Abstract
Osteogenesis imperfecta (OI) is a genetic disorder that results in low bone mineral density and brittle bones. Most cases result from dominant mutations in the type I procollagen genes, but mutations in a growing number of genes have been identified that produce autosomal recessive forms of the disease. Among these include mutations in the genes SERPINH1 and FKBP10, which encode the type I procollagen chaperones HSP47 and FKBP65, respectively, and predominantly produce a moderately severe form of OI. Little is known about the biochemical consequences of the mutations and how they produce OI. We have identified a new OI mutation in SERPINH1 that results in destabilization and mislocalization of HSP47 and secondarily has similar effects on FKBP65. We found evidence that HSP47 and FKBP65 act cooperatively during posttranslational maturation of type I procollagen and that FKBP65 and HSP47 but fail to properly interact in mutant HSP47 cells. These results thus reveal a common cellular pathway in cases of OI caused by HSP47 and FKBP65 deficiency.
Introduction
Osteogenesis imperfecta (OI) is an osteochondrodysplasia characterized by bone fragility and skeletal deformity. In most cases, OI results from defects in the type I procollagen genes, COL1A1 and COL1A2, which affect the quantity or quality of type I collagen present in the bone extracellular matrix. Dominantly inherited structural mutations in the type I procollagen genes affect the folding and structure of the type I procollagen trimer, leading to increased posttranslational modification (overmodification) of the resulting molecules. The overmodified molecules are often poorly secreted and accumulate in the endoplasmic reticulum (ER), leading to an unfolded protein response (1,2).
During the past decade, recessively inherited forms of OI, many of which result from mutations in genes implicated in type I procollagen synthesis, have been identified (3–5). A related group of these genes includes CRTAP, LEPRE1 and PPIB, which together encode the prolyl 3-hydroxylase complex, responsible for the hydroxylation of a single proline residue (P986) in the α1 (I) chain of type I procollagen. Mutations in the genes in this complex result in a mild degree of posttranslational overmodification, indicating that the complex acts during folding of the type I procollagen trimer (6–8).
Recessively inherited mutations in SERPINH1 or FKBP10, the genes which encode the type I procollagen chaperones HSP47 and FKBP65, respectively, result in moderately severe forms of OI characterized by osteopenia, bone fragility and skeletal deformities (9,10). SERPINH1 encodes HSP47, a chaperone located in the ER that appears to preferentially recognize and help maintain the folded state of the type I procollagen trimer (11,12). Two OI mutations have been reported in SERPINH1, one in dachshunds (13) and one in a single human case of moderately severe OI (9). In both instances, homozygosity for a missense mutation corresponding to the main functional domain of the protein, a serine-type endopeptidase inhibitor domain recognizing Xaa–Arg–Gly-containing procollagen sequences (14–16), was identified. Cultured dermal fibroblasts from the human case synthesized type I procollagen of normal electrophoretic mobility, suggesting that HSP47 acts after procollagen folding and therefore subsequent to the activity of the prolyl 3-hydroxylation complex (9). In contrast, homozygosity for an Hsp47-null allele in mouse is embryonically lethal at around E10 owing to extreme connective tissue fragility (17). Mouse cells lacking HSP47 are not able to cleave type I procollagen to its mature form, producing a defective procollagen that accumulates in Golgi compartments. These findings suggest that Hsp47 is essential for synthesis and secretion of type I procollagen (17,18).
FKBP10 encodes FKBP65, another type I procollagen chaperone resident in the ER. Null or missense mutations scattered throughout the gene (10,19–22) lead to two distinct phenotypes; moderately severe OI and OI with contractures (Bruck syndrome) (19,23,24). Similar to HSP47-mutant fibroblasts, fibroblasts with FKBP65 defects synthesize type I procollagen chains without posttranslational overmodification. This suggests that FKBP65 also acts after the prolyl 3-hydroxylation complex and at a similar stage during type I procollagen maturation as HSP47.
We have identified a familial case of OI caused by homozygosity for a mutation in SERPINH1 that decreases the level of HSP47 protein and has a secondary effect on the level of FKBP65, tying the activities of these two type I procollagen chaperones together. The two chaperones interact and together with type I procollagen are mislocalized within abnormal vesicles in cultured cells from OI cases with mutations in either gene, supporting a connection between the functions of these two proteins during type I procollagen biosynthesis. Abnormal intracellular trafficking and formation of vesicles in OI cases with defective HSP47 or FKBP65 suggest commonalities in the cellular mechanisms in this form of OI.
Results
Clinical findings
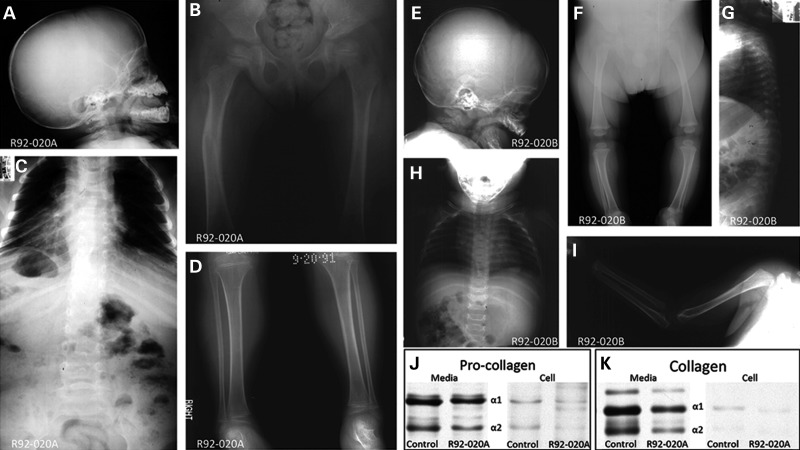
Two affected siblings (International Skeletal Dysplasia Registry reference numbers R92-020A and B), offspring of unaffected third cousin parents, were diagnosed with a moderately severe form of OI at the ages of 4 years and 6 months, respectively. No fractures occurred during their first few months of life, but radiographs showed generalized osteopenia (Fig. 1A–I), a large anterior fontanel and wormian bones in the skull (Fig. 1A and E), coxa valga, mild femoral bowing (Fig. 1B and F), reduced thorax size (Fig. 1C and H) and scoliosis with compression fractures in the vertebrae (Fig. 1G). Hyperextensibility was noted in the fingers, knees and hips. Blue sclerae were not observed, nor was there either dentinogenesis imperfecta or hearing loss. Type I procollagen synthesis and secretion by cultured dermal fibroblasts from one of the siblings (R92-020A) was indistinguishable from control cells (Fig. 1J and K). Recurrence, parental consanguinity and normal type I procollagen biosynthesis suggested that a recessive form of OI was segregating in the family.
