Abstract
Partial hepatectomy (PH) induces robust hepatic regenerative and metabolic responses that are considered to be triggered by humoral factors. The aim of the study was to identify plasma protein factors that potentially trigger or reflect the body’s immediate-early responses to liver mass reduction. Male C57BL/6 mice were subjected to sham operation, 70% PH, or 90% PH. Blood was collected from the inferior vena cava at 20, 60, and 180 minutes after surgery. Using a label-free quantitative mass spectrometry-based proteomics approach, we identified 399 proteins exhibiting significant changes in plasma expression between any two groups. Of the 399 proteins, 167 proteins had multiple unique sequences and high peptide ID confidence (>90%) and were defined as priority 1 proteins. A group of plasma proteins largely associated with metabolism is enriched after 70% PH. Among the plasma proteins that respond to 90% PH are a dominant group of proteins that are also associated with metabolism and one known cytokine (platelet factor 4). Ninety percent PH and 70% PH induces similar changes in plasma protein profile. Our findings enable us to gain insight into the immediate-early response of plasma proteins to liver mass loss. Our data support the notion that increased metabolic demands of the body after massive liver mass loss may function as a sensor that calibrates hepatic regenerative response.
Keywords: Partial hepatectomy, liver regeneration, plasma proteomics
Introduction
Partial hepatectomy (PH) induces hepatic regenerative and metabolic responses. An important aspect of the studies on liver regeneration is identification of the signals that trigger the initiation, progression, and termination of hepatic regeneration. Very early observations from the studies with parabiotic rats and ectopic transplants of hepatocytes provided a critical clue to the source of the signals. When rats were joined in pairs by parabiotic circulation, PH on one rat of the pair induced hepatic regenerative response in the other rat [1]. Moreover, PH on orthotopic liver caused a proliferative response in hepatic tissue or isolated hepatocytes transplanted extrahepatically into the host [2]. These studies convincingly indicated that the signals are humoral factors transmitted by the blood. Since then, a number of such factors, including IL-6, TNFα, HGF, and EGF, have been discovered. These factors are produced intra- and/or extrahepatically and exert various distinct or overlapping effects on liver regeneration [3–6]. For instance, proinflammatory cytokines IL-6 and TNFα participate in the induction of an early priming response, which renders hepatocytes competent to respond to growth factors. As potent mitogens, HGF and EGF stimulate hepatocyte replication. However, it is still unknown whether there are any immediate-early humoral factors that are responsible for the initial triggering of liver regeneration. Because many of the discovered factors appear in the circulation during liver regeneration, blood samples should be valuable in the design of studies aimed at answering this question. PH also induces the hepatic metabolic response [7–17]. It has been proposed that the increased metabolic demands placed on hepatocytes of the regenerating liver are linked to the machinery needed for hepatocyte proliferation and may function as a sensor that calibrates the regenerative response according to body demands [4]. Thus, blood samples should be valuable in finding humoral factors that reflect the systemic metabolic response to liver mass loss. Several groups have analyzed protein profiles in regenerating livers with proteomic approaches [18–24]. Those studies provided significant insights into the proteome of regenerating liver and identified proteins that are implicated in the regulation of liver regeneration. The aim of the present study was to identify blood-borne proteins that potentially trigger or reflect the body’s initial responses to liver resection. The availability of a powerful quantitative proteomic approach enabled us to pursue the aim by profiling immediate-early response plasma proteins in liver regeneration.
The widely used 70% PH was chosen for our study because hepatic regenerative response in this model can be precisely timed and is not accompanied by major cellular injury and inflammation [6, 25]. Ninety percent PH causes high mortality for unknown reasons. However, evidence indicates that the hepatic regenerative response is proportional to the extent of liver mass loss [1]. Thus, 90% PH was also utilized to (1) determine whether 90% PH induces changes in the immediate-early plasma proteomic profile that are similar to those from 70% PH, (2) consolidate the findings from the 70% PH model, and (3) identify plasma proteins associated with the extent of liver mass reduction. We used a label-free quantitative proteomics approach (LFQP) to profile the global protein expression in mouse plasma samples collected at three time points (20, 60 and 180 minutes) after sham operation, 70% PH, or 90% PH.
Materials and Methods
Mice and PH
C57BL/6 male mice were purchased from The Jackson Laboratory (Bar Harbor, Maine) and housed in plastic cages at 22 ± 1 °C on a 12-hour/12-hour light/dark cycle with light on from 6:00 am to 6:00 pm. Standard rodent chow and water were provided ad libitum throughout the entire feeding period. Six-month-old male mice were subjected to sham operation, 70% PH, or 90% PH. Standard 70% liver resection was performed following the procedure previously described by others and us [15, 26]. Ninety percent hepatectomy was performed by removing all hepatic lobes except for the caudate lobe. In the 70% and 90% PH procedures, each lobe to be surgically removed was individually ligated at its root. Surgery time for each mouse was determined for the collection of blood at 20, 60, or 180 minutes after surgery and between 3:00 pm and 4:00 pm to avoid the circadian clock-associated variations in plasma protein concentrations. Before blood collection, intra-abdominal inspection was conducted under anesthesia of isoflurane for mice that underwent 90% PH. Mice that showed congestion of the intestinal tract and portal system, which occasionally occurs as a sign of portal hypertension following 90% PH, were excluded from the experiment. Three to five mice were used per time point per surgery group. Blood was drawn from the inferior vena cava with the S-Monovette Blood Collection System with dried potassium EDTA (Sarstedt AG & Co, Nümbrecht, Germany) under anesthesia of isoflurane. Subsequently, blood was centrifuged for 10 minutes at 2,000 x g at room temperature to remove blood cells. The supernatant was transferred into an Eppendorf tube and then centrifuged for 15 minutes at 2,500 x g at room temperature to separate the platelets from the plasma. The plasma was transferred into an Eppendorf tube and stored at −80°C until use. All of the animal experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Protocols for the care and use of animals were approved by the Indiana University-Purdue University Indianapolis Animal Care and Use Committee.
Sample preparation for plasma proteomics analysis
Fifty microliters of plasma proteins were denatured in lysis buffer containing 8 M urea and 10 mM dithiothreitol (DTT) as previously described [27]. Prior to denaturing, high-abundance plasma proteins were depleted with a Sigma Seppro Mouse Affinity Column (Sigma-Aldrich, St. Louis, MO, USA), and protein concentrations were measured by Bradford assay [28]. The resulting protein extracts were reduced by triethylphosphine, alkylated by iodoethanol, and digested by trypsin [29]. Tryptic peptides were filtered through ultra-free MC 0.45-μm filters via centrifugation before they were applied to the high-performance liquid chromatography (HPLC) system. To assess the stability of the HPLC system and mass spectrometry (MS) instrument, chicken lysozyme (0.5 ng chicken lysozyme per μg protein extract) was spiked into to each sample before tryptic digestion as an internal reference for quality assessment and quality control.
Liquid Chromatography-Tandem Mass Spectrometry (LC/MS-MS)
Trypic peptides were injected randomly onto the X-Bridge C18 column (Waters, 2.1 mm X 100 mm) in the Thermo-Fisher Scientific Surveyor HPLC system (Waltham, MA). For peptide elution, a linear gradient from 5 to 40% acetonitrile (in water with 0.1% formic acid) was developed over 150 minutes at 50°C at a flow rate of 200 μL/min, and effluent was electro-sprayed into the LTQ mass spectrometer (Thermo-Fisher Scientific). Blanks were run prior to and between the sample runs to ensure that there was no significant signal from solvents or the column and that there was no ‘carry-over’. Data were collected in “Triple Play” (MS scan, Zoom scan, and MS/MS scan) mode. A proprietary algorithm was applied to filter and analyze the acquired data [30]. Database searches were performed with both the Sequest™ and X!Tandem algorithms [31]. The International Protein Index (IPI) Mouse database (V. 3.60) was used.
