Abstract
Glutathione S-transferase P1 (GSTP1), an enzyme involved in detoxification process, is frequently inactivated in prostate cancer due to epigenetic modifications. Through in silico analysis we identified a subset of miRNAs that are putative targets in regulating GSTP1. miRNAs are small endogenous non-coding RNA that are critical regulators of various physiologic and pathologic processes and their level of expression may play a precise role in early diagnosis and prognosis of cancer. These small molecules have been detected in a wide variety of human biological specimens including blood, serum, urine, ejaculate and tissues, which could be utilized as clinically useful biomarker in early detection and prognosis of prostate cancer. The chapter summarizes the current knowledge about miRNA involved in GSTP1 regulation in prostate cancer and their potential as useful biomarkers of disease for early detection and prognosis, along with challenges and limitations in this development.
Keywords: miRNA, GSTP1, prostate cancer, epigenetics, oxidative DNA damage
Introduction
Prostate cancer remains the most common form of cancer in males in the United States [1]. According to an estimate by the American Cancer Society, in 2014 approximately 233,000 new cases of prostate cancer will be diagnosed and about 29,480 men will die from this disease [1, 2]. Early diagnosis of tumor and timely detection of prostate cancer progression following either surgery or radiation therapy are critical for its effective and beneficial clinical outcome. The widespread use of serum-based total prostate-specific antigen (PSA) has led to detection of prostate cancer at a potentially curable stage however, its use as a screening tool remains controversial due to the absence of a true tPSA cutoff-point for identifying prostate cancer risk [3, 4]. First, approximately one-third of prostate cancers detected at a PSA level at or above 4ng/mL have already spread to the prostate capsule or beyond, and approximately 15% of men with a PSA level <4ng/mL have prostate cancer that is detectable by needle biopsy [5-7]. Specifically, the Prostate Cancer Prevention Trial reported that in men who underwent an empiric biopsy at PSA levels of <=0.5ng/mL, 0.6–1.0ng/mL, 1.1–2.0ng/mL, 2.1–3.0ng/mL, and 3.1–4.0ng/mL, the prostate cancer detection rates were 6.6%, 10.1%, 17.0%, 23.9%, and 26.9%, respectively [8]. Using current recommended guidelines to determine the need for confirmatory biopsy, false positive rates of 55-75% and false negative rates of at least 15% have been reported which limit the sensitivity and specificity of serum total PSA test as an effective population-based cancer detection tool [9, 10]. More recent concepts to improve specificity include use of age-adjusted PSA, PSA velocity, volume-adjusted PSA, and percent free PSA concentration [11, 12]. However, diagnostic accuracy and predictive values for tests using quantitative serum PSA assays remain controversial. There is a need of more sensitive and specific biomarker to detect prostate cancer.
Glutathione S-transferases
Glutathione S-transferases (GSTs) comprise of a multi gene enzymes family of phase II detoxifying enzymes of the xenobiotic metabolism [13]. The members of this family of dimeric enzymes are identified on the basis of their substrate specificity and amino acid sequences. GSTs catalyze the reactions in which reduced glutathione is conjugated to toxic oxidizing compounds. These compounds are produced either due to normal cellular activity of the cell or due to exposure of cells to xenobiotics and environmental pollutants such as carcinogens, pesticides, drugs and to endogenous molecules [14]. This conversion significantly detoxifies them by reducing their ability to react to cellular macromolecules. GSTs are ubiquitously present in every cell and in every living species examined, including both eukaryotes and in prokaryotes. Though, most of these enzymes are composed of cytosolic proteins, a small family of microsomal and mitochondrial (kappa) GSTs are also characterized. GSTs are considered as cell housekeepers due to their ability to detoxify both endogenous as well as exogenous cell substances. In some mammalian and rodents organs, cytosolic GSTs can constitute as high as 4-10% of cytosolic proteins. Soluble cytosolic GSTs exist as dimeric protein with an active site composed of two distinct functional groups including i) hydrophilic catalytically independent active G-site which binds to glutathione, and ii) physiological substrate of GSTs and an adjacent H-site which provides a hydrophobic environment for binding of electrophilic substrates with diverse structures [15, 16]. While G-site, which is in the amino terminal domain, is highly conserved among GSTs due to its high specificity for GSH, the H- site, which is in the carboxy-terminal domain, can be very divergent among GSTs, exhibiting broad and variable specificity to substrate binding [17]. GSTs catalyze the conjugation of reduced glutathione (GSH) via a sulfhydryl group to electrophilic centers on substrates with variable binding specificity. This activity detoxifies several reactive endogenously produced molecules such as α, β-unsaturated keto prostaglandins (i.e. PGA2), endogenous fatty acid oxidation products including 4-hydroxy-2-nonenal, peroxidized lipids and xenobiotics [18].
The mammalian GST super-family consists of seven classes of cytosolic GSTs which possess amino acid sequence similarity, substrate specificity and immunological cross-reactivity [16, 17]. They are named as GSTα (alpha)/GSTA, GSTμ (mu)/GSTM, GSTπ (pi)/GSTP, GSTσ (sigma)/GSTS, GSTθ (theta)/GSTT, GSTω (omega)/GSTO, and GSTζ (zeta)/GSTZ [18-20]. As functional GST enzymes are dimeric and the GSTα (GSTA) and GSTμ (GSTM) can form heterodimers in addition to homodimers, the number of isoenzymes in each class is large. One of the highly conserved classes of cytoplasmic GST is glutathione S-transferase Pi (GSTP1) that was found to be the predominant isoenzyme. It is mapped to chromosome 11q13 and the genes of this class are about 3kb long and contained seven exons. GSTP1 protects cells from cytotoxic and carcinogenic agents [21]. It is expressed at variable levels in different cell types in normal tissues and its altered activity and expression has been found to play an important role in determining susceptibility to different type of cancers, inflammatory disorders, asthma and neurodegenerative disorders [22-24]. The role of pi-class GST (GSTP1) is of particular interest in cancer biology. In humans, early loss of GSTP1 has been associated with cancer initiation and progression and suggested to possess tumor suppressor functions [25-27]. For example, GSTP−/− mice display a strong tendency to develop skin papillomas and lung cancer following carcinogen exposure, and loss of GSTP markedly enhances colon tumorigenesis in Apc (Min) mice [28-30]. On the contrary, over-expression of pi-class GST has been associated with tumor progression and drug resistance [31, 32]. GSTP1 over- expression has been reported in many human tumors, and has been shown to be correlated with advanced stage, disease aggressiveness, drug resistance, and poor survival [32]. A vast majority of human tumor cell lines over-expresses GSTP1, including cells selected in vitro for resistance to agents used for chemotherapy. In fact in 58 of the 60 human tumor cell lines used in the Drug Screen Program of the National Cancer Institute, GSTP1 was found to be the predominant isoenzyme (as high as 2.7% of the total cytosolic protein). A significant quantitative correlation among enzyme activity, protein and mRNA were shown particularly in those cell lines selected for resistance to alkylating agents [33]. Such comparable correlation was much less apparent for over-expression of GSTA and GSTPM.