Figure 1.
Clinical findings and collagen studies. (A–I) Radiographic analysis for patient R92-020A (A–D) at age of 4 and sibling R92-020B (E–I) at age 6 months. (J and K) Electrophoretic mobility of type I procollagen (J) and collagen (K) from cultured fibroblasts and media from case R92-020A. The α1 and α2 type I (pro)collagen chains are identified.
OI results from homozygosity for a mutation in SERPINH1
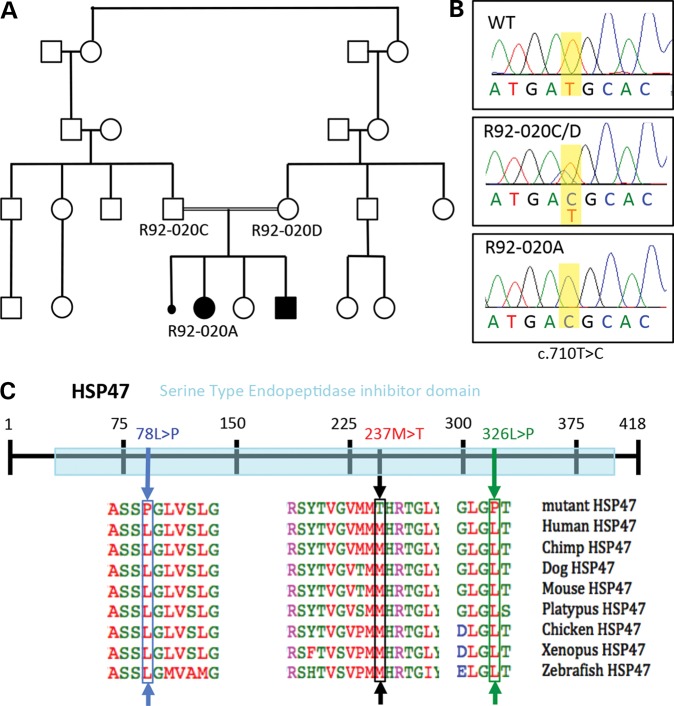
To determine the etiology of OI in the family (R92-020), the exon sequences of a set of known OI-associated genes were sequenced in DNA derived from R92-020A using a multiplex amplification and high-throughput sequencing approach. The genes analyzed included COL1A1, COL1A2, PLOD2, CRTAP, LEPRE1, PPIB, FKBP10, SERPINH1 and SERPINF1. Homozygosity for a single nucleotide variant in SERPINH1 (c.710T>C) was identified, predicting a single amino acid substitution in the protein (p.237Met>Thr; Fig. 2). The parents were carriers of the sequence change, consistent with autosomal recessive inheritance; DNA from the similarly affected sibling was not available. Sequence alignment of vertebrate HSP47 proteins showed that methionine 237 (M237) is highly conserved (Fig. 2C), supporting its requirement for the normal function of the protein. M237 is located in the major functional domain of HSP47, the serine-type endopeptidase inhibitor domain responsible for its chaperone function in the folding of fibrillar procollagen molecules. The previously reported missense mutations in HSP47-producing OI in dogs [green arrow in Fig. 2C; (13)] and humans [blue arrow in Fig. 2C; (9)] were also located in this domain. The genetic data are therefore consistent with the identified HSP47 missense change-producing OI in the family.
Figure 2.
Recessive missense mutations in SERPINH1 change a highly conserved HSP47 residue. (A) Pedigree of the family showing two of four affected siblings (R92-020A and B) and a phenotypically undescribed miscarriage. (B) Chromatograms showing the sequence of position c. 710 in SERPINH1 (in yellow) annotated as a thymine (T) in the WT allele and cytosine (C) in R92-020A and the carrier parents. (C) Representation of HSP47 protein with the serine-type endopeptidase inhibitor domain in light blue. Protein alignment of HSP47 sequences from several vertebrate species show high conservation of the residues with OI-related mutations. The R92-020A mutation, resulting in the missense change at amino acid 237, is labeled with a black box and arrow. The previously described human OI case with a missense mutation in another highly conserved residue (L87) is shown in blue, and the dachshund (dog) OI change at residue 326L is shown in green.
SERPINH1 mutation destabilizes HSP47 and FKBP65 at the protein level
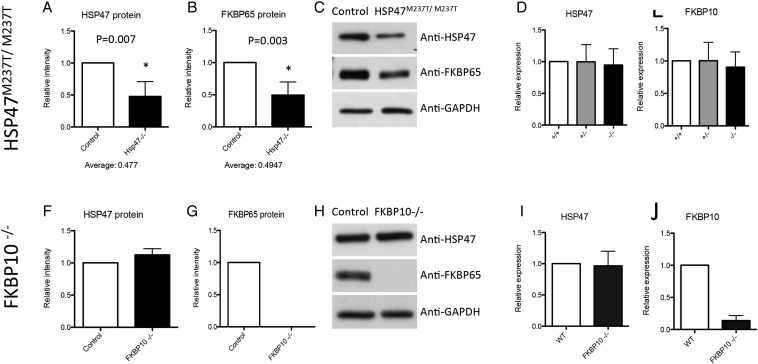
To determine the effect of the SERPINH1 variant on HSP47 synthesis, we analyzed cell lysates of cultured dermal fibroblasts by western blot. Compared with control cells, HSP47 in the mutant cells was reduced by ∼50% (Fig. 3A and C), suggesting that the mutation decreases the stability of the abnormal protein. To further test this finding, we used protein sequence analysis by liquid chromatography–mass spectrometry (LC-MS) to determine the percentage of wild-type versus mutated HSP47 in protein isolated from cells of one of the carrier parents. This analysis showed that 13% of the total HSP47 was derived from the mutant allele (data not shown), again suggesting that the mutant protein is unstable. At the RNA level, the amount of HSP47 transcript was similar in cells from controls, carriers and the OI case (Fig. 3D), indicating that decreased HSP47 protein was not the result of reduced transcription or mRNA stability.
Figure 3.
SERPINH1 mutation causes a reduction in HSP47 and FKBP65 protein levels. (A–C) Protein levels for HSP47 and FKBP65 in HSP47M237T/M237T cells show a decrease in both proteins. (D and E) Transcriptional levels of HSP47 and FKBP65 do not change in HSP47M237T/M237T cells as measured by qRT-PCR. (F–H) Protein levels in FKBP10−/− cells show no change for HSP47 and absence of FKBP65. (I and J) Transcriptional levels for FKBP10−/− cells do not show changes in HSP47 and confirm lack of FKBP10 transcript.
