Abstract
In this article, we will discuss the biochemistry of mitosis in eukaryotic cells. We will focus on conserved principles that, importantly, are adapted to the biology of the organism. It is vital to bear in mind that the structural requirements for division in a rapidly dividing syncytial Drosophila embryo, for example, are markedly different from those in a unicellular yeast cell. Nevertheless, division in both systems is driven by conserved modules of antagonistic protein kinases and phosphatases, underpinned by ubiquitin-mediated proteolysis, which create molecular switches to drive each stage of division forward. These conserved control modules combine with the self-organizing properties of the subcellular architecture to meet the specific needs of the cell. Our discussion will draw on discoveries in several model systems that have been important in the long history of research on mitosis, and we will try to point out those principles that appear to apply to all cells, compared with those in which the biochemistry has been specifically adapted in a particular organism.
During mitosis, cells undergo a series of dramatic changes that is coordinated in both time and space by biochemical mechanisms (e.g., antagonistic protein kinases and phosphatases, as well as ubiquitin-mediated proteolysis).
The aim of mitosis is to separate the genome and ensure that the two daughter cells inherit an equal and identical complement of chromosomes (Yanagida 2014). To achieve this, eukaryotic cells completely reorganize their microtubules to build a mitotic spindle that pulls apart the sister chromatids after the cohesin complexes are cut (see Cheeseman 2014; Hirano 2015; Reber and Hyman 2015; Westhorpe and Straight 2015) and, subsequently, use the actin cytoskeleton to divide the cell into two (cytokinesis) (see D’Avino et al. 2015). In some cells, such as in budding and fission yeasts, the spindle is built within the nucleus (closed mitosis), whereas in others, the nuclear envelope breaks down and the condensed chromosomes are captured by microtubules in the cytoplasm (open mitosis). This difference in the spatial organization of the mitotic cell has ramifications for the machinery controlling mitosis. In particular, the breakdown of the nuclear compartment disrupts the guanosine triphosphate (GTP)–guanosine diphosphate (GDP) gradient of the small GTPase called Ran. Ran usually controls nuclear-cytoplasmic transport through the importin chaperones; Ran-GDP in the cytoplasm promotes binding to nuclear transport substrates, whereas Ran-GTP in the nucleus promotes their dissociation (Güttler and Görlich 2011). As a result of nuclear envelope breakdown (NEBD), another Ran-GTP gradient is generated around the chromosomes, to which the RCC1 GTP-exchange factor binds (Clarke 2008). This Ran-GTP gradient is important for the interaction between microtubules and chromosomes because the high Ran-GTP levels around chromosomes promote the dissociation between the importin β chaperone and its binding partners, several of which help to stabilize or nucleate microtubules (Carazo-Salas et al. 1999; Kalab et al. 1999; Gruss et al. 2001; Wilde et al. 2001; Yokoyama et al. 2008).
The dramatic reorganization of the cell at mitosis must be coordinated in both time and space. There are several key temporal events: entry to mitosis, sister chromatid separation, and mitotic exit, and these are effectively made unidirectional by the biochemical machinery. We will discuss the biochemistry behind each of these temporal events, in turn, but it is important to emphasize that the control mechanisms are also spatially organized. Our understanding of this spatial organization has improved dramatically with advances in the technology to detect gradients of activity in cells, and this has revealed the importance of local gradients of antagonistic protein kinases and phosphatases, GTP-binding protein regulators, and ubiquitin ligases and deubiquitylases, to name only a few of the more prominent examples (reviewed in Pines and Hagan 2011).
ENTRY TO MITOSIS
Cyclin B–Cyclin-Dependent Kinase (CDK): The Main Switch
Entry to mitosis appears to have switch-like properties in most organisms studied; once a cell is committed to mitosis, there is no going back (see Fig. 1). But how the machinery is made switch-like does differ between systems. Nevertheless, the core of the switch seems to be the same in all cells: the activation of the cyclin B–Cdk1 mitotic protein kinase (Nurse 1990). A combination of genetics, cell biology, and biochemistry led to the identification of this protein kinase as the key regulator of mitosis, which is conserved from yeast to man (Doree 2002). The kinase is a heterodimer composed of a Cdk subunit and an activating cyclin subunit. Both are members of multigene families, and, usually, the B-type cyclins are essential to trigger mitosis. (One exception is Drosophila, in which the A-type cyclin is most important [Knoblich and Lehner 1993; Jacobs et al. 1998].) The mitotic cyclin–Cdk also binds a small protein cyclin-kinase subunit (Cks). Cks has a conserved anion-binding site (Bourne et al. 1995), which allows it to bind phosphoserine/phosphothreonine, and thereby retain interactions with substrates phosphorylated by the cyclin–Cdk. This allows the cyclin–Cdk to hyperphosphorylate its substrate by phosphorylating even low affinity sites, and this has been shown to be important in the activation of substrates, such as Xenopus Cdc25 and the anaphase-promoting complex or cyclosome (APC/C) (see below) (Patra et al. 1999) and in the degradation of cyclin A (Wolthuis et al. 2008; Di Fiore and Pines 2010).
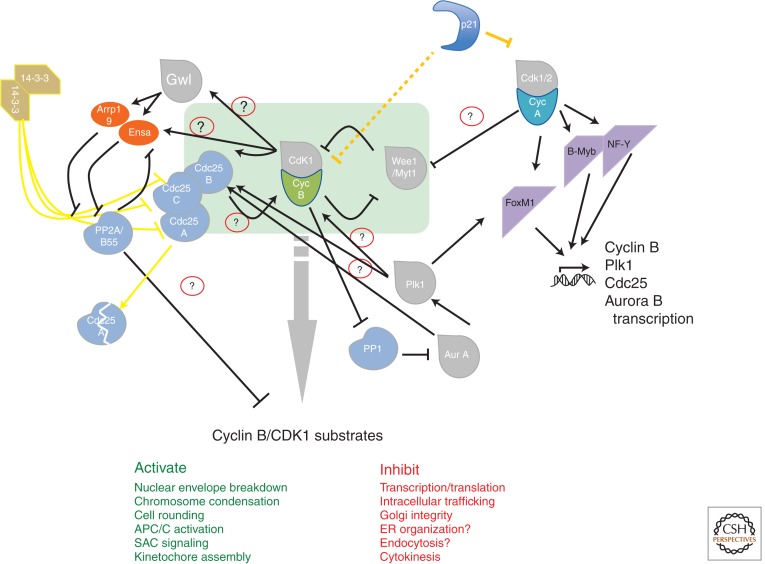
Figure 1.
Mitotic entry network. The core regulatory module is indicated as a green rectangle. Dark gray lines indicate the interactions during a nonperturbed mitotic cell cycle. Yellow lines indicate interaction on DNA damage or stress-activated G2 checkpoint signaling. Question marks in red circles indicate interactions that are contentious or not proven. Green and red colored text indicates processes that are stimulated and inhibited by cyclin B/Cdk1 during mitosis, respectively. Question marks indicate processes in which the direct involvement of cyclin B/Cdk1 is not clear or not proven. APC, Anaphase-promoting complex; CDK, cyclin-dependent kinase; ER, endoplasmic reticulum; SAC, spindle assembly checkpoint.
The crystal structures of a number of different cyclins, Cdks, and cyclin–Cdk complexes have been solved (Brown et al. 1995, 1999; Jeffrey et al. 1995; Russo et al. 1996; Pavletich 1999). These show that the Cdk subunit is completely inactive as a monomer for two reasons. First, the structure of the ATP-binding pocket in the amino-terminal lobe coordinates ATP in the wrong conformation for β-γ bond hydrolysis; second, the “T-loop,” which contains an activatory phosphorylation site, obscures the substrate binding cleft. Cyclin binding rectifies both defects by altering the structure of the Cdk amino-terminal lobe to remodel the ATP-binding site and draw the T-loop down and away from the substrate-binding cleft. When the T-loop is phosphorylated by Cdk-activating kinase (CAK), it interacts more strongly with a basic patch on the carboxy-terminal lobe of the Cdk, and this, then, is the fully active state.
