SUMMARY
T cell receptor (TCR) cross-reactivity between major histocompatibility complex II (MHCII)-binding self and foreign peptides could influence the naïve CD4+ T cell repertoire and autoimmunity. We found that nonamer peptides that bind to the same MHCII molecule only need to share five amino acids to cross-react on the same TCR. This property was biologically relevant since systemic expression of a self peptide reduced the size of a naïve cell population specific for a related foreign peptide by deletion of cells with cross-reactive TCRs. Reciprocally, an incompletely deleted naïve T cell population specific for a tissue-restricted self peptide could be triggered by related microbial peptides to cause autoimmunity. Thus, TCR cross-reactivity between similar self and foreign peptides can reduce the size of certain foreign peptide-specific T cell populations, and may allow T cell populations specific for tissue-restricted self peptides to cause autoimmunity after infection.
INTRODUCTION
Naïve T cell populations vary in size over several orders of magnitude (Alanio et al., 2010; Campion et al., 2014; Flesch et al., 2010; Kotturi et al., 2008; Kwok et al., 2012; Legoux et al., 2010; Moon et al., 2007; Obar et al., 2008; Schmidt et al., 2011; Su et al., 2013; Tan et al., 2011). It remains unclear, however, which factors account for this variability. The chemistry of the interaction between certain major histocompatibility complex bound peptides (p:MHC) and the T cell antigen receptor (TCR) is one important determinant (Chu et al., 2010; Turner et al., 2005). Alternatively, foreign-p:MHC-specific T cell populations with a high degree of cross-reactivity for self-p:MHC encountered during thymic development may be pared by negative selection. This possibility, however, has been difficult to assess because the rules for TCR cross-reactivity have not been determined.
Recent progress, however, has been made in this area for p:MHCII-specific TCRs expressed by CD4+ T cells (Birnbaum et al., 2014; Lucca et al., 2014; Su et al., 2013). Peptides of up to 20 amino acids bind to MHCII molecules via a nine amino acid core sequence (Rudolph et al., 2006). Certain amino acids within the core nonamer, often at positions 1, 4, 6 and 9, anchor the peptide by fitting into discrete pockets in the MHCII groove (Painter and Stern, 2012). The amino acids at positions 2, 3, 5, and 8 generally point up and out of the MHCII binding groove. Complementarity determining region (CDR) 1 and 2 of the TCR interact with the MHCII molecule, while CDR3 interacts mainly with the upward pointing residues in the peptide (Marrack et al., 2008). Using soluble TCRs to probe yeast-displayed MHCII-bound peptide libraries, Birnbaum, et al. (Birnbaum et al., 2014) found that a single TCR could bind dozens of peptides with similar TCR contact amino acids but very different MHCII anchor amino acids. If TCR specificity is determined by only 4 of the 9 amino acids in a peptide, then 1 in 160,000 (204) MHCII-bound peptides will have the same TCR contact amino acids and thus bind to the same TCR. Because a mammalian proteome can theoretically produce many more than 160,000 different MHCII-binding peptides, any one MHCII-bound foreign peptide could have several self-peptide homologs. These homologs could reduce the size of foreign peptide-specific populations by causing clonal deletion. Since different foreign peptides could have different numbers of self-peptide homologs, the corresponding naïve cell populations could undergo different degrees of selection and vary in size for this reason.
We tested this premise in C57BL/6 (B6) mice, which express the I-Ab MHCII molecule. We confirmed that TCR specificity for peptides bound to I-Ab (p:I-Ab) depended primarily on four TCR contact amino acids and was relatively independent of the nature of the MHCII anchor residues. This type of TCR cross-reactivity reduced the size of T cell populations specific for foreign peptides that had many self-peptide homologs. In addition, an incompletely deleted naive population specific for a tissue-restricted self peptide could be primed with I-Ab-bound bacterial peptides that matched the self peptide at TCR contact amino acids. Thus, TCR cross-reactivity between similar self and foreign peptides influences the composition of the foreign peptide-specific T cell repertoire via clonal deletion, and accounts for the capacity of microbial peptides to trigger autoimmunity to a self peptide.
RESULTS
Foreign-p:I-Ab-specific naïve T cell populations vary in size
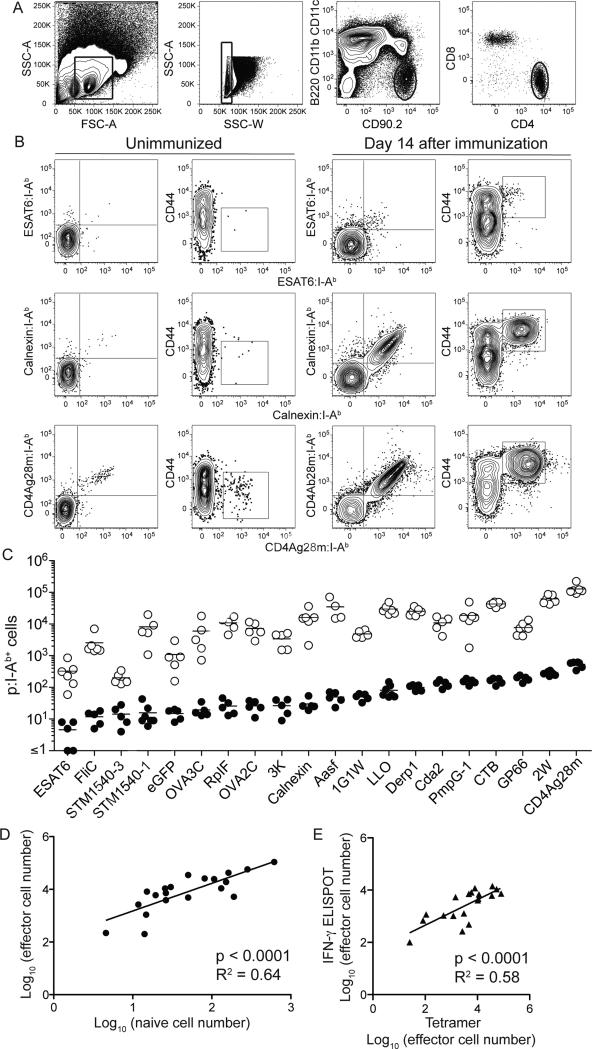
Naïve CD4+ T cell populations specific for 3 different I-Ab-bound foreign (non-mouse) peptides vary in size in B6 mice (Moon et al. 2007). We enumerated many more populations to estimate the normal range and to set the stage for assessment of the effect of clonal deletion by related self (mouse) peptides. A p:MHCII tetramer-based cell enrichment and flow cytometry strategy (Moon et al., 2009; Moon et al., 2007) was used to identify naïve CD4+ T cell populations in B6 mice specific for the first 20 different I-Ab-bound foreign peptides listed in Table 1. In each case, cells from the secondary lymphoid organs were stained with a pair of I-Ab tetramers containing the same peptide but labeled with different fluorochromes to maximize the TCR-specificity of the assay (Stetson et al., 2002; Tubo et al., 2013). Tetramer-bound cells were then labeled with magnetic beads, enriched on magnetized columns, and stained with antibodies specific for informative surface markers. The tetramer-bound cells in the bound fraction were detected by gating on lymphocyte-sized, single cells that expressed CD90.2 but not B220, CD11b, or CD11c, and CD4 but not CD8 (Figure 1A). Most double tetramer-binding cells had the CD44lo phenotype of naïve cells, indicating that these mice, which were housed in a specific pathogen-free facility, had no prior exposure to these epitopes (Figure 1B, left columns). The absolute number of naïve CD4+ cells ranged from fewer than an average of 5 ESAT6:I-Ab+ cells per mouse to over 500 CD4Ag28m:I-Ab+ cells per mouse (Figure 1C, filled circles).
Table 1.