Normal hydroxylation of Pro986 of the α1(I) chain (7), the lack of posttranslational overmodification of type I procollagen in HSP47-mutant cells and the preference of HSP47 for triple-helical molecules (11) are consistent with HSP47 acting on folded procollagen after the prolyl 3-hydroxylation complex (6–8). Cells derived from cases of recessive OI owing to mutations in FKBP10, which encodes another type I procollagen chaperone resident in the ER, also synthesize type I procollagen that is normally posttranslationally modified and have normal hydroxylation of Pro986 of the α1(I) chain (10,21,22,24). This correlation suggested the hypothesis that HSP47 and FKBP65 might act within the ER at a similar stage of type I procollagen biosynthesis. In addition, expression of the two genes is correlated in the UGET co-expression database (25) showing a mean correlation coefficient of 0.53 (Supplementary Material, Table S1). To determine whether the HSP47 deficiency might affect FKBP65, we measured the level of the protein in HSP47M237T/M237T cells. There was a similar decrease in the amount of FKBP65 as observed for HSP47 (Fig. 3B and C), suggesting that the abnormal activity and/or reduced levels of HSP47 protein had a concomitant effect on FKBP65 in the mutant cells. As for the HSP47 transcript, the expression level of FKBP10 was similar in control, carrier and HSP47-mutant cells (Fig. 3E), eliminating decreased transcription or transcript instability as the cause of decreased FKBP65 protein abundance in the presence of abnormal HSP47.
To determine whether the reciprocal reduction in both HSP47 and FKBP65 protein levels would also be present in cells from an OI case with a mutation in FKBP10, we analyzed proteins synthesized by cells derived from a case of recessive OI homozygous for a FKBP10-null mutation (c.831_832insC) (10). This mutation produces a translational frameshift (p.Gly278ArgfsX295) and has been demonstrated to result in complete absence of FKBP65 protein. In these cells, the level of HSP47 protein was similar in FKBP65-mutant and control cells (Fig. 3F–H), indicating that the loss of FKBP65 does not destabilize HSP47. As expected, there were no changes in the transcript level of HSP47, in accord with the preservation of HSP47 protein levels in the FKBP10-mutant cells (Fig. 3I and J).
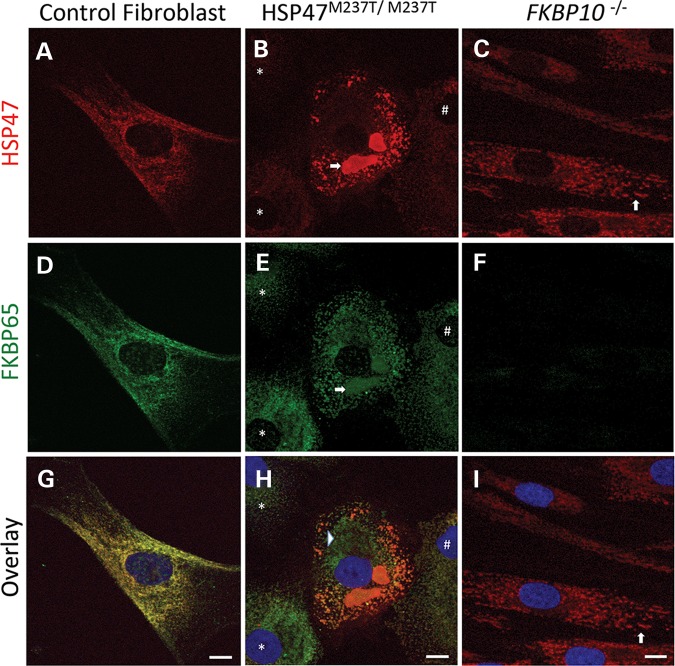
Mutant HSP47 is localized within punctate vesicles that are distinct from the main ER compartment
Under normal conditions, HSP47 and FKBP65 proteins are localized to the ER. To determine whether HSP47 and FKBP65 properly localized in mutant cells, we used immunofluorescence with antibodies against the two proteins. Control fibroblasts showed a reticular staining patterning typical of the ER organelle for both proteins, co-localizing them within the ER (Fig. 4A, D and G). HSP47-mutant cells showed a more rounded morphology, significantly different than that observed in fibroblasts (Fig. 4B, E and H). As expected, in HSP47-mutant cells, there was reduced HSP47 protein in most cells: 80% of cells had low HSP47 expression located in the ER, 10% had almost complete loss of signal and 10% had similar signal intensity to WT cells (Fig. 4B, E and H). In addition, HSP47 showed a punctate staining pattern differing from the reticular pattern observed in control cells. The abnormal staining pattern was characterized by the presence of vesicle-like structures, which labeled with anti-HSP47 antibody (Fig. 4B). Among cells containing a significant amount of HSP47, many cells showed a massive accumulation of HSP47 protein in the form of vacuolar-like compartments (arrow in Fig. 4B). In all HSP47-mutant cells, FKBP65 also showed a punctate pattern, partially co-localizing with HSP47 (Fig. 4E and H). FKBP65 was also detected in the large vacuolar-like structures containing HSP47 (arrow in Fig. 4E). Apart from the structures in which the two proteins co-localized, most of which were located at the periphery of the cells, FKBP65 labeling was also present in the main ER compartment where HSP47 staining was largely absent (arrowhead in Fig. 4H).
Figure 4.
HSP47M237T M237T and FKBP10−/− cells show protein accumulation in vesicles. Immunofluorescence against HSP47 (red) and FKBP65 (green) of control cells (A, D, and G), HSP47M237T/M237T (B, E and H) and FKBP65 (C, F and I) show co-localization of these proteins and their presence in massive vesicles in HSP47-mutant cells (arrows in B and E) and ER dilatation in FKBP10−/− (arrows in C and I). Arrow, asterisk and hash in B, E and H show variability of HSP47 signal in HSP47M237T M237T cells. Asterisk shows almost absence of HSP47 protein and hash shows a cell with low HSP47 expression. Bars represent 10 µm.
As expected, immunofluorescence analysis of FKBP10-mutant cells showed complete absence of FKBP65 (Fig. 4C, F and I). As predicted by the western data, the overall amount of HSP47 appeared to be similar to control cells, but it did not show the typical ER distribution (Fig. 4C and I, arrows). In these cells, HSP47 presented a punctate pattern similar to that observed in HSP47M237T/M237T cells, and ∼10% of the cells showed ER dilatation in the form of vesicles (Fig. 4C and I, arrows). These data show that although FKBP10−/− cells express a normal amount of HSP47, the absence of FKBP65 results in aggregation of HSP47 in vesicles that are not observed in control cells.
