Abstract
Muscle, heart and liver were analyzed in a male subject who succumbed to HSD10 disease. Respiratory chain enzyme analysis and BN-PAGE showed reduced activities and assembly of complexes I, III, IV, and V. The mRNAs of all RNase P subunits were preserved in heart and overexpressed in muscle, but MRPP2 protein was severely decreased. RNase P upregulation correlated with increased expression of mitochondrial biogenesis factors and preserved mitochondrial enzymes in muscle, but not in heart where this compensatory mechanism was incomplete. We demonstrate elevated amounts of unprocessed pre-tRNAs and mRNA transcripts encoding mitochondrial subunits indicating deficient RNase P activity. This study provides evidence of abnormal mitochondrial RNA processing causing mitochondrial energy failure in HSD10 disease.
Keywords: MHBD disease, HSD10 disease, RNase P, MRPP2, RNA processing
1. Introduction
2-methyl-3-hydroxy-butyryl-CoA dehydrogenase (MHBD) deficiency was initially described as an inborn error of isoleucine metabolism with characteristic findings in urine organic acids including 3-hydroxy-2-methylbutyrate and tiglylglycine (Zschocke et al., 2000). In the classical form of the disease, males often present following a precipitating event with lethargy, poor feeding and biochemical findings such as lactic acidosis, hypoglycemia, and hyperammonemia (Zschocke, 2012). Divergent from other organic acidemias, patients with MHBD deficiency have a period of normal development followed by progressive neurodegeneration typically occurring between 6 and 18 months of age. The progressive nature of the disease is characterized by visual and hearing impairments, neurologic dysfunction, a movement disorder and severe cardiomyopathy (García-Villoria et al., 2009; Korman, 2006; Zschocke, 2012).
Although a limited number of families with MHBD deficiency have been identified, a heterogeneous phenotype has emerged including rare neonatal (Perez-Cerda et al., 2005) and late onset forms (Olpin et al., 2002), and females with variable degrees of static psychomotor retardation (Ensenauer et al., 2002; Perez-Cerda et al., 2005; Rauschenberger et al., 2010; Zschocke, 2012). Regardless of the age of presentation, the primary manifestations in MHBD deficiency involve the central nervous system and cardiac dysfunction. Furthermore, the severity of the disease in MHBD deficiency does not correlate with the residual 2-methyl-3-hydroxybutyryl-CoA dehydrogenase enzyme activity suggesting a different mechanism leads for the neurodegenerative course (Rauschenberger et al., 2010).
Purification and analysis of the MHBD enzyme from bovine liver led to identification of the X-linked gene HSD17B10, and patients with MHBD deficiency were subsequently identified to have missense mutations in HSD17B10 (Ofman et al., 2003). HSD17B10 encodes a multifunctional enzyme that can oxidize a number of fatty acid, alcohol, steroid, and branched-chain amino acid substrates (Sriram et al., 2005). The HSD17B10 protein enzymatic function has been referred to as 3-hydroxyacyl-CoA dehydrogenase type 2 (HADH2), amyloid β-peptide-binding alcohol dehydrogenase (ABAD), short chain 3-hydroxyacyl-CoA dehydrogenase (SCHAD), and MHBD (Yang et al., 2005). Recognition of amyloid-binding affinity of HSD17B10 has also implicated the gene product in the pathology of Alzheimer’s disease (Chen and Yan, 2007, 2010).
Despite recognition of the HSD17B10 protein’s involvement in various metabolic pathways, the phenotype of MHBD deficiency is not consistent with impaired isoleucine or branched-chain fatty acid degradation. In 2008, the HSD17B10 gene product was also identified as a component of the mitochondrial RNase P complex (Holzmann et al., 2008). Mitochondrial DNA transcription generates large polycistronic RNAs, which have to be cleaved into its components of mitochondrial mRNAs, tRNAs, and rRNAs. Mitochondrial RNase P initiates this posttranscriptional processing by cleaving the mtDNA-encoded precursor RNA at the 5’ start site of tRNAs. Subsequent cleavage at the 3’ end by RNase Z releases the interspersed pre-tRNA into mature tRNAs and releases the adjacent protein-coding mRNAs of electron transport chain subunits for complexes I, III, IV, and V (Holzmann et al., 2008; Rorbach and Minczuk, 2012; Rossmanith et al., 1995).
Human mitochondrial RNase P is comprised of three protein subunits, MRPP1, MRPP2, and MRPP3, encoded by the genes RG9MTD1, HSD17B10, and KIAA0391 respectively (Holzmann et al., 2008; Sanchez et al., 2011), in contrast to other RNase P enzymes which are ribonucleoproteins. Moreover, MRPP1 together with MRPP2 also functions as a tRNA:m1R9 methyltransferase, a functionality for which HSD17B10 protein is necessary (Vilardo et al., 2012). Methylation of the N9 of both adenosine and guanosine occurs in 19 of 22 mitochondrial tRNAs, without which the tRNAs are unstable and ineffective. MRPP2, interacting with GRFS1, colocalizes in the mitochondrial RNA granules, a substructure inside the mitochondria for RNA processing (Jourdain et al., 2013). The discovery that the MRPP2 subunit is encoded by HSD17B10 has linked MHBD deficiency with mitochondrial gene expression, providing a possible mechanism by which these gene mutations cause a primary mitochondrial disorder.
Here we present data from human tissues that demonstrate abnormal mitochondrial RNA processing in MHBD deficiency, resulting in impaired mitochondrial respiration. We propose that the abnormal mtRNA transcript processing results in a mitochondrial bioenergetics failure which causes central nervous system dysfunction, cardiac failure, lactic acidosis and death in affected males.
2. Case report
The patient was mildly encephalopathic on the first day of life with lactic acidosis of 20 mmol/L (reference range 0.5–2.0 mmol/L), and mild hyperammonemia (166 µM). Additional laboratory studies included an acylcarnitine profile with elevated tiglylcarnitine (C5:1) and 3-hydroxy-isovalerylcarnitine (C5-OH) and urine organic acids with increased 2-methyl-3-hydroxybutyrate, tiglylglycine, and lactate without 2-methylacetoacetate suggestive of 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency. Sequencing of the HSD17B10 gene revealed a hemizygous mutation, c.740A>G ; p.N247S which had been reported previously in patients with MHBD deficiency (Perez-Cerda et al., 2005).
The patient was clinically stable over the first two months of life, but required gastrostomy tube placement for feeding difficulties. A brain MRI obtained at 5 days of age revealed restricted diffusion in the peri-rolandic white matter, and an EEG obtained on DOL 6 showed a mild dysmaturity without evidence for clinical or electrographic seizures. During the first six months, the infant attained appropriate developmental milestones, and was clinically stable. Echocardiogram at 4 days of age revealed a tiny patent ductus arteriosus, small foramen ovale, normal right and left ventricular size and systolic function, and a small pericardial effusion. At 3 months of age, only a tiny patent ductus arteriosus was noted on echocardiogram, and B-type natriuretic peptide was normal.
At six months of age, the child developed a mild gastroenteritis with poor oral intake. He presented the next morning with cardiac failure and Kussmaul breathing. Laboratory studies revealed metabolic acidosis, lactic acidosis, ketosis, hyperkalemia, and very elevated B-type natriuretic peptide (3235 ng/ml, normal < 99). Echocardiogram revealed severely depressed biventricular function with left ventricular shortening fraction of 18% (normal >30%) and biplane ejection fraction of 31% (normal 63–77%). With intensive care supportive measures, he demonstrated slight improvement in his cardiac status, but he developed recurrent and poorly tolerated episodes of tachyarrhythmia, and lactic acidosis triggered by a reduction in his sedation, by warming, or by weaning of ventilator support. These episodes twice required cardiopulmonary resuscitation. Laboratory studies showed anemia, thrombocytopenia, hypocalcemia, hyponatremia, elevated transaminases, and prolonged PT and PTT times. In light of the child’s overall poor prognosis, life-sustaining therapies were withdrawn and the patient died.
3. Methods and Materials
3.1 Pathology and Tissue Procurement
With consent, a metabolic autopsy was performed 45 minutes after death and heart muscle, skeletal muscle, and liver samples were flash frozen and stored at −80°C until analysis. Unfortunately, fibroblasts did not grow. For electron microscopy, tissues were fixed in glutaraldehyde, post-fixed with osmium tetroxide, embedded in epoxy resin, and 80 nm ultrathin sections were post-stained with uranyl acetate and lead citrate. Images were acquired using a Hitachi H-7650 transmission electron microscope at 60 kV accelerating voltage (Hitachi High-Technologies Corp., Tokyo, Japan). Control heart tissue was obtained from age-matched left ventricle tissue donated to the Colorado Multiple Institutional Review Board-approved Heart Transplant Tissue Bank at the University of Colorado. Controls were childhood donor hearts from individuals with normal left ventricular ejection fraction not transplanted for technical reasons, collected in the operating room at the time of explant and flash frozen. Control liver tissues were from transplant donors on resection for reduced size transplantation. Control muscle samples were excess material from biopsies of patients ultimately believed to be unaffected by a metabolic disorder.