Cyclin binding and CAK phosphorylation can be used to control entry to mitosis directly if a threshold of cyclin–Cdk activity must be reached to push cells into mitosis. The validity of this “threshold” hypothesis has been elegantly shown in genetically engineered fission yeast (Coudreuse and Nurse 2010), and the regulation of Cdk activity by local cyclin levels alone appears to be used to control mitosis in systems, such as rapidly dividing early Drosophila embryos (Su et al. 1998; McCleland et al. 2009). In other systems, however, the mitotic cyclin–Cdk complex accumulates in an inactive form through phosphorylation on a conserved tyrosine residue and, in some systems, the adjacent threonine residue, both in the ATP-binding site of the Cdk. Phosphorylation on these sites prevents the transfer of phosphate to a substrate (Atherton-Fessler et al. 1993). Subsequently, this complex can be rapidly activated by dephosphorylating the inhibitory sites to drive cells into mitosis. The Wee1 family of protein kinases, which inhibit the cyclin–Cdks and the antagonistic Cdc25 phosphatase family, are both conserved through yeast and animal evolution (although, perhaps, not in plants [Francis 2011]). The ability to control the key regulator of mitotic entry by phosphorylation/dephosphorylation allows the integration of signals from different pathways to control the timing of mitosis. Which pathways converge on the Wee1 and Cdc25 regulators depends on the biology of the organism. For example, in budding yeast, instead of entry to mitosis (Amon et al. 1992), it is used earlier in the cell cycle to arrest cells should synthesis of the bud be perturbed (the morphogenesis checkpoint) (Lew and Reed 1995). In many systems, unreplicated or damaged DNA activates “checkpoint” pathways that tip the balance in favor of Wee1 and against Cdc25 to prevent mitosis (reviewed in Langerak 2011). This can be achieved by stabilizing Wee1-like kinases and destabilizing or inhibiting Cdc25 phosphatases. Fission yeast cells control the size at which they divide; therefore, the Wee1-Cdc25 axis can be regulated by the target of rapamycin (TOR) pathway that responds to the nutritional state of the cell (Petersen and Nurse 2007; Atkin et al. 2014). The controls on the timing of mitosis in other systems are much less clear. Aside from the negative regulation by DNA damage, a number of factors have been implicated, including negative regulation by stress and, in animal cells, positive regulation by the other important mitotic cyclin, cyclin A. Cyclin A–Cdk can promote mitosis both through activating the transcription of other mitotic regulators (Laoukili et al. 2005) and, posttranscriptionally, in a much less well-defined manner (Furuno et al. 1999).
The balance between Wee1 and Cdc25 activities has been shown to impose switch-like behavior on the entry to mitosis in a number of systems. This is most clearly illustrated in the early Xenopus embryo, in which both Wee1 and Cdc25 are ultrasensitive substrates of cyclin B–Cdk itself (Kim and Ferrell 2007; Trunnell et al. 2011). Taking Wee1 first, cyclin B–Cdk1 phosphorylates Wee1 on two different sets of phosphorylation sites. The first set of sites that are phosphorylated do not affect Wee1 kinase activity, but serve to buffer the system such that cyclin B–Cdk1 activity must reach a critical threshold before it can phosphorylate the second set of sites. These inhibit Wee1 kinase activity, thereby setting up a double-negative feedback loop because the reduction in Wee1 activity allows more cyclin B–Cdk1 to be activated (Kim and Ferrell 2007). A similar, positive feedback loop is also set up by cyclin B–Cdk1, which phosphorylates and activates Cdc25 (Kumagai and Dunphy 1992; Trunnell et al. 2011). Together, these feedback loops ensure that once cyclin B1–Cdk1 activity passes a threshold, activation rapidly goes to completion. This mechanism of rapid cyclin B–Cdk1 activation has recently been shown to be particularly important to coordinate entry to mitosis in the very large (1 mm diameter) Xenopus egg, in which an initial, local activation of cyclin B–Cdk1 rapidly self-propagates to activate the kinase throughout the cell within a few minutes (Chang 2013).
Whether this rapid, self-propagating switch applies in the much smaller cells of other organisms is unclear. For example, in human cells, Wee1 does not appear to be inhibited through phosphorylation by cyclin–Cdks; instead, phosphorylation targets Wee1 for proteolysis through generating a phosphodegron (Watanabe et al. 1995). Similarly, although one Cdc25 isoform is activated by phosphorylation in mammalian cells, this particular isoform (Cdc25C) is not essential in the mouse (Chen et al. 2001; Ferguson et al. 2005). In contrast, the essential Cdc25A isoform (Lee et al. 2009) is not activated, but made more stable after phosphorylation by cyclin B–Cdk1 (Mailand et al. 2002); this generates a much slower positive feedback loop because it is limited by the rate of translation of Cdc25A. Moreover, in mammalian cells, two different measures of cyclin B1–Cdk1 activation, immunofluorescence staining of fixed cells (Lindqvist et al. 2007) and a Förster resonance energy transfer (FRET)–based sensor in living cells (Gavet and Pines 2010b), indicate that cyclin B1–Cdk1 is activated on a time scale of ∼30 min. This slower increase in cyclin B–Cdk1 levels seems to be important in coordinating mitotic entry because it triggers different events at dissimilar thresholds; cell rounding and the nuclear import of cyclin B–Cdk are the first events to be initiated, and APC/C activation and NEBD are the last (Gavet and Pines 2010a,b).
Nevertheless, there is evidence for the local activation of cyclin B–Cdk1 in other systems. The strongest evidence is in fission yeast, in which cyclin B–Cdk is first activated on the spindle pole bodies (SPBs), the organelles that nucleate microtubules to form the mitotic spindle. Locally activated cyclin B–Cdk recruits another mitotic regulator, Plo1 (a member of the conserved Polo-like kinase family, which plays key roles in mitosis in all systems), to the SPB. Once recruited to the SPB, Plo1 triggers the growth of the cell and sets the timing for mitotic commitment (Grallert et al. 2013). Elements of this control appear to be conserved; there is evidence that human cyclin B–Cdk1 is first activated on centrosomes (Jackman et al. 2003), the equivalent organelles to SPBs. Moreover, mitotic timing is strongly influenced by Plk1, an essential member of the Polo-like kinase family in mammalian cells, because inhibiting Plk1 in human cells delays mitosis by several hours (Lenart et al. 2007). It is not yet clear exactly how human Plk1 regulates the timing of mitosis. Acting together with cyclin B–Cdk1, it generates a phosphodegron on Wee1, which is recognized by the b-TrCP Skp1-Cul1-F-box protein (SCF) ubiquitin ligase (Watanabe et al. 1995), but whether this is sufficient to explain its requirement is not clear.
Plk1 has also been proposed to help trigger mitosis by promoting the rapid movement of cyclin B1 into the nucleus before NEBD (Toyoshima-Morimoto et al. 2001). This is a conserved feature of mitosis in animal cells in which the B-type cyclins have a nuclear export sequence in their amino terminus (Hagting et al. 1998; Toyoshima et al. 1998; Yang et al. 1998) and an ill-defined nuclear import (Yang et al. 1998; Takizawa et al. 1999; Jackman et al. 2002); as a result, the cyclin B–Cdk complex constantly shuttles between the nucleus and the cytoplasm in interphase, but the bulk of the protein is cytoplasmic. Coincident with its activation ∼20 min before NEBD (Lindqvist et al. 2007; Gavet and Pines 2010b), a large fraction of cyclin B–Cdk1 moves quickly into the nucleus (Pines and Hunter 1991; Ookata et al. 1992; Hagting et al. 1998), and it was reported that Plk1 is required for this by inhibiting the nuclear export of human cyclin B1–Cdk1 (Toyoshima-Morimoto et al. 2002). More recent studies, however, showed that cyclin B1–Cdk1 activation is sufficient to promote its own rapid movement into the nucleus in the absence of Plk1 activity (Gavet and Pines 2010a).
