring of in vivo biological processes. Anal Chem. 2011;84(2):1189–92. doi: 10.1021/ac202833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wurdinger T, Badr C, Pike L, de Kleine R, Weissleder R, Breakefield XO, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5(2):171–3. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badr CE, Hewett JW, Breakefield XO, Tannous BA. A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS One. 2007;2(6):e571. doi: 10.1371/journal.pone.0000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiramatsu N, Kasai A, Hayakawa K, Nagai K, Kubota T, Yao J, et al. Secreted protein-based reporter systems for monitoring inflammatory events: critical interference by endoplasmic reticulum stress. J Immunol Methods. 2006;315(1):202–7. doi: 10.1016/j.jim.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Tannous BA, Kim D-E, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11(3):435–43. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Santos EB, Yeh R, Lee J, Nikhamin Y, Punzalan B, Punzalan B, et al. Sensitive in vivo imaging of T cells using a membrane-bound Gaussia princeps luciferase. Nat Med. 2009;15(3):338–44. doi: 10.1038/nm.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derzelle S, Duchaud E, Kunst F, Danchin A, Bertin P. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl Environ Microbiol. 2002;68(8):3780–9. doi: 10.1128/AEM.68.8.3780-3789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meighen EA. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55(1):123–42. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Contag CH, Contag PR, Mullins JI, Spilman SD, Stevenson DK, Benaron DA. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18(4):593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 49.Francis KP, Joh D, Bellinger-Kawahara C, Hawkinson MJ, Purchio TF, Contag PR. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect Immun. 2000;68(6):3594–600. doi: 10.1128/IAI.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Min J-J, Nguyen VH, Kim H-J, Hong Y, Choy HE. Quantitative bioluminescence imaging of tumor-targeting bacteria in living animals. Nat Protoc. 2008;3(4):629–36. doi: 10.1038/nprot.2008.32. [DOI] [PubMed] [Google Scholar]
- 51.Min J-J, Kim H-J, Park JH, Moon S, Jeong JH, Hong Y-J, et al. Noninvasive real-time imaging of tumors and metastases using tumor-targeting light-emitting Escherichia coli. Mol Imag Biol. 2008;10(1):54–61. doi: 10.1007/s11307-007-0120-5. [DOI] [PubMed] [Google Scholar]
- 52.Mezzanotte L, Que I, Kaijzel E, Branchini B, Roda A, Löwik C. Sensitive dual color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase. PLoS One. 2011;6(4):e19277. doi: 10.1371/journal.pone.0019277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Najjar A, Zhang S, Rabinovich B, Willerson JT, Gelovani JG, et al. Molecular imaging of mesenchymal stem cell mechanistic insight into cardiac repair after experimental myocardial infarction. Circ Cardiovasc Imaging. 2012;5(1):94–101. doi: 10.1161/CIRCIMAGING.111.966424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mezzanotte L, Aswendt M, Tennstaedt A, Hoeben R, Hoehn M, Löwik C. Evaluating reporter genes of different luciferases for optimized in vivo bioluminescence imaging of transplanted neural stem cells in the brain. Contrast Media Mol Imaging. 2013;8(6):505–13. doi: 10.1002/cmmi.1549. [DOI] [PubMed] [Google Scholar]
- 55.Leng L, Wang Y, He N, Wang D, Zhao Q, Feng G, et al. Molecular imaging for assessment of mesenchymal stem cells mediated breast cancer therapy. Biomaterials. 2014;35(19):5162–70. doi: 10.1016/j.biomaterials.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakajima Y, Kimura T, Sugata K, Enomoto T, Asakawa A, Kubota H, et al. Multicolor luciferase assay system: one-step monitoring of multiple gene expressions with a single substrate. Biotechniques. 2005;38(6):891. doi: 10.2144/05386ST03. [DOI] [PubMed] [Google Scholar]
