Abstract
Ultraviolet (UV) radiation from sunlight is a major etiologic factor for skin cancer, the most prevalent cancer in the U.S., as well as premature skin aging. In particular, UVB radiation causes formation of specific DNA damage photoproducts between pyrimidine bases. These DNA damage photoproducts are repaired by a process called nucleotide excision repair, also known as UV-induced DNA repair. When left unrepaired, UVB-induced DNA damage leads to accumulation of mutations, predisposing people to carcinogenesis as well as to premature aging. Genetic loss of nucleotide excision repair leads to severe disorders, namely, xeroderma pigmentosum (XP), trichothiodystrophy (TTD) and Cockayne syndrome (CS), which are associated with predisposition to skin carcinogenesis at a young age as well as developmental and neurological conditions. Regulation of nucleotide excision repair is an attractive avenue to preventing or reversing these detrimental consequences of impaired nucleotide excision repair. Here we review recent studies on molecular mechanisms regulating nucleotide excision repair by extracellular cues and intracellular signaling pathways, with a special focus on the molecular regulation of individual repair factors.
INTRODUCTION
Ultraviolet (UV) radiation from sunlight is a major etiologic factor for skin cancer, the most prevalent cancer in the U.S. (1–6), as well as premature skin aging. UV radiation is classified into 3 types based on the wavelength- UVA (315–400 nm), UVB (280–315 nm) and UVC (100–280 nm) (7, 1). All UVC is blocked by the ozone layer, preventing it from reaching the surface of the earth (1). UVB forms only about 5% of all UV radiation reaching the earth’s surface, which effectively causes DNA damage (8, 9, 2). UVA forms about 95% of all UV radiation entering the earth, but is weaker than UVB in terms of causing DNA damage (8, 10, 2, 11).
UVB and UVC are absorbed directly by DNA, causing the formation of thymine dimers, mainly cyclobutane pyrimidine dimers (CPD) and pyrimidine (6-4) pyrimidone photoproducts (6-4PP) (2, 5). UVA exposure also causes thymine dimers; in addition, it leads to generation of reactive oxygen species (ROS) via photosensitizing reactions, and thus indirectly causes oxidative DNA damage lesions (2, 12, 11).
In humans and mice UV-induced CPD and 6-4PP lesions are repaired by nucleotide excision repair (NER), the most versatile DNA repair system. NER eliminates a wide variety of helix-distorting base lesions induced by environmental carcinogenic sources, including UV and air pollutants (13–20). Even though a primitive, more efficient DNA repair mechanism involving photolyases has been identified, it is absent in humans (20, 21). When NER is defective and the damage is left unrepaired, it leads to various disorders including xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothiodystrophy (TTD) (Table 1) (17, 16, 22). These disorders are characterized by increased carcinogenesis in various organs, developmental and immunological defects, neuronal and retinal degeneration, and aging (Table 1) (17, 16, 22). Defective NER predisposes affected individuals to carcinogenesis in the skin, brain, and lungs, and sensitizes mice to carcinogenesis in the skin, lungs, and liver (23, 24, 17, 25–27). Even though the versatile NER pathway can correct bulky nucleotide adducts distorting the DNA structure from a variety of environmental carcinogens, it is crucial for correction of UV-induced DNA photoproducts in the skin, since NER defective patients have high propensity to develop sunlight exposure induced skin cancer (27). Patients with defective NER manifest a 2,000–10,000 fold increase in risk of skin cancer, have a significantly lower age of onset of skin cancer compared to the general population, and have skin cancer as the most common cause of death as compared to other internal cancers (27). This establishes the most significant association of NER defects with UV-associated skin cancer amongst all cancers. Essential NER factors have been identified, including xeroderma pigmentosum complementation group A–G (XPA-XPG) and cockayne syndrome group A (CSA) and B (CSB) (17, 16, 22).
Table 1.
Disorders associated with defective NER (See Ref (17)).
| Disease due to defective NER | Genes causing disorder | Characteristics |
|---|---|---|
| Xeroderma pigmentosum(XP) | XPA, XPB, XPC, XPD, XPE, XPF, XPG, XPV (Xeroderma Pigmentosum Variant) | Sunlight exposure predisposes to various cancers, especially squamous cell carcinoma, basal cell carcinoma and melanoma skin cancer. |
| Trichothiodystrophy (TTD) | XPB, XPD, TTDN1 (TTD non- photosensitive 1 protein or M- phase-specific PLK1- interacting protein) and TTDA (general transcription factor IIH, polypeptide 5 (GTF2H5)) | Brittle, sulfur-deficient hair and ichthyosis, mental retardation |
| Cockayne syndrome (CS) | CSA, CSB, XPD and XPG | Developmental and neurological disorders, decreased lifespan |
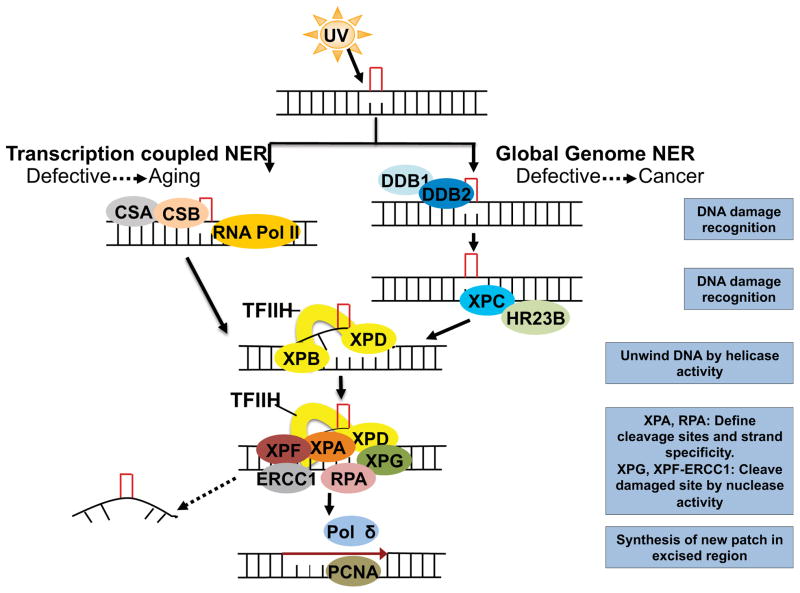
There are two main types of NER: global genome nucleotide excision repair (GG-NER) and transcription coupled nucleotide excision repair (TC-NER) (16, 17). GG-NER is mainly responsible for removing most of the CPD and 6-4PP damage in non-transcribed regions, whereas TC-NER does the same in regions under active transcription in the genome (16, 17). These two pathways differ in their damage recognition, but the following steps are the same in both pathways (Figure 1). In GG-NER, XPE (also known as DNA damage binding protein 2, or DDB2) and XPC first bind to the damage site and are responsible for UV-induced DNA damage recognition, in the heterodimeric complex with DDB1 (DNA damage binding protein 1) and HR23B, respectively (16, 17). For TC-NER, CSA and CSB proteins mediate recognition of UV-induced DNA damage in actively transcribed regions by relieving the stalled RNA polymerase II (RNA pol II) at these sites (16, 17).
Figure 1.
Sequential assembly of molecular players to remove UV-induced CPD and 6-4PP DNA damage lesions in global genome nucleotide excision repair (GG-NER) and transcription coupled nucleotide excision repair (TC-NER).
Following recognition, the rest of the NER pathway is the same for both the GG-NER and the TC-NER pathways (16, 17) (Figure 1). Upon recognition of the DNA damage, XPB and XPD, which form part of the transcription factor II H (TFIIH) complex, unwind the DNA through their helicase activity (16, 17) (Figure 1). XPA and RPA (replication protein) define the cleavage sites and strand specificity, to which XPG (also known as excision repair cross-complementation group 5, ERCC5) and the nuclease complex XPF-ERCC1 (excision repair cross-complementation group 1) bind to cleave the damaged site, followed by its excision (16, 17). The excised portion is replaced with a newly synthesized patch with the help of proliferating-cell nuclear antigen (PCNA) and replicative polymerase (Pol) δ (16, 17) (Figure 1).
Unrepaired DNA damage leads to replication fork breakdown and subsequent genomic instability during cell division, since regular high fidelity DNA polymerases cannot synthesize past DNA lesions (28–31). Translesion synthesis (TLS) polymerases can bypass the DNA damage and allow DNA replication to continue (28, 32–38). Even though TLS polymerases were originally considered low fidelity polymerases contributing to mutagenesis, recent advances suggest that TLS could be error-prone or error-free in a damage-specific and polymerase-specific manner (31, 28, 39–43).
Translesion synthesis across unrepaired CPD lesions is mediated by Pol η (initiation and extension) in an error-free manner (28, 43, 41, 44–46). When Pol η is absent, for example in XP-V patients, a two-step process involving initiation (TLS Pol ι,κ and/or an unknown polymerase) and extension (TLS Pol ζ or κ) mediates translesion synthesis across the CPD lesion in an error-prone manner (28, 47, 48). TLS across 6-4PP lesions is carried out by Pol ζ in an error-free manner, and alternatively by Pol η or Pol ι in an error-prone manner (49, 42). Since TLS is potentially mutagenic, regulating the comparatively error-free NER would be more desirable (50, 31).