The exact function of the striking change in cyclin B–Cdk localization in prophase is not yet clear. Altering the nuclear:cytoplasmic ratio could contribute to a spatially regulated positive feedback loop (Santos et al. 2012), but it also appears to be required to synchronize events in the nucleus and cytoplasm (Gavet and Pines 2010a). Indeed, Erich Nigg has called cyclin B–Cdk the “workhorse” of mitosis because it phosphorylates a very large number of substrates, including structural proteins as well as other mitotic regulators (Nigg 1993; Errico et al. 2010; Pagliuca et al. 2011). Thus, this kinase has an important role in restructuring the mitotic cell. This large number of substrates has made defining the myriad roles of cyclin B–Cdk in mitosis very challenging; it has been implicated in reshaping the microtubule cytoskeleton, intermediate filament and actin cytoskeletons, chromosome architecture, and the membrane compartments, promoting NEBD, and regulating the timing of anaphase and cytokinesis (reviewed in Nigg 1991, 1993; Errico et al. 2010). In many of these roles, cyclin B–Cdk activity is coordinated with other key mitotic kinases, notably the Plk and Aurora kinases.
The relation between cyclin B–Cdk and members of the Plk family is particularly intimate. The mitotic Plk (Plk1 in human cells) has a conserved phosphoserine/phosphothreonine-binding domain in its carboxyl terminus that is called the Polo box (Yaffe et al. 1997). This domain particularly favors Ser-phospho-Ser or Ser-phospho-Thr motifs, and an examination of Plk1-binding partners reveals enrichment for proteins, in which this dipeptide is followed by a proline (Lowery et al. 2007). Ser-Pro and Thr-Pro are the minimal motifs recognized by the Cdks; thus, for many substrates, the actions of the cyclin B–Cdk and Plk1 kinases are coordinated through cyclin B–Cdk phosphorylation creating a Plk1-binding site. Note that this, again, emphasizes the importance of the spatial relationship between the kinases and their substrates. This coordination is important in cells with an open mitosis where cyclin B–Cdk1 and Plk kinases are both required for complete disassembly of the pore (Muhlhausser and Kutay 2007).
Spatial control is also evident in the regulation of Aurora kinase activity. Like the Polo family, this conserved family of kinases was first identified in Drosophila. In animal cells, the Aurora A kinase is particularly important in forming the mitotic spindle through regulating microtubule nucleation from the centrosomes (see Reber and Hyman 2015). In vertebrates, the TPX2 protein also plays an important role by activating Aurora A through suppressing its dephosphorylation by the PP1 phosphatase (Bayliss et al. 2003) and recruiting Aurora A to microtubules (Wittmann et al. 2000; Kufer et al. 2002). The TPX2–Aurora A complex is then regulated by the PP6 phosphatase, which dephosphorylates the activatory T-loop phosphorylation on Aurora A (Zeng et al. 2010). The Aurora kinases, subsequently, have a crucial role in correcting erroneous microtubule attachments to the kinetochores, which depends on a spatial gradient set up at the centromere between Aurora (specifically, Aurora B in animal cells) and its antagonistic phosphatase, PP1 (see Cheeseman 2014). Once cells begin anaphase, Aurora B moves from the centromeres to the microtubules to coordinate cytokinesis (see D’Avino et al. 2015), and this change in localization depends on the decline in cyclin B–Cdk kinase and ubiquitin modification by Cul3-containing ubiquitin ligases, which make centromere-bound Aurora B a substrate for the p97 AAA ATPase (Sumara et al. 2007).
Inhibiting Protein Phosphatases
Most of the effort in mitosis research has focused on these essential protein kinases (cyclin B–Cdk, Plk, and Aurora), but, recently, the importance of controlling the antagonistic phosphatases has returned to view (reviewed in Bollen et al. 2009). Genetic screens in fission yeast (Kinoshita et al. 1990) and small interfering RNA (siRNA) screens in Drosophila cells (Chen et al. 2007) had identified the PP1 and PP2A families of phosphatases as playing important roles in mitosis, particularly in anaphase and mitotic exit. In addition, budding yeast depends on Cdc14 to dephosphorylate Cdk substrates (Ser-Pro and Thr-Pro motifs) to promote mitotic exit (reviewed in D’Amours and Amon 2004), a role that does not appear to be conserved in fission yeast or animal cells (Mocciaro et al. 2010; reviewed in Mocciaro and Schiebel 2010). It was a biochemical study using Xenopus extracts that underlined the importance of controlling phosphatases for mitotic entry. Mochida et al. showed that activating cyclin B–Cdk kinase was not sufficient to drive cells into mitosis; a member of the PP2A phosphatase family that antagonized cyclin B–Cdk1 had to be inactivated too (Mochida et al. 2009). A flurry of papers followed, which rapidly revealed that another feedback loop was at work (Gharbi-Ayachi et al. 2010; Mochida et al. 2010; Rangone et al. 2011; Blake-Hodek et al. 2012; Williams et al. 2014); more importantly, they revealed how a specific PP2A complex could be inhibited without affecting other complexes. Briefly, the Scant/Greatwall kinase is activated by cyclin–Cdk as cells enter mitosis (Blake-Hodek et al. 2012), and this phosphorylates a small, abundant protein called ENSA/ARPP19 (Gharbi-Ayachi et al. 2010; Mochida et al. 2010). Phospho-ENSA binds specifically to one type of PP2A complex (B55) as a competitive substrate inhibitor (Gharbi-Ayachi et al. 2010; Mochida et al. 2010; Williams et al. 2014). Phospho-ENSA is then slowly dephosphorylated by PP2A, which allows PP2A to reactivate once cyclin–Cdk and Greatwall kinase activity decline later in mitosis (Williams et al. 2014).
This was a particularly important mechanism to decipher because there are only a few protein phosphatase catalytic subunits in the cell, and these have very broad substrate specificity. Their exquisite substrate selection is conferred by incorporating the catalytic subunit into one of many different complexes, each containing a specific targeting subunit (reviewed in Virshup and Shenolikar 2009). This subunit recruits the phosphatase complex to a particular place, often at a specific time in the cell cycle, and also influences substrate binding. An inhibitor that only recognized the catalytic subunit would inhibit multiple different complexes all at the same time, which would not allow for regulation at the spatial and temporal levels that we are beginning to discover in cells. By generating a small molecule inhibitor that targets a specific subcomplex, the cell is able to inhibit only a subpopulation of the PP2A complexes, in both a spatially and temporally controlled manner.
Greatwall and ENSA, and their kinase substrate relationship, are conserved from yeast through to man, but they are adapted to the particular needs of the organism. Thus, it is required for Xenopus extracts to enter mitosis, whereas mammalian cells are able to enter mitosis in the absence of Greatwall, but cannot maintain the mitotic state (Manchado et al. 2010). This may be because ENSA can be phosphorylated directly by cyclin–Cdk complexes (Okumura et al. 2014), and this is sufficient to allow cells to enter mitosis. By contrast, in budding yeast, the Greatwall-ENSA homologs (Rim15 and Igo1 and -2) help to localize and maintain PP2A activity, which itself promotes mitotic entry by antagonizing the inhibitory effect of the Wee1 homolog, Swe1 (Juanes et al. 2013).
PP1 is the other highly abundant phosphatase in cells. As discussed above, in large part, the catalytic subunit of PP1 is regulated by recruiting it to specific proteins at precise times. These proteins have docking sites for PP1 with a core consensus motif of (R/K-V-X-F), (F-x-x-R/K-x-R/K), or (RRVTW) (Bollen et al. 2010; Heroes et al. 2013). In addition, there is a global inhibition of PP1 activity as cells enter mitosis because cyclin B–Cdk phosphorylates the catalytic subunit of PP1 at an inhibitory site on its carboxyl terminus (Wu et al. 2009).