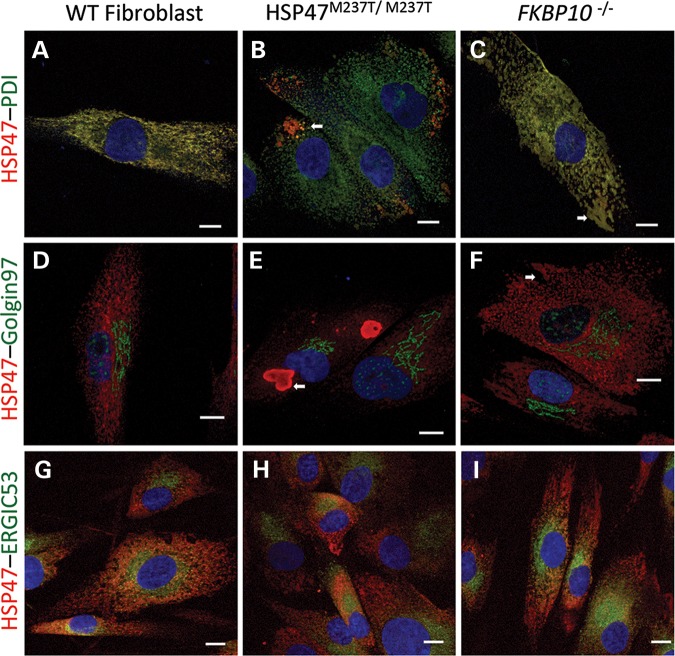
To examine the nature of the vesicles, we compared the localization of HSP47 and FKBP65 proteins with ER and Golgi markers. In control fibroblasts, HSP47 co-localized completely with the ER marker PDI (Fig. 5A). In HSP47M237T/M237T cells, HSP47 was barely present within the ER and the vesicle-like structures containing the bulk of the HSP47 were partially stained with the PDI antibody, particularly in regions immediately adjacent to the ER compartment, suggesting a possible origin of the vesicles in the ER (arrow in Fig. 5B). In FKBP10-mutant cells, HSP47 co-localized with the ER marker (Fig. 5C), including in an abnormal dilated portion of the ER that labeled with both antibodies (arrow in Fig. 5C). In the Golgi compartment, all three cell lines, control, HSP47M237T/M237T and FKBP10−/−, showed that there was no co-localization between HSP47 and Golgin97 antibodies (Fig. 5D–F). In addition, vesicles accumulating HSP47 and dilated ER in mutant cells were physically separated from the Golgi compartment (arrows in Fig. 5E and F). To determine whether the vesicle-like structures containing HSP47 were related to the Golgi apparatus, we used the ERGIC53 antibody, which labels the ER–Golgi intermediate compartment, the vesicles traveling from the ER to the Golgi. Similar to what was observed in control cells, both mutant cell lines, HSP47M237T/M237T and FKBP10−/−, showed HSP47 labeling in ERGIC53-labeled vesicles closer to the ER (Fig. 5G–I), suggesting that while some HSP47 transits beyond the ER, it is not found in the Golgi and is therefore likely to be recycled from the ERGIC to the ER. Because the co-localization of ERGIC53 and HSP47 was not observed in the large vesicles in HSP47-mutant cells, these vesicles might originate directly from the ER without involvement of the ERGIC.
Figure 5.
HSP47 accumulates in ER-related vesicles in OI-mutant cells. Immunofluorescence of HSP47 (red) and cell compartments (green): PDI for ER (A–C), Golgin97 for Golgi apparatus (D–F) and ERGIC53 for ER–Golgi intermediate compartment (G–I). Control (A, D and G), HSP47M237T M237T (B, E and H) and FKBP10−/− cells (C, F and I). White arrows identify vesicles accumulating HSP47. Bars represent 10 µm.
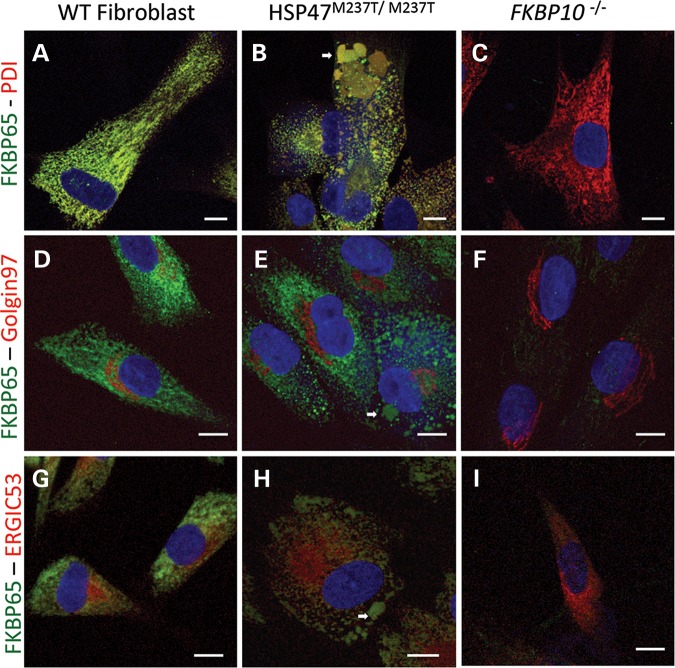
Similar results were observed when we performed immunofluorescence to localize FKBP65. In control cells, FKBP65 co-localized completely with PDI (Fig. 6A), consistent with its normal ER location. In HSP47M237T/M237T cells, FKBP65 was localized to the ER (Fig. 6B) as well as to the large HSP47-containing particles (arrow in Fig. 6B), with no staining of the Golgi and little staining of the ERGIC (Fig. 6E and H). Similar to the HSP47 staining, these data suggest that the Golgi is not involved in the generation of the large HSP47-containing vesicles. As expected, the FKBP10−/− cells did not show any signal with anti-FKBP65 antibody and markers for the ER, Golgi and ERGIC were normally distributed (Fig. 6C, F and I). In control cells, the lack of staining of the ERGIC compartment (Fig. 6G) suggests that FKBP65 is localized and primarily functions within the ER.
Figure 6.
FKBP65 accumulates in ER-related vesicles in OI-mutant cells. Immunofluorescence of FKBP65 (green) and cell compartments (red): PDI for ER (A–C), Golgin97 for Golgi apparatus (D–F) and ERGIC53 for ER–Golgi intermediate compartment (G–I). Control (A, D and G), HSP47M237T/M237T (B, E and H) and FKBP10−/− cells (C, F and I). White arrows identify vesicles accumulating FKBP65. Bars represent 10 µm.