Hence it is important to understand how NER is regulated, which could be exploited to prevent or ameliorate pathologies associated with defective NER. In this review, we summarize recent advances regarding the regulators of individual factors involved in NER, emphasizing the molecular regulation of NER through controlling the expression or activity of NER proteins.
XPC REGULATION
XPC is an indispensable factor for initial recognition of bulky DNA damage in non-transcribed regions (17, 16, 22). Loss of XPC function inhibits UV-induced CPD and 6-4PP DNA lesion repair, leading to accumulation of mutations upon replication, and increased cancer risk with UV exposure (17, 16, 22). Being such an important protein, XPC is regulated at multiple levels: genetic (mutations and polymorphisms), transcriptional, and post-translational, in addition to regulation under specific conditions like immunosuppression.
XPC polymorphisms
In spite of having normal NER, the Lys939Gln polymorphism in XPC has been associated with various cancers, indicating that NER-independent function of XPC is also important for cancer (51, 52). The XPC intron 11-5C/A SNP causes a reduction in DNA repair capacity due to a change in the frequency of alternatively spliced XPC mRNA.(53)
Promoter methylation of XPC
Recent studies have shown that XPC promoter methylation is increased by BRAFV600E (V600E mutant V-Raf Murine Sarcoma Viral Oncogene Homolog B), leading to decreased XPC mRNA levels and reduced DNA repair capacity (54) (Figure 2). This may play an important role in promoting spontaneous as well as UVB-induced melanomagenesis (54). XPC promoter hypermethylation is also associated with reduced XPC expression in lung tumors from patients (55). In lung cancer cell lines, the XPC promoter region, which consists of 17 CpG islands, was shown to be associated with XPC hypermethylation, leading to XPC repression (55).
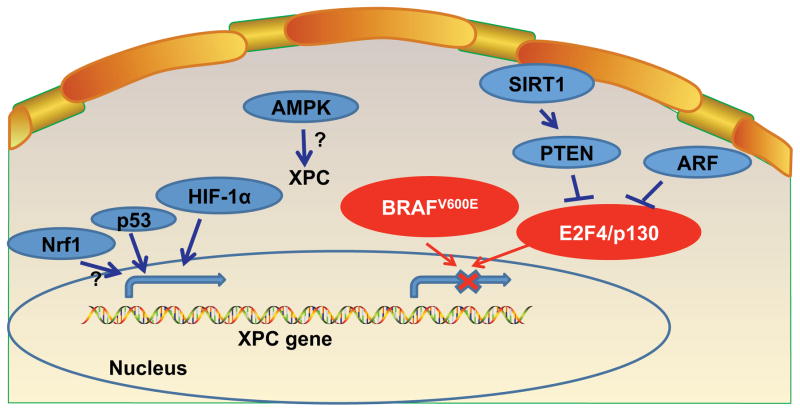
Figure 2.
Transcriptional Regulation of XPC.
Transcriptional regulation of XPC
Various distinct transcription factors, namely p53, nuclear factor erythroid 2-related factor 1 (Nrf1, also called NFE2L1), Hypoxia-inducible factor-1 alpha (HIF-1α), AMP-activated protein kinase (AMPK), E2F transcription factor 4 (E2F4), and 130 kDa retinoblastoma-associated protein (p130), have been found to regulate XPC expression, and non-transcription factors also have been found to act via some of these transcription factors to modulate XPC expression (Figure 2).
The p53 tumor suppressor signaling is well known to regulate XPC to enhance NER (56). In HaCaT keratinocytes, Nrf1 promotes CPD repair by increasing XPC expression (57). Nrf1 increases XPC expression via increasing glutathione availability (57). HIF-1α also contributes to increased XPC transcription after UVB (58). In mouse skin and normal human epidermal keratinocytes, the AMPK pathway promotes UVB-induced DNA repair by increasing XPC expression (59). Under growth arrest conditions, E2F4 and p130 repressors were found to bind the XPC promoter region in a genome-wide binding screen (60). In MEF cells, ARF was shown to reduce the binding of the E2F4-p130 repressor complex to XPC promoter, thus leading to increased expression of XPC (61, 62). ARF was shown to be required for efficient NER of UVC-induced CPD and 6-4PP lesions, due to its function of regulating XPC expression (62).
Loss of phosphatase and tensin homolog (PTEN), an important tumor suppressor, was shown to inhibit CPD repair and to a lesser extent 6-4PP repair in vitro (in HaCaT cells) and in vivo (in mouse epidermis) via decreasing XPC protein levels, by regulating XPC transcription (63). In in vitro cell culture models (MEFs and human keratinocytes), sirtuin 1 (SIRT1) inhibition impairs CPD and 6-4PP repair by increasing XPC transcription (64). SIRT1 increases XPC transcription by activating PTEN through its deacetylase activity, which inhibits phosphorylation of AKT and impairs the nuclear localization of p130 transcriptional repressor (64).
XPC regulation by post-translational modifications and protein-protein interactions
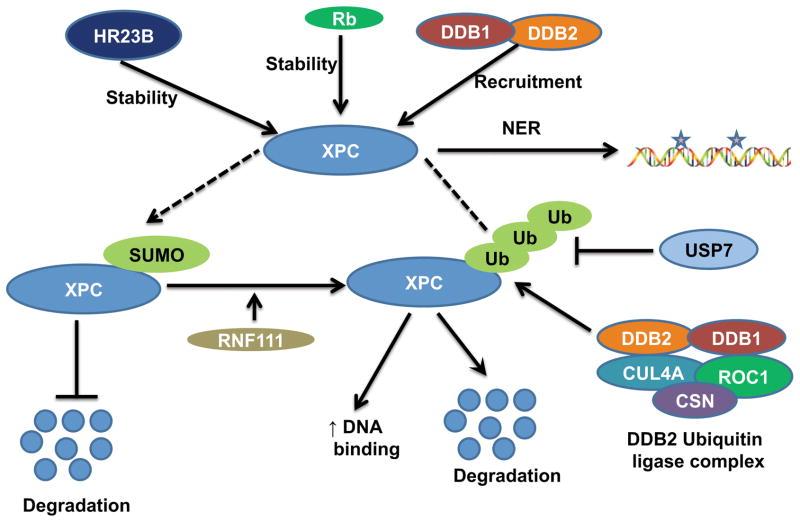
XPC can be regulated by two types of post-translational modifications: ubiquitylation and sumoylation (65). XPC can be polyubiqutinated by the UV-DDB E3 ligase complex consisting of DDB2, DDB1 (DNA damage binding protein 1), Cul4A, and several other proteins (66–69) (Figure 3). Ubiquitination of XPC increases XPC binding to DNA (68). Other studies also report that the UV-DDB complex mediates targeting of the XPC-HR23B complex to the site of CPD DNA damage (70, 71). Additionally, it was shown that DDB2 is necessary for degradation of XPC after UV-induced DNA photoproduct formation, but DDB1 and Cul4A, which are members of the same UV-DDB complex, inhibit XPC degradation upon UVC-induced DNA damage (72). After UV-induced DNA photoproduct formation, degradation of XPC was shown to be necessary for recruiting XPG at DNA damage sites and thus for efficient NER (72). XPC deubiquitination by ubiquitin-specific protease 7 (USP7) prevents XPC degradation and promotes NER (73). Sumoylation of XPC after UVC-induced DNA damage has also been shown to prevent its degradation (65). The K655 site on XPC is critical for sumoylation, as well as for degradation of XPC (72). RING finger protein 111 (RNF111) can polyubiquitinate sumoylated XPC to promote XPC’s binding to damaged DNA (65, 74).
Figure 3.
Post-translational regulation of XPC.
In addition, XPC interacts with other proteins to regulate its protein stability. For example, XPC-binding protein Rad23 (yeast homolog of HR23B) was shown to stabilize XPC (75, 76) (Figure 3). Inhibition of the proteasome pathway or overexpression of Rad23 increases the stability of Rad4 (yeast homolog of XPC) (77). The authors further show that Rad23 significantly impacts NER capacity via two independent but simultaneous mechanisms (77, 78). With p53-null cell lines, retinoblastoma protein RB was shown to increase the half-life/stability of XPC protein to enhance NER, through a direct interaction with XPC (79).
XPC regulation by immunosuppressants in organ transplant recipients
Organ transplant patients are at high risk of developing skin cancer (80). These skin cancers have long been attributed to the immunosuppressive therapy post-transplant, and the level of immunosuppression affects the development of skin cancer (80, 81). However, cyclosporin A (CsA), an immunosuppressant used for organ transplant recipients, promotes UVB-induced skin carcinogenesis in an immunosuppression-independent manner by (i) impairing DNA repair by suppressing XPC transcription, and (ii) impairing checkpoint and DNA damage response by upregulating CypA (82). Other reports have also shown that CsA inhibits NER in fibroblasts and lymphoblasts (83, 84). In contrast to keratinocytes, in fibroblasts CsA inhibits NER by reducing XPA and XPG but not XPC (85). In keratinocytes, the immunosuppressants tacrolimus and mycophenolate mofetil reduce UVB-caused DNA damage repair and apoptosis, and tacrolimus also impairs UVB-mediated checkpoint signaling, and thus may promote skin cancer in both an immunosuppression-dependent and -independent manner (86).