SISTER CHROMATID SEPARATION
Once cells have entered mitosis, their progress through division is largely regulated by ubiquitin-mediated proteolysis, which works by both altering the local balance of protein kinases and phosphatases, and through activating separase, an essential protease that cleaves the cohesin rings holding sister chromatids (Siomos et al. 2001) and centrioles (Schöckel et al. 2011) together. Crucially, the ubiquitin ligase that controls mitosis is itself controlled by a checkpoint mechanism that detects whether kinetochores are properly attached to the mitotic spindle. This is essential to the proper segregation of sister chromatids and, thus, genomic stability.
Ubiquitin-mediated proteolysis is facilitated by E2 enzymes that carry the ubiquitin and an E3 enzyme, or ubiquitin ligase, which binds the E2 and the substrate to bring them in close proximity to each other (reviewed in Hershko and Ciechanover 1998). The E2 then transfers the ubiquitin to the nearest available lysine, which can be on the substrate or an ubiquitin moiety already conjugated to the substrate. Thus, chains of ubiquitin are built up on the substrate, and specific types of chains are recognized by receptors on the 26S proteasome (Pickart 2001). Once a polyubiquitylated substrate binds to the proteasome, it is unfolded by ATPases in the cap of the proteasome and fed into the catalytic cylinder of the proteasome in which the peptide chain is cleaved by multiple proteases (Ciechanover 1994).
The key E3 or ubiquitin ligase that controls mitosis is called the APC/C (reviewed in Barford 2011; Pines 2011), and this works with two E2 enzymes that work in tandem. One E2, Ubc4 in budding yeast, UbcH10, or UbcH5 in animal cells adds the first ubiquitins to the substrate, and a second E2, Ubc1 (budding yeast), or Ube2S (animal) extends the ubiquitin chain (Rodrigo-Brenni and Morgan 2007; Garnett et al. 2009; Williamson et al. 2011). The E2 binds to one of the subunits of the APC/C, APC11. The APC/C has ∼15 subunits, the exact number depends on the organism, and is able to recognize a large number of different proteins, but each at a specific phase of mitosis or the subsequent G1 phase (see Fig. 2) (reviewed in Pines 2006, 2011).
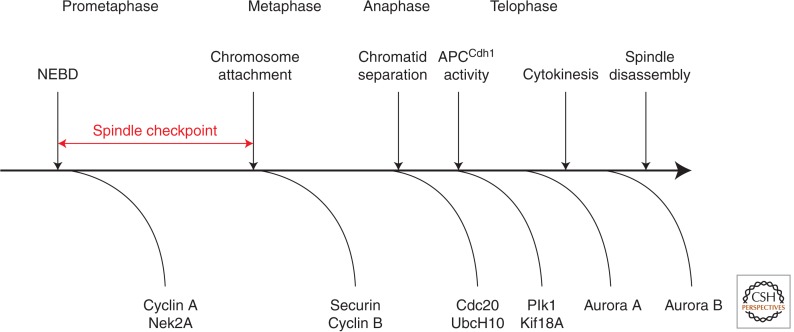
Figure 2.
APC/C-mediated proteolysis in mitosis. Time line of progress through mitosis with the times at which some important APC/C substrates are degraded indicated below the line. The spindle checkpoint prevents the degradation of all the indicated APC/C substrates, except cyclin A and Nek2A. The exchange of Cdc20 for Cdh1 in anaphase is required for the degradation of Aurora A, but not for some other substrates, including Plk1 and Cdc20. NEBD, nuclear envelope breakdown.
The APC/C recognizes its substrates through several different degrons, and recent structural data have begun to reveal how these degrons are recognized. The destruction box (D-box) was the first APC/C degron identified (Glotzer et al. 1991), and this ∼10-amino-acid motif binds to a bipartite receptor composed of the APC10 subunit and a second coactivator protein, Cdc20 (Passmore et al. 2003; Passmore and Barford 2005; Matyskiela and Morgan 2009; Buschhorn et al. 2011; da Fonseca et al. 2011; Izawa and Pines 2011; He et al. 2013). Cdc20 is replaced by Cdh1 later in mitosis in somatic cell cycles. Cdc20 and Cdh1 bind reversibly to the APC/C and, in doing so, activate it by changing its conformation to reposition APC11 to recruit the E2 enzyme in closer proximity to the bound substrate (Chang et al. 2014). Cdc20 and Cdh1 are WD40 family members and the amino-terminal residues of the D-box bind between blades 1 and 7 of the WD40 domain; the carboxy-terminal residues bind to APC10 (He et al. 2013). A second degron, Lys-Glu-Asn, called the KEN-box, binds to the top surface of the WD40 domain (Kraft et al. 2005; Buschhorn et al. 2011; Chao et al. 2012).
The APC/C is activated in mitosis, coincident with NEBD in metazoan cells (den Elzen and Pines 2001; Geley et al. 2001; Wolthuis et al. 2008) and this depends on cyclin B–Cdk activity, but exactly how this works is not clear. Cyclin B–Cdk phosphorylates the APC/C and this enhances the binding of Cdc20, but may not be essential for APC/C activity (Rudner and Murray 2000). Cyclin B–Cdk also phosphorylates Cdc20, but here the effect is more complicated. Some Cdk sites may activate Cdc20, whereas others may inhibit Cdc20 (Listovsky et al. 2000; Kraft et al. 2003; Labit et al. 2012); therefore, it is likely that there is an important interplay between cyclin B–Cdk phosphorylation and Cdc20 or APC/C-associated phosphatase(s), which control APC/C activation.
That cyclin B–Cdk activates the APC/C poses a problem for the cell because cells require cyclin B–Cdk activity to remain in mitosis, yet cyclin B is an APC/C substrate and its degradation inactivates its partner Cdk. This would severely limit the time that a cell could remain in mitosis to build a spindle and then correctly attach all its chromosomes. The solution to this problem is the spindle assembly checkpoint (SAC), which detects any kinetochores that are not correctly attached to microtubules (Hoyt et al. 1991; Li and Murray 1991; Li and Nicklas 1995; Rieder et al. 1995), and prevents the APC/C from recognizing cyclin B and another key substrate, securin (reviewed in Lara-Gonzalez et al. 2012). Although the mechanism of the SAC has not been completely elucidated, the key principles are known. Unattached kinetochores recruit a set of checkpoint proteins that were first identified by genetic screens in yeast (Hoyt et al. 1991; Li and Murray 1991). These are the MAD and BUB proteins, plus the MPS1 kinase. Our current understanding is that the recruitment of (at least) MAD1, MAD2, and BUB1 to unattached kinetochores catalyzes the assembly of an APC/C inhibitor (De Antoni et al. 2005) called the mitotic checkpoint complex (MCC) (Sudakin et al. 2001) through facilitating the binding between MAD2 and CDC20 (see Fig. 3) (De Antoni et al. 2005; Kulukian et al. 2009). The MCC is composed of the MAD2, BUBR1, and BUB3 proteins that are tightly bound to CDC20 (Sudakin et al. 2001; Chao et al. 2012). This complex inactivates CDC20 and prevents it from activating the APC/C because MAD2 binds to a motif on CDC20 that is required to bind the APC/C (Chao et al. 2012; Izawa and Pines 2012). A KEN box in the BUBR1 protein further inactivates CDC20 by binding to the top surface of CDC20, thereby acting as a pseudosubstrate inhibitor (Burton and Solomon 2007; Chao et al. 2012). There must be further mechanisms by which the MCC inhibits the APC/C, however, because the MCC subsequently binds to the APC/C.