Sequence of peptides in I-Ab tetramers Each amino acid sequence contained in an I-Ab tetramer used in this study is listed. Numbers 1 through 9 above the sequences indicate the nonamer core predicted by Zhu et al. (Zhu et al., 2003) and Liu et al. (Liu et al., 2002) for each peptide with putative TCR contacts at P2, 5, 7, and 8 indicated by asterisks at the top and I-Ab anchors at P1, 4, 6, 7, and 9 denoted with carets at the bottom. The # signs refer to peptides that were locked into the I-Ab molecule in the indicated register by a disulfide trap strategy (Stadinski et al., 2010). The ESAT6 peptide is from the ESAT6 protein of Mycobacterium tuberculosis (Reiley et al., 2010); the FliC, STM1540-3, and STM1540-1 peptides are from the flagellin or STM1540 proteins of Salmonella enterica serovar Typhimurium (Maybeno et al., 2012; McSorley et al., 2000); the OVA2C and OVA3C peptides are from chicken ovalbumin (Moon et al., 2011; Robertson et al., 2000); the RpIF, Aasf, and PmpG-1 peptides are from ribosomal protein L6, anti-anti-sigma factor, and polymorphic membrane protein G of Chlamydia muridarum (Li and McSorley, 2013); the LLO peptide is from the listeriolysin O protein of Listeria monocytogenes (Geginat et al., 2001; Pepper et al., 2011); the CTB peptide is from the cholera toxin B subunit of Vibrio cholerae (Cong et al., 1996); the GP66 peptide is from glycoprotein 1 of Lymphocytic choriomeningitis virus (Oxenius et al., 1995); the CD4Ag28m peptide is from a putative transmembrane protein of Toxoplasma gondii (Grover et al., 2012); the Derp1 peptide is from the Der fl protein of Dermatophagoides farinae (Hoyne et al., 1994); the MOG peptide is from mouse myelin oligodendrocyte glycoprotein (Mendel et al., 1995); the calnexin peptide is from the calnexin protein of Blastomyces dermatitidis; the Cda2 peptide is from the chitin deacetylase 2 protein of Cryptococcus neoformans; the eGFP peptide is from enhanced green fluorescent protein. The 2W (Rees et al., 1999), 3K (Rees et al., 1999), 2W109, and 1G1W (Chu et al., 2010) peptides are variants of the E alpha peptide from the I-E MHCII molecule (Murphy et al., 1992).
| Peptide | Position (P) 123456789 * * ** |
|---|---|
| ESAT6 | QQWNFAGIEAAASA |
| FliC | VQNRFNSAITNLGNT |
| STM1540-3# | VYYTTYAPQAT |
| STM1540-1# | YTTYAPQATSA |
| eGFP | HDFFKSAMPEGYVQE |
| OVA3C# | GHAAHAEINEA |
| RplF | FVSPAAHII |
| OVA2C# | QAVHAAHAEIN |
| 3K | EAQKAKANKAVDKA |
| Calnexin | LVVKNPAAHHAIS |
| AasF | VSSPAVQES |
| 1G1W | EAGGALANWAVDSA |
| LLO# | NEKYAQAYPNVS |
| Derp1 | CQIYPPNVNKI |
| Cda2 | HQYMTALSNEVVF |
| PmpG-1 | YVDPAAAGG |
| CTB | NNKTPHAIAAIS |
| GP66 | DIYKGVYQFKSV |
| 2W | EAWGALANWAVDSA |
| CD4Ag28m | VEIHRPVPGTA |
| 2W109# | EYWGPLPNWV |
| MOG# | GWYRSPFSRVVVHLY |
Figure 1. Relationship between p:I-Ab-specific CD4+ T cell population sizes in unimmunized and immunized mice.
(A) Gates used to identify lymphocyte-sized, non-doublet, CD90.2+ B220− CD11b− CD11c−, CD4+ T cells.
(B) Contour plots of double tetramer staining (far left) or CD44 expression (left) for CD4+ T cells from spleen and lymph nodes identified as in (A) from individual unimmunized B6 mice following enrichment with the indicated p:I-Ab tetramers labeled with streptavidin-PE (X- axis) or streptavidin-APC (Y-axis). The plots on the right represent similar staining for populations in spleen and draining lymph nodes 14 days after subcutaneous injection with 10 μg of the relevant peptide emulsified in CFA.
(C) Total number of tetramer-binding CD4+ T cells per mouse for I-Ab-bound peptides in individual unimmunized (open circles) or immunized (filled circles) mice identified as in (B). Horizontal bars indicate mean values. The depicted results were pooled from 11 independent experiments.
(D) Linear correlation between the average log10 naive and effector cell number for each of the populations identified in (C).
(E) Linear correlation between the average log10 effector cell number obtained by tetramer staining (X-axis) or ELISPOT (Y-axis), 14 days after subcutaneous injection of 19 peptides from (C) emulsified in CFA. The depicted results were from 1 experiment with the mean of 3 values from individual mice for each peptide for the tetramer enrichment assay and the mean of 2 values from individual mice for the ELISPOT assay.
Mice were then immunized with the 20 peptides emulsified in complete Freund's adjuvant (CFA) to test the hypothesis that naïve cell number predicts the magnitude of the effector cell response (Jenkins and Moon, 2012). Immunization caused the relevant p:I-Ab specific T cells to expand and induce CD44 expression 2 weeks after immunization (Figure 1B, right columns). The number of effector cells corresponded to the number of naïve cells in most cases (Figure 1C). Naïve cell number predicted 64% of the variance in the number of effector cells generated in response to peptide immunization across the 20 CD4+ T cell populations (Figure 1D).
The interferon-γ Enzyme-Linked ImmunoSpot (ELISPOT) assay (Streeck et al., 2009) was then used to confirm the effector cell population size measurements with a test that did not depend on tetramer binding. B6 mice were immunized with a pool of 19 peptides emulsified in CFA. Two weeks later the spleen and lymph nodes from individual mice were assayed by p:I-Ab tetramer-based cell enrichment or ELISPOT. A highly significant correlation was observed between the numbers of effector cells in the p:I-Ab-specific populations detected by each assay (Figure 1E). This result validated the use of tetramer-based cell enrichment to measure the sizes of p:I-Ab-specific populations and solidified the conclusion that the magnitude of the effector cell response is related to the size of the relevant naïve cell population. The lack of a perfect correlation between naïve and effector cell numbers, however, indicates that other factors influence the magnitude of the primary immune response such as how well or long a peptide binds MHCII or engages regulatory T cells (Sant et al., 2013).
Determining the TCR contact amino acids in nonamer core peptides
To determine if clonal deletion based on TCR cross-reactivity contributed to the variation in naïve population size, it was necessary to identify the TCR contact amino acids in the foreign peptides. An I-Ab-peptide binding motif (Liu et al., 2002; Zhu et al., 2003) was used to identify the most likely nonamer cores for the foreign peptides shown in Table 1. It was not necessary to identify the core for 5 of the peptides, because they were locked into the I-Ab molecule in the register shown in Table 1 using a disulfide trap strategy (Stadinski et al., 2010).