REGULATION OF XPE/DDB2
As an essential factor in DNA damage recognition, DDB2 is regulated at both transcriptional and post-translational levels. In response to UVC, DDB2 transcription is regulated by p53 in human cells, but not in mice (87, 88). In addition, DDB2 activity is modulated by multiple pathways. UV irradiation was shown to cause constitutive photomorphogenesis 9 (COP9) signalosome (CSN) dissociation from the DDB2 complex (89, 69). This in turn increased the ubiquitin-ligase activity of DDB2 (90, 69). Upon being polyubiquitinated by the DDB complex itself, DDB2 signals for its degradation by proteasomes and loses its DNA binding activity (68). Poly (ADP)-ribosylation (PARylation) of DDB2 was shown to inhibit DDB2 ubiquitination and degradation, to allow DDB2 more time to mediate chromatin modification (91, 92). In addition to DDB2 complex-mediated chromatin regulation, DDB2 itself can regulate NER through chromatin remodeling by (i) Poly (ADP-ribose) polymerase 1 (PARP-1) mediated PARylation of chromatin, and (ii) recruiting chromatin remodeler Amplified in Liver Cancer 1 (ALC1) (91, 93–95).
REGULATION of CSA and CSB
As an ubiquitin-ligase in TC-NER, CSA activity is decreased by UV irradiation via rapid association of COP9 signalosome (CSN) with CSA (69). CSA was shown to ubiquitinate CSB to target it for degradation via the ubiquitin proteasome pathway, ultimately aiding the reinitiating of transcription after DNA repair (96). UVSSA (UV-sensitive syndrome protein) was shown to recruit USP7 to deubiquitinate and stabilize CSB, thus opposing CSA-mediated ubiquitination and degradation of CSB (97, 98).
REGULATION OF RNA pol II
Following UV-induced DNA photoproduct formation at actively transcribed genes, stalled RNA pol II recruits CSA and CSB (99–102). RNA pol II is then polyubiquitylated by CSA- and CSB-complex, which leads to the degradation of RNA pol II in the proteasomes to allow the recruitment of downstream NER factors (99–102). Alternatively, RNA pol II degradation factor 1 (Def1) can mediate degradation of RNA pol II via ubiquitination independent of TC-NER (103). In addition, the VHS (Vps-27, Hrs and STAM) domain of UVSSA was found to be essential for ubiquitination and dephosphorylation of RNA pol II and for efficient TC-NER, and this ubiquitination of RNA pol II does not target RNA pol II for degradation (104). The authors also suggest that since UVSSA interacts with TFIIH, UVSSA probably ubiquitinates RNA pol II by recruiting TFIIH, helping to remove stalled RNA pol II from the damage sites to allow the TC-NER factors access to the DNA damage (104).
XPD REGULATION
XPD functions at the merging point of the GG-NER and TC-NER pathways. XPD is a part of the TFIIH complex, and along with XPB serves to unwind the recognized DNA damage via its helicase activity. In keratinocytes, immediately after UVB irradiation, XPD normally undergoes a small decrease, followed by its upregulation (58).
XPA REGULATION
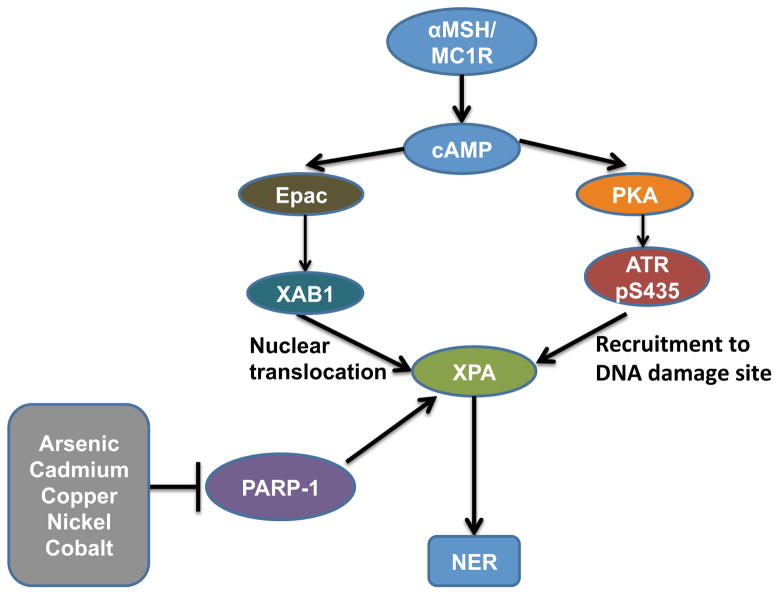
XPA level and activity are regulated through various mechanisms under physiological and stress conditions. XPA level is increased by deficiency in toll-like receptor 4 (TLR4) (105). In keratinocytes, αMSH/MC1R complex (α-Melanocyte-Stimulating Hormone/Melanocortin 1 Receptor complex) enhances the GTPase activity of XPA-binding protein 1 (XAB1), which in turn induces nuclear translocation of XPA, thus regulating NER for UVB-induced DNA damage via a pigmentation-independent mechanism (106) (Figure 4). PKA-mediated ATR phosphorylation enhances ATR-XPA interaction and rapid recruitment of XPA to DNA damage sites, ultimately promoting NER and reducing UVB or UVC-caused mutations (107). In addition, PARP-1 directly interacts with XPA to enhance NER (108). Arsenic is known to inhibit NER, and zinc was shown to protect against the detrimental effects of arsenic on DNA damage (109–112). In cell-based systems, XPA and PARP-1 (both zinc finger proteins) were shown to be molecular targets for arsenite (113–116). Cadmium, copper, nickel and cobalt also inhibited XPA by decreasing binding of XPA to UVC-damaged DNA (117). The mechanism of NER impairment by metals needs to be further investigated.
Figure 4.
Molecular Regulation of XPA.
REGULATION OF RPA and PCNA
RPA binds to the unwound DNA damage site and to XPA. XPA and RPA together define the cleavage sites and strand specificity for the downstream nucleases. After excision of the damaged DNA by the nucleases, PCNA promotes filling of the gap by DNA synthesis. Transcription of RPA and PCNA may be regulated by E2F1 (E2F Transcription Factor 1) and E2F4, which were found to bind to the promoter region of RPA3 and PCNA genes (118). E2F4 and p130 repressors, under growth arrest conditions, were also found to bind to the PCNA promoter region in a genome wide binding screen (60).
REGULATION OF XPG/ERCC5
XPG/ERCC5 participates in cleavage and excision of the damaged DNA lesion via its endonuclease activity. XPG level and activity are regulated by transcription and protein-protein interaction, respectively. In H23 or H460 human lung adenocarcinoma cell lines, CCAAT/enhancer-binding protein gamma (CEBPG) was shown to increase ERCC5 transcription (119) (Figure 5). Human interferon β (IFN-β) treatment was shown to increase XPG mRNA levels in fibroblasts isolated from CSA and CSB patients with defects in TC-NER (120). IFN-β-mediated upregulation of XPG could be a possible mechanism for IFN-β-mediated resistance to UVC-induced cell death, in a TC-NER-independent manner (121). Other mechanisms may also have a role in the effect of IFN-β and remain to be determined.
Figure 5.
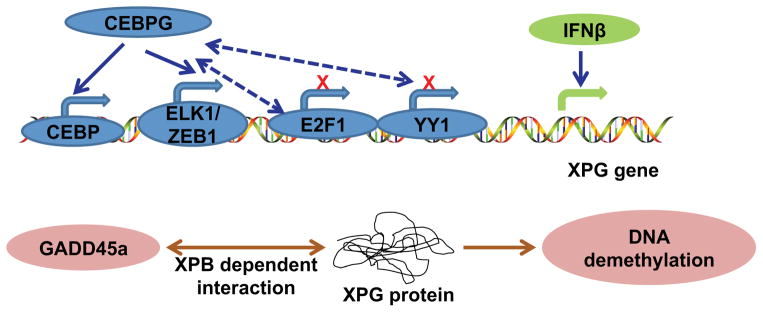
Molecular regulation and interactions of XPG.
Gadd45 (Growth Arrest And DNA-Damage-Inducible) has been shown to improve NER (122, 123). Gadd45a directly interacts with XPG to cause active DNA demethylation (124). Gadd45a-mediated DNA demethylation probably stimulates DNA repair via XPG and XPB, due to the association between DNA repair and DNA demethylation (124, 125). The precise mechanism remains to be elucidated.