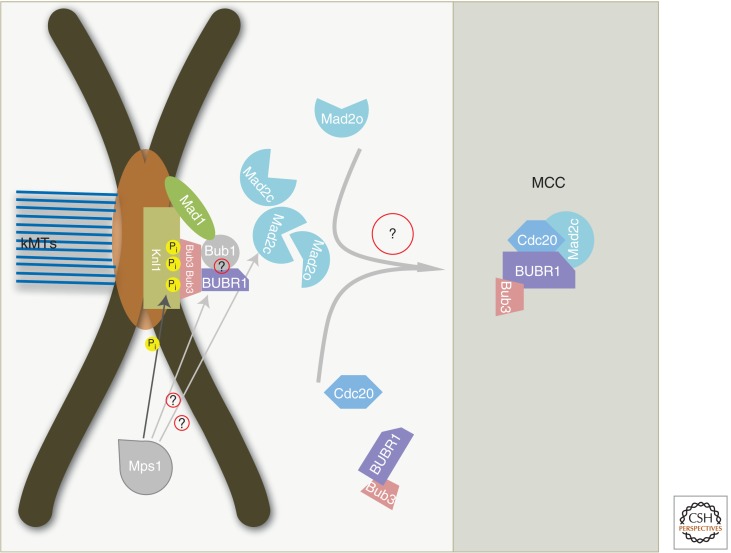
Figure 3.
Simplified model for how the mitotic checkpoint complex (MCC) may be generated at the kinteochore. The Bub1, Bub3, BubR1, Mad1, and Mad2 SAC proteins bind to unattached kinetochore, depending on Mps1 activity. Bub3/Bub1 and Bub3/BubR1 bind Knl1 after phosphorylation by Mps1. Mad1 binds too tightly to one molecule of Mad2 in its closed conformation (Mad2c). The Mad1–Mad2c complex recruits a molecule of Mad2 in its open conformation (Mad2o), and catalyzes a conformational change to Mad2c to enable it to bind Cdc20. Question marks indicate protein–protein interactions and other aspects of MCC assembly that are still incompletely understood.
Despite inhibiting CDC20, the MCC is unable to prevent the destruction of at least two APC/C substrates: cyclin A and Nek2A (den Elzen and Pines 2001; Geley et al. 2001; Hames et al. 2001). How Nek2A escapes inhibition is not yet clear, but depends on its ability to bind directly to the APC/C through its carboxy-terminal dipeptide Ile-Arg motif (Hayes et al. 2006). How cyclin A escapes is now a little clearer; it also depends on the ability of cyclin–Cdk to bind to the APC/C, which it does through the anion-binding Cks subunit that binds phosphorylated APC/C (Wolthuis et al. 2008; Di Fiore and Pines 2010). The second requirement is a direct interaction with CDC20 mediated by the amino terminus of cyclin A (Wolthuis et al. 2008; Di Fiore and Pines 2010). Moreover, cyclin A is able to compete with one of the MCC components, BUBR1, for binding to CDC20 (Di Fiore and Pines 2010). Thus, even while the SAC is generating the MCC to inhibit anaphase, cyclin A can bind sufficient CDC20 to target itself for ubiquitylation by the APC/C. Whether cyclin A has to be degraded in prometaphase is not clear, but a recent study found that persistent cyclin A–Cdk activity in prometaphase could perturb proper sister chromatid separation by altering kinetochore–microtubule interactions (Kabeche and Compton 2013).
The SAC continues to inhibit the APC/C until the last kinetochore attaches to the mitotic apparatus (Rieder et al. 1995). At this point, the SAC is turned off (metaphase), and very quickly after this, the APC/C begins to target cyclin B and securin for degradation (Clute and Pines 1999; Hagting et al. 2002; Collin et al. 2013; Dick and Gerlich 2013). It is still not understood how inactivation of the SAC and activation of the APC/C are so closely coupled; in particular, this implies that the MCC must have a very short half-life, which is difficult to reconcile with the potency of the SAC that can inhibit the APC/C when only a few kinetochores are unattached (Rieder et al. 1995; Collin et al. 2013; Dick and Gerlich 2013). But recent attention has focused on mutual inhibition between the MCC and the APC/C (Reddy et al. 2007), which requires a particular APC/C subunit, APC15 (Mansfeld et al. 2011; Foster and Morgan 2012; Uzunova et al. 2012). In animal cells, a second important player for MCC disassembly is the p31comet protein (Habu et al. 2002; Teichner et al. 2011; Westhorpe et al. 2011), which has recently been implicated in targeting an AAA ATPase to the MCC to initiate its disassembly (Eytan et al. 2014; Wang et al. 2014).
The cell separates its sister chromatids (anaphase) a few minutes after the APC/C begins to degrade cyclin B and securin. This is because both cyclin B–Cdk and securin inhibit the separase protease (Stemmann et al. 2001; Holland and Taylor 2006; Huang et al. 2008) that, once active, cleaves the Scc1 subunit of the cohesin complex, which holds the two sister chromatids together (see Hirano 2015). In animal cells, most of the cohesin complex on chromosome arms is removed by the Plk1 kinase (Hauf et al. 2005), but cohesin complexes remain at the centromere protected by the Shugoshin protein, which recruits an antagonistic PP2A phosphatase (Kitajima et al. 2006; Riedel et al. 2006; Tang et al. 2006). This illustrates the importance of spatial control of phosphatase activity because, clearly, this complex must not be inactivated as cells enter mitosis, but remain active until all the chromosomes have attached to the mitotic spindle.
Mitotic Exit
Anaphase is, effectively, the point of no return in mitosis. After the sister chromatids have separated, the cell is committed to exit mitosis because cyclin B–Cdk levels drop below the level required to keep cells in mitosis, and the cell has to coordinate the reshaping of its subcellular architecture to execute cytokinesis and return to the interphase state. Here, APC/C-mediated proteolysis plays some role, notably in degrading Plk1 and the Aurora kinases (Lindon and Pines 2004; Floyd et al. 2008), but the major part is played by phosphatases. In budding yeast, Cdc14 is important and is activated when the cyclin–Cdk1 levels drop below a threshold that allows Cdc14 to be released from a nuclear inhibitor by a pathway that also requires separase (reviewed in Stegmeier and Amon 2004; Rock and Amon 2009). In most other systems, the PP1 and PP2A complexes are most clearly implicated in controlling anaphase and mitotic exit (Kinoshita et al. 1990; Vagnarelli et al. 2011; Wurzenberger et al. 2012).
Global PP1 activity begins to increase in anaphase as cyclin B–Cdk activity drops because PP1 can autoactivate by removing the inhibitory phosphatase on its carboxyl terminus (Wu et al. 2009). PP1 is targeted to a number of structures in anaphase to help to reshape the subcellular architecture during mitotic exit. For example, Repo-Man protein targets PP1 to chromosomes in anaphase to coordinate chromosome decondensation (Vagnarelli et al. 2011), and the AKAP149 protein targets PP1 to the nuclear envelope to allow nuclear lamin reassembly (Steen et al. 2000). The drop in cyclin–Cdk activity levels in anaphase indirectly reactivates PP2A complexes because Greatwall kinase depends on cyclin–Cdk for its activity. As Greatwall activity declines, so does the rate at which phospho-ENSA is generated, thereby shifting the balance toward PP2A dephosphorylating and inactivating ENSA (Cundell et al. 2013; Williams et al. 2014). Reactivation of PP1, PP2A, and other phosphatases in a spatially and temporally coordinated fashion reshapes the cell to its interphase architecture and resets it to begin the next cycle of DNA replication, or to choose the alternative fates of quiescence or differentiation.
CONCLUDING REMARKS
Many years of research in a variety of powerful model systems has revealed the conserved core of the biochemistry of mitosis, but a number of important questions still remain. We still do not know what finally triggers mitosis in a normally dividing cell. Clearly, there are a number of signaling pathways that are integrated at this decision, but whether cells finally enter mitosis simply when all the negative inputs are turned off, or whether there is a key positive signal(s) that gives the final push, or whether mitosis is initiated once the balance tips between positive and negative feedback loops is unclear. Moreover, it is likely that different signaling pathways can inform the decision in different organisms and, indeed, cell types; when and where an epithelial cell divides differs from a fibroblast or a hepatocyte, for example.