- 57.Kitayama Y, Kondo T, Nakahira Y, Nishimura H, Ohmiya Y, Oyama T. An in vivo dual-reporter system of cyanobacteria using two railroad-worm luciferases with different color emissions. Plant Cell Physiol. 2004;45(1):109–13. doi: 10.1093/pcp/pch001. [DOI] [PubMed] [Google Scholar]
- 58.Kidd S, Caldwell L, Dietrich M, Samudio I, Spaeth EL, Watson K, et al. Mesenchymal stromal cells alone or expressing interferon-β suppress pancreatic tumors in vivo, an effect countered by anti-inflammatory treatment. Cytotherapy. 2010;12(5):615–25. doi: 10.3109/14653241003631815. [DOI] [PubMed] [Google Scholar]
- 59.Abe K. Therapeutic potential of neurotrophic factors and neural stem cells against ischemic brain injury. J Cereb Blood Flow Metab. 2000;20(10):1393–408. doi: 10.1097/00004647-200010000-00001. [DOI] [PubMed] [Google Scholar]
- 60.Koelling S, Miosge N. Stem cell therapy for cartilage regeneration in osteoarthritis. Expert Opin Biol Ther. 2009;9(11):1399–405. doi: 10.1517/14712590903246370. [DOI] [PubMed] [Google Scholar]
- 61.Lee DS. Optical imaging for stem cell differentiation to neuronal lineage. Nucl Med Mol Imag. 2012;46(1):1–9. doi: 10.1007/s13139-011-0122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sunwoo M, Yun H, Song S, Ham J, Hong J, Lee JE, et al. Mesenchymal stem cells can modulate longitudinal changes in cortical thickness and its related cognitive decline in patients with multiple system atrophy. Front Aging Neurosci. 2014;6:118. doi: 10.3389/fnagi.2014.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cihova M, Altanerova V, Altaner C. Stem cell based cancer gene therapy. Mol Pharm. 2011;8(5):1480–7. doi: 10.1021/mp200151a. [DOI] [PubMed] [Google Scholar]
- 64.Devine SM, Cobbs C, Jennings M, Bartholomew A, Hoffman R. Mesenchymal stem cells distribute to a wide range of tissues following systemic infusion into nonhuman primates. Blood. 2003;101(8):2999–3001. doi: 10.1182/blood-2002-06-1830. [DOI] [PubMed] [Google Scholar]
- 65.van der Bogt KE, Sheikh AY, Schrepfer S, Hoyt G, Cao F, Ransohoff KJ, et al. Comparison of different adult stem cell types for treatment of myocardial ischemia. Circulation. 2008;118(14 suppl 1):S121–9. doi: 10.1161/CIRCULATIONAHA.107.759480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang JJ, Liu ZQ, Zhang JM, Wang H-b HSY, Liu JF, et al. Real-time tracking of adipose tissue-derived stem cells with injectable scaffolds in the infarcted heart. Heart Vessel. 2013;28(3):385–96. doi: 10.1007/s00380-012-0275-0. [DOI] [PubMed] [Google Scholar]
- 67.Kim JE, Ahn B-C, Lee HW, Hwang MH, Shin SH, Lee SW et al. In vivo monitoring of survival and proliferation of hair stem cells in a hair follicle generation animal model. Mol Imag. 2013;12(5). [PubMed]
- 68.Cao F, Wagner RA, Wilson KD, Xie X, Fu J-D, Drukker M, et al. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS One. 2008;3(10):e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caspi O, Huber I, Kehat I, Habib M, Arbel G, Gepstein A, et al. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J Am Coll Cardiol. 2007;50(19):1884–93. doi: 10.1016/j.jacc.2007.07.054. [DOI] [PubMed] [Google Scholar]
- 70.Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, Habib IHM, et al. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res. 2007;100(2):263–72. doi: 10.1161/01.RES.0000257776.05673.ff. [DOI] [PubMed] [Google Scholar]
- 71.Kutschka I, Kofidis T, Chen IY, von Degenfeld G, Zwierzchoniewska M, Hoyt G, et al. Adenoviral human BCL-2 transgene expression attenuates early donor cell death after cardiomyoblast transplantation into ischemic rat hearts. Circulation. 2006;114(1 supp):I-I174–80. doi: 10.1161/CIRCULATIONAHA.105.001370. [DOI] [PubMed] [Google Scholar]