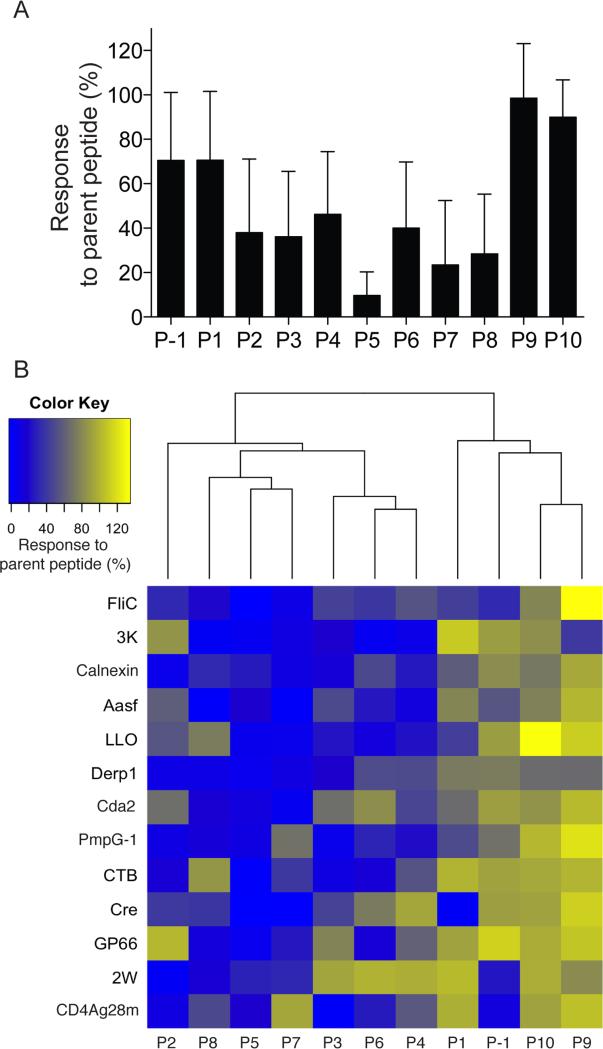
If the I-Ab binding nonamer cores shown in Table 1 are correct, then P5 should be the most important amino acid for TCR recognition because the CDR3s of TCRs generally interact with this amino acid (Marrack et al., 2008). Sets of 11 peptides, each 11 amino acids long and containing an alanine substitution at P-1 to P10, were prepared for each of 13 I-Ab-binding foreign parent peptides to test this possibility. All alanine-substituted peptides were predicted to bind I-Ab because alanine is a favorable residue for all of the pockets in this MHCII molecule (Liu et al., 2002; Zhu et al., 2003). Eleven amino acid peptides were used because it was not clear whether amino acids flanking the nonamer core could serve as additional TCR contacts (Huseby et al., 2005). Mice were primed with the parent peptides in CFA, and 2 weeks later, CD4+ T cells from the draining lymph nodes were stimulated with the parent peptide used for priming or the alanine substituted peptides. The T cell response was then measured by ELISPOT. Alanine substitutions at the predicted P5 residues reduced reactivity by parent peptide-primed T cells by an average of 90% (Figure 2A). Substitutions at P2 or P8 also reduced T cell reactivity by at least 65% as predicted for TCR contact residues (Huseby et al., 2005). In contrast, substitutions at P1 or P9, as well as the flanking amino acids, reduced T cell reactivity on average less than 30%. These results are consistent with P2, 5, and 8 being TCR contact residues for these I-Ab-bound peptides.
Figure 2. Alanine substitution analysis of CD4+ T cell responses to known foreign epitopes.
(A) Eleven amino acid versions of the indicated peptides (nonamer cores shown Table 1 plus P-1 and P10, nonamer core WFPAEPEDV for Cre) with single alanine substitutions at the indicated positions were tested as stimulators of parent peptide-primed CD4+ T cells in an ELISPOT assay. The % of the response to the parent peptide was determined by dividing the number of spots stimulated by the alanine substituted peptide by the number of spots stimulated by the parent peptide and multiplying by 100. The bars represent the average (± SD, n = 3 mice) values for the parent peptides shown in (B) from 2 independent experiments. (B) Unsupervised clustering of the % of response to the parent peptide after stimulation with alanine-substituted peptides for each of the 13 indicated parent peptides.
Substitutions at I-Ab anchor residues P4, 6, and 7 reduced reactivity by parent peptide-primed T cell populations by at least 50%. Because there was some variability in the effect of alanine substitution across the 13 peptides as evidenced by the large standard deviations in Figure 2A, an unsupervised clustering of the alanine scan data was performed as an independent way to identify the important TCR contacts (Figure 2B). P2, 5, and 8 clustered and had the largest effects on TCR recognition. P7 was also in this cluster. P3, 4, and 6 formed another cluster with smaller effects, while P-1, 1, 9 and 10 clustered for the smallest effects on TCR specificity. Based on these results, it is likely that P2, 5, 7, and 8 are the main TCR contacts for I-Ab-bound peptides. P7 may serve as both a TCR contact and an I-Ab anchor because it occupies a shallow pocket allowing its side chain to protrude for recognition by TCRs (Zhu et al., 2003). P3 may also be in this category. P4 and P6 could have affected TCR specificity indirectly by altering the positions of the upward pointing residues since it is likely that P4 and P6 are buried in the I-Ab molecule (Zhu et al., 2003) and thus unavailable for direct interaction with the TCR.
TCR contact amino acid conservation predicts p:MHCII cross-reactivity within polyclonal CD4+ T cell populations
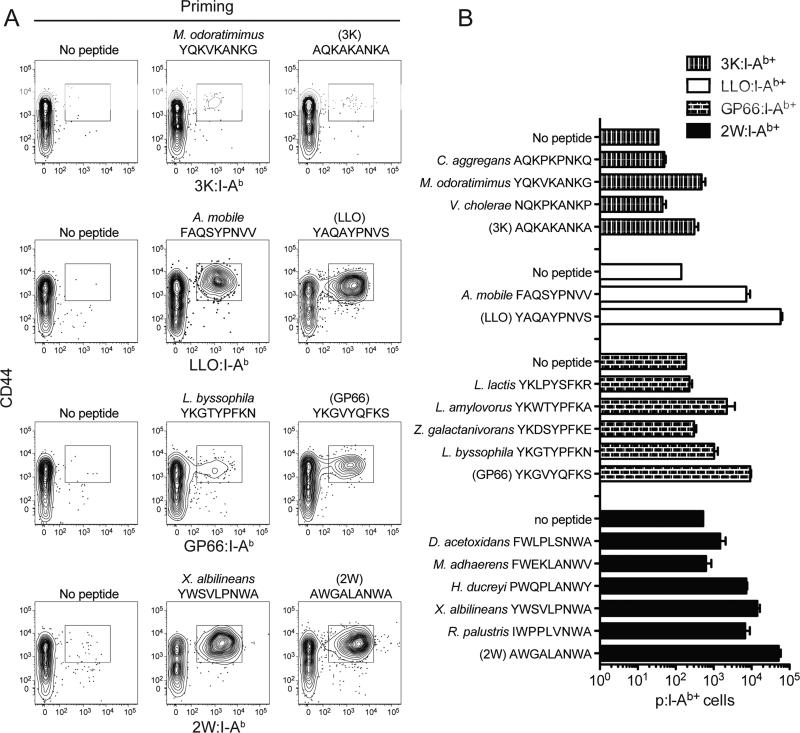
The knowledge that P2, 5, 7, and 8 are the main TCR contacts for I-Ab-bound peptides made it possible to test the limits of TCR cross-reactivity in this system. Over a 1,000 bacterial proteomes were searched for predicted I-Ab-binding nonamer peptides (Zhu et al., 2003) that matched immunogenic peptides from listeriolysin O (LLO) of Listeria monocytogenes, glycoprotein 1 of Lymphocytic choriomeningitis virus (GP66), or 3K or 2W variants of an MHCII peptide at P2, 5, 7, and 8 but at most one of the P1, 4, 6, or 9 residues. Three such peptides were selected for 3K, 1 for LLO, 4 for GP66, and 5 for 2W. B6 mice were then primed with the parent peptides or the corresponding variant peptides, and the number of responding CD4+ T cells was measured with 3K:I-Ab, LLO:I-Ab, GP66:I-Ab, or 2W:I-Ab tetramers. Mice primed 11 days earlier with 3K, LLO, GP66, or 2W peptides in CFA but not CFA alone had large populations of CD44high tetramer-binding cells (Figure 3A). One of 3 3K, 1 of 1 LLO, 2 of 4 GP66, and 4 of 5 2W variant peptides caused expansion of CD44hi CD4+ T cells capable of binding the relevant parent tetramer (Figure 3B). Thus, 8 of 13 (62%) I-Ab-binding peptides sharing 4 TCR contact amino acids but at most one I-Ab anchor residue exhibited TCR cross-reactivity.
Figure 3. Conservation of TCR contact amino acids predicts p:I-Ab cross-reactivity.
(A) Contour plots showing parent p:I-Ab tetramer staining (indicated on the Y-axes) of spleen and lymph node cells from mice primed 11 days earlier with CFA alone (no peptide), homologous bacterial peptide, or parental peptide. CD44hi tetramer-binding cells are indicated in the rectangular gates.
(B) Average number of 3K:I-Ab+, LLO:I-Ab+, GP66:I-Ab+, or 2W:I-Ab+ CD4+ T cells 11 days after priming with the indicated peptides emulsified in CFA (± SEM, n = 3 mice) from 1 experiment, which was representative of another.