Glutathione S-transferase-pi and prostate cancer
One of the most common epigenetic alterations described in human prostate cancer is the loss of expression of the glutathione-S-transferase-pi (GSTP1) which occurs in vast majority (>90%) of prostate tumors regardless of grade or stage [34]. Studies in human prostate tumor specimens and cancer cell lines have shown that GSTP1 gene is silenced due to epigenetic modifications [35]. Importantly, loss of GSTP1 function appears to be the characteristic of prostatic intraepithelial neoplasia (PIN) and proliferative inflammatory atrophy (PIA) lesions, throughout to represent prostate cancer precursors [36]. It has been proposed that GSTP1 is a caretaker gene, protecting cells against genomic damage mediated by oxidants and electrophiles from inflammation or dietary exposures [37]. Reports suggest that loss of GSTP1 shifts the prooxidant–antioxidant balance towards an oxidative state, resulting in increased inflammation and oxidative stress to prostate epithelial cells [38]. Studies have suggested age-related structural changes in the DNA of prostate tissue which is likely a result of oxidative damage induced by hydroxyl radicals [39]. Age-related oxidative DNA damage and increased accumulation of 8-oxo-2′- deoxyguanosine (8-OHdG) have been shown to be more pronounced in prostate neoplasms than in benign prostate tissue [40]. We have recently demonstrated that chronic intraprostatic inflammation causes premalignant and malignant changes in prostatic epithelium which may be due at least in part to accumulation of oxidative DNA products as a result of loss of GSTP1 expression in prostate epithelial cells [38, 41]. Since GSTP1 is epigenetically silenced in early stage prostate cancer, the elements of epigenetic GSTP1 regulation could serve as better biomarker for detection and prognosis of prostate cancer.
Epigenetics and gene regulation
Gene expression is intricately regulated through the epigenetic modifications such as DNA methylation, post-translational modifications of histone proteins, and transcriptional regulation of gene expression by non-coding regulatory microRNA [42, 43]. Numerous studies have demonstrated that the regulatory sequences near the GSTP1 gene are commonly inactivated by DNA hypermethylation during early stages of prostate carcinogenesis [44, 45]. Extensive methylation of deoxycytidine nucleotides distributed throughout the 5′CpG island region of GSTP1 is not detected in benign prostate tissue, but has been detected in high-grade intraepithelial neoplasia (HGPIN), prostate adenocarcinoma in the tissue and fluids including plasma, serum prostatic ejaculates and urine specimens [46-48].
Histone modification is closely associated with DNA methylation in prostate cancer [49]. Studies have demonstrated that class I histone deacetylases (HDACs) are frequently over-expressed in prostate cancer [50]. Studies demonstrate that HDAC1 contributes to aggressive tumor behavior and poor prognosis whereas HDAC2 expression is associated with shortened relapse free survival time in prostate cancer patients [51]. HDAC1-3 are highly expressed in prostate cancer and in corresponding HGPIN lesions coincide with the loss of GSTP1 expression in tumor specimens [51, 52]. Li et al. demonstrated that inhibition of HDAC1 by maspin, a tumor suppressor serpin, increases GSTP1 expression in human prostate cancer cells endorsing that HDAC1 plays a critical role in maspin-mediated GSTP1 re- expression [53].
Non-coding RNAs have emerged as a new class of key regulators of genes [54, 55]. miRNAs are short (∼20-24 nucleotides) non-coding RNAs that regulate gene expression mostly by facilitating the cleavage of target mRNA in plants [56]. Interestingly, miRNAs affect the expression of their target gene mostly by translational repression in animals. miRNAs target mRNAs by imperfect complimentary base- pairing to the 3′ untranslated region (3′ UTR) to downregulate target's protein synthesis either by deadenylation of the targeted message or by repressing the translation at the actively translating ribosomes [57, 58]. Currently, miRBase has a compilation of 2588 mature human miRNAs from human genome assembly (GRCh 38) to the GenBank [59]. These many miRNA are predicted to target > 45,000 sites that account for >60% of human genes.
Numerous studies in various organismal systems show that miRNAs play important roles in cellular processes such as development, differentiation, proliferation, apoptosis and metabolism [60]. Furthermore, strong evidence demonstrates that aberration in miRNAs expression and their targeting activities have been implicated in human diseases, including cancers [61]. Aberrant and deregulated expression of miRNAs has been identified in stages of carcinogenesis, development of resistance to therapeutics and in metastasis [61-63]. In addition, differential expression of miRNAs appears to play a significant role in the prognosis of various cancers including prostate cancer [61, 64, 65]. Studies of miRNAs show that over 50% of the miRNA genes are coded in the human genome at or near sites of frequent deletion and amplification as well as at CpG island methylation [66]. It is also becoming increasingly clear that miRNAs are transcriptionally silenced in various human cancers by epigenetic mechanisms including hypermethylation. These features of miRNA suggesting that deregulated expression of miRNAs play important role in tumorigenesis in nearly all types of cancers and may be linked with specific clinic-pathological parameters, risk, aggressiveness, staging and disease outcome [67-69].
Potential regulation of GSTP1 by miRNAs
Several computational and experimental approaches have been used to identify miRNAs and their targeting genes [70, 71]. For this study we used computational approaches to identify miRNAs potentially targeting and regulating GSTP1 expression (Table 1). These miRNA include miR-133a/b, miR- 144/144*, miR-153-1/2 and miR-590-3p/5p. In this chapter a short description of these miRNA and their involvement in cancer and GSTP1 regulation is highlighted.
Table 1. miRNAs potential to target GSTP1.