3.2 Analysis of mitochondrial respiratory chain proteins and enzyme activities
Respiratory chain enzyme activities for complexes I, II, II+III, III, IV, and citrate synthase were assayed in post 600 g supernatants spectrophotometrically on a Cary 300 spectrophotometer at 30° Celsius in heart, muscle and liver as described with some modifications (Rahman et al., 1996). For complex II, the reaction was started with the addition of 20 µM coenzyme Q1. For complex III, the sample was incubated with buffer and then the reaction was initiated by the addition of both decylbenzylquinol and oxidized cytochrome c. For complex IV, the sample was incubated with buffer and then the reaction was generated by the addition of reduced cytochrome c. Protein content of each sample was determined by Bradford assay (Bio-Rad Laboratories, Inc). For complexes I, II, II+III, and citrate synthase, enzyme activities were calculated as initial rates (nmol/min). For complexes III and IV, enzyme activities were calculated as first order rate constants derived within the first minute of the reaction (Fersht, 1985; SMITH, 1955). All activities were normalized to the total protein content of each sample and expressed as ratios over the activity of citrate synthase and of complex II. Normal ranges were determined from 25 muscle control samples, 16 control livers and, 18 control hearts. A control sample is concurrently analyzed with each assay. In control samples, the natural log of the ratios was normally distributed and the results were then expressed as Z-scores. Isolated mitochondrial membrane fractions of muscle, liver, and heart samples were analyzed by blue native polyacrylamide gel electrophoresis (BN-PAGE) with in-gel activity stain as described (Smet et al., 2005; Van Coster et al., 2001). In this assay, defects of transcription or translation can be identified by the presence for complex V of additional bands of lower molecular weight representing incomplete assembly (Smet et al., 2009).
3.3 RNA isolation and real-time quantitative PCR (RT-qPCR)
RNA was extracted from the patient’s heart, liver and skeletal muscle, as well as from 5 control pediatric hearts and 5 control pediatric muscle specimens. Isolation was performed using the mirVana™ kit (Ambion, Austin, TX, USA) according to manufacturer’s recommendation, and RNA quality was determined by Agilent Bioanalyzer 2100 nucleic acid analysis. Patient liver RNA was of poor quality and was not used in further analysis. Reverse transcription of RNAs to generate cDNAs was performed using the miScript Reverse Transcription Kit (Qiagen, Inc) according to manufacturer’s recommendations. RT-qPCR was performed on an Applied Biosystems 7500 Fast system using primers specific for seven mtDNA-encoded tRNA and mRNA cDNAs and the miScript SYBR Green PCR Kit (Qiagen, Inc). Primer pairs were designed for the three RNAse P subunits, for mitochondrial biogenesis factors, for pre-RNAs (tRNAs and mRNAs for RCE subunits) that span the cleavage sites, and for sites within processed, mature RNA transcripts (Supplemental material Table 1 and Supplemental Figure 1). All RT-qPCR reactions were performed in triplicate and normalized to 18S expression in the tissue. Melting curves were analyzed to ensure specificity of primer pairs.
3.4 Western Blot Analysis
Mitochondrial proteins were analyzed from a post-600 g crude mitochondrial homogenate loading 5 µg protein for separation on a 5–20% gradient polyacrylamide gel for separation with sodium dodecyl sulfate (SDS-PAGE), followed by Western blotting using the appropriate primary antibody and then subsequently incubating with horse radish peroxidase (HRP)-conjugated secondary antibody, and detection by chemiluminesence. The mitochondrial matrix protein citrate synthase was detected using an antibody that was a gift from Jessie M. Cameron, Genetics and Genome Research Program, Hospital for Sick Children, Toronto, Canada. The inner mitochondrial membrane protein adenine nucleotide translocator (ANT1) and the outer mitochondrial membrane protein porin were detected with an antibody obtained from Mitosciences (Eugene, OR, USA). The MRPP2 protein was detected using an antibody from Abcam (Cambridge, MA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. In addition, assembly of mitochondrial complexes was further evaluated by Western blotting after BN-PAGE of 10 µg mitochondrial protein, and probed with an antibody against ATP synthase 5A subunit (Mitosciences, MS502).
3.5 Northern Blot Analysis
Northern blot for tRNA-Met in total RNA from control and patient heart was performed using 500 ng total RNA isolated from tissues. Total RNA was resolved on 6% TBE-urea polyacrylamide gel and transferred to Hybond N+ Nylon. The membrane was incubated with a 5’-32P labeled oligonucleotide probe antisense to tRNA-Met (Supplementary Material Table I) overnight at 42°C in UltraHyb-oligo (Invitrogen), washed with 6XSSC, 0.1% SDS at 42°C for 15 minutes and exposed to a storage phosphor screen for 24 hours.