Protein identification
Proteins were classified from priority 1 [highest identification (ID) confidence] to priority 4 (lowest ID confidence) based on the protein identification quality. The confidence in the protein ID is increased with 1) increased peptide ID confidence and 2) a greater number of identified distinct amino acid sequences. The “peptide ID confidence” [ID quality of the amino acid sequence(s)] of the “best peptide” (the peptide with the highest peptide ID confidence) was used to assign each protein to a “high” (between 90 and 100% ID confidence), “moderate” (between 75 and 89% ID confidence), or “low” (less than 75% ID confidence) ID category, and all low category proteins were discarded before quantification. Proteins were also categorized based on the number of distinct amino acid sequences that were identified. High category proteins were considered priority 1 if multiple (≥2) unique peptide sequences with 90–100% ID confidence were identified; otherwise, they were ranked as priority 2. Moderate category proteins were considered priority 3 if multiple (≥2) unique sequences with 75–89% ID confidence were identified; otherwise, they were ranked as priority 4. The X!Tandem [31] and SEQUEST algorithms were used for amino acid sequence ID as previously described [30]. Briefly, each algorithm compared the observed peptide MS/MS spectrum and theoretically derived spectrums from the database to assign quality scores that were combined with other predictors in a proprietary algorithm to assign the overall score, “% ID confidence,” to each peptide.
Protein quantification
The quantification of proteins was performed as previously reported [32]. Raw files were obtained from the LTQ mass spectrometer and retention time was used to align all extracted ion chromatograms. Relative abundance was determined by the normalized area under the curve (AUC) for each individually aligned peak from each sample. The limit of detection of protein concentration for the methods and instruments used in the study is about 100 to 200 ng/mL.
Statistical analysis
ANOVA was used to detect significant changes in protein expression among groups. Randomization of the order of measurement and “quantile normalization” were used to eliminate technical bias and normalize the data [33]. A log2 scale (a one-unit difference on this log scale is equivalent to a two-fold change) was used for normalization. A p value was acquired from the ANOVA model to estimate the false positive rate. The p value was transformed to a q value by proprietary statistical methods to estimate the false discovery rate. The false discovery rate was controlled at 5% (< 0.05) by fixing the q value threshold. A change in protein expression between any two groups with a q value < 0.05 was defined as a “significant change” or “differential expression”. For each protein, a separate ANOVA model was fit with the PROC MIXED function in SAS software (Version 9) (SAS Institute, Cary, NC):
where Log2 (Intensity) is the protein intensity based on the weighted average of the quantile normalized log base 2 peptide intensities; Group Effect is the fixed effects (not random) caused by the experimental conditions or treatments that are being compared; and Sample Effect (nested within group) is the random effects from individual biological samples and sample preparation. Positive fold changes (FC), when the mean treated group ≥ mean control group, were computed from the means on the AUC scale (antilog): FC = mean treated group/mean control group. Negative FCs, when mean control group > mean treated group, were computed from the means on the AUC scale (antilog): FC = mean treated group/mean control group. Absolute (positive) values of the FCs were computed. The median percent coefficient of variation (% CV) for each priority level was determined by dividing the standard deviation (SD) by the mean on the AUC scale and is given on a percent scale.
Western blot analysis
Plasma samples were separated by polyacrylamide gel electrophoresis under reducing conditions. Proteins from the gels were electrophoretically transferred to polyvinylidene difluoride membranes. Antibodies against liver fatty acid binding protein (L-FABP) (Cell Signaling Technology, Danvers, MA), betaine homocysteine methyltransferase 2 (BHMT2) (GeneTex, Irvine, CA), fructose 1,6-bisphosphatase 1 (FBPase-1) (Santa Cruz Biotechnology, Santa Cruz, CA), selenium binding protein 1 (SELEBP1) (Aviva Systems Biology, San Diego, CA), and albumin (Abcam, Cambridge, MA) were used as probes. Immune complexes were detected using the enhanced chemiluminescence system (Pierce, Rockford, IL).
Enzyme-linked immunosorbent assay (ELISA) of plasma IL-6
BD OptEIA ELISA kit (Cat# 550950, BD Biosciences, San Jose, CA) was used to detect IL-6 levels in the plasma samples according to the manufacturer’s instruction. Five microliter of each plasma sample was added to each reaction. The absorbance was measured at 450 nm by BioTek synergy HT plate reader. Wavelength correction was performed by subtracting the optical density reading at 570 nm from the reading at 450 nm for each reaction.
Results
Proteomic profiling
Chicken lysozyme, an internal reference of the technical variation, did not display any significant changes in plasma concentrations between any two groups. The maximum fold change among any two group comparisons was 1.188 (an 18.8% change) (Supplementary Figure 1). This result indicated the reliability of the plasma sample preparation and the stability of the HPLC and MS instruments.
The overall findings from the global plasma protein analysis are summarized in Table 1. Among a total of 866 plasma proteins that were identified and quantified, 207 had multiple unique sequences with high peptide ID confidence and were listed as priority 1 proteins. Significant changes (q < 0.05) between any two groups were observed for 399 plasma proteins, of which 167 were priority 1 proteins.
Table 1.
Summary of all identified proteins
| Protein Priority | Peptide ID Confidence | Multiple Sequences Quantified | Number of Proteins | Number of Significant Changes | Maximum Absolute Fold Change | Median % Coeffecient of Variation |
|---|---|---|---|---|---|---|
| 1 | High | Yes | 207 | 167 | 5.69 | 15.53 |
| 2 | High | No | 205 | 93 | 9.18 | 25.32 |
| 3 | Moderate | Yes | 26 | 9 | 2.05 | 17.09 |
| 4 | Moderate | No | 428 | 130 | 12.40 | 29.05 |
|
| ||||||
| Overall | 866 | 399 | 12.40 | 24.34 | ||
To discover the proteins with significant changes associated with PH types and time points after surgery, pair-wise comparisons were performed between three surgery groups (70% PH vs. sham, 90% PH vs. sham, and 90% PH vs. 70% PH) at each time point (20, 60, and 180 minutes) (Table 2). The number of plasma proteins that exhibited significant alterations increased temporally, from 5 (at 20 minutes) to 57 (60 minutes) and further to 90 (180 minutes) in response to 70% PH; from 35 (at 20 minutes) to 52 (60 minutes) and further to 199 (180 minutes) in response to 90% PH. The temporal patterns of plasma protein expression allowed us to identify immediate-early response plasma proteins. When the 90% and 70% PH groups were compared, 1 protein at 20 minutes, no proteins at 60 minutes, and 20 proteins at 180 minutes were found to change significantly (Table 2). The data indicate that 90% PH and 70% PH induced similar changes in the plasma protein profile, especially during the first hour post-surgery. These 21 proteins that exhibited PH type-dependent alterations may be associated with the extent of liver mass loss. The priority, ID, annotation, mean protein intensity, fold change, and q value of all identified proteins that exhibited at least 1.5-fold changes with a q value < 0.05 are listed in Supplementary Table 1.
Table 2.