Once the decision has been made (or the balance toward mitosis shifted), the cell undergoes a remarkably quick reorganization of most of its subcellular architecture, on the mechanics of which we have very little understanding. Mass spectrometry screens have identified a myriad of phosphorylation sites in mitosis, but how these cause interphase structures to disassemble and mitotic structures to assemble is a highly complex problem. In silico modeling of the behavior of mitotic spindle components derived from studies with purified proteins have indicated that the mitotic spindle can self-organize from just a few key proteins (see Reber and Hyman 2015), but we have a long way to go to understand the mitotic cell as a whole.
Progress through mitosis depends critically on ubiquitin-mediated proteolysis directed by the APC/C, subject to control by the SAC. Our knowledge of how the APC/C works is advancing at a rapid rate, but it is not yet clear how it can recognize the right protein at the right time, especially in anaphase and after, nor how unattached kinetochores generate the MCC and negatively couple this to microtubule attachment. Similarly, the close temporal coupling between the APC/C and the SAC depends on the biochemistry of MCC assembly and disassembly, which has not yet been clarified. What is needed here are more quantitative data on molecule numbers, and association and dissociation constants, to inform models of how a few unattached kinetochores can generate sufficient MCC to keep the APC/C in check, but still allow rapid APC/C activation once the SAC is turned off.
Finally, understanding the coordination of events during mitotic exit is clearly as great a challenge as understanding mitotic entry, but our knowledge lags behind the wealth of data generated on mitotic kinases and entry. The recent focus on protein phosphatases and their global and local control is already beginning to illuminate this crucial but relatively neglected aspect of cell division.
Footnotes
Editors: Mitsuhiro Yanagida, Anthony A. Hyman, and Jonathon Pines
Additional Perspectives on Mitosis available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Amon A, Surana U, Muroff I, Nasmyth K 1992. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature 355: 368–371. [DOI] [PubMed] [Google Scholar]
- Atherton-Fessler S, Parker LL, Geahlen RL, Piwnica-Worms H 1993. Mechanisms of p34cdc2 regulation. Mol Cell Biol 13: 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin J, Halova L, Ferguson J, Hitchin JR, Lichawska-Cieslar A, Jordan AM, Pines J, Wellbrock C, Petersen J 2014. Torin1-mediated TOR kinase inhibition reduces Wee1 levels and advances mitotic commitment in fission yeast and HeLa cells. J Cell Sci 127: 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barford D 2011. Structural insights into anaphase-promoting complex function and mechanism. Philos Trans R Soc Lond B Biol Sci 366: 3605–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R, Sardon T, Vernos I, Conti E 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell 12: 851–862. [DOI] [PubMed] [Google Scholar]
- Blake-Hodek KA, Williams BC, Zhao Y, Castilho PV, Chen W, Mao Y, Yamamoto TM, Goldberg ML 2012. Determinants for activation of the atypical AGC kinase Greatwall during M phase entry. Mol Cell Biol 32: 1337–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M, Gerlich DW, Lesage B 2009. Mitotic phosphatases: From entry guards to exit guides. Trends Cell Biol 19: 531–541. [DOI] [PubMed] [Google Scholar]
- Bollen M, Peti W, Ragusa MJ, Beullens M 2010. The extended PP1 toolkit: Designed to create specificity. Trends Biochem Sci 35: 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y, Arvai AS, Bernstein SL, Watson MH, Reed SI, Endicott JE, Noble ME, Johnson LN, Tainer JA 1995. Crystal structure of the cell cycle-regulatory protein suc1 reveals a β-hinge conformational switch. Proc Natl Acad Sci 92: 10232–10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Garman EF, Wakatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson LN 1995. The crystal structure of cyclin A. Structure 3: 1235–1247. [DOI] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Lawrie AM, Morris MC, Tunnah P, Divita G, Johnson LN, Endicott JA 1999. Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. J Biol Chem 274: 8746–8756. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ 2007. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev 21: 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, Stark H, Peters JM 2011. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol 18: 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400: 178–181. [DOI] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D 2014. Molecular architecture and mechanism of the anaphase-promoting complex. Nature 513: 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D 2012. Structure of the mitotic checkpoint complex. Nature 484: 208–213. [DOI] [PubMed] [Google Scholar]
- *.Cheeseman IM 2014. The kinetochore. Cold Spring Harb Perspect Biol 6: a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Hurov J, White LS, Woodford-Thomas T, Piwnica-Worms H 2001. Absence of apparent phenotype in mice lacking Cdc25C protein phosphatase. Mol Cell Biol 21: 3853–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Archambault V, Kar A, Lio P, D’Avino PP, Sinka R, Lilley K, Laue ED, Deak P, Capalbo L, et al. 2007. Multiple protein phosphatases are required for mitosis in Drosophila. Curr Biol 17: 293–303. [DOI] [PubMed] [Google Scholar]
- Ciechanover A 1994. The ubiquitin-proteasome proteolytic pathway. Cell 79: 13–21. [DOI] [PubMed] [Google Scholar]
- Clute P, Pines J 1999. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol 1: 82–87. [DOI] [PubMed] [Google Scholar]
- Collin P, Nashchekina O, Walker R, Pines J 2013. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat Cell Biol 15: 1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudreuse D, Nurse P 2010. Driving the cell cycle with a minimal CDK control network. Nature 468: 1074–1079. [DOI] [PubMed] [Google Scholar]
- Cundell MJ, Bastos RN, Zhang T, Holder J, Gruneberg U, Novák B, Barr FA 2013. The BEG (PP2A-B55/ENSA/Greatwall) pathway ensures cytokinesis follows chromosome separation. Mol Cell 52: 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PCA, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D 2011. Structures of APC/CCdh1 with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature 470: 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, Amon A 2004. At the interface between signaling and executing anaphase—Cdc14 and the FEAR network. Genes Dev 18: 2581–2595. [DOI] [PubMed] [Google Scholar]
- *.D’Avino PP, Giansanti MG, Petronczki M 2015. Cytokinesis in animal cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol 15: 214–225. [DOI] [PubMed] [Google Scholar]
- den Elzen N, Pines J 2001. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol 153: 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick AE, Gerlich DW 2013. Kinetic framework of spindle assembly checkpoint signalling. Nat Cell Biol 15: 1370–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Pines J 2010. How cyclin A destruction escapes the spindle assembly checkpoint. J Cell Biol 190: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Deshmukh K, Tanaka Y, Pozniakovsky A, Hunt T 2010. Identification of substrates for cyclin dependent kinases. Adv Enzyme Regul 50: 375–399. [DOI] [PubMed] [Google Scholar]
- Eytan E, Wang K, Miniowitz-Shemtov S, Sitry-Shevah D, Kaisari S, Yen TJ, Liu S-T, Hershko A 2014. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and pp31comet. Proc Natl Acad Sci 111: 12019–12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AM, White LS, Donovan PJ, Piwnica-Worms H 2005. Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol Cell Biol 25: 2853–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd S, Pines J, Lindon C 2008. APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr Biol 18: 1649–1658. [DOI] [PubMed] [Google Scholar]
- Foster SA, Morgan DO 2012. The APC/C subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Mol Cell 47: 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D 2011. A commentary on the G2/M transition of the plant cell cycle. Ann Bot 107: 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno N, den Elzen N, Pines J 1999. Human cyclin A is required for mitosis until mid-prophase. J Cell Biol 147: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR 2009. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol 11: 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J 2010a. Activation of cyclin B1–Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol 189: 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J 2010b. Progressive activation of cyclin B1–Cdk1 coordinates entry to mitosis. Dev Cell 18: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T 2001. APC/C-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol 153: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi-Ayachi A, Labbé J-C, Burgess A, Vigneron S, Strub J-M, Brioudes E, Van-Dorsselaer A, Castro A, Lorca T 2010. The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330: 1673–1677. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138. [DOI] [PubMed] [Google Scholar]
- Grallert A, Patel A, Tallada VA, Chan KY, Bagley S, Krapp A, Simanis V, Hagan IM 2013. Centrosomal MPF triggers the mitotic and morphogenetic switches of fission yeast. Nat Cell Biol 15: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell 104: 83–93. [DOI] [PubMed] [Google Scholar]