3.6 Mutation Modeling Analysis
The mutation was modeled as a dimer on the high-resolution protein structure in the Protein Databank number 2023, and active site, and some known clinically severe and mild mutations shown as reference (Rauschenberger et al., 2010).
4. Results
4.1 Pathology findings
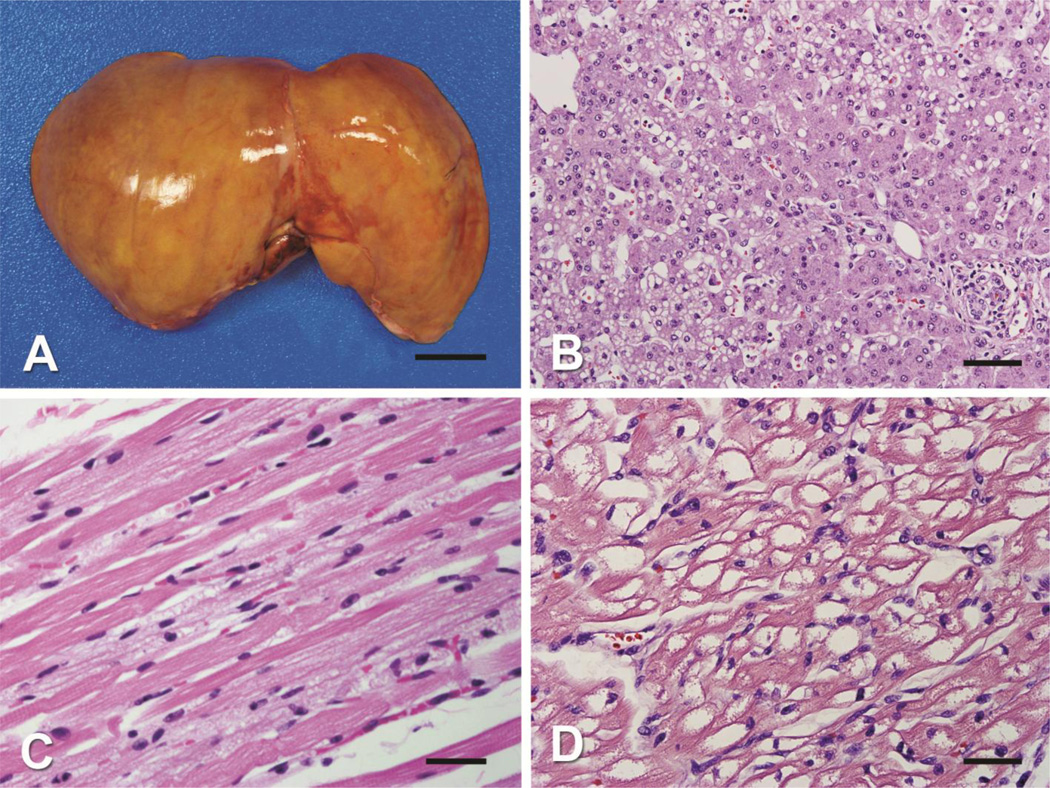
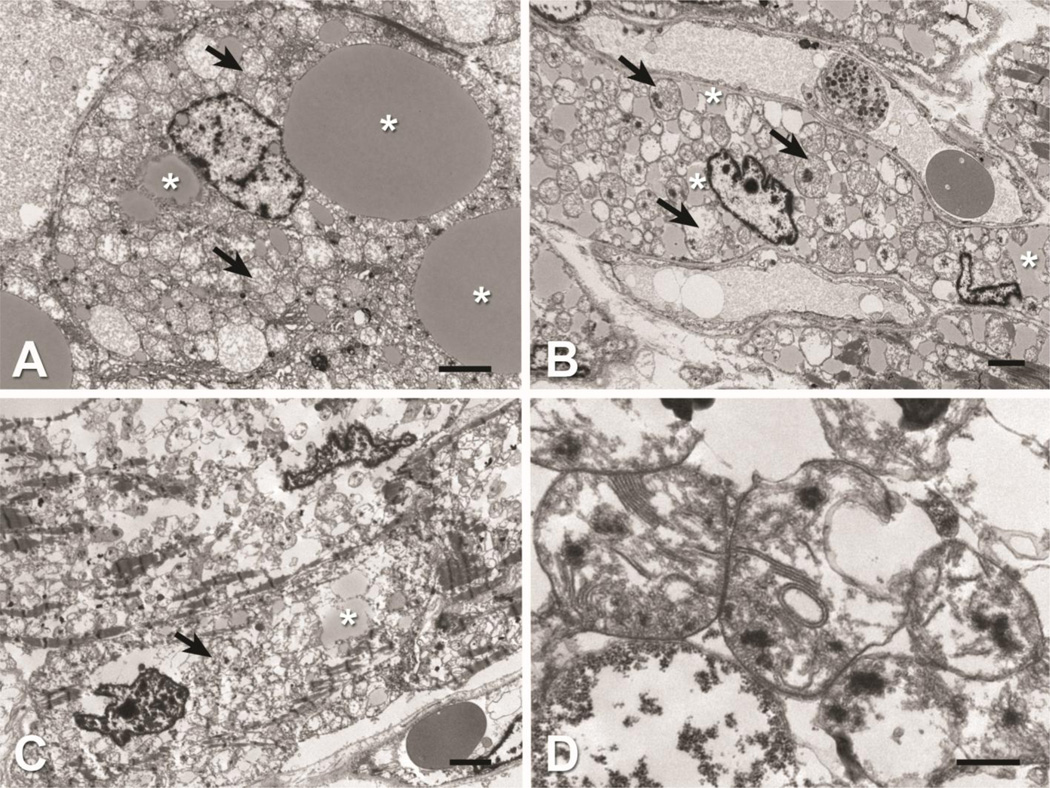
At autopsy, the infant had hepatomegaly (368 g, expected for age 259 g +/− 54 g) and cardiomegaly (120 g, expected for age 40 g +/− 8 g) with a small amount (17 mL) of serous pericardial effusion (Figure 1A). There was extensive vacuolation of tissues, and accumulation of lipid on oil-red O staining in muscle, heart, and in liver, the latter seen primarily in zone 2 as both macro- and microvesicular steatosis (Figures 1B, C, D). The brain weight was normal for age at 735 g and without evidence of demyelination or apparent abnormality. Transmission electron microscopy of liver, skeletal muscle, and heart shows disrupted mitochondrial architecture with strongly increased number of mitochondria and abnormal cristae structure in addition to increased accumulations of intracellular lipid within each of the tissues often displacing normal cellular structures (Figure 2). Enlarged mitochondria with macrovesicular and microvesicular lipid droplets are evident in hepatocytes (Figure 2A). Intracellular lipid, enlarged mitochondria, and paucity of contractile elements are evident in skeletal muscle (Figure 2B). Heart tissue showed severely diminished contractile elements, lipid accumulation, and numerous morphologically abnormal mitochondria (Figure 2C, 2D). Brain tissue showed spongy myelinopathy with myelin pallor and astrocytosis, most prominent in the cerebral cortex at the gray-white matter junction, and to a lesser extent in the brain stem tegmentum and globus pallidus. No inflammation, myelin breakdown, neuronal loss, or microcalcification were identified.
Figure 1. Pathology of tissues of HSD10 patient.
A) A gross photograph of the enlarged liver shows diffuse yellow discoloration (scale bar = 2.5 cm). B) The hepatocytes in zone 2 show micro- and macrovesicular steatosis (H&E stain, original magnification 40X objective, scale bar 100 µm). C) The skeletal muscle shows numerous cytoplasmic vacuoles (H&E stain, original magnification 40X objective, scale bar = 35 µm). D) The cardiomyocytes show extensive cytoplasmic vacuoles (H&E stain, original magnification 40X objective, scale bar = 35 µm).
Figure 2. Electron microscopy of tissues of HSD10 patient.
Transmission electron microscopy show disrupted mitochondrial architecture (arrows) and increased accumulations of intracellular lipid (asterisks) within liver, heart and skeletal muscle tissue. A) Liver: Hepatocyte containing enlarged mitochondria and macrovesicular and microvesicular lipid droplets. B) Skeletal muscle: Intracellular lipid, enlarged mitochondria, and lack of contractile elements within myocyte. C) Heart: Diminished contractile elements and lipid accumulations. D) Heart: High magnification of abnormal mitochondria observed within cardiomyocyte. Note stacked and occasional circular cristae. Scale Bars: A, B, C = 2 µm; D = 0.5 µm.
4.2 Mitochondrial Respiration and Assembly is Disrupted in HSD10 Disease
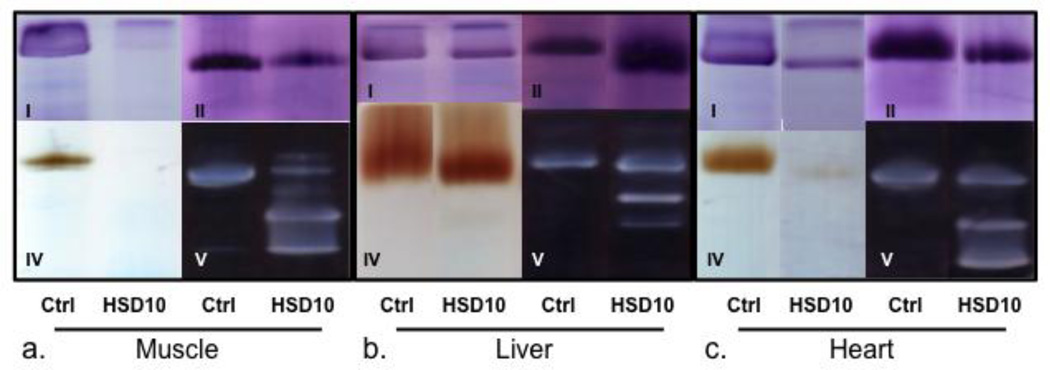
In the clinically most affected tissue, heart, the activities of the respiratory chain enzyme complexes I, III, and IV were severely decreased, whereas the activities of complex II and of citrate synthase were within the range established from tissues of control subjects (Table 1). In muscle, the activities of complex I, III, and IV were also decreased, but the activities of citrate synthase and to a lesser extent, of complex II were increased compared to control subjects. In liver tissue, only the activity of complex I was reduced. However, the activity of citrate synthase was strongly increased, and of complex II was also significantly increased. As a result, the ratio of complex III over these nuclear encoded enzyme activities was below normal. Similarly, on BN-PAGE analysis, the activities of complexes I and IV were decreased to nearly absent in both heart and muscle, but still preserved in liver (Figure 3A, 3B, 3C). In all tissues, extra bands of lower molecular weight of complex V were prominently present indicating substantial amounts of incompletely assembled complex V lacking the incorporation of mitochondrial DNA encoded subunits. The presence of incompletely assembled subunits of complex V was confirmed on Western blot using an antibody against ATP5A (Supplemental Material Figure 2).The presence of these bands in complex V has been shown to occur in patients with defects in mitochondrial transcription or translation (Smet et al., 2009).
Table 1.