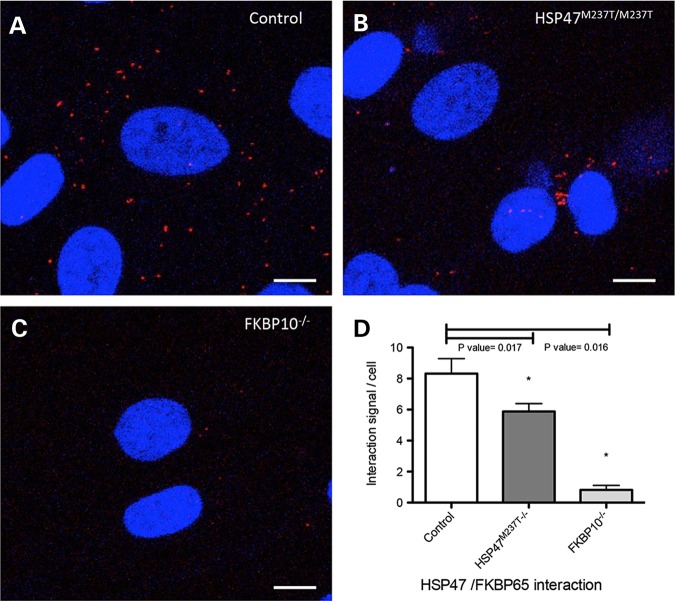
To determine whether co-localization between HSP47 and FKBP65 corresponds with the physical interaction between these proteins, we performed a Proximity Ligation Assay. This technique allows the identification of in-situ interactions of two proteins at endogenous levels (26). As observed in control fibroblasts, interaction of these two proteins was observed as single dots in the cytoplasm (Fig. 7A). The signal was also observed in HSP47-mutant cells; however, the number of interaction was reduced and clustered in a narrow area as opposed to the more dispersed appearance in controls (Fig. 7B and D). As expected, FKBP10-mutant cells showed an almost complete lack of signal owing to the absence of the FKBP65 protein (Fig. 7C). These results suggest that HSP47 and FKBP65 are interacting or working in a very close proximity and that abnormalities in either protein lead to abnormal trafficking, which drags a significant fraction of the complex into vesicles.
Figure 7.
In situ localization of the interaction between HSP47 and FKBP65. Interaction of endogenous chaperones was measured by Proximity Ligation Assay (PLA) in Control (A), HSP47M237T/M237T (B) and FKBP10−/− fibroblasts. (C). Significant reduction was observed in HSP47 and FKBP10-mutant cells (* in D). Bars represent 10 µm.
Vesicles with defective HSP47 and FKBP65 proteins contain type I procollagen
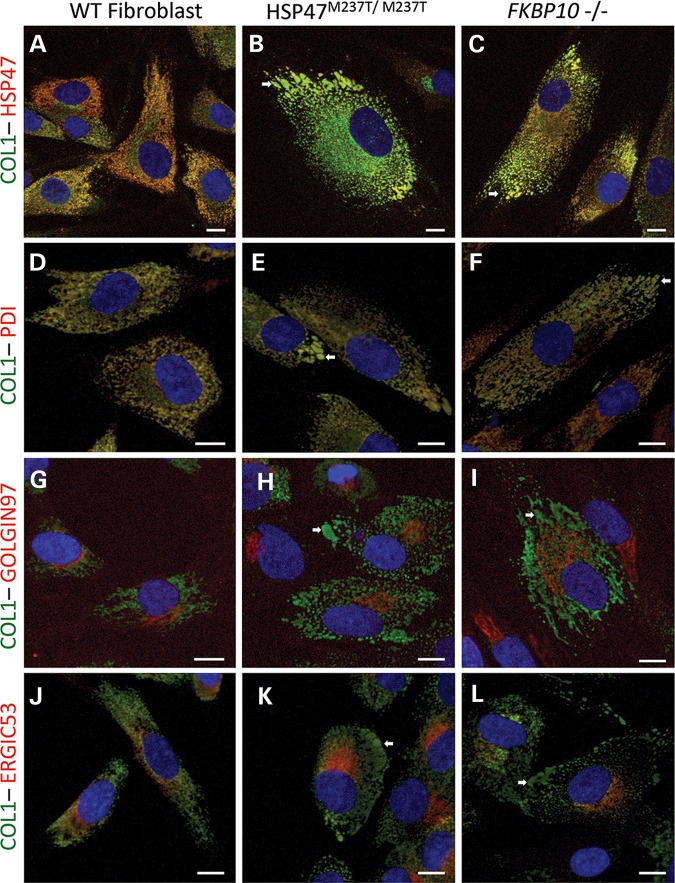
As HSP47 and FKBP65 chaperone fibrillar procollagen molecules during their posttranslational processing in the ER, we performed experiments to determine whether type I procollagen was present in the abnormal vesicles together with HSP47 and FKBP65. As expected, co-localization of HSP47 and type I procollagen was observed in the main ER compartment in control cells (Fig. 8A and D). Also as expected, type I procollagen was in the ERGIC and Golgi compartments where HSP47 is not present, consistent with the separation of the two proteins upon exit from the ER (Fig. 8G and J). In HSP47M237T/M237T cells, type I procollagen maintained its normal localization in the ER and Golgi (Fig. 8B, E, H and K) but was also found in the HSP47-accumulating vesicles (arrow in Fig. 8B, E, H and K) suggesting that type I procollagen exits the ER with mutant HSP47 and without trafficking through the Golgi. A similar pattern was found in FKBP10−/− cells, with presence of type I procollagen in the ER, ERGIC and Golgi, as well as in the dilated ER vesicles containing HSP47 (Fig. 8C, F, I and L). The presence of type I procollagen in the abnormal vesicles suggests that some type I procollagen molecules are not being processed normally through the Golgi and might be recycled or removed along with the defective chaperones.
Figure 8.
Type I procollagen is present in ER-related abnormal vesicles in OI-mutant cells. Immunofluorescence of COL1 (green) and HSP47 and cell compartments (red): HSP47 (A–C), PDI for ER (D–F) Golgin97 for Golgi apparatus (G–I) and ERGIC53 for ER–Golgi intermediate compartment (J–L). Control (A, D, G and J), HSP47M237T/M237T (B, E, H and K) and FKBP10−/− cells (C, F, I and L). White arrows identify vesicles accumulating HSP47 and type I procollagen. Bars represent 10 µm.