ERCC1 REGULATION
XPF-ERCC1 dimer participates in cleavage and excision of the damaged DNA lesion via its endonuclease activity. ERCC1 mutations contributing to NER disorders have rarely been found. A patient with a homozygous Exon 7 mutation in ERCC1 is reported to have CS symptoms, and a patient with ERCC1 deficiency had developmental failure and a mild defect in NER (126, 127). Ercc1 knockout mice have similar critical developmental disorders and neonatal lethal characteristics (128). Deletion of Smad4 (SMAD family member 4) decreases ERCC1 transcription to cause defective CPD repair via reduced Snail expression, leading to increased UVA and UVB-induced SCC in murine models with keratinocyte-specific Smad4 loss (129).
OTHER NER REGULATORY PATHWAYS
Melanin, MSH, and Melanocortin
In melanocytes with functional MC1R, total melanin and eumelanin contents (MC and EC) were found to be inversely proportional to CPD damage (130). Additionally, melanocytes with loss of MC1R function have higher UVB-induced CPD damage and lesser repair of these lesions, independent of their total melanin and eumelanin content (130).
MSH and adrenocorticotropic hormone (ACTH) have been shown to activate MC1R, leading to increased repair capacity of UVB-induced DNA photoproducts and decreased ROS generation (131–134). Forskolin was also shown to have effects similar to those of αMSH on UV-induced repair of DNA photoproducts, due to forskolin-mediated activation of the cAMP pathway, a downstream pathway common to melanocortins (131, 135, 136). Tetrapeptide and tripeptide analogs of melanocortin, containing a modified αMSH core with N capped groups and C terminal modifications respectively, also enhance CPD repair, by activating MC1R as MSH does (131, 137, 138).
Chromatin modification
Chromatin modification could be an important regulator of NER, since chromatin in an open conformation facilitates the binding of DNA repair factors to the damaged DNA (67, 139, 140, 99). In cell extracts and reconstituted human excision nuclease systems, with reconstituted nucleosomes, the NER rate was inhibited to only 10% of that in naked DNA (141). The repair kinetics for acetylaminofluorene-guanine (AAF-G) adduct is increased by the SWI/SNF complexes (SWItch/sucrose nonfermentable) through multiple mechanisms (142–152). In addition, by increasing the access of NER factors to nucleosomal DNA, NER can be increased by ACF (Asymmetric Crying Facies), an ATP-utilizing chromatin assembly and remodeling factor (153–155).
The DDB2 complex also facilitates NER by carrying out ubiquitination of H2A, H3, and H4 histone proteins, indicating DDB2’s role as a chromatin remodeler to allow the NER factors access to damaged DNA lesions (93–95). Histone acetylation mediates chromatin unfolding even after lesion detection, which is important for efficient NER. Indeed, histone acetyltransferases, such as GCN5 (general control of amino-acid synthesis 5), have been shown to be involved in this process (156–159). p53 was also shown to mediate whole genome relaxation to facilitate lesion detection and NER (159). The UV-DDB complex is also able to associate with GCN5 and p300, suggesting another probable mechanism of DDB-mediated chromatin regulation to facilitate NER (160–162).
E2F1 has been shown to facilitate NER by recruiting GCN5 at sites of UVC- or UVB-induced DNA photoproducts (158). GCN5 mediates histone H3 Lysine 9 (H3K9) acetylation to allow increased access of NER factors to the damaged DNA (158). The S29A mutation (S29 in mice, equivalent to human S31) in E2F1 hinders E2F1 recruitment and E2F1-mediated recruitment of GCN5 and H3K9 acetylators at damage sites, reducing access of the NER factors XPC and XPA to the DNA damage lesions (163). After removal of DNA damage, restarting of transcription is enhanced by rapid removal and exchange of H2A and H2B at UVC-induced DNA photoproducts, and by placement of H3.3 histone (164–166). In yeast, the loss of H2A histone variant HTZ1 (H2A.Z) inhibits UVC-induced CPD damage removal (167). HTZ1 promotes CPD repair by recruiting GCN5, leading to histone H3K9/K14 acetylation and increasing Rad14 (ortholog of XPA) binding to damaged DNA (167). Through their histone acetylase activity, p300 and CREB Binding Protein (CBP) redundantly lead to DDB2 recruitment to CPD lesions in compacted chromatin regions, facilitating repair of UVC-induced CPD lesions (168). p300 phosphorylation at S1834 is seminal for efficient CPD repair through facilitating DDB2 recruitment to the CPD lesion (168).
CONCLUSION AND PERSPECTIVES
UV-induced DNA repair, or NER, essentially removes DNA damage by inevitable environmental factors like solar UVB radiation and air pollutants. NER is vital to maintaining genomic integrity to protect animals and humans from skin, lung and brain cancer as well as neurological and developmental disorders, and thus justifiably has multiple factors and signaling mechanisms for its regulation. Recent studies have demonstrated that UV-induced DNA repair is regulated at multiple levels including transcriptional modulation and posttranslational modifications. Both extracellular cues and intracellular signaling regulate UV-induced DNA repair capacity. These regulations are achieved through regulating the availability or activity of individual repair factors, or modifying chromatin structure. Better understanding of the NER regulation can elucidate new opportunities to enhance the NER capacity and therefore improve our ability to prevent cancer initiation and progression, as both processes involve genetic mutations and/or genomic instability.
Acknowledgments
We apologize to those investigators whose work could not be directly referenced owing to space limitations. Work in the authors’ laboratory was supported by NIH/NIEHS grants ES016936 (YYH), the American Cancer Society (ACS) grant RSG-13-078-01 (YYH), the University of Chicago Cancer Research Center (P30 CA014599), the CTSA (NIH UL1 TR000430), and the University of Chicago Friends of Dermatology Endowment Fund.
ABBREVIATIONS
- 6-4PP
pyrimidine (6-4) pyrimidone photoproducts
- αMSH/MC1R complex
α-Melanocyte-Stimulating Hormone/Melanocortin 1 Receptor complex
- AAF-G
acetylaminofluorene-guanine
- ACF
Asymmetric Crying Facies
- ACTH
Adrenocorticotropic Hormone
- AICAR
5-aminoimidazole-4-carboxamide ribotide
- Akt
V-Akt Murine Thymoma Viral Oncogene Homolog
- ALC1
Amplified in Liver Cancer 1
- AMPK
AMP-activated protein kinase
- ARF
Alternative reading frame
- ATM
Ataxia Telangiectasia Mutated
- ATR
Ataxia Telangiectasia and Rad3 Related
- BCC
basal cell carcinoma
- BRAFV600E
V600E mutant V-Raf Murine Sarcoma Viral Oncogene Homolog B
- Camp
cyclic AMP
- CBP
CREB Binding Protein
- CEBPG
CCAAT/enhancer-binding protein gamma
- CPD
cyclobutane pyrimidine dimmers
- CS
Cockayne syndrome
- CsA
Cyclosporin A
- CSA
Cockayne syndrome group A
- CSB
Cockayne Syndrome group B
- CSN
constitutive photomorphogenesis 9 (COP9) signalosome
- Cul4A
Cullin 4A
- DDB1
Damage-Specific DNA Binding Protein 1
- DDB2
Damage-Specific DNA Binding Protein 2
- Def1
RNA polymerase II degradation factor 1
- DOT1L
DOT1-like protein
- DP1
DRTF polypeptide 1
- E2F1
E2F Transcription Factor 1
- E2F4
E2F transcription factor 4
- Epac
Exchange protein activated by cyclic AMP
- ERCC1
excision repair cross-complementation group 1
- ERCC5
excision repair cross-complementation group 5
- Gadd45
Growth Arrest And DNA-Damage-Inducible
- GCN5
general control of amino-acid synthesis 5
- GG-NER
global genome nucleotide excision repair
- H2A.Z
H2A histone variant HTZ1
- H3K9
Histone H3 Lysine 9
- HIF-1α
Hypoxia-inducible factor-1 alpha
- HIRA
HIR (Histone Cell Cycle Regulation Defective) Homolog A
- HMGN1
High Mobility Group Nucleosome Binding Domain 1
- HR23B
RAD23 Homolog B
- HRE
hypoxia response element
- IFN-β
interferon β
- MC and EC
total melanin and eumelanin contents
- MFA2
Mating Factor A
- MSH
Melanocyte Stimulating Hormone
- NAP1L1
Nucleosome Assembly Protein 1-Like 1
- NAP1L4
Nucleosome Assembly Protein 1-Like 4
- NEDD8
neural precursor cell expressed, developmentally down-regulated 8
- NER
nucleotide excision repair
- NOX1
NADPH Oxidase 1
- Nrf1
also called NFE2L1, nuclear factor erythroid 2-related factor 1
- p130
130 kDa retinoblastoma-associated protein
- p38 MAPK
p38 mitogen-activated protein kinases
- PARP-1
Poly (ADP-ribose) polymerase 1
- PARylation
Poly (ADP)-ribosylation
- PCNA
proliferating-cell nuclear antigen
- PI3K
Phosphatidylinositol-3-kinase
- PKA
Protein Kinase A
- Pol
polymerase
- PTEN
phosphatase and tensin homolog
- R4B
Rad4 binding domain
- RB
retinoblastoma protein
- RNA pol II
RNA polymerase II
- RNF111
RING finger protein 111
- ROC1
Regulator of Cullins 1
- ROS
reactive oxygen species
- RPA
replication protein A
- RPA3
Replication protein A3
- SCC
squamous cell carcinoma
- SHM
somatic hypermutation
- SIRT1
sirtuin 1
- Smad4
SMAD family member 4
- SWI/SNF
SWItch/sucrose nonfermentable
- TC-NER
transcription coupled nucleotide excision repair
- TFIIH
transcription factor II H
- TLS
Translesion synthesis
- TTD
trichothiodystrophy
- UBL
Ubiquitin-like domain
- USP7
Ubiquitin-specific-processing protease 7
- UV
Ultraviolet
- UVSSA
UV-sensitive syndrome protein
- VHS
Vps-27, Hrs and STAM domain
- XAB1
XPA-binding protein 1
- XP
xeroderma pigmentosum
- XPA
xeroderma pigmentosum, complementation group A
- XPB
xeroderma pigmentosum, complementation group B
- XPC
xeroderma pigmentosum, complementation group C
- XPD
xeroderma pigmentosum, complementation group D
- XPE
xeroderma pigmentosum, complementation group E
- XPF
xeroderma pigmentosum, complementation group F
- XPG
xeroderma pigmentosum, complementation group G
- YY1
Yin Yang 1
- ZEB1
Zinc Finger E-Box Binding Homeobox 1
Biographies
Palak Shah is currently pursuing a PhD in Molecular Pathogenesis and Molecular Medicine under the guidance of Dr. Yu-Ying He at the University of Chicago. She is also simultaneously pursuing an MS in Translational Sciences as part of the Howard Hughes Medical Institute PhD/MS Translational Training Program (TTP) offered at the University of Chicago. She received her B.Tech in Pharmaceutical Chemistry and Technology in 2010 from the Institute of Chemical Technology, Mumbai, India. Next she obtained Masters in Biotechnology in 2012 from the University of Pennsylvania. She is the recipient of the Dean’s International Student Fellowship from the University of Chicago in 2013. Her research interests are in genomic instability dependent and independent pathways in skin cancer.