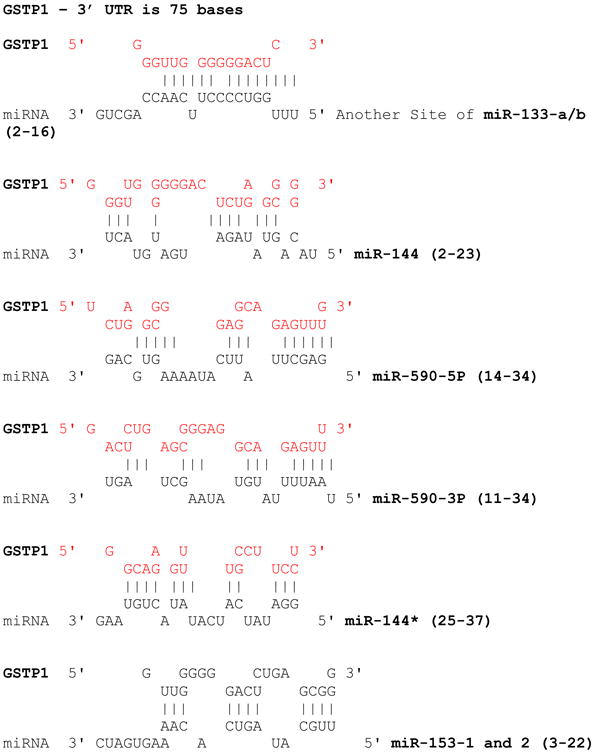
|
miR-133a/b
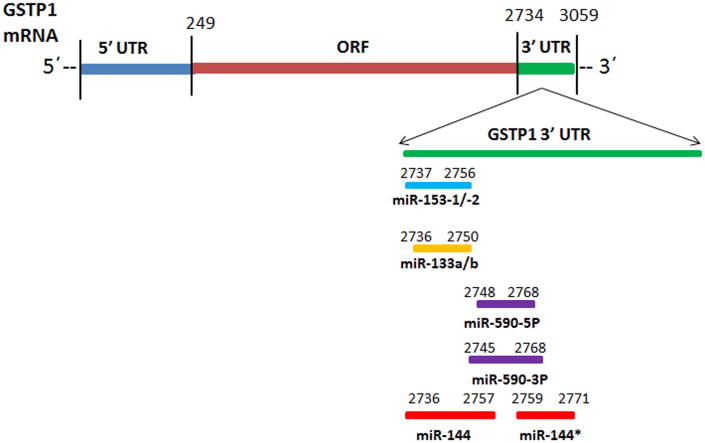
A search for TargetScan Human database identified miR-133a/b as a candidate miRNA regulator of GSTP1 by potentially targeting nucleotides 2736-2750 in the 3′UTR (Figure 1). The miR-133a1/a2-3p and miR-133b are transcribed from chromosome number 18, 20 and 6, respectively [72]. miR-133a and miR-133b differ only in one base at the 3′-end of the molecule (G→A). This position is furthest away from the seed region which is essential for miR:target interaction and its resultant effect on target mRNA translational repression. Therefore, it is likely that miR-133a and miR-133b will perform similar, if not, identical cellular function by regulating the expression of a common pool of target genes [73]. Interestingly, GENECODIS analysis revealed that miR-133a and miR-133b might supplement each other in many cancer pathways [74]. GSTP1 displays one single miR-133a and miR-133b binding site in its 3′UTR region (Figure 1). Downregulation of miR133a/b has been reported in various human cancers including head and neck squamous cell carcinoma, esophageal squamous cell carcinoma, non-small cell lung cancer, bladder, cervical, gastric and colorectal cancer [75-79]. Over-expression of miR-133a/b has been shown to inhibit tumor cell proliferation and induce apoptosis in various human cancer cell types [78, 80]. Specifically, miR-133b has been found to be involved in the regulation of cell death through death receptor-mediated apoptosis in human prostate cancer PC-3 cells [81]. It is a target of androgen receptor (AR) and required for androgen's mediated stimulation in LNCaP cells, and is also known to regulate CDC2L5, PTPRK, RB1CC1, and CPNE3 in prostate cancer cell lines [82]. Other putative targets of miR-133a/b are the Ras homolog gene family member A (RHOA), cell division cycle 42 (CDC42), FSCN1, LASP1 and c-MET oncogenic genes [83]. Association of miR-133b with prostate cancer progression and its potential as a diagnostic marker is also demonstrated by its downregulation observed in prostate secretion samples from patients, it has greater power (AUC 0.950) than PSA (AUC 0.463) to distinguish prostate cancer from benign prostate hyperplasia [84, 85].
Figure 1.
Schematic representation of miR-133-a/b, miR-153-1/2, miR-590-3P/5P and miR-144/144* target sites on GSTP1 3′UTR.
miR-144/miR-144*
The gene encoding miR-144 is located on chromosome 17 and has a passenger strand (miR- 144*). In silico analysis of miRNA-target mRNA prediction algorithm revealed single miR-144 and a miR- 144* binding sites in the GSTP1 3′UTR region with Watson-Crick match at miRNA positions 2736-2757 and 2759-2771, respectively. These sites raise the possibility that miR-144/144* are involved in the regulation of GSTP1 expression and possibly in detoxification aspect of the gene function. MiR-144 is dysregulated and involved in the many human tumors including osteosarcoma, mesothelioma, gastric and nasopharyengeal carcinoma [86-89]. Deregulation of miR-144 in colorectal cancer cells has been shown to activate mammalian target of rapamycin (mTOR) signaling and its down-regulation was associated with poor prognosis [90]. miR-144 expression level has been shown to be significantly decreased in bladder cancer, and its down-regulation increased bladder cancer cell proliferation by targeting Histone-lysine N-methyltransferase-EZH2 [91]. Similarly, gastric cancer stage IV patients also exhibit diminished expression of miR-144, where is promotes upregulation of ZFX proteins and subsequent cancer progression [88]. Notably, it has been demonstrated that the expression of miR-144 was reduced in thyroid cancer [92]. Recently, a comprehensive meta-analysis of miRNA expression microarray data sets revealed that miR-144 was downregulated in hepatocellular carcinoma, lung cancer and prostate cancer [93-95]. However, no information about the function or molecular mechanism(s) of miR-144 regulating GSTP1 has been reported. miR-144 was found to be over-expressed in high Gleason score (8 and 9) prostate tissue samples [94, 95], moreover miR-144* was significantly upregulated when compared to miR-144 in both metastatic and non-metastatic tumor xenograft models [95].