Respiratory chain enzyme activities in heart, skeletal muscle, and liver tissues
| HEART: | Activity (controls) | SD | Ratio /CS | SD | Ratio/complex II | SD |
|---|---|---|---|---|---|---|
| Complex I | 2.4 (139.9–335.8) | −14.3* | 2 (140–377) | −14.6* | 8 (517–1069) | −16.4* |
| Complex II | 316.8 (175.7–392.8) | +0.5 | 315 (227–357) | +0.9 | NA | NA |
| Complex III | 4.3 (15.5–189.6) | −3.0* | 4 (12–174) | −2.6* | 13 (43–566) | −2.7* |
| Complex II-III | 69.2 (175.3–570.7) | −3.1* | 69 (185–464) | −4.1* | 218 (714–1620) | −4.5* |
| Complex IV | 1.6 (12.0–27.5) | −8.0* | 2 (12–32) | −8.2* | 5 (42–101) | −8.7* |
| Citrate synthase | 1004.7 (492.8–1451.9) | +0.1 | NA | NA | NA | NA |
| MUSCLE: | ||||||
| Complex I | 3.0 (23.6–74.8) | −7.5* | 3 (98–271) | −9.7* | 26 (285–767) | −9.4* |
| Complex II | 115.2 (49.0–133.4) | +0.9 | 112 (251–573) | −2.3* | NA | NA |
| Complex III | 2.5 (5.7–31.4) | −2.7 | 2 (19–172) | −3.0* | 22 (45–369) | −2.8* |
| Complex II-III | 17.4 (34.2–107.6) | −3.3 | 17 (172–472) | −0.5 | 151 (549–1226) | −5.1* |
| Complex IV | 0.3 (1.1–3.8) | −4.9 | 0 (4–23) | −4.3* | 3 (11–68) | −4.6* |
| Citrate synthase | 1024.5 (159.8–353.3) | +5.3* | NA | NA | NA | NA |
| LIVER: | ||||||
| Complex I | 9.3 (14.4–56.0) | −2.5* | 8 (162–730) | −4.2* | 19 (68–252) | −3.2* |
| Complex II | 492.6 (174.7–309.8) | +3.1* | 399 (2304–3311) | −8.5* | NA | NA |
| Complex III | 13.2 (13.8–27.6) | −0.7 | 11 (128–315) | −3.1* | 27 (50–118) | −1.5 |
| Complex II-III | 130.5 (10.8–107.3) | +1.4 | 106 (138–1062) | −1.4 | 265 (62–383) | +0.6 |
| Complex IV | 1.3 (0.5–3.2) | −0.3 | 1 (6–35) | −0.5 | 3 (3–13) | −0.3 |
| Citrate synthase | 1235.7 (59.5–109.3) | +9.5* | NA | NA | NA | NA |
CS, citrate synthase; NA, not applicable; SD, standard deviation
SD represents how many SD the activity or ratio is above or below the mean established in tissues of control subjects.
SD of >2 is considered significant
Figure 3. Assembly and activity of respiratory chain enzyme complexes on blue native polyacrylamide gel.
Blue Native polyacrylamide gel from a.) muscle, b.) liver and c.) heart tissue showing activity staining for complexes I, II, IV and V as indicated. A) Little complex I or IV are appreciated in muscle, with incomplete complex V assembly as evidenced by lower molecular weight bands but with preserved complex II. B) Increased activity of complex II and incomplete complex V assembly in liver. C) Low levels of complex I and IV, with incomplete complex V assembly in heart. Ctrl- control tissue, HSD10- patient tissue.
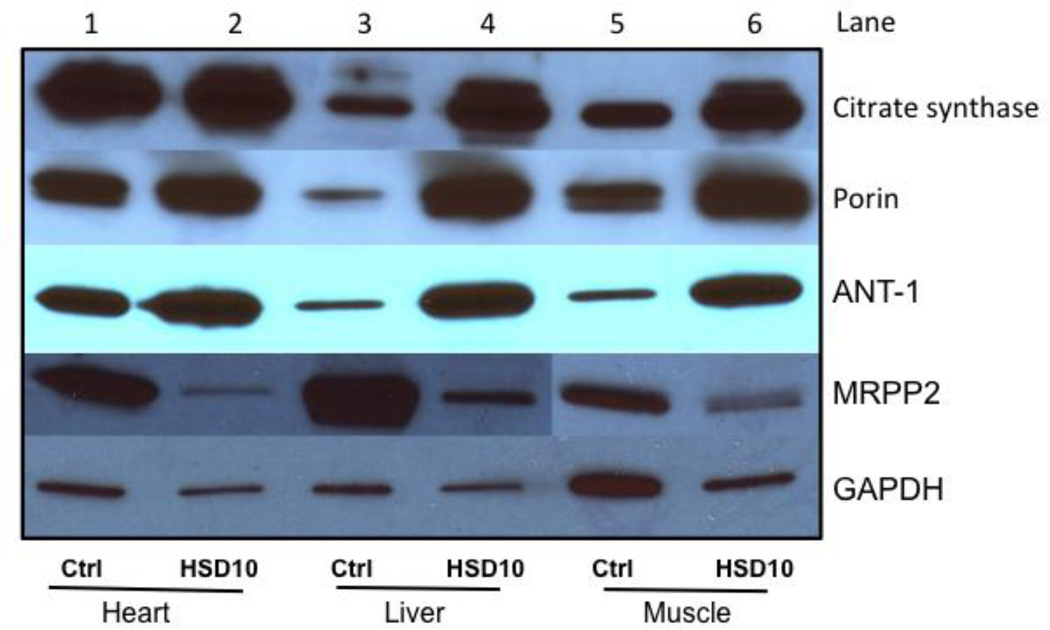
The increase in the enzyme activity of the mitochondrial matrix enzyme citrate synthase in liver and muscle, but not heart, corresponded to a similar increase in the amount of its protein on Western blot, which showed a strong increase compared to control tissue in liver and muscle, but not heart (Figure 4). Similarly, the inner mitochondrial membrane protein adenine nucleotide translocator (ANT-1), and the outer mitochondrial membrane protein porin were increased compared to controls in liver and muscle, but to a lesser extent in heart (Figure 4).
Figure 4. Western blot analysis of mitochondrial proteins and MRPP2 protein expression.
Mitochondrial proteins are separated on a gradient sodium dodecyl sulfate polyacrylamide gel and Western blotted prior to detection. GAPDH = glyceraldehyde 3-phosphate dehydrogenase is shown as loading control. ANT1 = adenine nucleotide translocator, MRPP2 = HSD10 protein
4.3 Relative expression of RNase P subunits is not perturbed in HSD10 disease
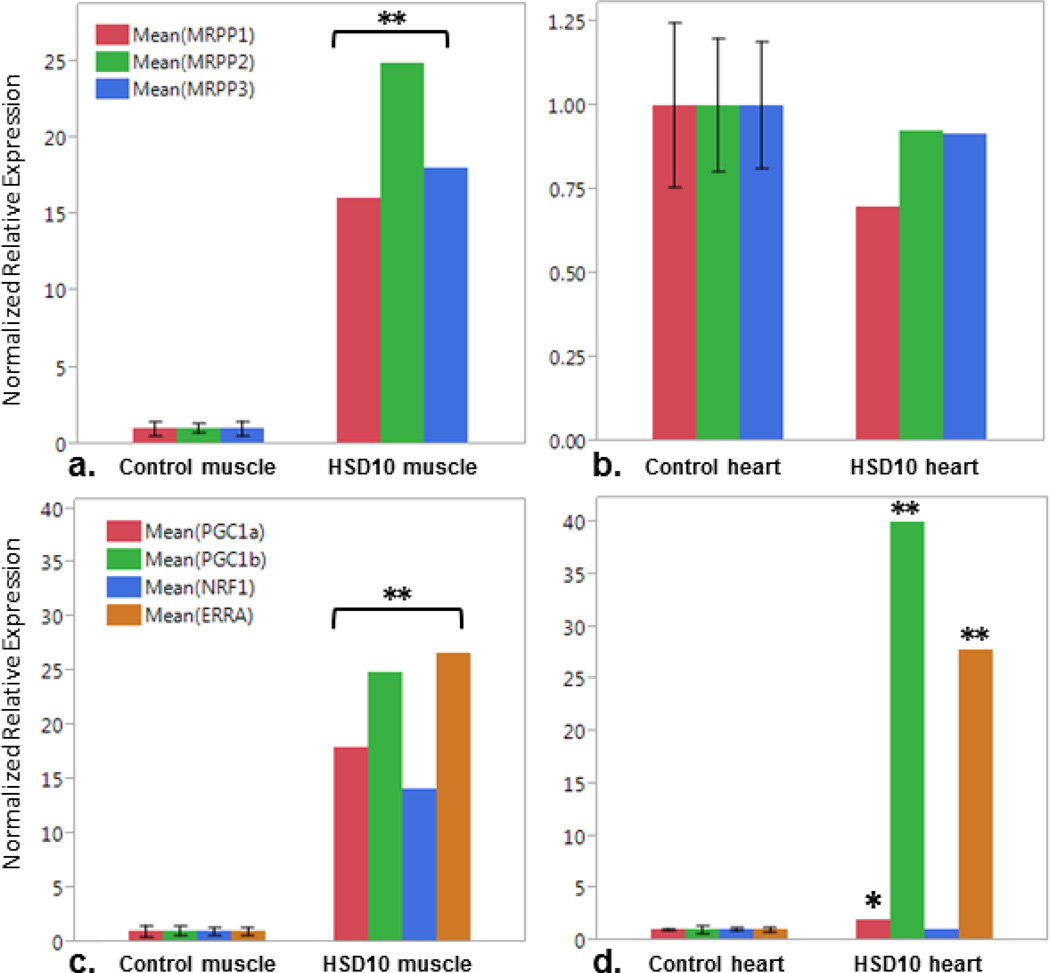
We first confirmed that sufficient RNase P was expressed in the tissues for which we had RNA of sufficient quality for analysis available, heart and skeletal muscle. The mRNA of the subunits MRPP1, MRPP2 and MRPP3, encoded by RG9MTD1, HSD17B10, and KIAA0391 was quantified by RT-qPCR and compared to expression in tissues from five control subjects (Figure 5). In skeletal muscle, there was significantly higher abundance of the RNA of each RNase P subunit in the patient tissue compared to controls; RG9MTD1 showed 16-fold, HSD17B10 25-fold, and KIAA0391 18-fold greater expression (Figure 5a). In contrast, expression of each subunit RNA was not significantly different in heart tissue from the patient when compared to controls (Figure 5b). These data confirm that the HSD17B10 transcript (MRPP2) as well as the other RNase P subunits MRPP1 and MRPP3 are expressed and stable in the tissues analyzed, and are in fact over-expressed in skeletal muscle.