Discussion
HSP47 function is essential for type I procollagen trafficking
A major function of the HSP47 chaperone is to assist in the proper folding of type I procollagen (27,28). Loss of Hsp47 in mice is embryonically lethal (17), revealing an essential role for the protein in early development. Type I procollagen trimers are synthesized in the Hsp47 knockout mice, and the resulting molecules are able to access the Golgi compartment, but they are unable to be cleaved at their amino-and carboxyl-termini (17,29). Thus, the absence of mature procollagen in the mice likely explains the lethal phenotype. In contrast, our data and previously published recessive OI cases with HSP47 missense mutations (9,13) demonstrate that mature type I collagen trimers can be synthesized, indicating that the mutant HSP47 retains partial function that allows synthesis, secretion and maturation of type I procollagen. While most of the procollagen is normally secreted, a fraction is translocated from the ER compartment to vesicles. Thus, HSP47 appears to have a dual function, first to assist with procollagen assembly in the ER and subsequently to assist in trafficking of trimeric molecules to the Golgi.
Hsp47 and FKBP65 cooperate in type I procollagen processing
Our experiments showed that protein levels of FKBP65 are reduced and mislocalized in HSP47M237T/M237T cells, identifying a relationship between the functions of the two molecules. While FKBP10−/− cells did not show a reciprocal effect on HSP47 protein levels, some HSP47 was mislocalized within the FKBP10-mutant cells, again suggesting that the functions of the two molecules are interdependent and that they cooperate in type I procollagen biosynthesis.
The precise mechanism by which FKBP65 is destabilized in HSP47M237T/M237T cells is not clear. One possibility is that the incorporation of defective HSP47 into vesicles leads to FKBP65 being dragged into the vesicles and degraded. The co-localization of the two proteins in abnormal ER vesicles suggests that HSP47 and FKBP65 are processed together in HSP47M237T/M237T cells. As both proteins participate in type I procollagen biosynthesis and type I procollagen is found in the abnormal vesicles in HSP47M237T/M237T and FKBP10−/− cells, perhaps the failure to properly process type I procollagen is the common event that leads to vesicle formation. This hypothesis supports the notion that there is an interaction among HSP47, FKBP65 and type I procollagen within the ER, which results in OI when it is disrupted. Co-immunoprecipitation experiments failed to demonstrate a direct interaction in either cultured fibroblast or bone lysates (data not shown), suggesting that the interaction is either indirect or that there is a direct interaction, which is either weak or transient. However, the Proximity Ligation Assay data suggest that HSP47 and FKBP65 physically interact in subcellular compartments that include the ER in control fibroblasts and the abnormal vesicles in HSP47-mutant cells. Furthermore, a reduction/absence of this functional interaction in the OI-mutant cells suggests that cooperation of these two proteins is failing during the synthesis of type I procollagen in these cells. We suggest that the similarity in phenotype between OI cases with mutations in either gene could be explained by the similar consequences on type I procollagen synthesis and the resulting cell biological effects.
Overall these data confirm that homozygosity for HSP47 missense mutations can produce moderately severe OI and establish an intracellular relationship between HSP47 and FKBP65, suggesting that they work together during type I procollagen biosynthesis. The data further show that mutant HSP47 leads to diminished FKBP65 protein. While loss of FKBP65 does not affect HSP47 levels, it does lead to HSP47 mislocalization. Co-localization of HSP47 with ERGIC proteins demonstrates that HSP47 transits with type I procollagen from the ER to the ERGIC. Abnormal HSP47 may contribute to the failure of type I procollagen to be properly trafficked to the Golgi, leading to the formation of intracellular vesicles. Failure of this cooperation alters type I procollagen trafficking and leads to its accumulation in ER-related vesicles These results thus reveal a common cellular pathway in cases of OI caused by HSP47 and FKBP65 deficiency.
Materials and Methods
Clinical information and DNA isolation
Patients and their family members were assessed under an approved human subjects protocol. Clinical information and radiographs were reviewed to determine the diagnostic findings in the affected individuals. DNA was isolated from blood or cultured fibroblasts via a standard manufacturer's protocol (QIAGEN).
Cell culture and analysis of collagenous proteins
Dermal fibroblast cultures were established from explanted skin biopsies from the cases (International Skeletal Dysplasia Registry reference numbers R92-020A and R93-188A and B) and the parents (R92-020C and R92-020D). Fibroblasts from affected individual and controls were plated at confluence (250 000 cells) in 35-mm tissue culture dishes in Dulbecco-Vogt Modified Eagle Medium supplemented with 10% FBS. For procollagen biosynthesis studies (30,31), confluent cells were starved of proline in serum-free medium supplemented with 50 mm ascorbic acid for 1 h and then labeled in serum-free medium with 100 mCi/ml [2,3,4,5-3H] proline. After labeling, cells were rinsed twice with complete medium with 50 mm ascorbic acid, and fresh chase medium was added that contained 12 mm proline. Then, the medium was decanted and the cell layer scraped into a separate tube in PBS. Protease inhibitors were added to both, and the collagenous proteins were precipitated with 50% v:v ethanol and carrier collagen in the presence of 15 mm iodoacetamide added to prevent new disulfide bond formation. Precipitated proteins were resuspended in SDS-sample buffer and separated on 5% SDS–PAGE under nonreducing conditions. All gels were fixed, treated with Enhance and dried. Radiolabeled bands were visualized by autoradiography.
Immunolocalization, immunoprecipitation and western blotting
Immunofluorescence experiments were performed using a Leica SP2 laser scanning confocal microscopy in the UCLA Advance Light Microscopy/Spectrometry core. Cultured fibroblasts were fixed in 4% PFA in PBS, then, washed and permeabilized with 0.1% Triton X-100 for 5 min, followed by blocking in 10% goat serum for 1 h. The primary antibody was incubated overnight at 4°C (mouse anti-HSP47, 1 : 100 Enzo Life Sciences M16.10A1; rabbit anti-FKBP65, 1 : 100 Proteintech 12172-1-AP; mouse anti-PDI 1 : 100 Abcam ab2792; rabbit anti-PDI 1 : 100 Santa Cruz H-160, mouse anti-Golgin97 1 : 200 Invitrogen A-21270; rabbit anti-Golgin97 1 : 100 Abcam 33701; mouse anti-ERGIC53, 1 : 1000 Enzo Life Sciences G1/93; rabbit anti-ERGIC53 1 : 200 Sigma E1031; rabbit anti-LC3 1 : 50 Cell Signaling 2775; rabbit anti-20S, 1 : 1500 Abcam ab22673; rabbit anti-COL1, 1 : 500 Abcam 34710). Fluorescent secondary antibody was incubated at a 1 : 1000 dilution for 1 h at room temperature antibody (alexa-fluor goat anti-mouse 488 and goat anti-rabbit 568 for all the samples), and DAPI at a 1 : 1000 dilution for 5 min at room temperature was applied before mounting.