- Güttler T, Görlich D 2011. Ran-dependent nuclear export mediators: A structural perspective. EMBO J 30: 3457–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habu T, Kim SH, Weinstein J, Matsumoto T 2002. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J 21: 6419–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagting A, Karlsson C, Clute P, Jackman M, Pines J 1998. MPF localisation is controlled by nuclear export. EMBO J 17: 4127–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagting A, den Elzen N, Vodermaier HC, Waizenegger IC, Peters J-M, Pines J 2002. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J Cell Biol 157: 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM 2001. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A–type D-box. EMBO J 20: 7117–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM 2005. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MJ, Kimata Y, Wattam SL, Lindon C, Mao G, Yamano H, Fry AM 2006. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat Cell Biol 8: 607–614. [DOI] [PubMed] [Google Scholar]
- He J, Chao WCH, Zhang Z, Yang J, Cronin N, Barford D 2013. Insights into degron recognition by APC/C coactivators from the structure of an Acm1–Cdh1 complex. Mol Cell 50: 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroes E, Lesage B, Görnemann J, Beullens M, Van Meervelt L, Bollen M 2013. The PP1 binding code: A molecular-lego strategy that governs specificity. FEBS J 280: 584–595. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A 1998. The ubiquitin system. Annu Rev Biochem 67: 425–479. [DOI] [PubMed] [Google Scholar]
- *.Hirano T 2015. Chromosome dynamics during mitosis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a015792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Taylor SS 2006. Cyclin-B1-mediated inhibition of excess separase is required for timely chromosome disjunction. J Cell Sci 119: 3325–3336. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Trotis L, Roberts BT 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66: 507–517. [DOI] [PubMed] [Google Scholar]
- Huang X, Andreu-Vieyra CV, York JP, Hatcher R, Lu T, Matzuk MM, Zhang P 2008. Inhibitory phosphorylation of separase is essential for genome stability and viability of murine embryonic germ cells. PLoS Biol 6: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Pines J 2011. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat Cell Biol 13: 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Pines J 2012. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J Cell Biol 199: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Kubota Y, den Elzen N, Hagting A, Pines J 2002. Cyclin A– and cyclin E–Cdk complexes shuttle between the nucleus and the cytoplasm. Mol Biol Cell 13: 1030–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Lindon C, Nigg EA, Pines J 2003. Active cyclin B1–Cdk1 first appears on centrosomes in prophase. Nat Cell Biol 5: 143–148. [DOI] [PubMed] [Google Scholar]
- Jacobs HW, Knoblich JA, Lehner CF 1998. Drosophila cyclin B3 is required for female fertility and is dispensable for mitosis like cyclin B. Genes Dev 12: 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP 1995. Structure of a cyclin A–CDK2 complex. Nature 376: 313–320. [DOI] [PubMed] [Google Scholar]
- Juanes MA, Khoueiry R, Kupka T, Castro A, Mudrak I, Ogris E, Lorca T, Piatti S 2013. Budding yeast Greatwall and endosulfines control activity and spatial regulation of PP2ACdc55 for timely mitotic progression. PLoS Genet 9: e1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L, Compton DA 2013. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature 502: 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M 1999. The ran GTPase regulates mitotic spindle assembly. Curr Biol 9: 481–484. [DOI] [PubMed] [Google Scholar]
- Kim SY, Ferrell JEJ 2007. Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell 128: 1133–1145. [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Ohkura H, Yanagida M 1990. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell 63: 405–415. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441: 46–52. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Lehner CF 1993. Synergistic action of Drosophila cyclins A and B during the G2-M transition. EMBO J 12: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters J-M 2003. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J 22: 6598–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM 2005. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell 18: 543–553. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Sillje HH, Korner R, Gruss OJ, Meraldi P, Nigg EA 2002. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol 158: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A, Han JS, Cleveland DW 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell 16: 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG 1992. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell 70: 139–151. [DOI] [PubMed] [Google Scholar]
- Labit H, Fujimitsu K, Bayin NS, Takaki T, Gannon J, Yamano H 2012. Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J 31: 3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH 2005. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 7: 126–136. [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS 2012. The spindle assembly checkpoint. Curr Biol 22: R966–R980. [DOI] [PubMed] [Google Scholar]
- Lee G, White LS, Hurov KE, Stappenbeck TS, Piwnica-Worms H 2009. Response of small intestinal epithelial cells to acute disruption of cell division through CDC25 deletion. Proc Natl Acad Sci 106: 4701–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of Polo-like kinase 1. Curr Biol 17: 304–315. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI 1995. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol 129: 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW 1991. Feedback control of mitosis in budding yeast. Cell 66: 519–531. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB 1995. Mitotic forces control a cell-cycle checkpoint. Nature 373: 630–632. [DOI] [PubMed] [Google Scholar]
- Lindon C, Pines J 2004. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol 164: 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, van Zon W, Karlsson Rosenthal C, Wolthuis RM 2007. Cyclin B1–Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol 5: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listovsky T, Zor A, Laronne A, Brandeis M 2000. Cdk1 is essential for mammalian cyclosome/APC regulation. Exp Cell Res 255: 184–191. [DOI] [PubMed] [Google Scholar]
- Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, Ong SE, Gammeltoft S, Carr SA, Yaffe MB 2007. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J 26: 2262–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Podtelejnikov AV, Groth A, Mann M, Bartek J, Lukas J 2002. Regulation of G2/M events by Cdc25A through phosphorylation-dependent modulation of its stability. EMBO J 21: 5911–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Guillamot M, de Cárcer G, Eguren M, Trickey M, García-Higuera I, Moreno S, Yamano H, Cañamero M, Malumbres M 2010. Targeting mitotic exit leads to tumor regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55α,δ phosphatase. Cancer Cell 18: 641–654. [DOI] [PubMed] [Google Scholar]
- Mansfeld J, Collin P, Collins MO, Choudhary J, Pines J 2011. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat Cell Biol 13: 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Morgan DO 2009. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell 34: 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, Farrell JA, O’Farrell PH 2009. Influence of cyclin type and dose on mitotic entry and progression in the early Drosophila embryo. J Cell Biol 184: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocciaro A, Schiebel E 2010. Cdc14: A highly conserved family of phosphatases with non-conserved functions? J Cell Sci 123: 2867–2876. [DOI] [PubMed] [Google Scholar]
- Mocciaro A, Berdougo E, Zeng K, Black E, Vagnarelli P, Earnshaw W, Gillespie D, Jallepalli P, Schiebel E 2010. Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J Cell Biol 189: 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S, Ikeo S, Gannon J, Hunt T 2009. Regulated activity of PP2A-B55 δ is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J 28: 2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S, Maslen SL, Skehel M, Hunt T 2010. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330: 1670–1673. [DOI] [PubMed] [Google Scholar]
- Muhlhausser P, Kutay U 2007. An in vitro nuclear disassembly system reveals a role for the RanGTPase system and microtubule-dependent steps in nuclear envelope breakdown. J Cell Biol 178: 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA 1991. The substrates of the cdc2 kinase. Semin Cell Biol 2: 261–270. [PubMed] [Google Scholar]
- Nigg EA 1993. Cellular substrates of p34cdc2 and its companion cyclin-dependent kinases. Trends Cell Biol 3: 296– 301. [DOI] [PubMed] [Google Scholar]
- Nurse P 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. [DOI] [PubMed] [Google Scholar]