Self p:MHCII cross-reactivity limits naïve population size and responsiveness to homologous foreign-p:MHCII
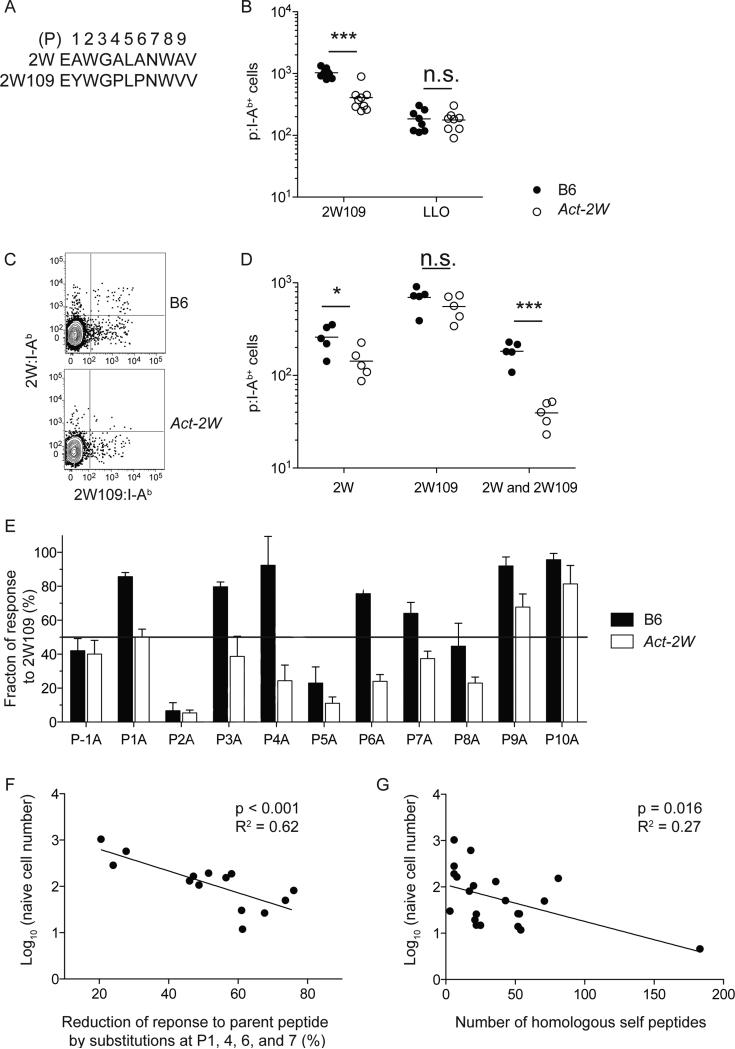
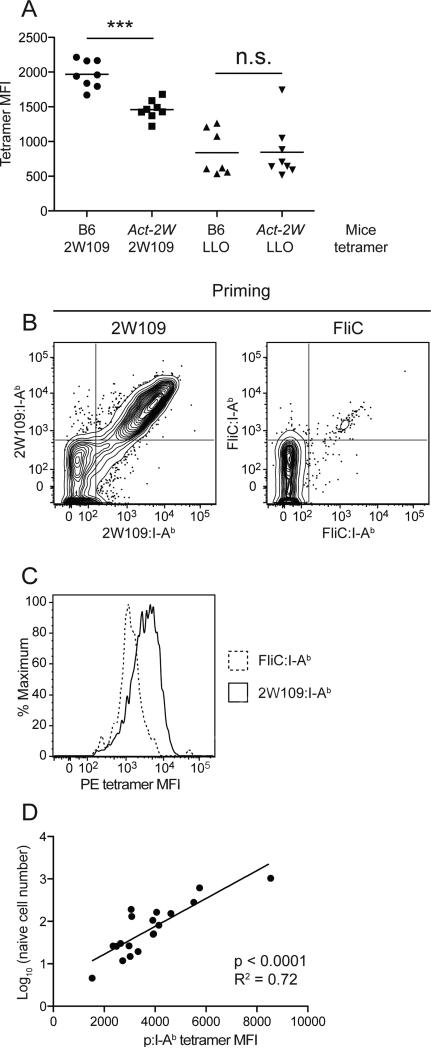
The results raised the possibility that TCR cross-reactivity between self and foreign peptides could lead to reductions in the sizes of foreign peptide-specific naïve populations because of clonal deletion. To determine if this phenomenon could occur, we examined Act-2W transgenic mice that express 2W as a self peptide under the control of the actin promoter (Moon et al., 2011) for the presence of T cells specific for a related peptide called 2W109, which has the same amino acids as 2W at P2, 3, 5, 7, and 8 but different residues at P1, 4, 6, and 9 (Table 1, Figure 4A). B6 mice had on average 1,000 2W109:I-Ab-specific naïve CD4+ T cells. In contrast, Act-2W mice contained a mean of 400 2W109:I-Ab-reactive cells (Figure 4B) suggesting that 600 2W109:I-Ab-specific cells were deleted in response to recognition of 2W:I-Ab. This difference was specific to 2W109:I-Ab since both strains had the same number of LLO:I Ab-specific naïve cells (Figure 4B).
Figure 4. Cross-reactivity with self-p:MHCII affects foreign-p:MHCII cell number.
(A) Sequence alignment of 2W and 2W109 peptides.
(B) Number of 2W109:I-Ab+ or LLO:I-Ab+ CD4+ T cells in the spleen and lymph nodes of unimmunized B6 (filled circles) or Act-2W (open circles) mice, enumerated by double tetramer staining as in Figure 1 (***p < 0.0001, n.s. = not significant). The depicted results were pooled from 3 independent experiments.
(C) Contour plots of 2W:I-Ab streptavidin-APC (Y-axis) versus 2W109:I-Ab streptavidin-PE (X-axis) tetramer staining for CD4+ T cells from spleen and lymph nodes from unimmunized B6 mice following enrichment with these tetramers.
(D) Number of CD4+ T cells in the spleen and lymph nodes of unimmunized B6 (filled circles) or Act-2W (open circles) mice that bound 2W:I-Ab tetramer only (upper left quadrants in (C)); 2W109:I-Ab tetramer only (bottom right quadrants in (C)), or both 2W:I-Ab and 2W109:I-Ab tetramers (upper right quadrants in (C)) following enrichment with these tetramers (***p < 0.0005, *p < 0.05, n.s. = not significant). The depicted results were from 1 experiment, which was representative of 2 others.
(E) Eleven amino acid versions of 2W109 peptides with single alanine substitutions at the indicated positions were tested as stimulators of 2W109-primed CD4+ T cells from B6 (filled bars) or Act-2W (open bars) mice in an ELISPOT assay. The % of the response to the 2W109 peptide was determined by dividing the number of spots stimulated by the alanine substituted peptide by the number of spots stimulated by the 2W109 peptide and multiplying by 100. The bars represent the average values (± SD, n = 3 mice) from 3 independent experiments.
(F) Linear regression analysis of the log10 naïve cell number versus the average % reduction of response to the parent peptide by alanine substitutions at I-Ab anchor residues at P1, P4, P6, and P7. For each point, the Y-axis value represents the mean from Figure 1C for an individual peptide and the X-axis value represents the mean for that peptide from Figure 2B.
(G) Linear regression analysis of the log10 naïve cell number versus the number of mouse peptides predicted to bind I-Ab and match each of the foreign peptides in Table 1 at a minimum of 4 out of 5 TCR contact positions (P2, P3, P5, P7, P8). For each point, the Y-axis value represents the mean from Figure 1C for an individual peptide. Spearman's Rho test, a rank order test that makes no assumptions about linearity, also revealed a significant correlation between these variables (p = 0.006), which was maintained even if the population with 183 self homologues was removed from the analysis (p = 0.02).