miR153-1/2
miR-153-1 and miR-153-2 are encoded in the intron of protein tyrosine phosphatase, receptor type N (PTPRN) gene on the chromosome 2 [96]. On other hand, miR-153-2 was found to be encoded in the intron of PTPRN 2 on chromosome 7 where the targeting site of miR-153-1/2 spans around nucleotides 2737-2756 in the GSTP1 3′UTR (Figure 1). In general, miR-153 has been observed to be over-expressed in tumors compared to normal tissue at higher levels in metastatic compared with non- metastatic tumors [97]. Studies conducted using various human prostate cancer cell lines demonstrate miR-153 represses PTEN expression to activate AKT kinase and downregulate the transcriptional activity of Forkhead box O (FOXO)1, leading to upregulation of the G1/S transitional promoter cyclin D1 and downregulation of the cyclin-dependent kinase (CDK) inhibitor p21 [98]. In contrast, miR-153 levels are shown to be reduced in human glioblastoma multiforme [99]. miR-153 induces apoptosis in these tumors by targeting Bcl-2 and Mcl-1, suggesting that miR-153 functions as a tumor suppressor [100]. Studies have revealed a tendency toward downregulation of miR-153 in relation to lymph node metastasis in ovarian epithelial tumors [101], and downregulation of miR-153 in high-risk medulloblastomas [99]. Therefore, miR-153 may function as a tumor suppressor or an oncogene depending on the tissue. However, no information about the function or molecular mechanism of miR-153-1/2 regulating GSTP1 has been reported.
miR-590-3p/5p
microRNA target databases, such as TargetScan and miRanda predicted that miR-590-3p/5p could regulate GSTP1 mRNA and/or protein expression miR-590-3p forms an 7mer at position 57-63 of GSTP1 3′UTR. Examination of the mRNA sequence of GSTP1 revealed that miR-590-3p/5p potential target sites are found at nucleotides 2748-2748 in the 3′UTR region (Figure 1). miR-590-3p/5p have been shown to be upregulated in some human cancers including hepatocellular carcinoma, clear cell renal cell carcinoma, myeloid leukemia and cervical cancer [102-105]. Upregulation of miR-590-5p has been shown to promote proliferation and invasion of clear cell renal cell carcinoma cells by downregulation of p21 (Waf1/Cip1) expression [103]. It is also shown to be differentially expressed in castrate-resistant prostate cancer compared to benign prostatic hyperplasia [106]. TargetScan predicted PTEN as the potential target genes of miR-590-3p/5p and was found to activate PI3K-AKT signaling pathway by down- regulating PTEN to promote AKT1-S473 phosphorylation [107], which may play an important role in the regulation of GSTP1 in prostate cancer.
Conclusion, Limitations and Future Direction
Measurement of serum PSA levels do not adequately detect prostate cancer or predict prognosis after definitive therapy. Therefore, there is a significant challenge to develop new non-invasive biomarker identification strategies to precisely detect and predict progression of prostate cancer. miRNAs are important regulators of oncogene and tumor suppressor genes that intercept various signaling pathways and pathological processes associated with tumorigenesis. Numerous publications have reported the potential of miRNA as a class of novel biomarkers in diagnosis and disease prognosis [63-69]. Our in silico analysis suggest that miRNA can exert their functions by potentially regulating GSTP1 expression and play an important role in prostate carcinogenesis (Table 1; Figure 1). With the identification of listed putative miRNAs that regulate GSTP1, a more detailed regulatory role may be deciphered which might be applicable in early detection and prognosis of prostate cancer.
However some limitations remain in their development and replacement to conventional biomarkers. This includes lack of established endogenous miRNA control to normalize for miRNA levels in body fluids. In this regard, U6 small nuclear RNA is frequently utilized to normalize miRNA levels, however its unstable nature and degradation in serum does not qualify it as an ideal standard control. Another limitation is the release of miRNA into body fluids, and its functional role and consequences remain unknown and/or limited inconsistencies in the analysis of the function of miRNA in prostate cancers. Furthermore, the exact cause-effect mechanism(s) has not been established for many miRNAs in prostate cancer despite significant efforts into such research. Therefore, further studies are needed to understand the role and consequences of miRNA in prostate cancer through novel high throughput techniques. Another caveat to consider is that the post-transcriptional gene regulation of miRNA as well as its target mRNA is further modulated by RNA binding proteins (RBPs). Numerous RBP have been characterized recently and many play a role in the both, miRNA and mRNA stabilities.
In conclusion, technical advancement in the detection of miRNAs regulating GSTP1 expression and function may have great promise as molecular biomarkers for prostate cancer. Further development and application of these miRNA-regulating GSTP1 assays to clinical specimens including blood, urine, ejaculate and prostate biopsy for cancer screening and early detection may have great promise as candidate clinical tests. A new series of studies critically assessing the predictive values of miRNAs in prostate cancer are needed.
Acknowledgments
The original work in SG laboratory outlined in this chapter is supported by the United States Public Health Service Grants R01CA108512, R01CA115491 and R03186179. Research in GCS laboratory is supported by the grants from Department of Defense W81XWH-11-10204 and National Science Foundation (MCB-0842606). We apologize to those investigators whose original work could not be cited owing to the space limitations.
Abbreviations
- 3′ UTR
3′ untranslated region
- 8-OHdG
8-oxo-2′-deoxyguanosine
- CDC42
cell division cycle 42
- CDK
cyclin-dependent kinase
- FOXO
Forkhead box O
- GSTP1
Glutathione S-transferase P1
- GSTs
Glutathione S-transferases
- HDAC
histone deacetylases
- HGPIN
high-grade intraepithelial neoplasia
- miRNA
microRNA
- mTOR
mammalian target of rapamycin
- PIA
proliferative inflammatory atrophy
- PIN
prostatic intraepithelial neoplasia
- PSA
prostate-specific antigen
- PTPRN
protein tyrosine phosphatase, receptor type N
- RBPs
RNA binding proteins
- RHOA
Ras homolog gene family member A
Footnotes
Conflict of Interest: Savita Singh, Girish C Shukla, and Sanjay Gupta declare that they have no conflict of interest.
Compliance with Ethics Guidelines: Human and Animal Rights and Informed Consent: This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as: ** Of major importance
- 1.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–71. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. 2014 at the web at: http://www.cancer.org.