Yu-Ying He is an Assistant Professor of Medicine at the University of Chicago. She received her PhD in Chemistry in 2000 from the Chinese Academy of Sciences in China. Then she received the Humboldt Research Fellowship working with Dr. Donat-P. Häder at the University of Erlangen-Nuerenburg in Germany. In 2001, she came to the NIEHS/NIH for her postdoctoral training with Dr. Colin Chignell. In 2007, she joined the faculty in the Department of Medicine at the University of Chicago. During her research career, Yu-Ying He has received several awards, including the first NIEHS Science Day Early Career Award, the NIH Fellows Award for Research Excellence (FARE), the American Skin Association Research Scholar Award, the American Cancer Society Research Scholar award, and the Outstanding New Environmental Scientist Award. Her research interests are in the genetic and environmental determinants of genomic stability using skin and skin cells as models.
References
- 1.Narayanan DL, Saladi RN, Fox JL. Ultraviolet radiation and skin cancer. International journal of dermatology. 2010;49:978–986. doi: 10.1111/j.1365-4632.2010.04474.x. [DOI] [PubMed] [Google Scholar]
- 2.D’Orazio J, Jarrett S, Amaro-Ortiz A, Scott T. UV Radiation and the Skin. Int J Mol Sci. 14:12222–12248. doi: 10.3390/ijms140612222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Program NT. Report on Carcinogens. 12. DIANE Publishing Company; 2011. [Google Scholar]
- 4.Breitbart EW, Greinert R, Volkmer B. Effectiveness of information campaigns. Progress in biophysics and molecular biology. 2006;92:167–172. doi: 10.1016/j.pbiomolbio.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 5.Cleaver JE, Crowley E. UV damage, DNA repair and skin carcinogenesis. Frontiers in bioscience : a journal and virtual library. 2002;7:d1024–1043. doi: 10.2741/A829. [DOI] [PubMed] [Google Scholar]
- 6.Diepgen TL, Mahler V. The epidemiology of skin cancer. The British journal of dermatology. 2002;146(Suppl 61):1–6. doi: 10.1046/j.1365-2133.146.s61.2.x. [DOI] [PubMed] [Google Scholar]
- 7.Lucas RWHO. Solar ultraviolet radiation : global burden of disease from solar ultraviolet radiation. World Health Organization; Geneva: 2006. [Google Scholar]
- 8.Hall BJ, Hall JC. Sauer’s Manual of Skin Diseases. Wolters Kluwer Health/Lippincott Williams & Wilkins; [Google Scholar]
- 9.Sinha RP, Hader DP. UV-induced DNA damage and repair: a review. Photochemical & photobiological sciences : Official journal of the European Photochemistry Association and the European Society for Photobiology. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 10.Kappes UP, Luo D, Potter M, Schulmeister K, Runger TM. Short- and long-wave UV light (UVB and UVA) induce similar mutations in human skin cells. J Invest Dermatol. 2006;126:667–675. doi: 10.1038/sj.jid.5700093. [DOI] [PubMed] [Google Scholar]
- 11.Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- 12.Rochette PJ, Therrien JP, Drouin R, Perdiz D, Bastien N, Drobetsky EA, Sage E. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003;31:2786–2794. doi: 10.1093/nar/gkg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual review of biochemistry. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 14.Niggli HJ, Rothlisberger R. Cyclobutane-type pyrimidine photodimer formation and induction of ornithine decarboxylase in human skin fibroblasts after UV irradiation. The Journal of investigative dermatology. 1988;91:579–584. doi: 10.1111/1523-1747.ep12477095. [DOI] [PubMed] [Google Scholar]
- 15.Vink AA, Berg RJ, de Gruijl FR, Roza L, Baan RA. Induction, repair and accumulation of thymine dimers in the skin of UV-B-irradiated hairless mice. Carcinogenesis. 1991;12:861–864. doi: 10.1093/carcin/12.5.861. [DOI] [PubMed] [Google Scholar]
- 16.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 17.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nature reviews. Genetics. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 18.Braithwaite E, Wu X, Wang Z. Repair of DNA lesions induced by polycyclic aromatic hydrocarbons in human cell-free extracts: involvement of two excision repair mechanisms in vitro. Carcinogenesis. 1998;19:1239–1246. doi: 10.1093/carcin/19.7.1239. [DOI] [PubMed] [Google Scholar]
- 19.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Molecular cell. 2010;37:702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jans J, Schul W, Sert YG, Rijksen Y, Rebel H, Eker AP, Nakajima S, van Steeg H, de Gruijl FR, Yasui A, Hoeijmakers JH, van der Horst GT. Powerful skin cancer protection by a CPD-photolyase transgene. Current biology : CB. 2005;15:105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Kneuttinger AC, Kashiwazaki G, Prill S, Heil K, Muller M, Carell T. Formation and Direct Repair of UV-induced Dimeric DNA Pyrimidine Lesions. Photochemistry and photobiology. 2013 doi: 10.1111/php.12197. [DOI] [PubMed] [Google Scholar]
- 22.Vogelstein B, Kinzler KW. The Genetic Basis of Human Cancer. McGraw-Hill, Medical Pub. Division; 2002. [Google Scholar]
- 23.Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim Biophys Acta. 2009;1790:963–969. doi: 10.1016/j.bbagen.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedberg EC. DNA Repair and Mutagenesis. ASM Press; 2006. [Google Scholar]
- 25.Friedberg EC, Bond JP, Burns DK, Cheo DL, Greenblatt MS, Meira LB, Nahari D, Reis AM. Defective nucleotide excision repair in xpc mutant mice and its association with cancer predisposition. Mutat Res. 2000;459:99–108. doi: 10.1016/s0921-8777(99)00068-3. [DOI] [PubMed] [Google Scholar]
- 26.Hengge UR, Emmert S. Clinical features of xeroderma pigmentosum. Advances in experimental medicine and biology. 2008;637:10–18. doi: 10.1007/978-0-387-09599-8_2. [DOI] [PubMed] [Google Scholar]
- 27.Bradford PT, Goldstein AM, Tamura D, Khan SG, Ueda T, Boyle J, Oh KS, Imoto K, Inui H, Moriwaki S, Emmert S, Pike KM, Raziuddin A, Plona TM, DiGiovanna JJ, Tucker MA, Kraemer KH. Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. Journal of medical genetics. 2011;48:168–176. doi: 10.1136/jmg.2010.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knobel PA, Marti TM. Translesion DNA synthesis in the context of cancer research. Cancer Cell Int. 11:39. doi: 10.1186/1475-2867-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nature reviews. Genetics. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 30.Stone JE, Kumar D, Binz SK, Inase A, Iwai S, Chabes A, Burgers PM, Kunkel TA. Lesion bypass by S. cerevisiae Pol zeta alone. DNA Repair (Amst) 2011;10:826–834. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell research. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 33.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 35.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase eta in the bypass of a (6-4) TT photoproduct. Molecular and cellular biology. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 38.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stallons LJ, McGregor WG. Translesion synthesis polymerases in the prevention and promotion of carcinogenesis. Journal of nucleic acids. 2010:2010. doi: 10.4061/2010/643857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 41.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase eta. Proc Natl Acad Sci U S A. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon JH, Prakash L, Prakash S. Error-free replicative bypass of (6-4) photoproducts by DNA polymerase zeta in mouse and human cells. Genes & development. 2010;24:123–128. doi: 10.1101/gad.1872810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yagi Y, Ogawara D, Iwai S, Hanaoka F, Akiyama M, Maki H. DNA polymerases eta and kappa are responsible for error-free translesion DNA synthesis activity over a cis-syn thymine dimer in Xenopus laevis oocyte extracts. DNA Repair (Amst) 2005;4:1252–1269. doi: 10.1016/j.dnarep.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 46.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 47.Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z. DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad Sci U S A. 2009;106:11552–11557. doi: 10.1073/pnas.0812548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Woodgate R, McManus TP, Mead S, McCormick JJ, Maher VM. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- 49.Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Verspuy J, Gali H, Haracska L, de Wind N. Mammalian polymerase zeta is essential for post-replication repair of UV-induced DNA lesions. DNA Repair (Amst) 2009;8:1444–1451. doi: 10.1016/j.dnarep.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 50.Araujo SJ, Wood RD. Protein complexes in nucleotide excision repair. Mutat Res. 1999;435:23–33. doi: 10.1016/s0921-8777(99)00042-7. [DOI] [PubMed] [Google Scholar]