A pairwise summary of the significant changes among groups (q < 0.05)
| Protein Priority | 20 min after PH
|
60 min after PH
|
180 min after PH
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 70% PH vs. Sham | 90% PH vs. Sham | 90% PH vs. 70% PH | 70% PH vs. Sham | 90% PH vs. Sham | 90% PH vs. 70% PH | 70% PH vs. Sham | 90% PH vs. Sham | 90% PH vs. 70% PH | |
| 1 | 3 | 8 | 1 | 48 | 48 | 0 | 60 | 115 | 11 |
| 2 | 2 | 7 | 0 | 2 | 1 | 0 | 21 | 46 | 4 |
| 3 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 2 | 0 |
| 4 | 0 | 19 | 0 | 7 | 3 | 0 | 8 | 36 | 5 |
|
| |||||||||
| Overall | 5 | 35 | 1 | 57 | 52 | 0 | 90 | 199 | 20 |
Characterization of identified proteins
To identify the proteins that showed robust changes in response to PH, priority 1 proteins that displayed at least two-fold changes identified in the 70% and 90% PH groups in comparison with the sham groups at each time point were chosen for further analysis (Table 3). At the earliest time point (20 minutes after surgery), only 1 protein, liver fatty acid binding protein (L-FABP), was identified in the 90% PH group. L-FABP is the only priority 1 protein that we found to be the most immediate-early response plasma protein in the 90% PH group. The elevation of L-FABP in plasma abundance lasted through the first three hours and was most dramatic (5.1-fold) at 180 minutes, although only a 1.72-fold change was observed at 60 minutes (Supplementary Table 1), following surgery. At 60 minutes post-PH, only 1 protein, betaine-homocysteine S-methyltransferase 2 (BHMT2), was found to be significantly increased in the plasma in response to both 70% and 90% PH. BHMT2 is the only most immediate-early response plasma protein that was found in both PH groups. Plasma expression of BHMT2 further increased at 180 minutes by 4.6- and 5.7-fold in response to 70% and 90% PH, respectively. At 180 minutes post-PH, plasma protein responses became robust. Nineteen common proteins between the 70% and 90% PH group were observed. The plasma concentrations of these common proteins were all increased with fold changes that ranged from 2.0 to 5.7 in comparison to the sham controls. Strikingly, all but 2 proteins (L-FABP and a putative uncharacterized protein) are metabolic enzymes. At 180 minutes following PH, 3 proteins were identified solely in the 70% PH group, whereas 25 proteins were observed solely in the 90% PH group. Interestingly, 12 proteins that were reduced in plasma abundance by at least 2-fold compared to the sham controls were found exclusively in the 90% PH group at 180 minutes after surgery. Five proteins (major urinary proteins 2, 3, and 5, major urinary protein gene family member 3, and Mup1 protein) of the 12 identified proteins belong to the major urinary protein family.
Table 3.
List of all identified priority 1 proteins (q < 0.05 with minimum two-fold change)
| Protein ID | 70% PH vs Sham | FC | Protein ID | 90% PH vs Sham | FC | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 20 min | IPI00120451.1 | Liver fatty acid binding protein | 2.3 | ||||
|
| |||||||
| 60 min | IPI00471239.2 | Betaine homocysteine S-methyltransferase 2 | 2.2 | IPI00471239.2 | Betaine homocysteine S-methyltransferase 2 | 2.3 | |
|
| |||||||
| 180 min | Common | IPI00471239.2 | Betaine homocysteine S-methyltransferase 2 | 4.6 | IPI00471239.2 | Betaine homocysteine S-methyltransferase 2 | 5.7 |
| IPI00121280.5 | Glutathione S-transferase Mu 7 | 3.8 | IPI00121280.5 | Glutathione S-transferase Mu 7 | 4.3 | ||
| IPI00228630.5 | Fructose 1,6-bisphosphatase 1 | 3.5 | IPI00228630.5 | Fructose 1,6-bisphosphatase 1 | 3.5 | ||
| IPI00130950.1 | Betaine homocysteine S-methyltransferase 1 | 3.4 | IPI00130950.1 | Betaine homocysteine S-methyltransferase 1 | 3.9 | ||
| IPI00555023.2 | Glutathione S-transferase P 1 | 3.0 | IPI00555023.2 | Glutathione S-transferase P 1 | 2.6 | ||
| IPI00283531.8 | Glutathione S-transferase P 2 | 2.8 | IPI00283531.8 | Glutathione S-transferase P 2 | 2.2 | ||
| IPI00127206.6 | Fructose bisphosphate aldolase B | 2.7 | IPI00127206.6 | Fructose bisphosphate aldolase B | 3.0 | ||
| IPI00467066.7 | Glycine N-methyltransferase | 2.6 | IPI00467066.7 | Glycine N-methyltransferase | 3.2 | ||
| IPI00122344.1 | Cystathionine gamma-lyase | 2.4 | IPI00122344.1 | Cystathionine gamma-lyase | 2.2 | ||
| IPI00554931.3 | 4-hydroxyphenylpyruvate dioxygenase | 2.4 | IPI00554931.3 | 4-hydroxyphenylpyruvate dioxygenase | 2.3 | ||
| IPI00230212.5 | Glutathione S-transferase Mu 1 | 2.4 | IPI00230212.5 | Glutathione S-transferase Mu 1 | 2.5 | ||
| IPI00310035.3 | Fumarylacetoacetase | 2.4 | IPI00310035.3 | Fumarylacetoacetase | 2.6 | ||
| IPI00653931.1 | Putative uncharacterized protein | 2.4 | IPI00653931.1 | Putative uncharacterized protein | 2.6 | ||
| IPI00120451.1 | Liver fatty acid binding protein | 2.2 | IPI00120451.1 | Liver fatty acid binding protein | 5.1 | ||
| IPI00319652.2 | Glutathione peroxidase 1 | 2.2 | IPI00319652.2 | Glutathione peroxidase 1 | 2.6 | ||
| IPI00314788.5 | Argininosuccinate lyase | 2.2 | IPI00314788.5 | Argininosuccinate lyase | 2.1 | ||
| IPI00331241.8 | Glutathione S-transferase A3 | 2.1 | IPI00331241.8 | Glutathione S-transferase A3 | 2.7 | ||
| IPI00117914.3 | Arginase 1 | 2.0 | IPI00117914.3 | Arginase 1 | 2.2 | ||
| IPI00626662.3 | Retinal dehydrogenase 1 | 2.0 | IPI00626662.3 | Retinal dehydrogenase 1 | 2.2 | ||
|
| |||||||
| Uncommon | IPI00623845.3 | Selenium binding protein 1 | 2.5 | IPI00469114.5 | Hemoglobin subunit alpha | 3.8 | |
| IPI00323816.5 | Selenium binding protein 2 | 2.5 | IPI00831055.2 | Beta globin | 3.6 | ||
| IPI00114330.3 | Homogentisate 1,2 dioxygenase | 2.1 | IPI00762198.2 | Beta globin | 3.5 | ||
| IPI00110658.1 | Putative uncharacterized protein | 3.5 | |||||
| IPI00316491.4 | Hemoglobin subunit beta 2 | 3.4 | |||||
| IPI00555059.2 | Peroxiredoxin 6 | 3.3 | |||||
| IPI00553333.2 | Hemoglobin subunit beta 1 | 3.1 | |||||
| IPI00130589.8 | Superoxide dismutase [Cu-Zn] | 2.8 | |||||
| IPI00222430.5 | Diazepam binding inhibitor isoform 1 | 2.5 | |||||
| IPI00129393.1 | Platelet factor 4 | 2.3 | |||||
| IPI00122684.2 | Enolase | 2.0 | |||||
| IPI00753038.1 | Sorbitol dehydrogenase | 2.0 | |||||
| IPI00336362.2 | Aldehyde dehydrogenase, cytosolic 1 | 2.0 | |||||
| IPI00120832.1 | Major urinary protein 3 | −2.2 | |||||
| IPI00762131.2 | 17 kDa protein | −2.2 | |||||
| IPI00761300.2 | 11 kDa protein | −2.2 | |||||
| IPI00118457.1 | Serum amyloid A-2 protein | −2.6 | |||||
| IPI00466399.1 | Major urinary protein 2 | −2.6 | |||||
| IPI00480401.3 | Major urinary protein gene family member 3 | −2.6 | |||||
| IPI00132542.1 | Putative uncharacterized protein | −2.6 | |||||
| IPI00930848.1 | Mup1 protein | −2.8 | |||||
| IPI00890309.1 | Hypothetical protein LOC100039008 | −2.8 | |||||
| IPI00115243.1 | Major urinary protein 5 | −2.8 | |||||
| IPI00649653.2 | Hypothetical protein LOC100039089 | −2.9 | |||||
| IPI00118455.1 | Serum amyloid A 1 protein | −3.0 | |||||
To find plasma proteins associated with the extent of liver mass reduction, the plasma protein profile of the 90% PH group was compared with that of the 70% PH group with less stringency (q < 0.05 with minimum change of 1.5-fold). None of the proteins were found at 20 and 60 minutes after PH. Even at 180 minutes, only 8 proteins were observed (Table 4). The data indicated that 90% PH and 70% PH induced similar plasma protein responses during the first three hours post-surgery. These 8 proteins may be associated with the extent of liver mass decrease.