- Okumura E, Morita A, Wakai M, Mochida S, Hara M, Kishimoto T 2014. Cyclin B–Cdk1 inhibits protein phosphatase PP2A-B55 via a Greatwall kinase-independent mechanism. J Cell Biol 204: 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Okano T, Tachibana K, Kishimoto T 1992. Relocation and distinct subcellular localization of p34cdc2–cyclin B complex at meiosis reinitiation in starfish oocytes. EMBO J 11: 1763–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca FW, Collins MO, Lichawska A, Zegerman P, Choudhary JS, Pines J 2011. Quantitative proteomics reveals the basis for the biochemical specificity of the cell-cycle machinery. Mol Cell 43: 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Barford D 2005. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep 6: 873–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, McCormack EA, Au SW, Paul A, Willison KR, Harper JW, Barford D 2003. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J 22: 786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra D, Wang SX, Kumagai A, Dunphy WG 1999. The Xenopus Suc1/Cks protein promotes the phosphorylation of G2/M regulators. J Biol Chem 274: 36839–36842. [DOI] [PubMed] [Google Scholar]
- Pavletich NP 1999. Mechanisms of cyclin-dependent kinase regulation: Structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol 287: 821–828. [DOI] [PubMed] [Google Scholar]
- Petersen J, Nurse P 2007. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol 9: 1263–1272. [DOI] [PubMed] [Google Scholar]
- Pickart CM 2001. Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533. [DOI] [PubMed] [Google Scholar]
- Pines J 2006. Mitosis: A matter of getting rid of the right protein at the right time. Trends Cell Biol 16: 55–63. [DOI] [PubMed] [Google Scholar]
- Pines J 2011. Cubism and the cell cycle: The many faces of the APC/C. Nat Rev Mol Cell Biol 12: 427–438. [DOI] [PubMed] [Google Scholar]
- Pines J, Hagan I 2011. The Renaissance or the cuckoo clock. Philos Trans R Soc Lond B Biol Sci 366: 3625–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J, Hunter T 1991. Human cyclins A and B are differentially located in the cell and undergo cell cycle dependent nuclear transport. J Cell Biol 115: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangone H, Wegel E, Gatt MK, Yeung E, Flowers A, Debski J, Dadlez M, Janssens V, Carpenter ATC, Glover DM 2011. Suppression of scant identifies Endos as a substrate of Greatwall kinase and a negative regulator of protein phosphatase 2A in mitosis. PLoS Genet 7: e1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Reber S, Hyman AA 2015. Emergent properties of the metaphase spindle. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW 2007. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446: 921–925. [DOI] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al. 2006. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441: 53–61. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol 130: 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JM, Amon A 2009. The FEAR network. Curr Biol 19: R1063–R1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO 2007. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 130: 127–139. [DOI] [PubMed] [Google Scholar]
- Rudner AD, Murray AW 2000. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol 149: 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Pavletich NP 1996. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol 3: 696–700. [DOI] [PubMed] [Google Scholar]
- Santos SD, Wollman R, Meyer T, Ferrell JEJ 2012. Spatial positive feedback at the onset of mitosis. Cell 149: 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöckel L, Möckel M, Mayer B, Boos D, Stemmann O 2011. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol 13: 966–972. [DOI] [PubMed] [Google Scholar]
- Siomos MF, Badrinath A, Pasierbek P, Livingstone D, White J, Glotzer M, Nasmyth K 2001. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr Biol 11: 1825–1835. [DOI] [PubMed] [Google Scholar]
- Steen RL, Martins SB, Tasken K, Collas P 2000. Recruitment of protein phosphatase 1 to the nuclear envelope by A-kinase anchoring protein AKAP149 is a prerequisite for nuclear lamina assembly. J Cell Biol 150: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Amon A 2004. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet 38: 203–232. [DOI] [PubMed] [Google Scholar]
- Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW 2001. Dual inhibition of sister chromatid separation at metaphase. Cell 107: 715–26. [DOI] [PubMed] [Google Scholar]
- Su TT, Sprenger F, DiGregorio PJ, Campbell SD, O’Farrell PH 1998. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev 12: 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 154: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Quadroni M, Frei C, Olma MH, Sumara G, Ricci R, Peter M 2007. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell 12: 887–900. [DOI] [PubMed] [Google Scholar]
- Takizawa CG, Weis K, Morgan DO 1999. Ran-independent nuclear import of cyclin B1–Cdc2 by importin β. Proc Natl Acad Sci 96: 7938–7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H 2006. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell 10: 575–585. [DOI] [PubMed] [Google Scholar]
- Teichner A, Eytan E, Sitry-Shevah D, Miniowitz-Shemtov S, Dumin E, Gromis J, Hershko A 2011. p31comet promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc Natl Acad Sci 108: 3187–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E 1998. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J 17: 2728–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima-Morimoto F, Taniguchi E, Shinya N, Iwamatsu A, Nishida E 2001. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature 410: 215–220. [DOI] [PubMed] [Google Scholar]
- Toyoshima-Morimoto F, Taniguchi E, Nishida E 2002. Plk1 promotes nuclear translocation of human Cdc25C during prophase. EMBO Rep 3: 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunnell NB, Poon AC, Kim SY, Ferrell JEJ 2011. Ultrasensitivity in the regulation of Cdc25C by Cdk1. Mol Cell 41: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova K, Dye BT, Schutz H, Ladurner R, Petzold G, Toyoda Y, Jarvis MA, Brown NG, Poser I, Novatchkova M, et al. 2012. APC15 mediates CDC20 autoubiquitylation by APC/CMCC and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol 19: 1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P, Ribeiro S, Sennels L, Sanchez-Pulido L, de Lima Alves F, Verheyen T, Kelly DA, Ponting CP, Rappsilber J, Earnshaw WC 2011. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev Cell 21: 328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup DM, Shenolikar S 2009. From promiscuity to precision: Protein phosphatases get a makeover. Mol Cell 33: 537–545. [DOI] [PubMed] [Google Scholar]
- Wang K, Sturt-Gillespie B, Hittle JC, Macdonald D, Chan GK, Yen TJ, Liu S-T 2014. Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J Biol Chem 289: 23928–23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Broome M, Hunter T 1995. Regulation of the human Wee1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J 14: 1878–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Westhorpe FG, Straight AF 2015. The centromere: Epigenetic control of chromosome segregation during mitosis. Cold Spring Harb Perspect Biol 7: a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS 2011. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci 124: 3905–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A, Lizarraga SB, Zhang L, Wiese C, Gliksman NR, Walczak CE, Zheng Y 2001. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat Cell Biol 3: 221–227. [DOI] [PubMed] [Google Scholar]
- Williams BC, Filter JJ, Blake-Hodek KA, Wadzinski BE, Fuda NJ, Shalloway D, Goldberg ML 2014. Greatwall-phosphorylated Endosulfine is both an inhibitor and a substrate of PP2A-B55 heterotrimers. Elife 3: e01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Banerjee S, Zhu X, Philipp I, Iavarone AT, Rape M 2011. Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Mol Cell 42: 744–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Wilm M, Karsenti E, Vernos I 2000. TPX2, A novel Xenopus MAP involved in spindle pole organization. J Cell Biol 149: 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthuis R, Clay-Farrace L, van Zon W, Yekezare M, Koop L, Ogink J, Medema R, Pines J 2008. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell 30: 290–302. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Guo JY, Tang W, Yang C-S, Freel CD, Chen C, Nairn AC, Kornbluth S 2009. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat Cell Biol 11: 644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C, Held M, Lampson MA, Poser I, Hyman AA, Gerlich DW 2012. Sds22 and Repo-Man stabilize chromosome segregation by counteracting Aurora B on anaphase kinetochores. J Cell Biol 198: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, et al. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science 278: 1957–1960. [DOI] [PubMed] [Google Scholar]
- *.Yanagida M 2014. The role of model organisms in the history of mitosis research. Cold Spring Harb Perspect Biol 6: a015768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bardes ES, Moore JD, Brennan J, Powers MA, Kornbluth S 1998. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev 12: 2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H, Gruss OJ, Rybina S, Caudron M, Schelder M, Wilm M, Mattaj IW, Karsenti E 2008. Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate. J Cell Biol 180: 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Bastos RN, Barr FA, Gruneberg U 2010. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J Cell Biol 191: 1315–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]