For immunoprecipitation, proteins were extracted with RIPA buffer and pulled down with anti-HSP47 (1 : 500 Enzo Life Sciences and Protein A/G Plus agarose, Santa Cruz sc-2003) antibody, overnight at 4°C. Immunoprecipitates were washed in PBS, collected and separated by SDS–PAGE. Gels were stained with Coomassie Blue, and parallel western blots were probed with HSP47 antibody to confirm the immunoprecipitation. The corresponding band stained with Coomassie blue was cut out for trypsin digestion and analysis by liquid chromatography–mass spectrometry (LC-MS) as described (6).
For western blot analyses, proteins lysates were separated by electrophoresis through a 10% SDS–polyacrylamide gel, transferred to PVDF membranes, blocked in 5% milk and probed with primary antibodies (anti-Hsp47 antibody, 1 : 500 Enzo Life Sciences M16.10A1; rabbit anti-FKBP65 1 : 5000, Proteintech 12172-1-AP; rabbit anti-GAPDH 1 : 2000 Cell Signaling 2118S). Peroxidase-conjugated secondary antibodies (Cell Signaling 7071 and 7072) were used and immunocomplexes identified by using the ECL detection reagent (Cell Signaling 7003). Mann–Whitney test was performed for statistic analysis using Prism software.
Quantitative RT-PCR
Total RNA was extracted from cultured dermal fibroblasts at 80% confluence using TRIZOL (Invitrogen 15596), treated with DNAase (Invitrogen 18068) and reversed-transcribed (Thermo Scientific K1621) according to the manufacturer's instructions. Quantitative PCR was performed with a real-time PCR detection system (Stratagene MX3005P) using SYBR green PCR master mix (Thermo Scientific K0222) and standard thermocycler conditions. PCR was performed in a total volume of 25 μl in at least three independent experiments. Each sample was analyzed for at least two housekeeping genes to normalize for RNA input amounts and to perform relative quantifications. Levels of transcripts in controls were set at 1. Melting curve analysis showed a single, sharp peak with the expected temperature melting for all samples. Primers were designed with pairs from multiple different exons wherever possible (Supplementary Material, Table S1).
Proximity Ligation Assay
In situ interactions were detected by Proximity Ligation Assay kit, Duolink (SIGMA, DUO92101) (26,32). Fibroblast cells were fixed and permeabilized as described for immunofluorescence. Antibody incubation and probe amplification were performed according to the manufacturer's instructions. As a negative controls we performed the proximity ligation assay in absence of one of the primary antibodies and on null allele cells for FKBP65 (see Supplementary Material, Fig. S1).
Supplementary Material
Funding
This work was supported in part by grants from the National Institutes of Health (RO1 AR062651, P01 HD070394, RO1 DE19567 and RO1 AR066124). The research was also supported by the NIH/National Center for Advancing Translational Science (NCATS) (UCLA CTSI Grant Number UL1TR000124) and funds from the Orthopaedic Hospital Research Center at UCLA, the Orthopaedic Institute for Children, the Joseph Drown Foundation, the Dreiseszun Family Foundation and the March of Dimes Foundation.
Supplementary Material
Acknowledgements
We thank the family for their participation in this study. Confocal laser microscopy was performed at the CNSI Advanced Light Microscopy/Spectroscopy Resource Facility at UCLA, supported with funding from NIH-NCRR shared resources grant (CJX1-443835-WS-29646) and NSF Major Research Instrumentation grant (CHE-0722519).
Conflict of Interest statement. None declared.
References
- 1.Murakami T., Saito A., Hino S.I., Kondo S., Kanemoto S., Chihara K., Sekiya H., Tsumagari K., Ochiai K., Yoshinaga K., et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat. Cell Biol. 2009;10:1205–1211. doi: 10.1038/ncb1963. [DOI] [PubMed] [Google Scholar]
- 2.Lisse T.S., Thiele F., Fuchs H., Hans W., Przemeck G.K., Abe K., Rathkolb B., Quintanilla-Martinez L., Hoelzlwimmer G., Helfrich M., et al. ER stress-mediated apoptosis in a new mouse model of osteogenesis imperfecta. PLoS Genet. 2008;2:e7. doi: 10.1371/journal.pgen.0040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forlino A., Cabral W.A., Barnes A.M., Marini J.C. New perspectives on osteogenesis imperfecta. Nat. Rev. Endocrinol. 2011;7:540–557. doi: 10.1038/nrendo.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byers P.H., Pyott S.M. Recessively inherited forms of osteogenesis imperfecta. Annu. Rev. Genet. 2012;46:475–497. doi: 10.1146/annurev-genet-110711-155608. [DOI] [PubMed] [Google Scholar]
- 5.Marini J.C., Blissett A.R. New genes in bone development: what's new in osteogenesis imperfecta. J. Clin. Endocrinol. Metab. 2013;98:3095–3103. doi: 10.1210/jc.2013-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morello R., Bertin T.K., Chen Y., Hicks J., Tonachini L., Monticone M., Castagnola P., Rauch F., Glorieux F.H., Vranka J., et al. CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127:291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 7.Cabral W.A., Chang W., Barnes A.M., Weis M., Scott M.A., Leikin S., Makareeva E., Kuznetsova N.V., Rosenbaum K.N., Tifft C.J., et al. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat. Genet. 2007;39:359–365. doi: 10.1038/ng1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dijk F.S., Nesbitt I.M., Zwikstra E.H., Nikkels P.G.J., Piersma S.R., Fratantoni S.A., Jimenez C.R., Huizer M., Morsman A.C., Cobben J.M., et al. PPIB mutations cause severe osteogenesis imperfecta. Am. J. Hum. Genet. 2009;85:521–527. doi: 10.1016/j.ajhg.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christiansen H.E., Schwarze U., Pyott S.M., AlSwaid A., Balwi A.l.M., Alrasheed S., Pepin M.G., Weis M.A., Eyre D.R., Byers P.H. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2010;86:389–398. doi: 10.1016/j.ajhg.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alanay Y., Avaygan H., Camacho N., Utine G.E., Boduroglu K., Aktas D., Alikasifoglu M., Tuncbilek E., Orhan D., Bakar F.T., et al. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2010;86:551–559. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macdonald J.R., Bächinger H.P. HSP47 binds cooperatively to triple helical type I collagen but has little effect on the thermal stability or rate of refolding. J. Biol. Chem. 2001;276:25399–25403. doi: 10.1074/jbc.M102471200. [DOI] [PubMed] [Google Scholar]