- 51.Khan SG, Metter EJ, Tarone RE, Bohr VA, Grossman L, Hedayati M, Bale SJ, Emmert S, Kraemer KH. A new xeroderma pigmentosum group C poly(AT) insertion/deletion polymorphism. Carcinogenesis. 2000;21:1821–1825. doi: 10.1093/carcin/21.10.1821. [DOI] [PubMed] [Google Scholar]
- 52.He J, Shi TY, Zhu ML, Wang MY, Li QX, Wei QY. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta-analysis. International journal of cancer. Journal international du cancer. 2013;133:1765–1775. doi: 10.1002/ijc.28089. [DOI] [PubMed] [Google Scholar]
- 53.Khan SG, Muniz-Medina V, Shahlavi T, Baker CC, Inui H, Ueda T, Emmert S, Schneider TD, Kraemer KH. The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res. 2002;30:3624–3631. doi: 10.1093/nar/gkf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo C, Sheng J, Hu MG, Haluska FG, Cui R, Xu Z, Tsichlis PN, Hu GF, Hinds PW. Loss of ARF sensitizes transgenic BRAFV600E mice to UV-induced melanoma via suppression of XPC. Cancer Res. 2013;73:4337–4348. doi: 10.1158/0008-5472.CAN-12-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu YH, Tsai Chang JH, Cheng YW, Wu TC, Chen CY, Lee H. Xeroderma pigmentosum group C gene expression is predominantly regulated by promoter hypermethylation and contributes to p53 mutation in lung cancers. Oncogene. 2007;26:4761–4773. doi: 10.1038/sj.onc.1210284. [DOI] [PubMed] [Google Scholar]
- 56.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc Natl Acad Sci U S A. 2002;99:12985–12990. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han W, Ming M, Zhao R, Pi J, Wu C, He YY. Nrf1 CNC-bZIP protein promotes cell survival and nucleotide excision repair through maintaining glutathione homeostasis. J Biol Chem. 287:18788–18795. doi: 10.1074/jbc.M112.363614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rezvani HR, Mahfouf W, Ali N, Chemin C, Ged C, Kim AL, de Verneuil H, Taieb A, Bickers DR, Mazurier F. Hypoxia-inducible factor-1alpha regulates the expression of nucleotide excision repair proteins in keratinocytes. Nucleic Acids Res. 38:797–809. doi: 10.1093/nar/gkp1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu CL, Qiang L, Han W, Ming M, Viollet B, He YY. Role of AMPK in UVB-induced DNA damage repair and growth control. Oncogene. 32:2682–2689. doi: 10.1038/onc.2012.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD. A common set of gene regulatory networks links metabolism and growth inhibition. Molecular cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 61.Datta A, Sen J, Hagen J, Korgaonkar CK, Caffrey M, Quelle DE, Hughes DE, Ackerson TJ, Costa RH, Raychaudhuri P. ARF directly binds DP1: interaction with DP1 coincides with the G1 arrest function of ARF. Molecular and cellular biology. 2005;25:8024–8036. doi: 10.1128/MCB.25.18.8024-8036.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominguez-Brauer C, Chen YJ, Brauer PM, Pimkina J, Raychaudhuri P. ARF stimulates XPC to trigger nucleotide excision repair by regulating the repressor complex of E2F4. EMBO reports. 2009;10:1036–1042. doi: 10.1038/embor.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ming M, Feng L, Shea CR, Soltani K, Zhao B, Han W, Smart RC, Trempus CS, He YY. PTEN positively regulates UVB-induced DNA damage repair. Cancer Res. 71:5287–5295. doi: 10.1158/0008-5472.CAN-10-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ming M, Shea CR, Guo X, Li X, Soltani K, Han W, He YY. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc Natl Acad Sci U S A. 107:22623–22628. doi: 10.1073/pnas.1010377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang TT, D’Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 67.Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harbor perspectives in biology. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 69.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 70.Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, Matsunaga T. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J Biol Chem. 2002;277:1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- 71.Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J Biol Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 72.Wang QE, Praetorius-Ibba M, Zhu Q, El-Mahdy MA, Wani G, Zhao Q, Qin S, Patnaik S, Wani AA. Ubiquitylation-independent degradation of Xeroderma pigmentosum group C protein is required for efficient nucleotide excision repair. Nucleic Acids Res. 2007;35:5338–5350. doi: 10.1093/nar/gkm550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He J, Zhu Q, Wani G, Sharma N, Han C, Qian J, Pentz K, Wang QE, Wani AA. Ubiquitin-specific Protease 7 Regulates Nucleotide Excision Repair through Deubiquitinating XPC Protein and Preventing XPC Protein from Undergoing Ultraviolet Light-induced and VCP/p97 Protein-regulated Proteolysis. The Journal of biological chemistry. 2014;289:27278–27289. doi: 10.1074/jbc.M114.589812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikstrom M, Choudhary C, Houtsmuller AB, Marteijn JA, Bekker-Jensen S, Mailand N. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. The Journal of cell biology. 2013;201:797–807. doi: 10.1083/jcb.201212075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng JM, Vermeulen W, van der Horst GT, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JH. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes & development. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okuda Y, Nishi R, Ng JM, Vermeulen W, van der Horst GT, Mori T, Hoeijmakers JH, Hanaoka F, Sugasawa K. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 2004;3:1285–1295. doi: 10.1016/j.dnarep.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 77.Ortolan TG, Chen L, Tongaonkar P, Madura K. Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway. Nucleic Acids Res. 2004;32:6490–6500. doi: 10.1093/nar/gkh987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Molecular cell. 1999;3:687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- 79.Hardy TM, Kumar MS, Smith ML. RB stabilizes XPC and promotes cellular NER. Anticancer Res. 30:2483–2488. [PubMed] [Google Scholar]
- 80.Euvrard S, Kanitakis J, Claudy A. Skin cancers after organ transplantation. N Engl J Med. 2003;348:1681–1691. doi: 10.1056/NEJMra022137. [DOI] [PubMed] [Google Scholar]
- 81.Kuschal C, Thoms KM, Schubert S, Schafer A, Boeckmann L, Schon MP, Emmert S. Skin cancer in organ transplant recipients: effects of immunosuppressive medications on DNA repair. Experimental dermatology. 2012;21:2–6. doi: 10.1111/j.1600-0625.2011.01413.x. [DOI] [PubMed] [Google Scholar]
- 82.Han W, Soltani K, Ming M, He YY. Deregulation of XPC and CypA by cyclosporin A: an immunosuppression-independent mechanism of skin carcinogenesis. Cancer Prev Res (Phila) 5:1155–1162. doi: 10.1158/1940-6207.CAPR-12-0185-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thoms KM, Kuschal C, Oetjen E, Mori T, Kobayashi N, Laspe P, Boeckmann L, Schon MP, Emmert S. Cyclosporin A, but not everolimus, inhibits DNA repair mediated by calcineurin: implications for tumorigenesis under immunosuppression. Experimental dermatology. 2011;20:232–236. doi: 10.1111/j.1600-0625.2010.01213.x. [DOI] [PubMed] [Google Scholar]
- 84.Kuschal C, Thoms KM, Mori T, Kobayashi N, Boeckmann L, Laspe P, Emmert S. Cyclosporin A, but not everolimus, inhibits DNA repair in human fibroblasts and lymphoblasts. International journal of clinical pharmacology and therapeutics. 2009;47:38–40. doi: 10.5414/cpp47038. [DOI] [PubMed] [Google Scholar]
- 85.Kuschal C, Thoms KM, Boeckmann L, Laspe P, Apel A, Schon MP, Emmert S. Cyclosporin A inhibits nucleotide excision repair via downregulation of the xeroderma pigmentosum group A and G proteins, which is mediated by calcineurin inhibition. Experimental dermatology. 2011;20:795–799. doi: 10.1111/j.1600-0625.2011.01320.x. [DOI] [PubMed] [Google Scholar]
- 86.Ming M, Zhao B, Qiang L, He YY. Effect of Immunosuppressants Tacrolimus and Mycophenolate Mofetil on the Keratinocyte UVB Response. Photochemistry and photobiology. 2014 doi: 10.1111/php.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nichols AF, Itoh T, Graham JA, Liu W, Yamaizumi M, Linn S. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J Biol Chem. 2000;275:21422–21428. doi: 10.1074/jbc.M000960200. [DOI] [PubMed] [Google Scholar]