Figure 5. Expression of RNase P subunits and mitochondrial biogesis factors.
RT-qPCR relative expression of RNAse P subunits, MRPP1, MRPP2 (encodes by HSD10 gene), and MRPP3, in a.) muscle and b.) heart tissue. Expression of mitochondrial biogenesis factors PGC1α and β, NRF1 and ERRα in c.) muscle and d.) heart. Comparison between controls (Ctrl, n=5) and patient (HSD10). (*p<0.05, **p<0.0005)
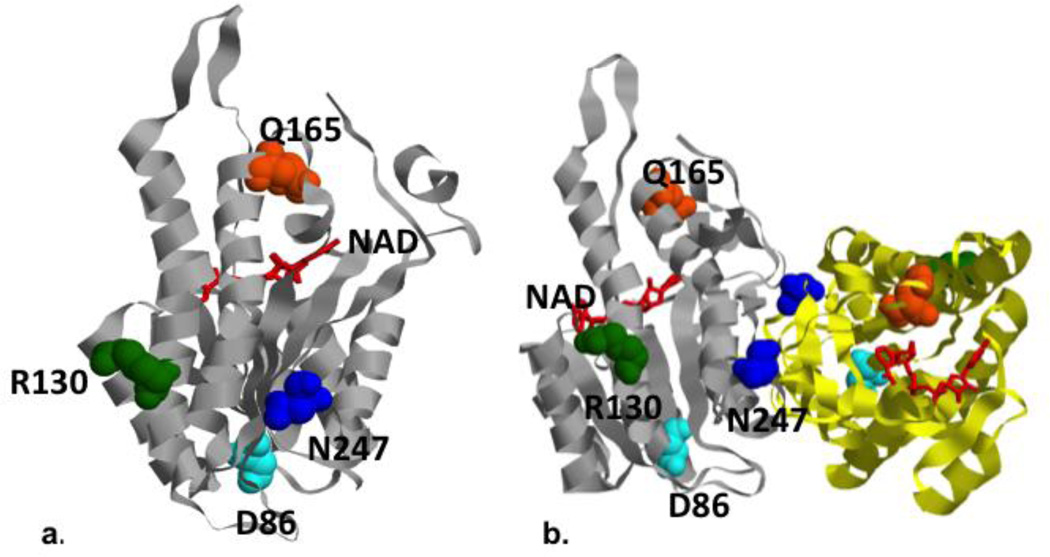
Despite the abundance of its mRNA, the HSD10 protein level (MRPP2) was severely decreased, but not absent, in all three tissues heart, muscle, and liver compared to multiple controls (Figure 4, one representative control shown). When modeled on the high resolution protein structure in the Protein Databank (Pdb ID: 2023), the mutated residue N247 is located near the dimerization domain, and away from the catalytically active cleft for the dehydrogenase function (Figure 6). It is close to the amino acid D86, which was mutated in a clinically severely affected patient with preserved MHBD enzymatic function, and away from the amino acid Q165 which was observed in a clinically mildly affected patient but with completely deficient MHBD enzyme activity (Rauschenberger et al., 2010).
Figure 6. Modeling of human HSD10/MRPP2 mutations.
Locations of mutated residues in the HSD10/MRPP2 protein, and protein dimmer Crystal structure of human HSD10 (pdb: 2023) with the locations of mutated residues highlighted. a.) The HSD10 patient’s mutation, Asn247, (shown in blue) is located on the same side of the enzyme as Arg130 (green) and Asp86 (light blue), away from the NAD (red) binding cleft and Gln165 (orange). b.) The mutations associated with a more severe phenotype (N247 and D86) are adjacent to the homodimerization domain (the third chain of the homodimer is shown in yellow ribbon).
4.4 Mitochondrial biogenesis is disrupted in heart in HSD10 disease
In order to understand why citrate synthase expression and activity are not increased in HSD10 heart compared to liver and muscle; and likewise why MRPP subunits are over-expressed in muscle but not heart, we evaluated the mRNA expression of the mitochondrial biogenesis factors, PGC1α, PGC1β, NRF1 and ERRα in heart and skeletal muscle and compared these to control tissues (Figure 5c, 5d). Each of these 4 transcription factors is highly overexpressed in muscle tissue (Figure 5c, p<0.0005). PGC1β and ERRα are highly overexpressed in HSD10 heart similar to what was seen in muscle (p<0.0005). In contrast, PGC1α is only modestly overexpressed in HSD10 heart, and NRF1 expression is not significantly different from controls (Figure 5d, PGC1α p<0.05).
4.5 Mitochondrial transcripts are abnormally processed in HSD10 disease
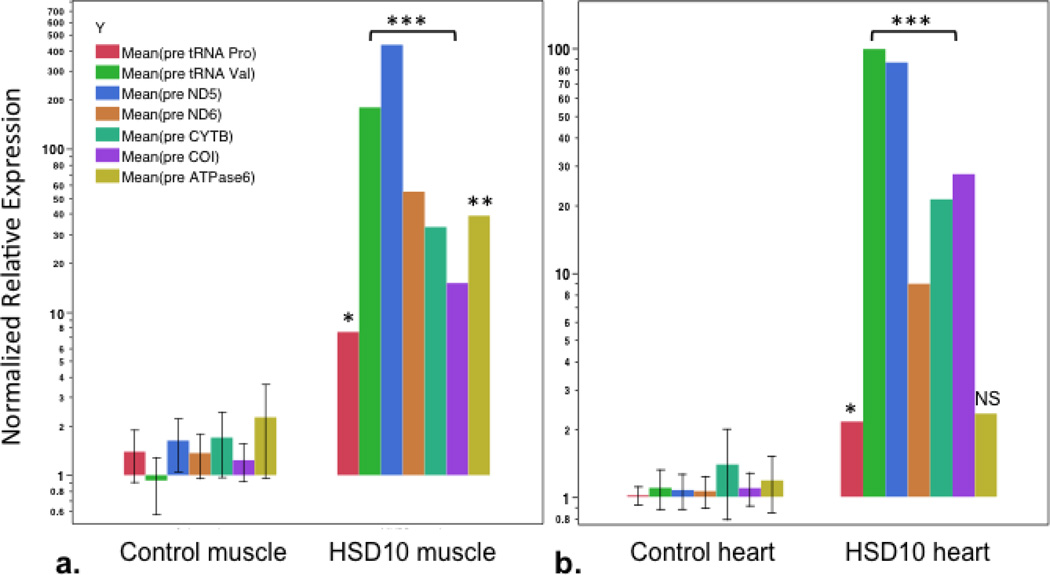
Assessment of mtDNA-encoded RNA transcript cleavage processing was conducted using RT-qPCR for specific mtDNA-encoded respiratory chain subunit mRNAs and tRNAs. Quantification of amplified cDNA sequence spanning cleavage start sites for the following transcripts was performed in skeletal muscle and heart tissues for the following genes: mt.tRNAVal (MT-TV), mt.tRNAPro (MT-TP), the NADH dehydrogenase complex I subunit ND5 (MT-ND5) and subunit ND6 (MT-ND6), cytochrome B (MT-CYB), cytochrome C oxidase subunit I (MT-COI), and mt.ATPase6 (MT-ATP6) (Figure 7, and supplementary figure 1). The quantity of unprocessed sequence that included the start sites for mt.tRNAVal, ND5, ND6, CYB, and COI, were highly overexpressed (15–400 fold higher levels; p values <0.0001) in tissues from the patient as compared to the unprocessed transcript in control muscle and heart tissues. Levels of transcript for mt.tRNAPro were also elevated to a lesser but still significant degree in both tissues (p<0.01), and the mt.ATPase6 transcript was present in levels 17-fold higher in muscle than in control tissue, but not significantly higher than control tissue in heart. The mtATPase6 is not processed by RNAase P at its start site and its level of unprocessed RNA was not different from controls in heart tissue illustrating specificity. These data indicate very high quantities of unprocessed mitochondrial RNA in tissues from the patient with HSD10 disease in comparison to tissues from control subjects.