- 3.Schröder FH, Roobol MJ. Defining the optimal prostate-specific antigen threshold for the diagnosis of prostate cancer. Curr Opin Urol. 2009;19:227–31. doi: 10.1097/MOU.0b013e328329a2d0. [DOI] [PubMed] [Google Scholar]
- 4.Catalona WJ, Loeb S. Prostate cancer screening and determining the appropriate prostate-specific antigen cutoff values. J Natl Compr Canc Netw. 2010;8:265–70. doi: 10.6004/jnccn.2010.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canby-Hagino E, Hernandez J, Brand TC, Troyer DA, Higgins B, Ankerst DP, Thompson IM, Leach RJ, Parekh DJ. Prostate cancer risk with positive family history, normal prostate examination findings, and PSA less than 4.0 ng/mL. Urology. 2007;70:748–52. doi: 10.1016/j.urology.2007.06.1105. [DOI] [PubMed] [Google Scholar]
- 6.Schröder FH, Raaijmakers R, Postma R, van der Kwast TH, Roobol MJ. 4-year prostate specific antigen progression and diagnosis of prostate cancer in the European Randomized Study of Screening for Prostate Cancer, section Rotterdam. J Urol. 2005;174:489–94. doi: 10.1097/01.ju.0000165568.76908.5c. [DOI] [PubMed] [Google Scholar]
- 7.Minardi D, Galosi AB, Recchioni A, Giammarco L, Polito M, Muzzonigro G. Diagnostic accuracy of percent free prostate-specific antigen in prostatic pathology and its usefulness in monitoring prostatic cancer patients. Urol Int. 2001;67:272–82. doi: 10.1159/000051003. [DOI] [PubMed] [Google Scholar]
- 8.Lucia MS, Darke AK, Goodman PJ, La Rosa FG, Parnes HL, Ford LG, Coltman CA, Jr, Thompson IM. Pathologic characteristics of cancers detected in The Prostate Cancer Prevention Trial: implications for prostate cancer detection and chemoprevention. Cancer Prev Res (Phila) 2008;1:167–73. doi: 10.1158/1940-6207.CAPR-08-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulmert D, Serio AM, O'Brien MF, Becker C, Eastham JA, Scardino PT, Björk T, Berglund G, Vickers AJ, Lilja H. Long-term prediction of prostate cancer: prostate-specific antigen (PSA) velocity is predictive but does not improve the predictive accuracy of a single PSA measurement 15 years or more before cancer diagnosis in a large, representative, unscreened population. J Clin Oncol. 2008;26:835–41. doi: 10.1200/JCO.2007.13.1490. [DOI] [PubMed] [Google Scholar]
- 10.Capitanio U, Perrotte P, Zini L, Suardi N, Antebi E, Cloutier V, Jeldres C, Shariat SF, Duclos A, Arjane P, Saad F, Montorsi F, Karakiewicz PI. Population-based analysis of normal Total PSA and percentage of free/Total PSA values: results from screening cohort. Urology. 2009;73:1323–7. doi: 10.1016/j.urology.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Catalona WJ, Southwick PC, Slawin KM, Partin AW, Brawer MK, Flanigan RC, Patel A, Richie JP, Walsh PC, Scardino PT, Lange PH, Gasior GH, Loveland KG, Bray KR. Comparison of percent free PSA, PSA density, and age-specific PSA cutoffs for prostate cancer detection and staging. Urology. 2000;56:255–60. doi: 10.1016/s0090-4295(00)00637-3. [DOI] [PubMed] [Google Scholar]
- 12.Basso D, Fogar P, Piva MG, Navaglia F, Mazza S, Prayer-Galetti T, Castellucci E, Pagano F, Plebani M. Total PSA, free PSA/total PSA ratio, and molecular PSA detection in prostate cancer: which is clinically effective and when? Urology. 2000;55:710–5. doi: 10.1016/s0090-4295(99)00596-8. [DOI] [PubMed] [Google Scholar]
- 13.Seidegård J, Ekström G. The role of human glutathione transferases and epoxide hydrolases in the metabolism of xenobiotics. Environ Health Perspect. 1997;105:791–9. doi: 10.1289/ehp.105-1470052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mannervik B, Danielson UH. Glutathione transferases-structure and catalytic activity. CRC Crit Rev Biochem. 1998;23:283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- 15.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong RN. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem Res Toxicol. 1997;10:2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 18.Mannervik B, Alin P, Guthenberg C, Jensson H, Tahir MK, Warholm M, Jörnvall H. Identification of three classes of cytosolic glutathione transferase common to several mammalian species: correlation between structural data and enzymatic properties. Proc Natl Acad Sci U S A. 1985;82:7202–6. doi: 10.1073/pnas.82.21.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer DJ, Coles B, Pemble SE, Gilmore KS, Fraser GM, Ketterer B. Theta, a new class of glutathione transferases purified from rat and man. Biochem J. 1991;274:409–14. doi: 10.1042/bj2740409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Board PG, Baker RT, Chelvanayagam G, Jermiin LS. Zeta, a novel class of glutathione transferases in a range of species from plants to humans. Biochem J. 1997;328:929–35. doi: 10.1042/bj3280929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mannervik B, Awasthi YC, Board PG, Hayes JD, Di Ilio C, Ketterer B, Listowsky I, Morgenstern R, Muramatsu M, Pearson WR. Nomenclature for human glutathione transferases. Biochem J. 1992;282:305–6. doi: 10.1042/bj2820305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Ilio C, Aceto A, Bucciarelli T, Angelucci S, Felaco M, Grilli A, Zezza A, Tenaglia R, Federici G. Glutathione transferase isoenzymes in normal and neoplastic human kidney tissue. Carcinogenesis. 1991;12:1471–75. doi: 10.1093/carcin/12.8.1471. [DOI] [PubMed] [Google Scholar]
- 23.Schroer KT, Gibson AM, Sivaprasad U, Bass SA, Ericksen MB, Wills-Karp M, Lecras T, Fitzpatrick AM, Brown LA, Stringer KF, Hershey GK. Downregulation of glutathione S-transferase pi in asthma contributes to enhanced oxidative stress. J Allergy Clin Immunol. 2011;128:539–48. doi: 10.1016/j.jaci.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohanec Grabar P, Logar D, Tomsic M, Rozman B, Dolzan V. Genetic polymorphisms of glutathione S-transferases and disease activity of rheumatoid arthritis. Clin Exp Rheumatol. 2009;27:229–36. [PubMed] [Google Scholar]
- 25.Fiolka R, Zubor P, Janusicova V, Visnovsky J, Mendelova A, Kajo K, Lasabova Z, Plank L, Danko J. Promoter hypermethylation of the tumor-suppressor genes RASSF1A, GSTP1 and CDH1 in endometrial cancer. Oncol Rep. 2013;30:2878–86. doi: 10.3892/or.2013.2752. [DOI] [PubMed] [Google Scholar]