Simultaneous staining with 2W:I-Ab and 2W109:I-Ab tetramers was then performed to directly assess TCR cross-reactivity as the basis for the smaller 2W109:I-Ab-specific population in Act-2W mice. B6 mice contained 260 naïve cells that bound 2W:I-Ab alone and this number was reduced by about half to 140 cells in Act-2W mice (Figure 4C) as previously described (Moon et al., 2011). In contrast, the number of cells in B6 and Act-2W mice that were specific for 2W109:I-Ab alone, 700 and 560 respectively was not significantly different. B6 mice also contained cells that bound both 2W:I-Ab and 2W109:I-Ab tetramers, proving the existence of TCRs that were cross-reactive on these p:I-Ab complexes. B6 mice contained a mean of 180 of these cells, while Act-2W mice contained only 40. By this method, 880 2W109:I-Ab-specific cells were detected in B6 mice and 600 in Act-2W mice, suggesting the loss of 280 cells in the latter strain. This reduction was smaller than the difference of 600 2W109:I-Ab-specific observed in B6 and Act-2W mice after staining with 2W109:I-Ab tetramer alone (Figure 4B). This discrepancy could have been related to under detection of targets due to competition between the 2W:I-Ab and 2W109:I-Ab tetramers or an inability to exclude false positive events in the single tetramer binding populations. In any case, these results demonstrated that B6 mice had two populations of 2W109:I-Ab-specific T cells, one that was cross-reactive on 2W:I-Ab and one that was not. The cross-reactive population was eliminated in Act-2W mice, leaving the population that was only specific for 2W109:I-Ab.
We next determined why the 2W109:I-Ab-specific cells that remained in Act-2W mice were not deleted. One possibility was that the shared TCR contact amino acids of I-Ab-bound 2W and 2W109 adopt slightly different conformations, and the T cells that remained in Act-2W mice were specific for the 2W109 but not the 2W conformation. If so, then the 2W109:I-Ab-specific cells in Act-2W mice would be expected to be more sensitive to substitutions at I-Ab anchor positions than 2W109:I-Ab-specific cells in B6 mice. The reactivity of 2W109:I-Ab-specific CD4+ T cells in B6 mice was reduced by at least 50% when stimulated with peptides containing alanine substitutions at P-1, 2, 5, or 8 (Figure 4E, black bars). In contrast, the reactivity of the 2W109:I-Ab-specific population in Act-2W mice was reduced by at least 50% when stimulated with peptides containing substitutions at P-1, 1, 2, 3, 4, 5, 6, 7, or 8 (Figure 4E, white bars). Thus, the 2W109:I-Ab-specific population that survived negative selection in the presence of 2W:IAb appeared to be specific for TCR contact amino acids in an I-Ab anchor residue-dependent fashion.
The unusual sensitivity of 2W109:I-Ab-specific T cells in Act-2W mice to I-Ab anchor substitutions suggested that this feature might be useful for identifying other foreign peptide-specific T cell populations that experienced clonal deletion. Indeed, small foreign peptide-specific populations tended to more sensitive to substitutions at IAb anchors P1, 4, 6, and 7 than large populations (Figure 4F).
A search was then performed for self-peptide homologs that could have contributed to deletion of clones from each of the foreign p:I-Ab-specific CD4+ T cell populations. A bioinformatic screen was developed to identify mouse peptides that were predicted to bind I-Ab and share TCR contact amino acids with the foreign peptides shown in Table 1 (Figure S1). The stringency of the search was reduced to include any combination of 4 out of 5 identical amino acid residues at P2, 3, 5, 7, and 8 to capture any self peptide that could have an effect. Mouse-peptide homologs were identified for all the foreign peptides, with a range of 3 to 183 (Table S1). A significant inverse correlation was observed between naïve CD4+ T cell population size and the number of self-peptide homologs (Figure 4G), although the r2 value was only 0.27. These results support the conclusion that TCR cross-reactivity on self-peptide homologs plays some role in determining the size of MHCII-bound foreign peptide-specific CD4+ T cell populations.
Small foreign p:MHCII-specific T cell populations contain cells with low affinity TCRs
The relatively weak correlation in Figure 4G spurred a search for additional evidence that clonal deletion sculpts naïve CD4+ T cell population size. Earlier work indicated that self p:MHCII-specific T cells that survived clonal deletion bound tetramer poorly due to expression of low affinity TCRs (Moon et al., 2011; Zehn and Bevan, 2006). Based on this observation, we determined whether tetramer binding intensity could be used to identify foreign p:MHCII-specific populations that experienced clonal deletion on a related self p:MHCII ligand. Indeed, as shown in Figure 5A, 2W109:I-Ab-specific naïve T cells in Act-2W mice bound 2W109:I-Ab tetramer less well than the comparable population in B6 mice. This difference was not observed for LLO:I-Ab-specific T cells in these strains indicating that it resulted as a consequence of 2W:I-Ab complexes in Act-2W mice. Thus, clonal deletion on a self p:MHCII ligand produced a smaller and lower affinity population of T cells specific for a related foreign p:MHCII ligand.
Figure 5. Small p:MHCII-specific T cell populations are comprised of cells with low affinity TCRs.
(A) Mean fluorescence intensity (MFI) for I-Ab tetramer staining of naïve CD4+ T cells from the indicated mice (***p < 0.0001, n.s. = not significant). The depicted results were pooled from 2 independent experiments.
(B) Contour plots of CD4+ T cells from spleen and draining lymph nodes of B6 mice 14 days after priming with 10 μg of the indicated peptide emulsified in CFA enriched with the indicated p:I-Ab tetramers labeled with streptavidin-PE or streptavidin-APC.
(C) Histogram overlays of 2W109:I-Ab streptavidin-PE or FliC:I-Ab streptavidin-PE tetramer binding for cells from the upper right quadrants of (B).
(D) Linear regression analysis of the log10 naïve cell number versus the tetramer MFI for 19 different p:I-Ab-specific populations from peptide-primed B6 mice. Each point represents values for an individual peptide. For each point, the Y-axis value represents the mean from Figure 1C for an individual peptide and the X-axis value the mean for that peptide from 2 independent experiments,
Based on this result, we determined whether the small foreign p:MHCII-specific naïve populations had this fingerprint of clonal deletion. Expanded populations of cells generated by peptide priming were analyzed to obtain accurate quantification of tetramer binding intensity. 2W109:I-Ab tetramer stained the large 2W109:I-Ab-specific population more intensely than the FliC:I-Ab tetramer stained the small FliC:I-Ab-specific population (Figure 5B and 5C). Analysis of the populations specific for 19 of the I-Ab-bound foreign peptides shown in Table 1 revealed a highly significant correlation between the number of cells in a given population and their tetramer binding intensity (Figure 5D). These results indicate that small foreign p:MHCII-specific naïve populations have lost cells with the highest affinity TCRs due to deletion of clones that also recognize self p:MHCII ligands.
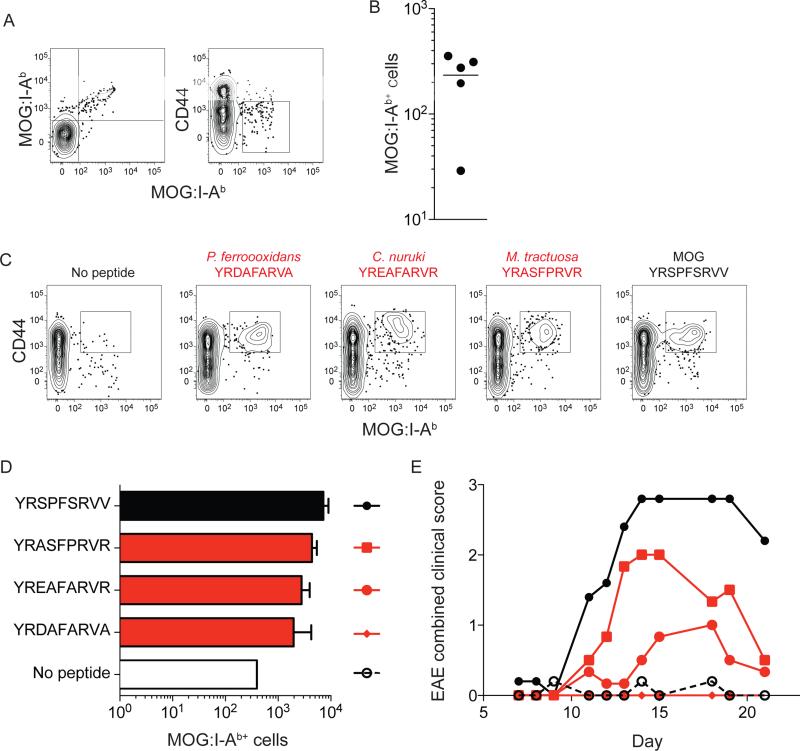
A tissue-restricted self p:MHCII ligand is recognized by a large naïve T cell population
The results indicated that negative selection on a self-peptide homolog could reduce the size of a foreign peptide-specific CD4+ T cell population. We considered the corollary case in which a self peptide-specific CD4+ T cell population that had not undergone complete deletion could be triggered to cause autoimmunity by a foreign peptide with the same TCR contact amino acids. This idea was tested with myelin oligodendrocyte glycoprotein (MOG) peptide 40-48, which causes experimental autoimmune encephalomyelitis (EAE) in B6 mice when injected with CFA and pertussis toxin (Miller et al., 2010). B6 mice that were not immunized with MOG contained about 250 MOG:IAb-specific CD44lo naïve CD4+ T cells (Figure 5A and B), which was at the high end of the range for I-Ab-bound foreign peptides (Figure 1B). Bacterial proteomes were then searched for peptides predicted to bind I-Ab and matching MOG at P1, 2, 5, 7, and 8 but differing at P3, 4, 6, and 9. Matching at P1 was maintained because alanine substitution experiments showed that P1 was critical for TCR recognition of MOG:I-Ab (data not shown). Numerous peptides fitting the criteria were identified and 3 were selected to immunize B6 mice. As shown in Figure 5C and D, all three peptides induced expansion of CD44high MOG:I-Ab tetramer-binding T cells although not as well as MOG itself. Two of the three bacterial peptides, the two that stimulated the most expansion of MOG:I-Ab-specific T cells, also caused EAE (Figure 5E). These results demonstrate that a large naïve population specific for a tissue-restricted self peptide can be activated by bacterial-peptide homologs to induce autoimmunity.