- 88.Tan T, Chu G. p53 Binds and activates the xeroderma pigmentosum DDB2 gene in humans but not mice. Molecular and cellular biology. 2002;22:3247–3254. doi: 10.1128/MCB.22.10.3247-3254.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 90.Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 91.Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, Hensbergen P, Deelder A, de Groot A, Matsumoto S, Sugasawa K, Thoma N, Vermeulen W, Vrieling H, Mullenders L. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. The Journal of cell biology. 2012;199:235–249. doi: 10.1083/jcb.201112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robu M, Shah RG, Petitclerc N, Brind’Amour J, Kandan-Kulangara F, Shah GM. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci U S A. 2013;110:1658–1663. doi: 10.1073/pnas.1209507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci U S A. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guerrero-Santoro J, Kapetanaki MG, Hsieh CL, Gorbachinsky I, Levine AS, Rapic-Otrin V. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 2008;68:5014–5022. doi: 10.1158/0008-5472.CAN-07-6162. [DOI] [PubMed] [Google Scholar]
- 95.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Molecular cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 96.Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A, Nakatani Y. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes & development. 2006;20:1429–1434. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JH, Demmers JA, Fousteri M, Vermeulen W, Marteijn JA. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nature genetics. 2012;44:598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 98.Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S, Tahara H, Neilan EG, Honma M, Nohmi T, Yasui A, Tanaka K. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nature genetics. 2012;44:593–597. doi: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
- 99.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 100.Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci U S A. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Molecular and cellular biology. 1999;19:6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 103.Woudstra EC, Gilbert C, Fellows J, Jansen L, Brouwer J, Erdjument-Bromage H, Tempst P, Svejstrup JQ. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415:929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- 104.Nakazawa Y, Sasaki K, Mitsutake N, Matsuse M, Shimada M, Nardo T, Takahashi Y, Ohyama K, Ito K, Mishima H, Nomura M, Kinoshita A, Ono S, Takenaka K, Masuyama R, Kudo T, Slor H, Utani A, Tateishi S, Yamashita S, Stefanini M, Lehmann AR, Yoshiura K, Ogi T. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nature genetics. 2012;44:586–592. doi: 10.1038/ng.2229. [DOI] [PubMed] [Google Scholar]
- 105.Ahmad I, Simanyi E, Guroji P, Tamimi IA, delaRosa HJ, Nagar A, Nagar P, Katiyar SK, Elmets CA, Yusuf N. Toll-like receptor-4 deficiency enhances repair of UVR-induced cutaneous DNA damage by nucleotide excision repair mechanism. The Journal of investigative dermatology. 2014;134:1710–1717. doi: 10.1038/jid.2013.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dong L, Wen J, Pier E, Zhang X, Zhang B, Dong F, Ziegler N, Mysz M, Armenta R, Cui R. Melanocyte-stimulating hormone directly enhances UV-Induced DNA repair in keratinocytes by a xeroderma pigmentosum group A-dependent mechanism. Cancer Res. 70:3547–3556. doi: 10.1158/0008-5472.CAN-09-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jarrett Stuart G, Horrell Erin MW, Christian Perry A, Vanover Jillian C, Boulanger Mary C, Zou Y, D’Orazio John A. PKA-Mediated Phosphorylation of ATR Promotes Recruitment of XPA to UV-Induced DNA Damage. Molecular cell. 54:999–1011. doi: 10.1016/j.molcel.2014.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 108.King BS, Cooper KL, Liu KJ, Hudson LG. Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair. J Biol Chem. 2012;287:39824–39833. doi: 10.1074/jbc.M112.393504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Okui T, Fujiwara Y. Inhibition of human excision DNA repair by inorganic arsenic and the co-mutagenic effect in V79 Chinese hamster cells. Mutat Res. 1986;172:69–76. doi: 10.1016/0165-1218(86)90108-4. [DOI] [PubMed] [Google Scholar]
- 110.Lee-Chen SF, Yu CT, Jan KY. Effect of arsenite on the DNA repair of UV-irradiated Chinese hamster ovary cells. Mutagenesis. 1992;7:51–55. doi: 10.1093/mutage/7.1.51. [DOI] [PubMed] [Google Scholar]
- 111.Hartwig A, Groblinghoff UD, Beyersmann D, Natarajan AT, Filon R, Mullenders LH. Interaction of arsenic(III) with nucleotide excision repair in UV-irradiated human fibroblasts. Carcinogenesis. 1997;18:399–405. doi: 10.1093/carcin/18.2.399. [DOI] [PubMed] [Google Scholar]
- 112.Cooper KL, King BS, Sandoval MM, Liu KJ, Hudson LG. Reduction of arsenite-enhanced ultraviolet radiation-induced DNA damage by supplemental zinc. Toxicology and applied pharmacology. 2013;269:81–88. doi: 10.1016/j.taap.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou X, Sun X, Cooper KL, Wang F, Liu KJ, Hudson LG. Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. J Biol Chem. 2011;286:22855–22863. doi: 10.1074/jbc.M111.232926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hartwig A, Pelzer A, Asmuss M, Burkle A. Very low concentrations of arsenite suppress poly(ADP-ribosyl)ation in mammalian cells. International journal of cancer. Journal international du cancer. 2003;104:1–6. doi: 10.1002/ijc.10911. [DOI] [PubMed] [Google Scholar]
- 115.Schwerdtle T, Walter I, Hartwig A. Arsenite and its biomethylated metabolites interfere with the formation and repair of stable BPDE-induced DNA adducts in human cells and impair XPAzf and Fpg. DNA Repair (Amst) 2003;2:1449–1463. doi: 10.1016/j.dnarep.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 116.Ding W, Liu W, Cooper KL, Qin XJ, de Souza Bergo PL, Hudson LG, Liu KJ. Inhibition of poly(ADP-ribose) polymerase-1 by arsenite interferes with repair of oxidative DNA damage. J Biol Chem. 2009;284:6809–6817. doi: 10.1074/jbc.M805566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Asmuss M, Mullenders LH, Eker A, Hartwig A. Differential effects of toxic metal compounds on the activities of Fpg and XPA, two zinc finger proteins involved in DNA repair. Carcinogenesis. 2000;21:2097–2104. doi: 10.1093/carcin/21.11.2097. [DOI] [PubMed] [Google Scholar]
- 118.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes & development. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Crawford EL, Blomquist T, Mullins DN, Yoon Y, Hernandez DR, Al-Bagdhadi M, Ruiz J, Hammersley J, Willey JC. CEBPG regulates ERCC5/XPG expression in human bronchial epithelial cells and this regulation is modified by E2F1/YY1 interactions. Carcinogenesis. 2007;28:2552–2559. doi: 10.1093/carcin/bgm214. [DOI] [PubMed] [Google Scholar]
- 120.Sugita K, Suzuki N, Higuchi Y, Kita K, Suzuki Y, Lehmann A. Enhancement of XPG mRNA expression by human interferon-beta in Cockayne syndrome cells. Mutat Res. 1998;408:67–72. doi: 10.1016/s0921-8777(98)00020-2. [DOI] [PubMed] [Google Scholar]
- 121.Sugita K, Suzuki N, Niimi H. Involvement of antipain-sensitive protease activity in the interferon-beta-induced UV-refractoriness of Cockayne syndrome fibroblasts. Mutat Res. 1996;357:177–181. doi: 10.1016/0027-5107(96)00098-x. [DOI] [PubMed] [Google Scholar]
- 122.Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, Kastan MB, O’Connor PM, Fornace AJ., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 123.Smith ML, Kontny HU, Zhan Q, Sreenath A, O’Connor PM, Fornace AJ., Jr Antisense GADD45 expression results in decreased DNA repair and sensitizes cells to u.v.-irradiation or cisplatin. Oncogene. 1996;13:2255–2263. [PubMed] [Google Scholar]
- 124.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 125.Le May N, Fradin D, Iltis I, Bougneres P, Egly JM. XPG and XPF endonucleases trigger chromatin looping and DNA demethylation for accurate expression of activated genes. Molecular cell. 2012;47:622–632. doi: 10.1016/j.molcel.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 126.Jaspers NG, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, Giglia-Mari G, Hoogstraten D, Kleijer WJ, Hoeijmakers JH, Vermeulen W. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. American journal of human genetics. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L, Li TS, Yoshiura K, Utani A, Hirano A, Yamashita S, Greenblatt D, Nardo T, Stefanini M, McGibbon D, Sarkany R, Fassihi H, Takahashi Y, Nagayama Y, Mitsutake N, Lehmann AR, Ogi T. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. American journal of human genetics. 2013;92:807–819. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McWhir J, Selfridge J, Harrison DJ, Squires S, Melton DW. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nature genetics. 1993;5:217–224. doi: 10.1038/ng1193-217. [DOI] [PubMed] [Google Scholar]