- 26.Gumy-Pause F, Pardo B, Khoshbeen-Boudal M, Ansari M, Gayet-Ageron A, Sappino AP, Attiyeh EF, Ozsahin H. GSTP1 hypermethylation is associated with reduced protein expression, aggressive disease and prognosis in neuroblastoma. Genes Chromosomes Cancer. 2012;51:174–85. doi: 10.1002/gcc.20941. [DOI] [PubMed] [Google Scholar]
- 27.Steinmann K, Sandner A, Schagdarsurengin U, Dammann RH. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep. 2009;22:1519–26. doi: 10.3892/or_00000596. [DOI] [PubMed] [Google Scholar]
- 28.Henderson CJ, Smith AG, Ure J, Brown K, Bacon EJ, Wolf CR. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc Natl Acad Sci U S A. 1998;95:5275–80. doi: 10.1073/pnas.95.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie KJ, Henderson CJ, Wang XJ, Vassieva O, Carrie D, Farmer PB, Gaskell M, Park K, Wolf CR. Glutathione transferase pi plays a critical role in the development of lung carcinogenesis following exposure to tobacco-related carcinogens and urethane. Cancer Res. 2007;67:9248–57. doi: 10.1158/0008-5472.CAN-07-1764. [DOI] [PubMed] [Google Scholar]
- 30.Ritchie KJ, Walsh S, Sansom OJ, Henderson CJ, Wolf CR. Markedly enhanced colon tumorigenesis in Apc(Min) mice lacking glutathione S-transferase Pi. Proc Natl Acad Sci U S A. 2009;106:20859–64. doi: 10.1073/pnas.0911351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruzza P, Rosato A, Rossi CR, Floreani M, Quintieri L. Glutathione transferases as targets for cancer therapy. Anticancer Agents Med Chem. 2009;9:763–77. doi: 10.2174/187152009789056895. [DOI] [PubMed] [Google Scholar]
- 32.Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369–75. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tew KD, Monks A, Barone L, Rosser D, Akerman G, Montali JA, Wheatley JB, Schmidt DE., Jr Glutathione-associated enzymes in the human cell lines of the National Cancer Institute Drug Screening Program. Mol Pharmacol. 1996;50:149–59. [PubMed] [Google Scholar]
- 34.Bostwick DG, Meiers I, Shanks JH. Glutathione S-transferase: differential expression of alpha, mu, and pi isoenzymes in benign prostate, prostatic intraepithelial neoplasia, and prostatic adenocarcinoma. Hum Pathol. 2007;38:1394–401. doi: 10.1016/j.humpath.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Maldonado L, Brait M, Loyo M, Sullenberger L, Wang K, Peskoe SB, Rosenbaum E, Howard R, Toubaji A, Albadine R, Netto GJ, Hoque MO, Platz EA, Sidransky D. GSTP1 Promoter Methylation Is Associated with Recurrence in Early Stage Prostate Cancer. J Urol. 2014;5347:3413–2. doi: 10.1016/j.juro.2014.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Bergh A, Damber JE. Morphological transition of proliferative inflammatory atrophy to high-grade intraepithelial neoplasia and cancer in human prostate. Prostate. 2009;69:1378–86. doi: 10.1002/pros.20992. [DOI] [PubMed] [Google Scholar]
- 37.Li G, Wang H, Liu AB, Cheung C, Reuhl KR, Bosland MC, Yang CS. Dietary carcinogen 2-amino-1- methyl-6-phenylimidazo[4,5-b]pyridine-induced prostate carcinogenesis in CYP1A-humanized mice. Cancer Prev Res (Phila) 2012;5:963–72. doi: 10.1158/1940-6207.CAPR-12-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Kanwal R, Pandey M, Bhaskaran N, Maclennan GT, Fu P, Ponsky LE, Gupta S. Protection against oxidative DNA damage and stress in human prostate by glutathione S-transferase P1. Mol Carcinog. 2014;53:8–18. doi: 10.1002/mc.21939. This manuscript presents the functional role of GSTP1 as ‘caretaker’ of oxidative stress and DNA damage in normal prostate epithelial cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malins DC, Johnson PM, Wheeler TM, Barker EA, Polissar NL, Vinson MA. Age-related radical- induced DNA damage is linked to prostate cancer. Cancer Res. 2001;61:6025–8. [PubMed] [Google Scholar]
- 40.Lockett KL, Hall MC, Clark PE, Chuang SC, Robinson B, Lin HY, Su LJ, Hu JJ. DNA damage levels in prostate cancer cases and controls. Carcinogenesis. 2006;27:1187–93. doi: 10.1093/carcin/bgi288. [DOI] [PubMed] [Google Scholar]
- 41.MacLennan GT, Eisenberg R, Fleshman RL, Taylor JM, Fu P, Resnick MI, Gupta S. The influence of chronic inflammation in prostatic carcinogenesis: a 5-year followup study. J Urol. 2006;176:1012–6. doi: 10.1016/j.juro.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 42.Kanwal R, Gupta S. Epigenetic modifications in cancer. Clin Genet. 2012;81:303–11. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed H. Promoter Methylation in Prostate Cancer and its Application for the Early Detection of Prostate Cancer Using Serum and Urine Samples. Biomark Cancer. 2010;2010:17–33. doi: 10.4137/BIC.S3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–86. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 46.Dumache R, Puiu M, Motoc M, Vernic C, Dumitrascu V. Prostate cancer molecular detection in plasma samples by glutathione S-transferase P1 (GSTP1) methylation analysis. Clin Lab. 2014;60:847–52. doi: 10.7754/clin.lab.2013.130701. [DOI] [PubMed] [Google Scholar]
- 47.elgado-Cruzata L, Hruby GW, Gonzalez K, McKiernan J, Benson MC, Santella RM, Shen J. DNA methylation changes correlate with Gleason score and tumor stage in prostate cancer. DNA Cell Biol. 2012;31:187–92. doi: 10.1089/dna.2011.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meiers I, Shanks JH, Bostwick DG. Glutathione S-transferase pi (GSTP1) hypermethylation in prostate cancer: review 2007. Pathology. 2007;39:299–304. doi: 10.1080/00313020701329906. [DOI] [PubMed] [Google Scholar]
- 49.Abbas A, Gupta S. The role of histone deacetylases in prostate cancer. Epigenetics. 2008;3:300–9. doi: 10.4161/epi.3.6.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waltregny D, North B, Van Mellaert F, de Leval J, Verdin E, Castronovo V. Screening of histone deacetylases (HDAC) expression in human prostate cancer reveals distinct class I HDAC profiles between epithelial and stromal cells. Eur J Histochem. 2004;48:273–90. [PubMed] [Google Scholar]