Figure 7. Quantification of pre-processed mitochondrial RNA transcripts.
RT-qPCR relative quantification of pre-processed mitochondrial RNA transcripts of tRNA Proline (mt.tRNAPro), tRNA Valine (mt.tRNAVal), NADH dehydrogenase 5 and 6 (ND5, ND6), Cytochrome B (CYTB), Cytochrome C oxidase-I (COI), and ATP synthase 6 (ATPase6) from a.) muscle and b.) heart; comparison between control (Ctrl) and patient (HSD10) tissues. Primer pairs on either side of 5’ start site for each gene. P values as noted: *p<0.01, **p<0.0005, ***p<0.0001, NS- not significant.
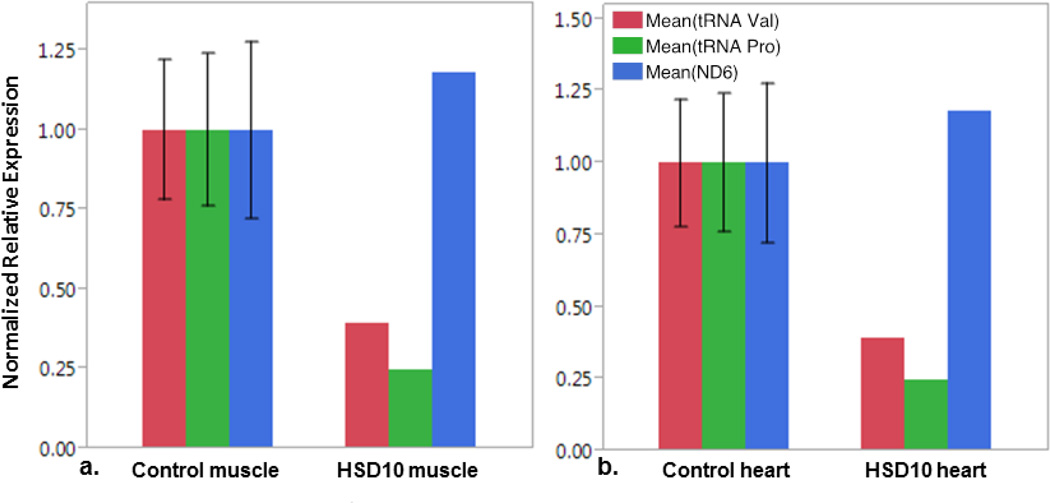
Primer pairs were designed to amplify sequence contained within the processed RNA fragments from some of the same mtDNA genes. These primer pairs amplify cDNA from both processed and unprocessed mitochondrial-encoded RNAs, and reflect total amounts of mitochondrial-encoded RNA in tissues. Expression of sequence within the tRNA valine, tRNA proline, and ND6 message were not significantly different in patient tissues (heart and skeletal muscle) compared to control tissues (Figure 8). These data indicate that mitochondrial RNA is present in tissues from a patient with HSD10 disease at a level that is similar to normal controls; however, a significant proportion of RNAs from the HSD10 patient must remain in an unprocessed form without cleavage at the 5’ site.
Figure 8. Quantification of total mitochondrial RNA transcripts.
RT-qPCR relative quantification of mitochondrial RNA transcripts tRNA Val, tRNA Pro, and NADH dehydrogenase 6 (ND6) as noted in legend in a.) muscle and b.) heart tissue from control subjects (Ctrl) and from the patient (HSD10). Primers sets anneal within gene exon, and therefor amplify both pre and processed transcripts. No statistically difference between controls and patient tissues.
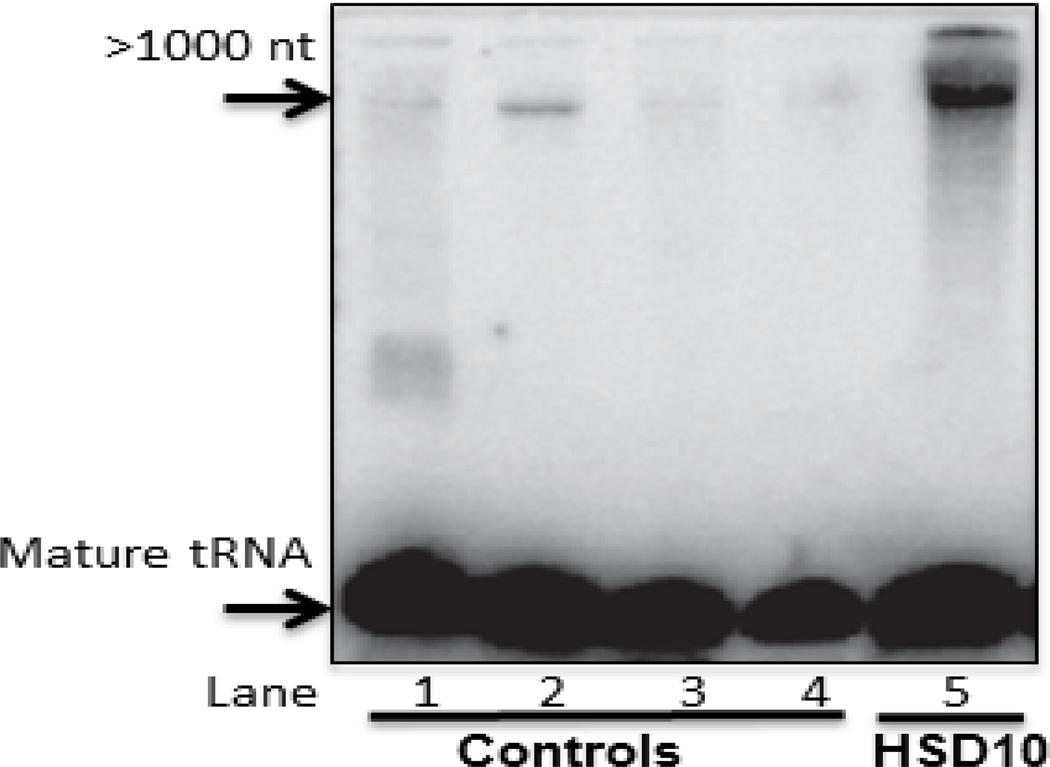
To further validate that mitochondrial RNA cleavage is disrupted in HSD10 disease a Northern blot was performed to look for mature tRNA-methionine (tRNA-Met), and look for evidence of unprocessed RNA in heart tissue from the HSD10 patient. As expected, only mature tRNA-Met is evident in heart tissue from control samples (Figure 9). In the HSD10 patient sample, there is abundant mature tRNA-Met in addition to a significant amount of higher molecular weight unprocessed RNA, greater than 1000 nucleotides in length.
Figure 9. Northern blot analysis of unprocessed mitochondrial tRNA in HSD10 patient heart.
Total RNA from heart tissue of four controls (Lanes 1–4) and HSD10 patient (Lane 5) was isolated and resolved on TBE-urea polyacrylamide gel and transferred to a Hybond N+ membrane and probed with an antisense oligonucleotide probe to tRNA-Met. A band of unprocessed tRNA present in the HSD10 heart sample is strongly increased over controls.
5. Discussion
The multi-functionality of the HSD17B10 gene product and its role in several metabolic pathways has led to a number of hypotheses for the mechanism of the disease originally named 2-methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency (Zschocke, 2012). The recent discovery that the HSD17B10 gene product also functions as a component of mitochondrial RNAse-P, a protein activity critical for mtDNA transcript processing, has provided a plausible link between the clinical course characterized by lactic acidosis, cardiomyopathy, liver dysfunction and neurodegeneration in most patients and the pathophysiology of the disease (Rauschenberger et al., 2010; Zschocke, 2012). Furthermore, Deutschmann et al have demonstrated that knockdown of HSD10 in HeLa cells disrupts processing of mtRNAs, in particular those encoded in the mitochondrial heavy strand (Deutschmann et al., 2014). Accumulation of pre-tRNAs was also demonstrated in skin fibroblasts from patients with HSD10 disease.