Table 4.
A list of the priority 1 proteins identified by comparision between 90% and 70% PH group at 180 minutes after surgery (q < 0.05 with minimum 1.5-fold change)
| Protein ID | 90% PH vs 70% PH | FC |
|---|---|---|
| IPI00111210.1 | Platelet basic protein | 2.47 |
| IPI00120451.1 | Liver Fatty acid-binding protein | 2.28 |
| IPI00222430.5 | Diazepam binding inhibitor isoform 1 | 2.00 |
| IPI00555059.2 | Peroxiredoxin-6 | 1.66 |
| IPI00130589.8 | Superoxide dismutase [Cu-Zn] | 1.56 |
| IPI00133456.1 | Regucalcin | 1.52 |
| IPI00762131.2 | 17 kDa protein | −1.52 |
| IPI00761300.2 | 11 kDa protein | −1.52 |
To define the major biological processes in which the identified plasma proteins are involved, the PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System (http://www.patherdb.org) was used to group all priority 1 proteins listed in Table 3 based on their biological functions. In this program, one protein could be categorized into multiple biological function groups. In the 70% PH group, a total of 22 proteins were analyzed, excluding 1 protein due to unknown function (Table 5). These 21 proteins were categorized into three main function groups: metabolic process, immune system response, and response to stimulus. Fourteen of the 21 proteins (66.67%) are associated with the metabolic process. In the 90% PH group, 44 proteins were input into the program, and 13 proteins were excluded due to unknown identity or function. Thus, a total of 31 proteins were analyzed (Table 6). As a result, the majority of the proteins (19 proteins, 61.3% of the 31 proteins) participate in the metabolic process. A less dominant group of proteins (13 proteins) are associated with immune system response. Ten proteins participate in response to stimulus. Taken together, these data demonstrate that, regardless of the extent of liver mass loss, most of the immediate-early response plasma proteins are involved in the metabolic process.
Table 5.
A summary of the biological functions of the priority 1 plasma proteins listed in 70% PH group in Table 3
| Biological Process | Number of Proteins | Protein List |
|---|---|---|
| Metabolic process | 14 | Fructose 1,6-bisphosphatase 1, Betaine homocysteine S-methyltransferase 1, Retinal dehydrogenase 1, Liver fatty acid-binding protein, Glutathione peroxidase 1, Cystathionine gamma-lyase, 4-hydroxyphenylpyruvate dioxygenase, Fumarylacetoacetase, Argininosuccinate lyase, Homogentisate 1,2-dioxygenase, Betaine-homocysteine S-methyltransferase 2, Fructose bisphosphate aldolase B, Arginase 1, Glutathione S-transferase A3 |
| Immune system response | 8 | Selenium binding protein 1, Selenium binding protein 2, Glutathione S-transferase P 2, Glutathione S-transferase A3, Glutathione S-transferase Mu 7, Glutathione S-transferase Mu 1, Glutathione S-transferase P 1, Glutathione peroxidase 1 |
| Response to stimulus | 6 | Glutathione S-transferase P 2, Glutathione S-transferase A3, Glutathione S-transferase Mu 7, Glutathione S-transferase Mu 1, Glutathione peroxidase 1, Glutathione S-transferase P 1 |
| System process | 1 | Glutathione S-transferase A3 |
| Transport | 1 | Glutathione S-transferase A3 |
| Cell communication | 1 | Liver fatty acid binding protein |
| Cellular Process | 1 | Liver fatty acid binding protein |
| Developmental Process | 1 | Liver fatty acid binding protein |
| Unknown | 1 | IPI00653931 |
Table 6.
A summary of the biological functions of the priority 1 plasma proteins listed in 90% PH group in Table 3
| Biological Process | Number of Proteins | Protein List |
|---|---|---|
| Metabolic process | 19 | Fructose1,6-bisphosphatase 1, Betaine homocysteine S-methyltransferase 1, Arginase 1 Retinal dehydrogenase 1, Liver fatty acid binding protein, OTTMUSG00000012492, Cystathionine gamma-lyase, 4-hydroxyphenylpyruvate dioxygenase, Fumarylacetoacetase, Argininosuccinate lyase, Major urinary protein 5, Betaine homocysteine S-methyltransferase 2, Fructose bisphosphate aldolase B, Glutathione peroxidase 1, Aldehyde dehydrogenase cytosolic 1, Sorbitol dehydrogenase, Glutathione S-transferase A3, Peroxiredoxin 6, Major urinary protein 3 |
| Immune system response | 13 | Glutathione S-transferase Mu 7, Glutathione S-transferase Mu 1, OTTMUSG00000012492, Glutathione S-transferase P 2, Major urinary protein 5, Glutathione S-transferase A3, Peroxiredoxin 6, Major urinary protein 3, Glutathione S-transferase P 1, Platelet factor 4, Glutathione peroxidase 1, Amyloid protein A, Serum amyloid A-1 protein |
| Response to stimulus | 10 | OTTMUSG00000012492, Glutathione S-transferase P 2, Major urinary protein 5, Platelet factor 4 Glutathione S-transferase A3, Glutathione S-transferase Mu 7, Glutathione S-transferase Mu 1, Glutathione peroxidase 1, Major urinary protein 3, Glutathione S-transferase P 1 |
| System process | 7 | Hemoglobin subunit alpha, OTTMUSG00000012492, Major urinary protein 5, Glutathione S-transferase A3, Hemoglobin subunit beta 1, Hemoglobin subunit beta 2, Major urinary protein 3 |
| Transport | 7 | Liver fatty acid binding protein, Hemoglobin subunit alpha, OTTMUSG00000012492, Major urinary protein 5, Hemoglobin subunit beta 1, Hemoglobin subunit beta 2, Major urinary protein 3 |
| Cell communication | 5 | Liver fatty acid-binding protein, Major urinary protein 5, Major urinary protein 3, OTTMUSG00000012492, Platelet factor 4 |
| Cellular Process | 5 | Liver fatty acid binding protein, OTTMUSG00000012492, Major urinary protein 5, Major urinary protein 3, Platelet factor 4 |
| Reproduction | 3 | OTTMUSG00000012492, Major urinary protein 5, Major urinary protein 3 |
| Apoptosis | 2 | Sorbitol dehydrogenase, Platelet factor 4 |
| Developmental Process | 2 | Liver fatty acid binding protein, Platelet factor 4 |
| Unknown | 13 | Putative uncharacterized protein (3), Beta globin (2), Diazepam binding inhibitor isoform 1, Major urinary protein 2, Major urinary protein gene family member 3, Mup1 protein, Enolase, Hypothetical protein LOC100039008, 17 kDa protein, 11 kDa protein |
Note: Number in parantheses shows the number of protein with same name but different protien ID.