- 51.Weichert W, Röske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M, Kristiansen G. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–10. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakagawa M, Oda Y, Eguchi T, Aishima S, Yao T, Hosoi F, Basaki Y, Ono M, Kuwano M, Tanaka M, Tsuneyoshi M. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Rep. 2007;18:769–74. [PubMed] [Google Scholar]
- 53.Li X, Kaplun A, Lonardo F, Heath E, Sarkar FH, Irish J, Sakr W, Sheng S. HDAC1 inhibition by maspin abrogates epigenetic silencing of glutathione S-transferase pi in prostate carcinoma cells. Mol Cancer Res. 2011;9:733–45. doi: 10.1158/1541-7786.MCR-10-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 55.Vislovukh A, Vargas TR, Polesskaya A, Groisman I. Role of 3′-untranslated region translational control in cancer development, diagnostics and treatment. World J Biol Chem. 2014;5:40–57. doi: 10.4331/wjbc.v5.i1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhuo Y, Gao G, Shi JA, Zhou X, Wang X. miRNAs: biogenesis, origin and evolution, functions on virus-host interaction. Cell Physiol Biochem. 2013;32:499–510. doi: 10.1159/000354455. [DOI] [PubMed] [Google Scholar]
- 57.Dalmay T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 2013;54:29–38. doi: 10.1042/bse0540029. [DOI] [PubMed] [Google Scholar]
- 58.Shruti K, Shrey K, Vibha R. Micro RNAs: tiny sequences with enormous potential. Biochem Biophys Res Commun. 2011;407:445–9. doi: 10.1016/j.bbrc.2011.03.058. [DOI] [PubMed] [Google Scholar]
- 59.New human genome assembly (GRCh38) released! On the web at: http://www.ncbi.nlm.nih.gov/news/12-23-2013-grch38-released/
- 60.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–39. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 61.Nikitina EG, Urazova LN, Stegny VN. MicroRNAs and human cancer. Exp Oncol. 2012;34:2–8. [PubMed] [Google Scholar]
- 62.Raisch J, Darfeuille-Michaud A, Nguyen HT. Role of microRNAs in the immune system, inflammation and cancer. World J Gastroenterol. 2013;19:2985–96. doi: 10.3748/wjg.v19.i20.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, Musolino C. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) Int J Oncol. 2012;41:1897–912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 64.Sita-Lumsden A, Dart DA, Waxman J, Bevan CL. Circulating microRNAs as potential new biomarkers for prostate cancer. Br J Cancer. 2013;108:1925–30. doi: 10.1038/bjc.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang YX, Gao WQ. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene. 2014;33:135–47. doi: 10.1038/onc.2013.54. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki H, Maruyama R, Yamamoto E, Kai M. Epigenetic alteration and microRNA dysregulation in cancer. Front Genet. 2013;4:258. doi: 10.3389/fgene.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–69. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 68.Sethi S, Ali S, Kong D, Philip PA, Sarkar FH. Clinical implication of microRNAs in molecular pathology. Clin Lab Med. 2013;33:773–86. doi: 10.1016/j.cll.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 69**.Fendler A, Jung K. MicroRNAs as new diagnostic and prognostic biomarkers in urological tumors. Crit Rev Oncog. 2013;18:289–302. doi: 10.1615/critrevoncog.2013007176. This paper highlights the latest findings on miRNA in tissue and body fluids of patients suffering from urological cancers and their clinical utility as biomarker. [DOI] [PubMed] [Google Scholar]
- 70**.Reyes-Herrera PH, Ficarra E. One decade of development and evolution of microRNA target prediction algorithms. Genomics Proteomics Bioinformatics. 2012;10:254–63. doi: 10.1016/j.gpb.2012.10.001. This manuscript summarizes a decade of evolution and development of computational algorithms for miRNA target prediction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watanabe Y, Tomita M, Kanai A. Computational methods for microRNA target prediction. Methods Enzymol. 2007;427:65–86. doi: 10.1016/S0076-6879(07)27004-1. [DOI] [PubMed] [Google Scholar]
- 72.Yu H, Lu Y, Li Z, Wang Q. microRNA-133: Expression, Function and Therapeutic Potential in Muscle Diseases and Cancer. Curr Drug Targets. 2014;15:817–28. doi: 10.2174/1389450115666140627104151. [DOI] [PubMed] [Google Scholar]
- 73.Ohanian M, Humphreys DT, Anderson E, Preiss T, Fatkin D. A heterozygous variant in the human cardiac miR-133 gene, MIR133A2, alters miRNA duplex processing and strand abundance. BMC Genet. 2013;14:18. doi: 10.1186/1471-2156-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moriya Y, Nohata N, Kinoshita T, Mutallip M, Okamoto T, Yoshida S, Suzuki M, Yoshino I, Seki N. Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. J Hum Genet. 2012;57:38–45. doi: 10.1038/jhg.2011.126. [DOI] [PubMed] [Google Scholar]
- 76.Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b:Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804–14. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 77.Wu D, Pan H, Zhou Y, Zhou J, Fan Y, Qu P. microRNA-133b downregulation and inhibition of cell proliferation, migration and invasion by targeting matrix metallopeptidase-9 in renal cell carcinoma. Mol Med Rep. 2014;9:2491–8. doi: 10.3892/mmr.2014.2116. [DOI] [PubMed] [Google Scholar]
- 78.Uchida Y, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Kawahara K, Nishiyama K, Seki N, Nakagawa M. MiR-133a induces apoptosis through direct regulation of GSTP1 in bladder cancer cell lines. Urol Oncol. 2013;31:115–23. doi: 10.1016/j.urolonc.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 79.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patron JP, Fendler A, Bild M, Jung U, Müller H, Arntzen MØ, Piso C, Stephan C, Thiede B, Mollenkopf HJ, Jung K, Kaufmann SH, Schreiber J. MiR-133b targets antiapoptotic genes and enhances death receptor-induced apoptosis. PLoS One. 2012;7:e35345. doi: 10.1371/journal.pone.0035345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin C, Zhang W. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep. 2012;27:1967–75. doi: 10.3892/or.2012.1711. [DOI] [PubMed] [Google Scholar]