We have shown that all the components of the RNase P are adequately expressed in the tissues we were able to examine at the mRNA level. In fact, there is extensive up-regulation of HSD10 message in muscle not seen in heart, similar to other mitochondrial proteins as discussed below. Despite the presence of message, the levels of MRPP2 protein are severely reduced but not absent in the patient’s tissues, likely decreasing its functional activity without abolishing activity. This is probably due to instability of the protein, which we propose may be due to the location of the mutation on the crystal structure near the multimerization domain. The mutated amino acid is close to amino acid D86 which was mutated in a clinically severely affected patient and with a great effect on mitochondrial integrity and cell survival, but with less effect on the MHBD dehydrogenase enzyme activity (Rauschenberger et al., 2010). In contrast, mutations adjacent to the NADH-binding domain are likely to have a greater effect on MHBD enzyme activity, with a lesser mitochondrial respiratory chain deficient phenotype. There are no reported nonsense or frameshift mutations, thus it is likely that total absence of expression is embryonic-lethal (Zschocke, 2012). Although we show that HSD10/MRPP2 protein is reduced in patient tissues, it is not absent and some level of MHBD activity and other enzyme activities such as RNase P is likely preserved explaining the presence of mature mitochondrial RNAs.
Next, we provided evidence that the reduced presence of HSD17B10 protein was associated with deficient mitochondrial RNA processing. Polycistronic mitochondrial RNA precursors of both future tRNA and protein-coding genes accumulated at ten to hundred fold levels in tissues from an affected patient, whereas the total amount of RNA products is unchanged, implying a reduction in the 5’-processed RNA products. This is compatible with defective functioning of RNase P. Similarly, RNAi silencing of MRPP2 had resulted in accumulation of mitochondrial pre-tRNA and reduction in processed mitochondrial tRNA. Such a shortage of mature tRNAs will result in a decreased synthesis rate of proteins from the coding mRNAs, which themselves have not been processed at the normal rate. The reduced availability of tRNAs will impair the translation process for all mitochondrial mRNAs. It was previously shown that reduction of RNase P activity by silencing MRPP1 decreased the amount of rRNA resulting in decreased ribosomal assembly (Holzmann et al., 2008) and resulted in a decreased protein synthesis rate (Sanchez et al., 2011). Thus, a decrease in tRNA and rRNA affects the processing of all mitochondrial DNA encoded proteins due to the requirement for translation, even though the processing of some mRNA occurs independent of RNase P. We see evidence of this in the processing of ATPase 6, which has an overlapping reading frame with ATPase 8, which is located 5’ to ATPase 6 and is devoid of an intervening tRNA. This site is not a target of RNAse P (Jourdain et al., 2013; Sanchez et al., 2011; Temperley et al., 2010). The processing of this site appears more intact, although it is still abnormal in muscle tissue. Overall, the result is a decrease in protein synthesis rate of all mitochondrial DNA encoded protein subunits of the respiratory chain. In addition to its role in RNase P activity, the HSD17B10 / MRPP2 protein is also required for mitochondrial tRNA:m1R9 methyltransferase activity (Vilardo et al., 2012). This activity is independent of HSD10 activity as a short-branched chain fatty acid dehydrogenase such as 2-methylbutyryl-CoA dehydrogenase, does not involve RNA binding, and is independent of the RNase-P activity. Lack of methylation of N9 on guanosine or adenosine disrupts folding and critically destabilizes mitochondrial tRNAs such as mt.tRNALys and would even further impair mitochondrial translation.
We indeed found evidence of decreased mitochondrial protein synthesis. Complex V showed the presence of subcomplexes that contained nuclear encoded subunits but lack mitochondrial DNA encoded subunits on both BN-PAGE with in gel activity stain and with Western blotting. This is typically observed when there is a differential rate of synthesis of the nuclear encoded subunits and the mitochondrial DNA encoded subunits (Smet et al., 2009). The enzyme activities and the amount of protein of the respiratory chain complexes that have mitochondrial DNA encoded subunits, I, III, and IV were severely decreased, particularly in heart, whereas the activity of the nuclear encoded subunits complex II and citrate synthase were not affected. Rather, there was a very striking increase compared to control subjects of complex II and particularly of citrate synthase activity in liver, moderately so in muscle, but not in heart, the most affected tissue.
The amount of proteins of the matrix (citrate synthase), of the inner mitochondrial membrane (adenine nucleotide translocator), and of the outer mitochondrial membrane (porin) was equally increased in liver and muscle. This suggests that there is increased mitochondrial biogenesis in liver and muscle in response to energy deficiency. Indeed several known major mitochondrial biogenesis factors were highly upregulated in muscle. This mitochondrial proliferation is likely the trigger for the observed upregulation of the RNase P-complex subunit transcription in skeletal muscle tissue. In all tissues, on pathology there was a striking increase in the number of mitochondria, consistent with increased biogenesis, and displacing the normal sarcomeric cell structure. This is very apparent in heart tissue, clinically the most severely affected tissue. In contrast, citrate synthase and complex II activities were not increased in this tissue nor were the level of other proteins including ANT1 and porin. The biogenesis response was still strong for PGC1β and ERRα in heart but muted for PGC1α and NRF1. Decreased expression of PGC1α has been shown previously to occur in pathologically (or decompensated) dilated cardiomyopathy (Abel and Doenst, 2011; Ahuja et al., 2013). This differential response of major mitochondrial biogenesis factors in the heart could be related to the observed loss of upregulation and the more severe enzyme deficiency in the heart tissue. This observation may explain the tissue specificity of organ dysfunction, with severe cardiomyopathy at a time where the liver and muscle function remained fairly preserved.
A detrimental effect of HSD10 deficiency on mitochondrial function and morphology has previously been demonstrated in RNAi experiments, in a Xenopus knock-down model of HSD10 disease, in a conditional HSD17B10 knock-out mouse, and in patient samples, further implicating mitochondrial dysfunction as a primary etiology of the disease. The mitochondrial ultrastructure was similarly disrupted with decreased mitochondrial cristae in disrupted Xenopus cells by antisense knock-down, in knock-out animal cell lines, and in some patient fibroblasts (Rauschenberger et al., 2010). The impairment of bioenergetics was shown by decreased 1-14C-pyruvate turnover following HSD17B10 antisense knock-down in Xenopus animal caps. Induction of apoptosis was present in both the Xenopus and the conditional knock-out mouse model (Rauschenberger et al., 2010).
Electron transport chain analysis has been previously reported in a number of patients with HSD10 disease (Table 2). Respiratory chain enzyme analysis in skeletal muscle of two males with MHBD deficiency (Ensenauer et al., 2002) revealed complex I deficiency and combined complex I and IV deficiency (Olpin et al., 2002). It was normal in the skin fibroblasts of two male patients with MHBD deficiency (Perez-Cerda et al., 2005; Sass et al., 2004). This discrepancy may be due to tissue specific expression of the HSD10 protein, or by differential effect in various tissues as demonstrated in this study. Discrepant respiratory chain activity may also relate to the progression of the disease, the location of the mutation, or sex of the patient. HSD10 is an X-linked gene, and respiratory chain enzymes were normal in a female with the same mutation (p.N247S) as the current report (Perez-Cerda et al., 2005), which may be the result of the presence of a normal allele and random X-inactivation.
Table 2.
MHBD Deficiency and Mitochondrial Enzyme Analysis
| Reference | Sex | Genotype | MHBD enzyme activity (controls) nmol/min/mg protein) |
RC Tissue | RC enzyme activity |
|---|---|---|---|---|---|
| Ensenaure et al, 2002 | M | c.388C>T (p.R130C) | 0.57 ± 0.17 (7.10 ± 0.83) | SM | Complex I deficiency |
| Olpin et al, 2002 | M | c.745G>C (p.E249Q) | 0.89 (7.3 ± 0.7) | SM | Complex I and IV deficiency |
| Sass et al, 2004 | M | c.388C>T (p.R130C) | 0 (1.06 ± 0.5) | SF | Normal RC activity |
| Perez-Cerda et al, 2005 | F | c.740A>G (p.N247S) | 3.81 (7.27 ± 1.16) | SM | Normal RC activity |
| Perez-Cerda et al, 2005 | M | c.740A>G (p.N247S) | 1.5 (7.27 ± 1.16) | SF | Normal RC activity |
| Current Report | M | c.740A>G (p.N247S) | 4.9± 0.5 (21.4 ± 0.1; 21.2 ± 0.2)a | SM Liver Heart |
Complex I, IV, V deficiency Complex I, IV, V deficiency Complex I, III, IV, V deficiency |
simultaneous controls
NA, not available; M, male; F, female; SM, skeletal muscle; SF, skin fibroblasts; RC, respiratory chain
In conclusion, our data demonstrate that deficiency of RNA processing due to abnormal RNase P activity plays a central role in the pathology of HSD10 disease. We propose that this is the primary mechanism of mitochondrial failure and of clinical outcome in severely affected patients. Patient tissues show remarkably elevated quantities of unprocessed pre-tRNAs, and elevated levels of unprocessed transcripts. Normal mitochondrial RNA cleavage and translation is necessary for complex I, III, IV and V assembly and function. We have provided evidence that abnormal mitochondrial RNA processing due to known HSD17B10 mutation, in this case a previously reported p.N247S missense change, leads to complex I, III, IV and V deficiency and consequent energy failure. This identifies HSD10 disease as only the third recognized disorder of mitochondrial RNA posttranscriptional processing after mitochondrial poly(A) polymerase deficiency and defects of RNase Z encoded by ELAC2 (Crosby et al., 2010; Haack et al., 2013). Finally, it must be noted that some patients with HSD10 disease do not exhibit a degenerative course and only present with increased 2-methyl-3-hydroxybutyric acid (Rauschenberger et al., 2010). This may be related to the location of the mutation, which in such patients affects the catalytic dehydrogenase site without disrupting any other functions further affirming the central role of mitochondrial RNA processing in the degenerative process. This makes counseling of newly affected patients more complex.