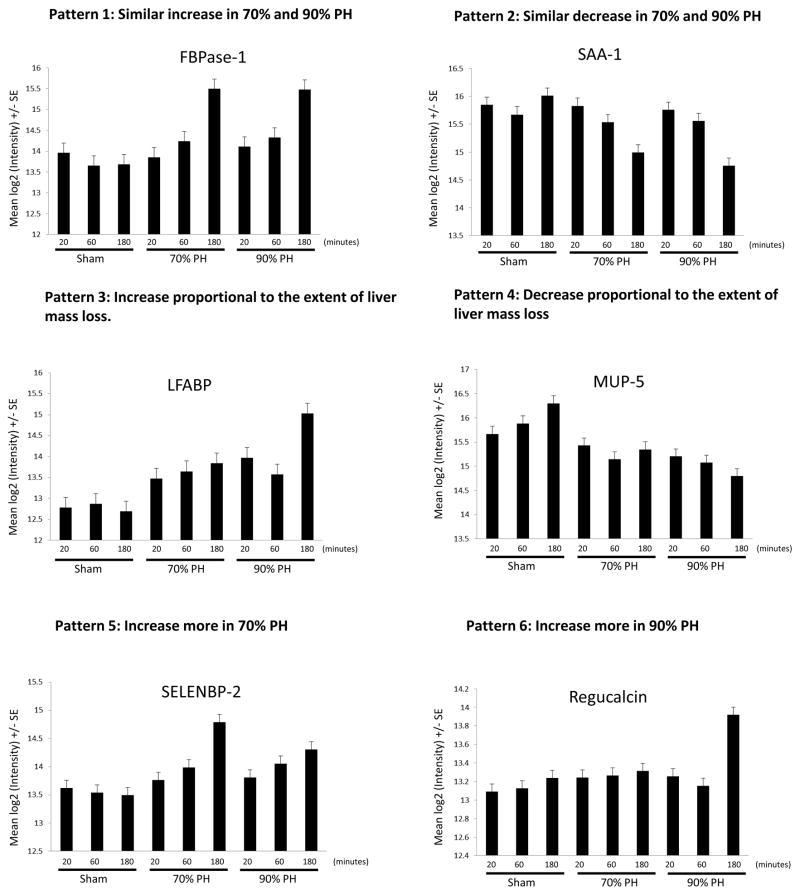
We found 6 typical patterns that can be used to monitor the alterations of plasma protein expression during the first three hours after PH. The representative proteins that exhibit those patterns are depicted in Figure 1. The identification of these patterns helps us to determine how a protein behaves in plasma expression after PH and whether such behavior depends on the extent of liver mass loss.
Fig. 1. Representative proteins showing typical patterns of plasma protein expression 20, 60, and 180 minutes after sham operation, 70% partial hepatectomy (PH), or 90% PH.
Six major patterns of plasma protein expression were noted and a representative protein displaying each pattern is presented in panels A to F. The mean Log2 intensity ± SE (y-axis) is shown for each protein (a difference of one unit is equivalent to a two-fold change). The x-axis is labeled with three experiment groups (sham operation group, 70% PH group, and 90% PH group) and three time points after surgery (20 minutes, 60 minutes, and 180 minutes).). FBPase-1, fructose 1,6-bisphosphatase 1; LFABP, liver fatty acid binding protein; MUP-5, major urinary protein 5; SAA-1, serum amyloid A-1 protein; SELENBP-2, selenium-binding protein 2.
To verify the global proteomic data, the plasma expression of several selected proteins was analyzed by western blotting (Figure 2). L-FABP plasma abundance increased slightly at 60 and 180 minutes following 70% PH and was highest at 180 minutes after 90% PH. BHMT2 plasma levels were elevated 60 minutes and 180 minutes after 70% PH and 90% PH in comparison with the sham controls. Plasma FBPase1 was only detected at 180 minutes after both types of PH. A PH-dependent increase in SELEBP1 plasma expression was most evident at 180 minutes post-70% PH. Collectively, the results revealed plasma expression patterns of these proteins that are similar to those demonstrated by quantitative proteomic analysis.
Fig. 2. Plasma protein levels of L-FABP, BHMT2, FBPase-1, and SELEBP1 after partial hepatectomy (PH).
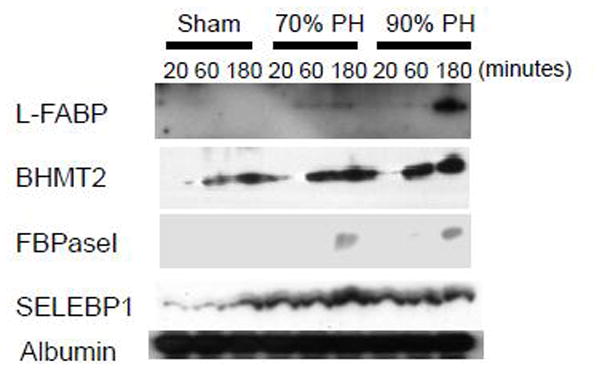
Blood was collected 20, 60 or 180 minutes after sham operation, 70% PH, or 90% PH. EDTA-plasma was prepared. Aliquots of plasma from each mouse per time point per surgery group were combined. One microliter of each combined plasma sample was subjected to western blotting with antibodies against the proteins indicated. Albumin protein levels were used as loading controls. L-FABP, liver fatty acid binding protein; BHMT2, betaine-homocysteine S-methyltransferase 2; FBPase-1, fructose 1,6-bisphosphatase 1; and SELEBP1, selenium binding protein 1.
Using ELISA assays, several groups demonstrated that PH induces increases in protein concentrations of several cytokines and growth factors, such as IL-6, TNFα, and HGF, in the circulation [34, 35]. However, those proteins were not detected in our plasma samples using the LFQP approach. To evaluate whether those proteins exist in our plasma samples, we quantified IL-6, a typical proinflammatory cytokine associated with liver regeneration, by ELISA assay in aliquots of plasma samples used in the proteomic analysis (Fig. 3). As a result, circulating IL-6 protein exhibited significant increases at 180 minutes after both 70% and 90% PH in comparison with sham controls. The average IL-6 concentration was 1,193 pg/mL in the blood at 180 minutes following 70% PH, resembling previous reports [34, 35]. At the same time point, IL-6 protein displayed higher magnitude of response to 90% PH compared with 70% PH, reaching 2,165 pg/mL. The result suggests that PH-induced enrichment of circulating IL-6 did not reach a level that can be detected by the proteomic approach.
Fig. 3. Plasma interleukin 6 (IL-6) levels after partial hepatectomy (PH).
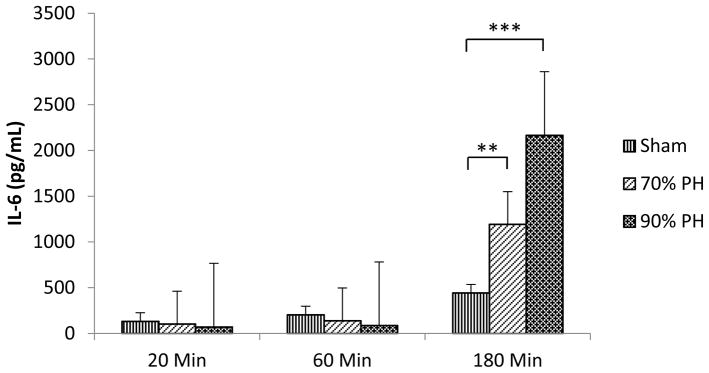
Mice were subjected to sham operation, 70% PH, or 90% PH. Blood was collected from the inferior vena cava at 20, 60, or 180 minutes after surgery. EDTA-plasma was prepared and used for IL-6 quantification by assay. **, P < 0.01; ***, P < 0.001 in comparison with sham operation controls (n= 3–5).