DISCUSSION
Work on multiple MHCII molecules has led to the conclusion that many TCRs cross-react on peptides with the same surface exposed amino acids but different MHCII-anchor residues (Birnbaum et al., 2014; Lucca et al., 2014; Su et al., 2013). Our results confirm this conclusion for the I-Ab MHCII molecule by showing that I-Ab-binding peptides sharing 4 putative TCR contact amino acids but at most one MHCII anchor amino acid with the parent peptide were immunogenic for T cells specific for the parent peptide. In addition, we found that this type of TCR cross-reactivity extended to negative selection as evidenced by the loss of cells with 2W:I-Ab and 2W109:I-Ab-specific TCRs in mice expressing the 2W peptide. These results suggested that many TCRs focus primarily on the TCR contact amino acids no matter how the peptide is anchored to MHCII. This conclusion is supported by structural biology studies showing that peptides that bind MHCII with different amino acids but have the same upward pointing amino acids orient those amino acids in very similar (but non-identical) ways, and are bound with the same footprint by cross-reactive TCRs (Birnbaum et al., 2014).
Some I-Ab-restricted TCRs could, however, distinguish peptides that shared upward pointing amino acids but had different I-Ab anchors as described for TCRs restricted by other MHC molecules (Kersh et al., 2001; Stewart-Jones et al., 2012). The 2W109:I-Ab-specific T cells that could not recognize the related 2W:I-Ab ligand fell in this category. It is unlikely that these TCRs directly recognized the anchor amino acids because their side chains probably do not protrude from groove of the I-Ab molecule (Zhu et al., 2003). Rather, it is likely that the different anchor amino acids in the two peptides produce two subtly different conformations of the identical TCR contact amino acids. B6 mice have T cells with TCRs that can distinguish the two conformations and others that cannot. Act-2W mice lacked the cross-reactive T cells, presumably because of clonal deletion on 2W:I-Ab complexes, but retained the ones specific for the conformation of TCR contacts that depends on the 2W109 MHCII anchors. It is therefore not surprising that changes to any of the anchor amino acids negated the reactivity of these T cells.
Several pieces of evidence indicated that naïve p:I-Ab-specific T cell populations of small size experienced extensive clonal deletion. Like the 2W109:I-Ab-specific T cells that survived clonal deletion in Act-2W mice, the cells in the small populations were sensitive to MHCII anchor substitutions and expressed lower affinity TCRs. These observations fit with work showing that T cells that develop under conditions of minimal clonal deletion have highly cross-reactive TCRs (Huseby et al., 2005) and T cells that survive clonal deletion have lower affinity TCRs (Moon et al., 2011; Zehn and Bevan, 2006). Thus, large naïve populations may be especially valuable to the host because they generate large numbers of effector cells expressing high affinity TCRs that are more tolerant to MHC-anchor variants of peptides produced by pathogens.
The inverse correlation between the number of cells in a foreign peptide-specific naïve T cell population and the number of predicted self peptides with similar TCR contacts is evidence that small populations are pared by deletion of clones with self p:MHC-specific TCRs. It is important to note, however, that this correlation was relatively weak. One reason for this could be that peptides that share TCR contact amino acids but have different MHCII anchor amino acids are often, but not always, recognized by the same TCRs. This uncertainty means that some of the mouse peptides that were predicted to cause negative selection probably do not. Another reason could be that many of the predicted mouse peptides are in parent proteins that are not expressed by thymic antigen-presenting cells or are not processed correctly to release the peptide. Additionally, other factors such as the chemistry of TCR contact amino acids likely influence naïve cell population size (Chu et al., 2010; Turner et al., 2005).
Our findings may also be relevant for certain forms for autoimmunity caused by tissue-restricted self peptides. We found that one such peptide from MOG was recognized by a large naïve T cell population. The number of MOG:I-Ab-specific T cells may be even larger than described here because some cells with low affinity TCRs are probably not detected by p:MHCII tetramer staining (Sabatino et al., 2011). The large size of the MOG:I-Ab-specific T cell population is consistent with earlier work (Ben-Nun et al., 2006) indicating that it experiences a low degree of clonal deletion. This may be the case because MOG or other mouse peptides with the same TCR contact amino acids are not abundantly displayed by I-Ab on thymic antigen-presenting cells. The MOG:I-Ab-specific T cell population may be especially prone to causing autoimmunity because its large size increases the likelihood that some if its members will be activated by a microbial mimic peptide. A similar situation may exist for humans as it has been shown that several microbial peptides stimulated myelin basic protein peptide:HLA-D specific T cell clones (Birnbaum et al., 2014; Wucherpfennig and Strominger, 1995). These studies suggest that a better understanding of TCR cross-reactivity and negative selection may make it possible to identify the infections that trigger autoimmunity.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 (B6) mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) or the National Cancer Institute Mouse Repository. Act-2W (Moon et al., 2011) mice were bred in our facilities. All mice were housed in specific pathogen-free conditions at the University of Minnesota and all experiments were conducted in accordance with institutional and federal guidelines.
p:MHCII tetramer production
pRMHa-3 vectors containing the alpha and beta chains of I-Ab under the control of the metallothionein promoter were constructed as previously described (Moon et al., 2007). Sequences encoding antigenic peptides (shown in Table 1) were fused to the N terminus of the I-Ab beta chain via a flexible polyglycine linker. Tetramers were produced in insect cells as previously described (Moon et al., 2007) with the exception that in some cases biotinylated p:I-Ab monomers were purified on a Pierce Monomeric Avidin UltraLink Resin (Thermo Scientific). Biotinylated p:I-Ab monomers were used to make tetramers with streptavidin-phycoerythrin (PE) or streptavidin-allophycocyanin (APC) (Prozyme).
Cell Enrichment and Flow Cytometry
Single cell suspensions were prepared from spleen and lymph nodes. Staining with p:IAb-streptavidin-PE and p:I-Ab-streptavidin-APC tetramers was for 1 hour at room temperature. Anti-PE and anti-APC magnetic beads were then added and bead-bound cells were enriched as previously described (Moon et al., 2007; Tubo et al., 2013). Cells contained in the bound fraction were stained on ice with antibodies specific for cell surface molecules and analyzed on an LSR II or Fortessa (Becton Dickinson) flow cytometer. Data were analyzed with FlowJo software.
Peptides
Peptides used for immunization or ELISPOT were purchased from Genscript. The peptides were 11 amino acids long with the putative nonamer core in the middle. The nonamer core sequences are shown Table 1 or the relevant figures. Peptides were emulsified in CFA and 0.05 ml of emulsion containing 10 μg of peptide was injected subcutaneously at the base of the tail.