- 129.Mitra D, Fernandez P, Bian L, Song N, Li F, Han G, Wang XJ. Smad4 loss in mouse keratinocytes leads to increased susceptibility to UV carcinogenesis with reduced Ercc1-mediated DNA repair. J Invest Dermatol. 2013;133:2609–2616. doi: 10.1038/jid.2013.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hauser JE, Kadekaro AL, Kavanagh RJ, Wakamatsu K, Terzieva S, Schwemberger S, Babcock G, Rao MB, Ito S, Abdel-Malek ZA. Melanin content and MC1R function independently affect UVR-induced DNA damage in cultured human melanocytes. Pigment Cell Res. 2006;19:303–314. doi: 10.1111/j.1600-0749.2006.00315.x. [DOI] [PubMed] [Google Scholar]
- 131.Abdel-Malek ZA, Ruwe A, Kavanagh-Starner R, Kadekaro AL, Swope V, Haskell-Luevano C, Koikov L, Knittel JJ. alpha-MSH tripeptide analogs activate the melanocortin 1 receptor and reduce UV-induced DNA damage in human melanocytes. Pigment Cell Melanoma Res. 2009;22:635–644. doi: 10.1111/j.1755-148X.2009.00598.x. [DOI] [PubMed] [Google Scholar]
- 132.Kadekaro AL, Kavanagh R, Kanto H, Terzieva S, Hauser J, Kobayashi N, Schwemberger S, Cornelius J, Babcock G, Shertzer HG, Scott G, Abdel-Malek ZA. alpha-Melanocortin and endothelin-1 activate antiapoptotic pathways and reduce DNA damage in human melanocytes. Cancer Res. 2005;65:4292–4299. doi: 10.1158/0008-5472.CAN-04-4535. [DOI] [PubMed] [Google Scholar]
- 133.Bohm M, Wolff I, Scholzen TE, Robinson SJ, Healy E, Luger TA, Schwarz T, Schwarz A. alpha-Melanocyte-stimulating hormone protects from ultraviolet radiation-induced apoptosis and DNA damage. J Biol Chem. 2005;280:5795–5802. doi: 10.1074/jbc.M406334200. [DOI] [PubMed] [Google Scholar]
- 134.Smith AG, Luk N, Newton RA, Roberts DW, Sturm RA, Muscat GE. Melanocortin-1 receptor signaling markedly induces the expression of the NR4A nuclear receptor subgroup in melanocytic cells. J Biol Chem. 2008;283:12564–12570. doi: 10.1074/jbc.M800480200. [DOI] [PubMed] [Google Scholar]
- 135.D’Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, Ito S, Fisher DE. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 136.Passeron T, Namiki T, Passeron HJ, Le Pape E, Hearing VJ. Forskolin protects keratinocytes from UVB-induced apoptosis and increases DNA repair independent of its effects on melanogenesis. J Invest Dermatol. 2009;129:162–166. doi: 10.1038/jid.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haskell-Luevano C, Hendrata S, North C, Sawyer TK, Hadley ME, Hruby VJ, Dickinson C, Gantz I. Discovery of prototype peptidomimetic agonists at the human melanocortin receptors MC1R and MC4R. J Med Chem. 1997;40:2133–2139. doi: 10.1021/jm960840h. [DOI] [PubMed] [Google Scholar]
- 138.Yang Y, Dickinson C, Haskell-Luevano C, Gantz I. Molecular basis for the interaction of [Nle4, D-Phe7]melanocyte stimulating hormone with the human melanocortin-1 receptor. J Biol Chem. 1997;272:23000–23010. doi: 10.1074/jbc.272.37.23000. [DOI] [PubMed] [Google Scholar]
- 139.Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA- reapir synthesis. Proc Natl Acad Sci U S A. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peterson CL, Almouzni G. Nucleosome dynamics as modular systems that integrate DNA damage and repair. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hara R, Mo J, Sancar A. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Molecular and cellular biology. 2000;20:9173–9181. doi: 10.1128/mcb.20.24.9173-9181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Molecular and cellular biology. 2002;22:6779–6787. doi: 10.1128/MCB.22.19.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ray A, Mir SN, Wani G, Zhao Q, Battu A, Zhu Q, Wang QE, Wani AA. Human SNF5/INI1, a component of the human SWI/SNF chromatin remodeling complex, promotes nucleotide excision repair by influencing ATM recruitment and downstream H2AX phosphorylation. Molecular and cellular biology. 2009;29:6206–6219. doi: 10.1128/MCB.00503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, Wani AA. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem. 2009;284:30424–30432. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Muftuoglu M, Selzer R, Tuo J, Brosh RM, Jr, Bohr VA. Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene. 2002;283:27–40. doi: 10.1016/s0378-1119(01)00870-8. [DOI] [PubMed] [Google Scholar]
- 146.Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 147.Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM, Jr, Bohr VA. Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res. 2002;30:782–793. doi: 10.1093/nar/30.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lake RJ, Geyko A, Hemashettar G, Zhao Y, Fan HY. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Molecular cell. 2010;37:235–246. doi: 10.1016/j.molcel.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Molecular and cellular biology. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cho I, Tsai PF, Lake RJ, Basheer A, Fan HY. ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS genetics. 2013;9:e1003407. doi: 10.1371/journal.pgen.1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Molecular cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 152.Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, Li L. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc Natl Acad Sci U S A. 2010;107:17274–17279. doi: 10.1073/pnas.1008388107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ura K, Araki M, Saeki H, Masutani C, Ito T, Iwai S, Mizukoshi T, Kaneda Y, Hanaoka F. ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. The EMBO journal. 2001;20:2004–2014. doi: 10.1093/emboj/20.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 155.Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes & development. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yu S, Teng Y, Waters R, Reed SH. How chromatin is remodelled during DNA repair of UV-induced DNA damage in Saccharomyces cerevisiae. PLoS genetics. 2011;7:e1002124. doi: 10.1371/journal.pgen.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA Repair (Amst) 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 158.Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011;39:1390–1397. doi: 10.1093/nar/gkq983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rubbi CP, Milner J. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. The EMBO journal. 2003;22:975–986. doi: 10.1093/emboj/cdg082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Datta A, Bagchi S, Nag A, Shiyanov P, Adami GR, Yoon T, Raychaudhuri P. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat Res. 2001;486:89–97. doi: 10.1016/s0921-8777(01)00082-9. [DOI] [PubMed] [Google Scholar]
- 161.Rapic-Otrin V, McLenigan MP, Bisi DC, Gonzalez M, Levine AS. Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res. 2002;30:2588–2598. doi: 10.1093/nar/30.11.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Molecular and cellular biology. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Biswas AK, Mitchell DL, Johnson DG. E2F1 responds to ultraviolet radiation by directly stimulating DNA repair and suppressing carcinogenesis. Cancer Res. 2014;74:3369–3377. doi: 10.1158/0008-5472.CAN-13-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, Fousteri M, Houtsmuller AB, Vermeulen W, Marteijn JA. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Molecular cell. 2013;51:469–479. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 165.Oksenych V, Zhovmer A, Ziani S, Mari PO, Eberova J, Nardo T, Stefanini M, Giglia-Mari G, Egly JM, Coin F. Histone methyltransferase DOT1L drives recovery of gene expression after a genotoxic attack. PLoS genetics. 2013;9:e1003611. doi: 10.1371/journal.pgen.1003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013;155:94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 167.Yu Y, Deng Y, Reed SH, Millar CB, Waters R. Histone variant Htz1 promotes histone H3 acetylation to enhance nucleotide excision repair in Htz1 nucleosomes. Nucleic Acids Res. 2013;41:9006–9019. doi: 10.1093/nar/gkt688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang QE, Han C, Zhao R, Wani G, Zhu Q, Gong L, Battu A, Racoma I, Sharma N, Wani AA. p38 MAPK- and Akt-mediated p300 phosphorylation regulates its degradation to facilitate nucleotide excision repair. Nucleic Acids Res. 2013;41:1722–1733. doi: 10.1093/nar/gks1312. [DOI] [PMC free article] [PubMed] [Google Scholar]