Discussion
Our study revealed a group of plasma proteins associated with the body’s immediate-early responses to massive liver mass loss. It is well established that 70% PH induces robust hepatic regenerative response. Thus, we anticipated that, with this model, cytokines or growth factors associated with inflammatory response, cell proliferation, or organ growth could be identified as protein factors that participate in triggering hepatic regeneration. Surprisingly, no such factors were found among all priority 1 proteins exhibiting at least 2-fold or even 1.5-fold changes during the first three hours after 70% PH (Table 3 and Supplementary Table 1). Notably, in the 90% PH model, only one cytokine, platelet factor 4 (PF4), was identified as a priority 1 protein at 180 minutes following surgery (Table 3). PF4 is involved in inflammation and wound healing [36]. Here we linked PF4 with liver regeneration. Further studies are needed to determine whether PF4 plays a role in mediating hepatic regenerative responses. Remarkably, in both the 70% and 90% PH models, the majority of the plasma proteins identified are catalytic enzymes (Table 3) and most of the proteins are associated with metabolism (Tables 5 and 6). Among the 22 and 37 priority 1 proteins with known identity in the 70% PH and 90% PH groups, 20 (90.9%) and 22 (59.5%) are catalyzing enzymes, respectively (Table 3). The findings suggest that systemic metabolic changes may dominate the most immediate-early responses of the body to liver mass reduction. Several lines of evidence implicate the connection of metabolic pathways with DNA replication during liver regeneration [7, 8, 11, 37–42]. PH or transplantation of reduced-size livers may lead to a hypermetabolic state [43]. Our findings support the notion that increased metabolic demands of the body may function as a sensor that calibrates the hepatic regenerative response [4]. This notion is further supported by a very recent report that demonstrates that pancreatic beta cell regeneration is controlled by glucose metabolism [44].
One intriguing question is how the enzymes identified in the current study are released into the blood stream. Most of the enzymes are known to be expressed in the liver. Increased concentrations of liver enzymes in the blood are often considered to be indicative of hepatic damage. However, it is well established that 70% PH triggers robust hepatic regenerative response without major cellular injury and is considered to be a clean model for studying liver regeneration [25]. Indeed, plasma levels of alanine aminotransferase (ALT) and sorbitol dehydrogenase (SDH), which are often used to evaluate liver injury, were not significantly changed during the first three hours, whereas plasma level of aspartate transaminase (AST), another liver injury index, was increased only by 1.61-fold at 180 minutes after 70% PH (Supplemental Table 1). The data indicate that liver injury was minimal in mice subjected to 70% PH within the first three hours after surgery. Similar plasma protein profiles between 70% PH and 90% PH are also indicative of minimal liver injury in mice subjected to 90% PH during the period. Furthermore, a comparison of the plasma protein profiles between the 70% PH group and the 90% PH group did not elicit any significant differences at each time point in the plasma concentrations of ALT, AST, and SDH (Supplemental Table 1). If hepatic cellular damage is the cause of increased concentrations of liver enzymes in the circulation, then we would see the appearance of a full panel of liver enzymes, including phase I metabolic enzymes that are abundant in hepatocytes, in the blood. However, none of the cytochrome P450 monooxygenases exhibited more than 1.5-fold PH-dependent changes at any time point during the first three hours after PH. At this juncture, we are not able to address the question of how metabolic enzymes are released into the blood without major liver injury after PH. We believe that the appearance of those enzymes in the circulation may reflect a hypermetabolic state in the remaining liver following PH.
A number of proteins identified in our studies are associated with lipid, amino acid, and glucose metabolism and phase II detoxification. L-FABP belongs to a family of small and highly conserved proteins that bind long-chain fatty acids and play important roles in fatty acid uptake, transport, and metabolism [45]. However, a lack of L-FABP has no apparent effect on liver regeneration, although hepatic fat accumulation is reduced [46]. Thus, a redundant mechanism should exist if L-FABP participates in inducing immediate-early response during liver regeneration. A group of identified proteins are associated with amino acid metabolism, including BHMT2, BHMT, glycine N-methyltransferase, cystathionine gamma-lyase, argininosuccinate lyase, 4-hydroxyphenylpyruvate dioxygenase, and fumarylacetoacetate hydrolase [47–51]. Rapid increases of these enzymes in plasma expression after PH may reflect the enhancement of amino acid metabolism in response to liver mass decrease. Two identified proteins associated with glucose metabolism are fructose 1,6-bisphosphatase 1 and fructose bisphosphatase aldolase B [52, 53], which might be indicative of the enhancement of hepatic gluconeogenesis to compensate for a blood glucose deficiency caused by PH. Plasma protein levels of 5 members of the glutathione S-transferase (GST) family (GST P1, P2, Mu1, Mu7, and A3) were elevated by 2.1- to 4.3- fold, regardless of the extent of liver mass loss (Table 3). GSTs are major phase II detoxification enzymes that also carry out a range of other functions, including steroid and leukotriene biosynthesis, peroxide degradation, and ligand binding and transport [54]. Thus, increases of those GSTs in plasma concentration may reflect hepatic metabolic response to PH to compensate for the reduced capacity of xenobiotic and endobiotic metabolism in the remaining liver. We observed that plasma expression of 5 major urinary proteins (MUPs) was decreased at 180 minutes after 90% PH (Table 3). MUPs are a family of proteins that contain a conserved β-barrel structure with a characteristic central hydrophobic pocket. They are secreted by the liver, are excreted into the urine, and function as regulators of pheromone signaling [55]. Very recent studies revealed a novel function of MUPs in regulating glucose and lipid metabolism. MUP1 suppresses hepatic gluconeogenesis and lipogenesis and promotes energy expenditure in skeletal muscle [56, 57]. Thus, the decrease of plasma expression of MUPs may stimulate hepatic glucose and lipid synthesis to meet the body’s metabolic needs after PH. Collectively, most of the proteins identified in our study are involved in a broad range of metabolic processes, which might reflect the hypermetabolic state of partially hepatectomized liver.
We found that 70% PH and 90% PH induced similar changes in the plasma protein profile within the first three hours after surgery. When these two PH groups were compared, only 8 priority 1 proteins displayed changes above 1.5-fold in the plasma concentration at 180 minutes after surgery, whereas none of the priority 1 proteins showed such changes at 20 and 60 minutes following surgery (Table 4). The data suggest that, irrespective of the percentage of liver mass loss, hepatectomy induces similar immediate-early responses in plasma protein profile. In contrast to rats who can survive from 90% PH, mice subjected to 90% PH usually die within 24 hours of surgery, most likely due to the impact of portal vein pressure on hepatic artery flow and/or insufficient liver metabolic capacity [25, 58]. However, in the study of parabiotic rats in pairs, total removal of the liver in one rat induced maximum regenerative response in the intact liver of the other rat in the pair [1]. This previous observation indicated that the removal of the entire liver can induce the strongest regenerative response. In line with this finding, our data suggest that the immediate-early response of the body to 90% PH is intact and hepatic regeneration failure caused by 90% PH may be due to the later events that occur after the initiation stage of liver regeneration.
Notably, proinflammatory cytokines and growth factors known to be associated with liver regeneration, including IL-6, TNFα, and HGF, are not in the list of the proteins detected in our study. It is known that mRNA expression of hepatic IL-6, TNFα, and HGF is upregulated within the first few hours after PH [35, 59]. However, none of the reported proteomic studies detected those proteins in the liver after PH [18–20, 22]. IL-6, TNFα, and HGF proteins were also not detectable by western blotting analysis in the remnant livers in the first three hours following PH in the current study (data not shown). We did not find any reports showing PH-dependent expression of these proteins in the liver by western blotting analysis. Thus, it is likely that the abundance of these proteins in regenerating livers is still too low to be detected by proteomic and immunoblotting approaches. Using ELISA assay, several groups demonstrated that the levels of circulating IL-6, TNFα, IL-1β and HGF are rapidly increased following PH [34, 35, 60–62]. Within the first four hours post-70% PH in mice, the highest concentrations of those proteins in the blood were 1,000 to 2,000 pg/mL for IL-6 [34, 35, 60], approximate 15 pg/mL for TNFα [34, 35], around 100 pg/mL for IL-1β [34], and about 1,250 pg/mL for HGF [34]. We quantified IL-6 protein in our plasma samples used for the proteomic study by ELISA assay. The average plasma IL-6 concentration elevated to1,193 pg/mL at 180 minutes after 70% PH (Fig. 3), which is within the reported range. At the same time point, 90% PH further increased the level of plasma IL-6 protein to 2,165 pg/mL (Fig. 3). However, IL-6, TNFα, and HGF were not detected in our plasma samples by western blotting analysis (data not shown). Therefore, plasma abundance of those proteins may not reach a level that can be detected by either proteomic approach used in the current study or immunoblotting measurement. Vice versa, the sensitivity of the proteomic approach may not allow for profiling very low-abundance proteins in the blood.