ELISPOT
Mice were immunized by injection of 10 μg of peptide or pools of peptides emulsified in CFA, as described above. Two weeks later, CD4+ T cells were isolated from the draining inguinal and para-aortic lymph nodes and purified by positive selection with anti-CD4 magnetic beads (Miltenyi Biotec). The cells were cultured for 24 hours with B6 splenocytes and candidate peptides at a final concentration of 20 μM and analyzed for the presence of IFN-γ-producing cells by ELISPOT. The ELISPOT assay was adapted from a published protocol (Streeck et al., 2009) and a commercially available kit (Merck Millipore). Individual peptides were assayed in triplicate.
Alanine scanning approach
Single alanine substitutions were made at P-1 to P10 of test peptides. In cases where A was in the native sequence, V was substituted, with the exception of P4 or P6, where P was substituted. This was to avoid shifting the I-Ab-binding register because P is a preferred amino acid at P4 and P6 based on the I:Ab-binding matrix (Zhu et al., 2003). The peptides were assayed by ELISPOT as described above. The results were analyzed using the publicly available script “heatmap.2” available for the statistical analysis program “R.”
Identification of bacterial peptides with the same TCR contact amino acids as 2W, GP66, LLO, 3K, or MOG peptides
The set of 1,632 complete bacterial proteomes was downloaded in fasta format from Uniprot (www.uniprot.org) using the publicly available batch retrieval perl script. Each of these proteomes was then parsed into all possible ten amino acid substrings (P-1 to P9) and I-Ab-binding scores were generated based on a published matrix (Zhu et al., 2003). The top 1% of predicted I-Ab-binding peptides from each bacterial proteome were then screened against 2W, GP66, LLO, 3K, or MOG for overlap at amino acid positions 2, 5, 7, and 8 using custom c++ scripts.
Identification of mouse peptides with the same TCR contact amino acids as known immunogenic peptides
The Mus musculus proteome was downloaded from the NCBI website (ftp://ftp.ncbi.nlm.nih.gov/genomes/M_musculus/protein/protein.fa.gz). All possible ten amino acid substrings were extracted from these peptide sequences and each substring was given an I-Ab-binding score based on a published matrix (Zhu et al., 2003). The top 5% of predicted I-Ab-binding mouse peptides were then screened against the sequences of the foreign peptides shown in Table 1 using custom c++ scripts to identify mouse peptides that shared 4 of 5 amino acids at positions 2, 3, 5, 7, and 8 with the foreign peptides.
EAE Induction and clinical evaluation
Mice were immunized subcutaneously with 100 μg of MOG peptide or 11 amino acid versions of homologous bacterial peptides emulsified in CFA containing Mycobacterium tuberculosis. In addition, the mice received 200 ng of pertussis toxin (List Biological Laboratories) intravenously on the day of, and 2 days after, peptide immunization. Individual animals were graded according to clinical severity as follows: grade 0, no abnormality; grade 1, limp tail; grade 2, limp tail and hind limb weakness; grade 3, partial hind limb paralysis; grade 4, complete hind limb paralysis; as previously described (Fife et al., 2000).
Statistical analysis
Two-tailed Student's t tests, linear regressions, and Spearman's Rho test were performed with Prism (Graphpad) software.
Supplementary Material
Figure 6. TCR cross-reactivity for foreign peptides similar to MOG:I-Ab can elicit EAE.
(A) Contour plots of double tetramer staining (left) or CD44 expression (right) for CD4+ T cells from spleen and lymph nodes from individual unimmunized B6 mice following enrichment with the MOG:I-Ab tetramers labeled with streptavidin-PE (X-axis) or streptavidin-APC (Y-axis).
(B) Total number of MOG:I-Ab double tetramer-binding CD4+ T cells in individual unimmunized mice from 2 independent experiments. Horizontal bar indicates the mean.
(C) Contour plots of clonal expansion and CD44 expression by MOG:I-Ab-specific cells 14 days after subcutaneous priming with 10 μg of the indicated peptides emulsified in CFA.
(D) Average number of MOG:I-Ab+ CD4+ T cells after injection of 10 μg of the indicated peptides (± SEM, n = 3 mice/group). The depicted results were from 1 experiment, which was representative of another.
(E) EAE course after subcutaneous injection of 100 μg of the peptides indicated in (D). The depicted results show mean disease score values (n = 5) for each peptide from 1 experiment, which was representative of another.
ACKNOWLEDGEMENTS
We thank J. Walter and A. Quade for technical assistance, S. McSorley for the Aasf:IAb, RpiF:I-Ab, and PmpG1:I-Ab tetramers, and all members of the Jenkins Lab for helpful discussions. Grants from the NIH (T32 GM008244 and F30 DK093242 to R.W.N., UL1 TR000114 to D.B., R01 AI107020 to J.J.M., R01 AI080275 to K.N., R01 AI105816 and R01 AI040996 to B.S.K., R01 AI093553 to M.W., P01 AI035296 to B.T.F., and R37 AI027998, R01 AI03916, P01 AI035296, and R01 AI103760 to M.K.J.) and the William F. Milton Fund (to J.J.M.) supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicting financial interests.
REFERENCES
- Alanio C, Lemaitre F, Law HK, Hasan M, Albert ML. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood. 2010;115:3718–3725. doi: 10.1182/blood-2009-10-251124. [DOI] [PubMed] [Google Scholar]
- Ben-Nun A, Kerlero de Rosbo N, Kaushansky N, Eisenstein M, Cohen L, Kaye JF, Mendel I. Anatomy of T cell autoimmunity to myelin oligodendrocyte glycoprotein (MOG): prime role of MOG44F in selection and control of MOG-reactive T cells in H-2b mice. Eur. J. Immunol. 2006;36:478–493. doi: 10.1002/eji.200535363. [DOI] [PubMed] [Google Scholar]
- Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, Ozkan E, Davis MM, Wucherpfennig KW, Garcia KC. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion SL, Brodie TM, Fischer W, Korber BT, Rossetti A, Goonetilleke N, McMichael AJ, Sallusto F. Proteome-wide analysis of HIV-specific naive and memory CD4(+) T cells in unexposed blood donors. J. Exp. Med. 2014;211:1273–1280. doi: 10.1084/jem.20130555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HH, Moon JJ, Kruse AC, Pepper M, Jenkins MK. Negative selection and peptide chemistry determine the size of naive foreign peptide-MHC class II-specific CD4+ T cell populations. J. Immunol. 2010;185:4705–4713. doi: 10.4049/jimmunol.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Bowdon HR, Elson CO. Identification of an immunodominant T cell epitope on cholera toxin. Eur. J. Immunol. 1996;26:2587–2594. doi: 10.1002/eji.1830261108. [DOI] [PubMed] [Google Scholar]
- Fife BT, Huffnagle GB, Kuziel WA, Karpus WJ. CC chemokine receptor 2 is critical for induction of experimental autoimmune encephalomyelitis. J. Exp. Med. 2000;192:899–905. doi: 10.1084/jem.192.6.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flesch IE, Woo WP, Wang Y, Panchanathan V, Wong YC, La Gruta NL, Cukalac T, Tscharke DC. Altered CD8(+) T cell immunodominance after vaccinia virus infection and the naive repertoire in inbred and F(1) mice. J. Immunol. 2010;184:45–55. doi: 10.4049/jimmunol.0900999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geginat G, Schenk S, Skoberne M, Goebel W, Hof H. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J. Immunol. 2001;166:1877–1884. doi: 10.4049/jimmunol.166.3.1877. [DOI] [PubMed] [Google Scholar]
- Grover HS, Blanchard N, Gonzalez F, Chan S, Robey EA, Shastri N. The Toxoplasma gondii peptide AS15 elicits CD4 T cells that can control parasite burden. Infect. Immun. 2012;80:3279–3288. doi: 10.1128/IAI.00425-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyne GF, Callow MG, Kuo MC, Thomas WR. Inhibition of T-cell responses by feeding peptides containing major and cryptic epitopes: studies with the Der p I allergen. Immunology. 1994;83:190–195. [PMC free article] [PubMed] [Google Scholar]
- Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Jenkins MK, Moon JJ. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J. Immunol. 2012;188:4135–4140. doi: 10.4049/jimmunol.1102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersh GJ, Miley MJ, Nelson CA, Grakoui A, Horvath S, Donermeyer DL, Kappler J, Allen PM, Fremont DH. Structural and functional consequences of altering a peptide MHC anchor residue. J. Immunol. 2001;166:3345–3354. doi: 10.4049/jimmunol.166.5.3345. [DOI] [PubMed] [Google Scholar]
- Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J. Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok WW, Tan V, Gillette L, Littell CT, Soltis MA, LaFond RB, Yang J, James EA, DeLong JH. Frequency of epitope-specific naive CD4(+) T cells correlates with immunodominance in the human memory repertoire. J. Immunol. 2012;188:2537–2544. doi: 10.4049/jimmunol.1102190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoux F, Debeaupuis E, Echasserieau K, De La Salle H, Saulquin X, Bonneville M. Impact of TCR reactivity and HLA phenotype on naive CD8 T cell frequency in humans. J. Immunol. 2010;184:6731–6738. doi: 10.4049/jimmunol.1000295. [DOI] [PubMed] [Google Scholar]
- Li LX, McSorley SJ. B cells enhance antigen-specific CD4 T cell priming and prevent bacteria dissemination following Chlamydia muridarum genital tract infection. PLoS pathogens. 2013;9:e1003707. doi: 10.1371/journal.ppat.1003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Dai S, Crawford F, Fruge R, Marrack P, Kappler J. Alternate interactions define the binding of peptides to the MHC molecule IA(b). Proc. Natl. Acad. Sci. USA. 2002;99:8820–8825. doi: 10.1073/pnas.132272099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucca LE, Desbois S, Ramadan A, Ben-Nun A, Eisenstein M, Carrie N, Guery JC, Sette A, Nguyen P, Geiger TL, et al. Bispecificity for myelin and neuronal self-antigens is a common feature of CD4 T cells in C57BL/6 mice. J. Immunol. 2014;193:3267–3277. doi: 10.4049/jimmunol.1400523. [DOI] [PubMed] [Google Scholar]
- Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu. Rev. Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maybeno M, Redeker A, Welten SP, Peters B, Loughhead SM, Schoenberger SP, Sette A, Arens R. Polyfunctional CD4+ T cell responses to immunodominant epitopes correlate with disease activity of virulent Salmonella. PloS one. 2012;7:e43481. doi: 10.1371/journal.pone.0043481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSorley SJ, Cookson BT, Jenkins MK. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J. Immunol. 2000;164:986–993. doi: 10.4049/jimmunol.164.2.986. [DOI] [PubMed] [Google Scholar]
- Mendel I, Kerlero de Rosbo N, Ben-Nun A. A myelin oligodendrocyte glycoprotein peptide induces typical chronic experimental autoimmune encephalomyelitis in H-2b mice: fine specificity and T cell receptor V beta expression of encephalitogenic T cells. Eur. J. Immunol. 1995;25:1951–1959. doi: 10.1002/eji.1830250723. [DOI] [PubMed] [Google Scholar]
- Miller SD, Karpus WJ, Davidson TS. Coligan John E., editor. Experimental autoimmune encephalomyelitis in the mouse. Current protocols in immunology. 2010 doi: 10.1002/0471142735.im1501s88. Chapter 15, Unit 15 11. [DOI] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Hataye J, Pagan AJ, Pepper M, McLachlan JB, Zell T, Jenkins MK. Tracking epitope-specific T cells. Nat. Protoc. 2009;4:565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Dash P, Oguin TH, 3rd, McClaren JL, Chu HH, Thomas PG, Jenkins MK. Quantitative impact of thymic selection on Foxp3+ and Foxp3-subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc. Natl. Acad. Sci. USA. 2011;108:14602–14607. doi: 10.1073/pnas.1109806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DB, Rath S, Pizzo E, Rudensky AY, George A, Larson JK, Janeway CA., Jr. Monoclonal antibody detection of a major self peptide-MHC class II complex. J. Immunol. 1992;148:3483–3491. [PubMed] [Google Scholar]
- Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenius A, Bachmann MF, Ashton-Rickardt PG, Tonegawa S, Zinkernagel RM, Hengartner H. Presentation of endogenous viral proteins in association with major histocompatibility complex class II: on the role of intracellular compartmentalization, invariant chain and the TAP transporter system. Eur. J. Immunol. 1995;25:3402–3411. doi: 10.1002/eji.1830251230. [DOI] [PubMed] [Google Scholar]
- Painter CA, Stern LJ. Conformational variation in structures of classical and non-classical MHCII proteins and functional implications. Immunol. Rev. 2012;250:144–157. doi: 10.1111/imr.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees W, Bender J, Teague TK, Kedl RM, Crawford F, Marrack P, Kappler J. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley WW, Shafiani S, Wittmer ST, Tucker-Heard G, Moon JJ, Jenkins MK, Urdahl KB, Winslow GM, Woodland DL. Distinct functions of antigen-specific CD4 T cells during murine Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. USA. 2010;107:19408–19413. doi: 10.1073/pnas.1006298107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JM, Jensen PE, Evavold BD. DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323-339 epitope. J. Immunol. 2000;164:4706–4712. doi: 10.4049/jimmunol.164.9.4706. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Sabatino J, Huang JJ, J., Zhu C, Evavold BD. High prevalence of low affinity peptide–MHC II tetramer–negative effectors during polyclonal CD4+ T cell responses J. Exp. Med. 2011;208:81–90. doi: 10.1084/jem.20101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant AJ, Chaves FA, Leddon SA, Tung J. The control of the specificity of CD4 T cell responses: thresholds, breakpoints, and ceilings. Front. Immunol. 2013;4:340. doi: 10.3389/fimmu.2013.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J, Neumann-Haefelin C, Altay T, Gostick E, Price DA, Lohmann V, Blum HE, Thimme R. Immunodominance of HLA-A2-restricted hepatitis C virus-specific CD8+ T cell responses is linked to naive-precursor frequency. J. Virol. 2011;85:5232–5236. doi: 10.1128/JVI.00093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc. Natl. Acad. Sci. USA. 2010;107:10978–10983. doi: 10.1073/pnas.1006545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Mohrs M, Mallet-Designe V, Teyton L, Locksley RM. Rapid expansion and IL-4 expression by Leishmania-specific naive helper T cells in vivo. Immunity. 2002;17:191–200. doi: 10.1016/s1074-7613(02)00363-1. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones GB, Simpson P, van der Merwe PA, Easterbrook P, McMichael AJ, Rowland-Jones SL, Jones EY, Gillespie GM. Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms. Proc. Natl. Acad. Sci. USA. 2012;109:3483–3492. doi: 10.1073/pnas.1207896109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck H, Frahm N, Walker BD. The role of IFN-gamma Elispot assay in HIV vaccine research. Nat. Protoc. 2009;4:461–469. doi: 10.1038/nprot.2009.7. [DOI] [PubMed] [Google Scholar]
- Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AC, La Gruta NL, Zeng W, Jackson DC. Precursor frequency and competition dictate the HLA-A2-restricted CD8+ T cell responses to influenza A infection and vaccination in HLA-A2.1 transgenic mice. J. Immunol. 2011;187:1895–1902. doi: 10.4049/jimmunol.1100664. [DOI] [PubMed] [Google Scholar]
- Tubo NJ, Pagan AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, Jenkins MK. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 2013;153:785–796. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SJ, Kedzierska K, Komodromou H, La Gruta NL, Dunstone MA, Webb AI, Webby R, Walden H, Xie W, McCluskey J, et al. Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations. Nat. Immunol. 2005;6:382–389. doi: 10.1038/ni1175. [DOI] [PubMed] [Google Scholar]
- Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehn D, Bevan MJ. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity. 2006;25:261–270. doi: 10.1016/j.immuni.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Rudensky AY, Corper AL, Teyton L, Wilson IA. Crystal structure of MHC class II I-Ab in complex with a human CLIP peptide: prediction of an I Ab peptide-binding motif. J. Mol. Biol. 2003;326:1157–1174. doi: 10.1016/s0022-2836(02)01437-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.