- 12.Kojima T., Miyaishi O., Saga S., Ishiguro N., Tsutsui Y., Iwata H. The retention of abnormal type I procollagen and correlated expression of HSP 47 in fibroblasts from a patient with lethal osteogenesis imperfecta. J. Pathol. 1998;184:212–218. doi: 10.1002/(SICI)1096-9896(199802)184:2<212::AID-PATH996>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 13.Drögemüller C., Becker D., Brunner A., Haase B., Kircher P., Seeliger F., Fehr M., Baumann U., Lindblad-Toh K., Leeb T. A missense mutation in the SERPINH1 gene in dachshunds with osteogenesis imperfecta. PLoS Genetics. 2009;5:e1000579. doi: 10.1371/journal.pgen.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koide T., Takahara Y., Asada S., Nagata K. Xaa-Arg-Gly triplets in the collagen triple helix are dominant binding sites for the molecular chaperone HSP47. J. Biol. Chem. 2002;277:6178–6182. doi: 10.1074/jbc.M106497200. [DOI] [PubMed] [Google Scholar]
- 15.Ono T., Miyazaki T., Ishida Y., Uehata M., Nagata K. Direct in vitro and in vivo evidence for interaction between Hsp47 protein and collagen triple helix. J. Biol. Chem. 2012;287:6810–6818. doi: 10.1074/jbc.M111.280248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yagi-Utsumi M., Yoshikawa S., Yamaguchi Y., Nishi Y., Kurimoto E., Ishida Y., Homma T., Hoseki J., Nishikawa Y., Koide T., et al. NMR and mutational identification of the collagen-binding site of the chaperone Hsp47. PLoS ONE. 2012;7:e45930. doi: 10.1371/journal.pone.0045930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagai N., Hosokawa M., Itohara S., Adachi E., Matsushita T., Hosokawa N., Nagata K. Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis. J. Cell Biol. 2000;150:1499–1506. doi: 10.1083/jcb.150.6.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagata K. HSP47 as a collagen-specific molecular chaperone: function and expression in normal mouse development. Semin. Cell Dev, Biol. 2003;14:275–282. doi: 10.1016/j.semcdb.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 19.Kelley B.P., Malfait F., Bonafé L., Baldridge D., Homan E., Symoens S., Willaert A., Elcioglu N., Van Maldergem L., Verellen-Dumoulin C., et al. Mutations in FKBP10 cause recessive osteogenesis imperfecta and bruck syndrome. J. Bone Miner. Res. 2011;26:666–672. doi: 10.1002/jbmr.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Setijowati E.D., Van Dijk F.S., Cobben J.M., van Rijn R.R., Sistermans E.A., Faradz S.M.H., Kawiyana S., Pals G. A novel homozygous 5 bp deletion in FKBP10 causes clinically Bruck syndrome in an Indonesian patient. Eur. J. Med. Genet. 2012;55:17–21. doi: 10.1016/j.ejmg.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Venturi G., Monti E., Carbonare L.D., Corradi M., Gandini A., Valenti M.T., Boner A., Antoniazzi F. A novel splicing mutation in FKBP10 causing osteogenesis imperfecta with a possible mineralization defect. Bone. 2012;50:343–349. doi: 10.1016/j.bone.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 22.Barnes A.M., Cabral W.A., Weis M., Makareeva E., Mertz E.L., Leikin S., Eyre D., Trujillo C., Marini J.C. Absence of FKBP10 in recessive type XI osteogenesis imperfecta leads to diminished collagen cross-linking and reduced collagen deposition in extracellular matrix. Hum. Mutat. 2012;33:1589–1598. doi: 10.1002/humu.22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaheen R., Al-Owain M., Sakati N., Alzayed Z.S., Alkuraya F.S. FKBP10 and Bruck syndrome: phenotypic heterogeneity or call for reclassification? Am. J. Hum. Genet. 2010;87:306–307. doi: 10.1016/j.ajhg.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarze U., Cundy T., Pyott S.M., Christiansen H.E., Hegde M.R., Bank R.A., Pals G., Ankala A., Conneely K., Seaver L., et al. Mutations in FKBP10, which result in Bruck syndrome and recessive forms of osteogenesis imperfecta, inhibit the hydroxylation of telopeptide lysines in bone collagen. Hum. Mol. Genet. 2012;22:1–17. doi: 10.1093/hmg/dds371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day A., Dong J., Funari V.A., Harry B., Strom S.P., Cohn D.H., Nelson S.F. Disease gene characterization through large-scale co-expression analysis. PLoS ONE. 2009;4:e8491. doi: 10.1371/journal.pone.0008491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G., Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 27.Nagata K. Hsp47: a collagen-specific molecular chaperone. Trends. Biochem. Sci. 1996;21:22–26. doi: 10.1016/0968-0004(96)80881-4. [DOI] [PubMed] [Google Scholar]
- 28.Nagata K., Saga S., Yamada K.M. Characterization of a novel transformation-sensitive heat-shock protein (HSP47) that binds to collagen. Biochem. Biophys. Res. Commun. 1988;153:428–434. doi: 10.1016/s0006-291x(88)81242-7. [DOI] [PubMed] [Google Scholar]
- 29.Masago Y., Hosoya A., Kawasaki K., Kawano S., Nasu A., Toguchida J., Fujita K., Nakamura H., Kondoh G., Nagata K. The molecular chaperone Hsp47 is essential for cartilage and endochondral bone formation. J. Cell Sci. 2012;125:1118–1128. doi: 10.1242/jcs.089748. [DOI] [PubMed] [Google Scholar]
- 30.Bonadio J., Holbrook K.A., Gelinas R.E., Jacob J., Byers P.H. Altered triple helical structure of type I procollagen in lethal perinatal osteogenesis imperfecta. J. Biol. Chem. 1985;260:1734–1742. [PubMed] [Google Scholar]
- 31.Bonadio J., Byers P.H. Subtle structural alterations in the chains of type I procollagen produce osteogenesis imperfecta type II. Nature. 1985;316:363–366. doi: 10.1038/316363a0. [DOI] [PubMed] [Google Scholar]
- 32.Weibrecht I., Leuchowius K.-J., Clausson C.-M., Conze T., Jarvius M., Howell W.M., Kamali-Moghaddam M., Söderberg O. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev Proteomics. 2010;7:401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.