- 82.Mo W, Zhang J, Li X, Meng D, Gao Y, Yang S, Wan X, Zhou C, Guo F, Huang Y, Amente S, Avvedimento EV, Xie Y, Li Y. Identification of novel AR-targeted microRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate cancer. PLoS One. 2013;8:e56592. doi: 10.1371/journal.pone.0056592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up- regulation of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med. 2009;180:713–9. doi: 10.1164/rccm.200903-0325OC. [DOI] [PubMed] [Google Scholar]
- 84.Guzel E, Karatas OF, Semercioz A, Ekici S, Aykan S, Yentur S, Creighton CJ, Ittmann M, Ozen M. Identification of microRNAs differentially expressed in prostatic secretions of patients with prostate cancer. Int J Cancer. 2014 doi: 10.1002/ijc.29054. [DOI] [PubMed] [Google Scholar]
- 85.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–38. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 86.Namløs HM, Meza-Zepeda LA, Barøy T, Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma -A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48:615–23. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]
- 88.Akiyoshi S, Fukagawa T, Ueo H, Ishibashi M, Takahashi Y, Fabbri M, Sasako M, Maehara Y, Mimori K, Mori M. Clinical significance of miR-144-ZFX axis in disseminated tumour cells in bone marrow in gastric cancer cases. Br J Cancer. 2012;107:1345–53. doi: 10.1038/bjc.2012.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang LY, Ho-Fun Lee V, Wong AM, Kwong DL, Zhu YH, Dong SS, Kong KL, Chen J, Tsao SW, Guan XY, Fu L. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis. 2013;34:454–63. doi: 10.1093/carcin/bgs346. [DOI] [PubMed] [Google Scholar]
- 90.Iwaya T, Yokobori T, Nishida N, Kogo R, Sudo T, Tanaka F, Shibata K, Sawada G, Takahashi Y, Ishibashi M, Wakabayashi G, Mori M, Mimori K. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33:2391–7. doi: 10.1093/carcin/bgs288. [DOI] [PubMed] [Google Scholar]
- 91.Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang Z, Qiu F, Lin J. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013;280:4531–8. doi: 10.1111/febs.12417. [DOI] [PubMed] [Google Scholar]
- 92.Swierniak M, Wojcicka A, Czetwertynska M, Stachlewska E, Maciag M, Wiechno W, Gornicka B, Bogdanska M, Koperski L, de la Chapelle A, Jazdzewski K. In-depth characterization of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98:E1401–9. doi: 10.1210/jc.2013-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zha W, Cao L, Shen Y, Huang M. Roles of Mir-144-ZFX pathway in growth regulation of non-small- cell lung cancer. PLoS One. 2013 Sep 16;8(9):e74175. doi: 10.1371/journal.pone.0074175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Watahiki A, Wang Y, Morris J, Dennis K, O'Dwyer HM, Gleave M, Gout PW, Wang Y. MicroRNAs associated with metastatic prostate cancer. PLoS One. 2011;6:e24950. doi: 10.1371/journal.pone.0024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95**.Walter BA, Valera VA, Pinto PA, Merino MJ. Comprehensive microRNA Profiling of Prostate Cancer. J Cancer. 2013;4:350–7. doi: 10.7150/jca.6394. This paper presents a comprehensive profiling of miRNA in prostate cancer from 37 patients whose prostate tissue were microdissected to obtain pure population of tumor cells, normal epithelium and adjacent stroma. This study provides lead in microRNA research in the development, progression and prognosis of prostate cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mandemakers W, Abuhatzira L, Xu H, Caromile LA, Hébert SS, Snellinx A, Morais VA, Matta S, Cai T, Notkins AL, De Strooper B. Co-regulation of intragenic microRNA miR-153 and its host gene Ia-2β: identification of miR-153 target genes with functions related to IA-2β in pancreas and brain. Diabetologia. 2013;56:1547–56. doi: 10.1007/s00125-013-2901-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu Q, Sun Q, Zhang J, Yu J, Chen W, Zhang Z. Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis. 2013;34:539–49. doi: 10.1093/carcin/bgs374. [DOI] [PubMed] [Google Scholar]
- 98.Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate. 2013;73:596–604. doi: 10.1002/pros.22600. [DOI] [PubMed] [Google Scholar]
- 99.Zhao S, Deng Y, Liu Y, Chen X, Yang G, Mu Y, Zhang D, Kang J, Wu Z. MicroRNA-153 is tumor suppressive in glioblastoma stem cells. Mol Biol Rep. 2013;40:2789–98. doi: 10.1007/s11033-012-2278-4. [DOI] [PubMed] [Google Scholar]
- 100.Xu J, Liao X, Wong C. Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer. 2010;126:1029–35. doi: 10.1002/ijc.24823. [DOI] [PubMed] [Google Scholar]
- 101.Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, An HJ. Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology. 2010;57:734–43. doi: 10.1111/j.1365-2559.2010.03686.x. [DOI] [PubMed] [Google Scholar]
- 102.Jiang X, Xiang G, Wang Y, Zhang L, Yang X, Cao L, Peng H, Xue P, Chen D. MicroRNA-590-5p regulates proliferation and invasion in human hepatocellular carcinoma cells by targeting TGF-β RII. Mol Cells. 2012;33:545–51. doi: 10.1007/s10059-012-2267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xiao X, Tang C, Xiao S, Fu C, Yu P. Enhancement of proliferation and invasion by MicroRNA-590-5p via targeting PBRM1 in clear cell renal carcinoma cells. Oncol Res. 2013;20:537–44. doi: 10.3727/096504013X13775486749335. [DOI] [PubMed] [Google Scholar]
- 104.Favreau AJ, Sathyanarayana P. miR-590-5p, miR-219-5p, miR-15b and miR-628-5p are commonly regulated by IL-3, GM-CSF and G-CSF in acute myeloid leukemia. Leuk Res. 2012;36:334–41. doi: 10.1016/j.leukres.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chu Y, Ouyang Y, Wang F, Zheng A, Bai L, Han L, Chen Y, Wang H. MicroRNA-590 promotes cervical cancer cell growth and invasion by targeting CHL1. J Cell Biochem. 2014;115:847–53. doi: 10.1002/jcb.24726. [DOI] [PubMed] [Google Scholar]
- 106.Jalava SE, Urbanucci A, Latonen L, Waltering KK, Sahu B, Jänne OA, Seppälä J, Lähdesmäki H, Tammela TL, Visakorpi T. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012;31:4460–71. doi: 10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- 107.Yang H, Zheng W, Zhao W, Guan C, An J. Roles of miR-590-5p and miR-590-3p in the development of hepatocellular carcinoma. Nan Fang Yi Ke Da Xue Xue Bao. 2013;33:804–11. [PubMed] [Google Scholar]