Our study demonstrated the value and robustness of the LFQP approach in evaluating hepatic regenerative and metabolic responses during liver regeneration by profiling plasma proteins. However, our study has several limitations. Fourteen priority 1 proteins (1 in the 70% PH group and 13 in the 90% PH group, Tables 5 & 6) that displayed at least 2-fold changes could not be identified and, thus, cannot be analyzed due to incomplete annotation of the mouse protein database. In addition, priority 2, 3, and 4 proteins were not included in our analysis because of low protein identification confidence. It is likely that some important proteins associated with hepatic regenerative and metabolic responses are among those unknown proteins and, thus, have not been identified or analyzed by our study. Moreover, our study suggests that the LFQP approach may not be sensitive enough to detect very low-abundance proteins in the blood.
In summary, using the proteomic approach and two PH models, we analyzed plasma protein profiles within the first three hours after PH. A group of immediate-early response plasma proteins was revealed in each PH model. A group of proteins largely associated with metabolism exhibits an increase in plasma abundance after 70% PH. Moreover, 90% PH induces plasma protein responses similar to 70% PH. A dominant group of proteins associated with metabolism and one known cytokine (platelet factor 4) significantly respond to 90% PH. Our findings suggest that systemic metabolic responses might be an important factor to consider in the future efforts on identifying the initial trigger of liver regeneration.
Supplementary Material
Acknowledgments
Grant: This work was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (7RO1DK07596).
Abbreviations
- PH
partial hepatectomy
- IL-6
interleukin 6
- TNF
tumor necrosis factor
- EGF
epidermal growth factor
- HGF
hepatocyte growth factor
- L-FABP
liver fatty acid binding protein
- BHMT
betaine-homocysteine S-methyltransferase
- FBPase
fructose 1,6-bisphosphatase
- SELEBP
selenium binding protein
- CV
coefficient of variation
- FC
fold change
- LFQP
label-free quantitative proteomics
References
- 1.Moolten FL, Bucher NL. Regeneration of rat liver: transfer of humoral agent by cross circulation. Science. 1967;158(798):272–4. doi: 10.1126/science.158.3798.272. [DOI] [PubMed] [Google Scholar]
- 2.Grisham JW, Leong GF, Hole BV. Heterotopic Partial Autotransplantation of Rat Liver: Technic and Demonstration of Structure and Function of the Graft. Cancer Res. 1964;24:1474–95. [PubMed] [Google Scholar]
- 3.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213(2):286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43(2 Suppl 1):S45–53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 5.Zimmermann A. Liver regeneration: the emergence of new pathways. Med Sci Monit. 2002;8(3):RA53–63. [PubMed] [Google Scholar]
- 6.Koniaris LG, et al. Liver regeneration. J Am Coll Surg. 2003;197(4):634–59. doi: 10.1016/S1072-7515(03)00374-0. [DOI] [PubMed] [Google Scholar]
- 7.Jiang YP, Ballou LM, Lin RZ. Rapamycin-insensitive regulation of 4e-BP1 in regenerating rat liver. J Biol Chem. 2001;276(14):10943–51. doi: 10.1074/jbc.M007758200. [DOI] [PubMed] [Google Scholar]
- 8.Volarevic S, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288(5473):2045–7. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 9.Tsukamoto H. Cytokine regulation of hepatic stellate cells in liver fibrosis. Alcohol Clin Exp Res. 1999;23(5):911–6. [PubMed] [Google Scholar]
- 10.Tsukada K, et al. Ribosomal change in liver after partial hepatectomy and acute stress. J Biol Chem. 1968;243(6):1152–9. [PubMed] [Google Scholar]
- 11.Goggin MM, et al. Rapamycin-sensitive induction of eukaryotic initiation factor 4F in regenerating mouse liver. Hepatology. 2004;40(3):537–44. doi: 10.1002/hep.20338. [DOI] [PubMed] [Google Scholar]
- 12.Tsukada K, et al. Relationship between the ribosomal alteration after partial hepatectomy and the increase in liver protein synthesis in vivo. J Biol Chem. 1968;243(6):1160–5. [PubMed] [Google Scholar]
- 13.Delgado-Coello B, et al. Cholesterol: recapitulation of its active role during liver regeneration. Liver Int. 2011;31(9):1271–84. doi: 10.1111/j.1478-3231.2011.02542.x. [DOI] [PubMed] [Google Scholar]
- 14.Shteyer E, et al. Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology. 2004;40(6):1322–32. doi: 10.1002/hep.20462. [DOI] [PubMed] [Google Scholar]
- 15.Dai G, et al. Pregnane X receptor is essential for normal progression of liver regeneration. Hepatology. 2008;47(4):1277–87. doi: 10.1002/hep.22129. [DOI] [PubMed] [Google Scholar]
- 16.Huang W, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312(5771):233–6. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 17.Anderson SP, et al. Delayed liver regeneration in peroxisome proliferator-activated receptor-alpha-null mice. Hepatology. 2002;36(3):544–54. doi: 10.1053/jhep.2002.35276. [DOI] [PubMed] [Google Scholar]
- 18.Strey CW, et al. Partial hepatectomy induced liver proteome changes in mice. Proteomics. 2005;5(1):318–25. doi: 10.1002/pmic.200400913. [DOI] [PubMed] [Google Scholar]
- 19.Guo F, et al. Proteomic analysis of the transition from quiescent to proliferating stages in rat liver hepatectomy model. Proteomics. 2006;6(10):3075–86. doi: 10.1002/pmic.200500322. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, et al. Liver proteome analysis of adaptive response in rat immediately after partial hepatectomy. Proteomics. 2007;7(23):4398–407. doi: 10.1002/pmic.200600913. [DOI] [PubMed] [Google Scholar]
- 21.Cao H, et al. Proteomic analysis of regenerating mouse liver following 50% partial hepatectomy. Proteome Sci. 2009;7:48. doi: 10.1186/1477-5956-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh HC, et al. Protein profilings in mouse liver regeneration after partial hepatectomy using iTRAQ technology. J Proteome Res. 2009;8(2):1004–13. doi: 10.1021/pr800696m. [DOI] [PubMed] [Google Scholar]
- 23.Li X, et al. Proteomics analysis of plasma membrane from liver sinusoidal endothelial cells after partial hepatectomy by an improved two-dimensional electrophoresis. Mol Cell Biochem. 2010;344(1–2):137–50. doi: 10.1007/s11010-010-0537-z. [DOI] [PubMed] [Google Scholar]
- 24.Yuan X, et al. Lipid metabolism and peroxisome proliferator-activated receptor signaling pathways participate in late-phase liver regeneration. J Proteome Res. 2011;10(3):1179–90. doi: 10.1021/pr100960h. [DOI] [PubMed] [Google Scholar]
- 25.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176(1):2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greene AK, Puder M. Partial hepatectomy in the mouse: technique and perioperative management. J Invest Surg. 2003;16(2):99–102. [PubMed] [Google Scholar]
- 27.Wang M, et al. Label-free mass spectrometry-based protein quantification technologies in proteomic analysis. Brief Funct Genomic Proteomic. 2008;7(5):329–39. doi: 10.1093/bfgp/eln031. [DOI] [PubMed] [Google Scholar]