- 72.Shi M, Li J, Liao L, Chen B, Li B, Chen L, et al. Regulation of CXCR4 expression in human mesenchymal stem cells by cytokine treatment: role in homing efficiency in NOD/SCID mice. Haematologica. 2007;92(7):897–904. doi: 10.3324/haematol.10669. [DOI] [PubMed] [Google Scholar]
- 73.Zhang D, Fan G-C, Zhou X, Zhao T, Pasha Z, Xu M, et al. Over-expression of CXCR4 on mesenchymal stem cells augments myoangiogenesis in the infarcted myocardium. J Mol Cell Cardiol. 2008;44(2):281–92. doi: 10.1016/j.yjmcc.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng Z, Ou L, Zhou X, Li F, Jia X, Zhang Y, et al. Targeted migration of mesenchymal stem cells modified with CXCR4 gene to infarcted myocardium improves cardiac performance. Mol Ther. 2008;16(3):571–9. doi: 10.1038/sj.mt.6300374. [DOI] [PubMed] [Google Scholar]
- 75.Wang H, Cao F, De A, Cao Y, Contag C, Gambhir SS, et al. Trafficking mesenchymal stem cell engraftment and differentiation in tumor-bearing mice by bioluminescence imaging. Stem Cells. 2009;27(7):1548–58. doi: 10.1002/stem.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang Y, Shah K, Messerli SM, Snyder E, Breakefield X, Weissleder R. In vivo tracking of neural progenitor cell migration to glioblastomas. Hum Gene Ther. 2003;14(13):1247–54. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- 77.Akbar S, Fazle M, Abe M, Yoshida O, Murakami H, Onji M. Dendritic cell-based therapy as a multidisciplinary approach to cancer treatment: present limitations and future scopes. Curr Med Chem. 2006;13(26):3113–9. doi: 10.2174/092986706778742882. [DOI] [PubMed] [Google Scholar]
- 78.Gao JQ, Okada N, Mayumi T, Nakagawa S. Immune cell recruitment and cell-based system for cancer therapy. Pharm Res. 2008;25(4):752–68. doi: 10.1007/s11095-007-9443-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Costa GL, Sandora MR, Nakajima A, Nguyen EV, Taylor-Edwards C, Slavin AJ, et al. Adoptive immunotherapy of experimental autoimmune encephalomyelitis via T cell delivery of the IL-12 p40 subunit. J Immunol. 2001;167(4):2379–87. doi: 10.4049/jimmunol.167.4.2379. [DOI] [PubMed] [Google Scholar]
- 80.Rabinovich BA, Ye Y, Etto T, Chen JQ, Levitsky HI, Overwijk WW, et al. Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proc Natl Acad Sci U S A. 2008;105(38):14342–6. doi: 10.1073/pnas.0804105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edinger M, Cao YA, Verneris MR, Bachmann MH, Contag CH, Negrin RS. Revealing lymphoma growth and the efficacy of immune cell therapies using in vivo bioluminescence imaging. Blood. 2003;101(2):640–8. doi: 10.1182/blood-2002-06-1751. [DOI] [PubMed] [Google Scholar]
- 82.Tang Q, Adams JY, Tooley AJ, Bi M, Fife BT, Serra P, et al. Visualizing regulatory T cell control of autoimmune responses in nonobese diabetic mice. Nat Immunol. 2005;7(1):83–92. doi: 10.1038/ni1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen VH, Zeiser R, Chang DS, Beilhack A, Contag CH, Negrin RS. In vivo dynamics of regulatory T-cell trafficking and survival predict effective strategies to control graft-versus-host disease following allogeneic transplantation. Blood. 2007;109(6):2649–56. doi: 10.1182/blood-2006-08-044529. [DOI] [PubMed] [Google Scholar]
- 84.Shin IJ, Shon S-M, Schellingerhout D, Park J-Y, Kim J-Y, Lee SK, et al. Characterization of partial ligation-induced carotid atherosclerosis model using dual-modality molecular imaging in ApoE knock-out mice. PLoS One. 2013;8(9):e73451. doi: 10.1371/journal.pone.0073451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee HW, Jeon YH, Hwang M-H, Kim J-E, T-i P, Ha J-H, et al. Dual reporter gene imaging for tracking macrophage migration using the human sodium iodide symporter and an enhanced firefly luciferase in a murine inflammation model. Mol Imag Biol. 2013;15(6):703–12. doi: 10.1007/s11307-013-0645-8. [DOI] [PubMed] [Google Scholar]