- 28.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 29.Hale JE, et al. A simplified procedure for the reduction and alkylation of cysteine residues in proteins prior to proteolytic digestion and mass spectral analysis. Anal Biochem. 2004;333(1):174–81. doi: 10.1016/j.ab.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 30.Higgs RE, et al. Estimating the statistical significance of peptide identifications from shotgun proteomics experiments. J Proteome Res. 2007;6(5):1758–67. doi: 10.1021/pr0605320. [DOI] [PubMed] [Google Scholar]
- 31.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–7. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 32.Higgs RE, et al. Comprehensive label-free method for the relative quantification of proteins from biological samples. J Proteome Res. 2005;4(4):1442–50. doi: 10.1021/pr050109b. [DOI] [PubMed] [Google Scholar]
- 33.Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Sgroi A, et al. Interleukin-1 receptor antagonist modulates the early phase of liver regeneration after partial hepatectomy in mice. PLoS ONE. 2011;6(9):e25442. doi: 10.1371/journal.pone.0025442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin S, et al. Enhanced liver regeneration in IL-10-deficient mice after partial hepatectomy via stimulating inflammatory response and activating hepatocyte STAT3. Am J Pathol. 2011;178(4):1614–21. doi: 10.1016/j.ajpath.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandercappellen J, Van Damme J, Struyf S. The role of the CXC chemokines platelet factor-4 (CXCL4/PF-4) and its variant (CXCL4L1/PF-4var) in inflammation, angiogenesis and cancer. Cytokine Growth Factor Rev. 2011;22(1):1–18. doi: 10.1016/j.cytogfr.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 37.Mead JE, et al. Induction of replicative competence (“priming”) in normal liver. Cancer Res. 1990;50(21):7023–30. [PubMed] [Google Scholar]
- 38.McGowan J, Atryzek V, Fausto N. Effects of protein-deprivation on the regeneration of rat liver after partial hepatectomy. Biochem J. 1979;180(1):25–35. doi: 10.1042/bj1800025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelsen CJ, et al. Amino acids regulate hepatocyte proliferation through modulation of cyclin D1 expression. J Biol Chem. 2003;278(28):25853–8. doi: 10.1074/jbc.M302360200. [DOI] [PubMed] [Google Scholar]
- 40.Martin KA, Blenis J. Coordinate regulation of translation by the PI 3-kinase and mTOR pathways. Adv Cancer Res. 2002;86:1–39. doi: 10.1016/s0065-230x(02)86001-8. [DOI] [PubMed] [Google Scholar]
- 41.Avruch J, et al. Recent advances in the regulation of the TOR pathway by insulin and nutrients. Curr Opin Clin Nutr Metab Care. 2005;8(1):67–72. doi: 10.1097/00075197-200501000-00010. [DOI] [PubMed] [Google Scholar]
- 42.Kim DH, Sabatini DM. Raptor and mTOR: subunits of a nutrient-sensitive complex. Curr Top Microbiol Immunol. 2004;279:259–70. doi: 10.1007/978-3-642-18930-2_15. [DOI] [PubMed] [Google Scholar]
- 43.Lehmann TG, et al. Minimizing oxidative stress by gene delivery of superoxide dismutase accelerates regeneration after transplantation of reduced-size livers in the rat. Liver Transpl. 2006;12(4):550–9. doi: 10.1002/lt.20632. [DOI] [PubMed] [Google Scholar]
- 44.Porat S, et al. Control of pancreatic beta cell regeneration by glucose metabolism. Cell Metab. 2011;13(4):440–9. doi: 10.1016/j.cmet.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Atshaves BP, et al. Liver fatty acid-binding protein and obesity. J Nutr Biochem. 2010;21(11):1015–32. doi: 10.1016/j.jnutbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newberry EP, et al. Altered hepatic triglyceride content after partial hepatectomy without impaired liver regeneration in multiple murine genetic models. Hepatology. 2008;48(4):1097–105. doi: 10.1002/hep.22473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mani S, Yang G, Wang R. A critical life-supporting role for cystathionine gamma-lyase in the absence of dietary cysteine supply. Free Radic Biol Med. 2011;50(10):1280–7. doi: 10.1016/j.freeradbiomed.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 48.Turner MA, et al. Human argininosuccinate lyase: a structural basis for intragenic complementation. Proc Natl Acad Sci U S A. 1997;94(17):9063–8. doi: 10.1073/pnas.94.17.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryle MJ, Hausinger RP. Non-heme iron oxygenases. Curr Opin Chem Biol. 2002;6(2):193–201. doi: 10.1016/s1367-5931(02)00302-2. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura K, et al. Animal models of tyrosinemia. J Nutr. 2007;137(6 Suppl 1):1556S–1560S. doi: 10.1093/jn/137.6.1556S. discussion 1573S–1575S. [DOI] [PubMed] [Google Scholar]
- 51.Pajares MA, Perez-Sala D. Betaine homocysteine S-methyltransferase: just a regulator of homocysteine metabolism? Cell Mol Life Sci. 2006;63(23):2792–803. doi: 10.1007/s00018-006-6249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalby AR, Tolan DR, Littlechild JA. The structure of human liver fructose-1,6-bisphosphate aldolase. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 11):1526–33. doi: 10.1107/s0907444901012719. [DOI] [PubMed] [Google Scholar]
- 53.van Poelje PD, Potter SC, Erion MD. Fructose-1, 6-bisphosphatase inhibitors for reducing excessive endogenous glucose production in type 2 diabetes. Handb Exp Pharmacol. 2011;(203):279–301. doi: 10.1007/978-3-642-17214-4_12. [DOI] [PubMed] [Google Scholar]
- 54.Oakley A. Glutathione transferases: a structural perspective. Drug Metab Rev. 2011;43(2):138–51. doi: 10.3109/03602532.2011.558093. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y, Rui L. Major urinary protein regulation of chemical communication and nutrient metabolism. Vitam Horm. 2010;83:151–63. doi: 10.1016/S0083-6729(10)83006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Jiang L, Rui L. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem. 2009;284(17):11152–9. doi: 10.1074/jbc.M900754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hui X, et al. Major urinary protein-1 increases energy expenditure and improves glucose intolerance through enhancing mitochondrial function in skeletal muscle of diabetic mice. J Biol Chem. 2009;284(21):14050–7. doi: 10.1074/jbc.M109.001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makino H, et al. A good model of hepatic failure after excessive hepatectomy in mice. J Surg Res. 2005;127(2):171–6. doi: 10.1016/j.jss.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 59.Zarnegar R, et al. Expression of hepatocyte growth factor mRNA in regenerating rat liver after partial hepatectomy. Biochem Biophys Res Commun. 1991;177(1):559–65. doi: 10.1016/0006-291x(91)92020-k. [DOI] [PubMed] [Google Scholar]
- 60.Vaquero J, et al. Toll-like receptor 4 and myeloid differentiation factor 88 provide mechanistic insights into the cause and effects of interleukin-6 activation in mouse liver regeneration. Hepatology. 2011;54(2):597–608. doi: 10.1002/hep.24420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindroos PM, Zarnegar R, Michalopoulos GK. Hepatocyte growth factor (hepatopoietin A) rapidly increases in plasma before DNA synthesis and liver regeneration stimulated by partial hepatectomy and carbon tetrachloride administration. Hepatology. 1991;13(4):743–50. [PubMed] [Google Scholar]
- 62.Patijn GA, et al. Hepatocyte growth factor induces hepatocyte proliferation in vivo and allows for efficient retroviral-mediated gene transfer in mice. Hepatology. 1998;28(3):707–16. doi: 10.1002/hep.510280317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.