Supplementary Material
Highlights.
A mutation at the dimer interface of HSD10 has resulted in decreased protein levels in the presence of increased mRNA message
A mutation in HSD10 causing reduced HSD10 protein results in deficient RNase P activity.
Deficient RNase P activity results in impaired mtDNA transcript processing with elevated amounts of unprocessed RNAs, which are necessary for mitochondrial translation.
Lack of upregulation of mitochondrial enzymes is associated with the most severe mitochondrial biochemical defect and fatal symptoms.
HS10 disease is the second recognized disorder of mitochondrial RNA processing
Acknowledgements
This work was supported in part by funds from a grant to K.C.C. from the NIH/NCATS Colorado CTSI (UL1 TR000154) and support in part for M.W.F. and J.V.H was provided from Miracles for Mito. Contents are the authors’ sole responsibility and do not necessarily represent official NIH views.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts to declare.
References
- Abel ED, Doenst T. Mitochondrial adaptations to physiological vs. pathological cardiac hypertrophy. Cardiovasc. Res. 2011;90:234–242. doi: 10.1093/cvr/cvr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja P, Wanagat J, Wang Z, Wang Y, Liem DA, Ping P, Antoshechkin IA, Margulies KB, MacLellan WR. Divergent Mitochondrial Biogenesis Responses in Human Cardiomyopathy. Circulation. 2013;127:1957–1967. doi: 10.1161/CIRCULATIONAHA.112.001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JX, Yan SD. Amyloid-beta-induced mitochondrial dysfunction. J. Alzheimers Dis. 2007;12:177–184. doi: 10.3233/jad-2007-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JX, Yan SS. Role of mitochondrial amyloid-beta in Alzheimer’s disease. J. Alzheimers Dis. 2010;2(20 Suppl.):S569–S578. doi: 10.3233/JAD-2010-100357. [DOI] [PubMed] [Google Scholar]
- Crosby AH, Patel H, Chioza BA, Proukakis C, Gurtz K, Patton MA, Sharifi R, Harlalka G, Simpson MA, Dick K, Reed JA, Al-Memar A, Chrzanowska-Lightowlers ZMA, Cross HE, Lightowlers RN. Defective mitochondrial mRNA maturation is associated with spastic ataxia. Am. J. Hum. Genet. 2010;87:655–660. doi: 10.1016/j.ajhg.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschmann AJ, Amberger A, Zavadil C, Steinbeisser H, Mayr JA, Feichtinger RG, Oerum S, Yue WW, Zschocke J. Mutation or knock-down of 17β-hydroxysteroid dehydrogenase type 10 cause loss of MRPP1 and impaired processing of mitochondrial heavy strand transcripts. Hum. Mol. Genet. 2014;23:3618–3628. doi: 10.1093/hmg/ddu072. [DOI] [PubMed] [Google Scholar]
- Ensenauer R, Niederhoff H, Ruiter JPN, Wanders RJA, Schwab KO, Brandis M, Lehnert W. Clinical variability in 3-hydroxy-2-methylbutyryl-CoA dehydrogenase deficiency. Ann. Neurol. 2002;51:656–659. doi: 10.1002/ana.10169. [DOI] [PubMed] [Google Scholar]
- Fersht A. Enzyme Structure and Mechanism. 2nd ed. W H Freeman & Co (Sd); 1985. [Google Scholar]
- García-Villoria J, Navarro-Sastre A, Fons C, Pérez-Cerdá C, Baldellou A, Fuentes-Castelló MA, González I, Hernández-Gonzalez A, Fernández C, Campistol J, Delpiccolo C, Cortés N, Messeguer A, Briones P, Ribes A. Study of patients and carriers with 2-methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency: difficulties in the diagnosis. Clin. Biochem. 2009;42:27–33. doi: 10.1016/j.clinbiochem.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Haack TB, Kopajtich R, Freisinger P, Wieland T, Rorbach J, Nicholls TJ, Baruffini E, Walther A, Danhauser K, Zimmermann FA, Husain RA, Schum J, Mundy H, Ferrero I, Strom TM, Meitinger T, Taylor RW, Minczuk M, Mayr JA, Prokisch H. ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2013;93:211–223. doi: 10.1016/j.ajhg.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Jourdain AA, Koppen M, Wydro M, Rodley CD, Lightowlers RN, Chrzanowska-Lightowlers ZM, Martinou J-C. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013;17:399–410. doi: 10.1016/j.cmet.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korman SH. Inborn errors of isoleucine degradation: a review. Mol. Genet. Metab. 2006;89:289–299. doi: 10.1016/j.ymgme.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Ofman R, Ruiter JPN, Feenstra M, Duran M, Poll-The BT, Zschocke J, Ensenauer R, Lehnert W, Sass JO, Sperl W, Wanders RJA. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency is caused by mutations in the HADH2 gene. Am. J. Hum. Genet. 2003;72:1300–1307. doi: 10.1086/375116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olpin SE, Pollitt RJ, McMenamin J, Manning NJ, Besley G, Ruiter JPN, Wanders RJA. 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency in a 23-year-old man. J. Inherit. Metab. Dis. 2002;25:477–482. doi: 10.1023/a:1021251202287. [DOI] [PubMed] [Google Scholar]
- Perez-Cerda C, García-Villoria J, Ofman R, Sala PR, Merinero B, Ramos J, García-Silva MT, Beseler B, Dalmau J, Wanders RJA, Ugarte M, Ribes A. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency: an X-linked inborn error of isoleucine metabolism that may mimic a mitochondrial disease. Pediatr. Res. 2005;58:488–491. doi: 10.1203/01.pdr.0000176916.94328.cd. [DOI] [PubMed] [Google Scholar]
- Rahman S, Blok RB, Dahl HH, Danks DM, Kirby DM, Chow CW, Christodoulou J, Thorburn DR. Leigh syndrome: clinical features and biochemical and DNA abnormalities. Ann. Neurol. 1996;39:343–351. doi: 10.1002/ana.410390311. [DOI] [PubMed] [Google Scholar]
- Rauschenberger K, Schöler K, Sass JO, Sauer S, Djuric Z, Rumig C, Wolf NI, Okun JG, Kölker S, Schwarz H, Fischer C, Grziwa B, Runz H, Nümann A, Shafqat N, Kavanagh KL, Hämmerling G, Wanders RJA, Shield JPH, Wendel U, Stern D, Nawroth P, Hoffmann GF, Bartram CR, Arnold B, Bierhaus A, Oppermann U, Steinbeisser H, Zschocke J. A non-enzymatic function of 17beta-hydroxysteroid dehydrogenase type 10 is required for mitochondrial integrity and cell survival. EMBO Mol Med. 2010;2:51–62. doi: 10.1002/emmm.200900055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J, Minczuk M. The post-transcriptional life of mammalian mitochondrial RNA. Biochem. J. 2012;444:357–373. doi: 10.1042/BJ20112208. [DOI] [PubMed] [Google Scholar]
- Rossmanith W, Tullo A, Potuschak T, Karwan R, Sbisà E. Human mitochondrial tRNA processing. J. Biol. Chem. 1995;270:12885–12891. doi: 10.1074/jbc.270.21.12885. [DOI] [PubMed] [Google Scholar]
- Sanchez MIGL, Mercer TR, Davies SMK, Shearwood A-MJ, Nygård KKA, Richman TR, Mattick JS, Rackham O, Filipovska A. RNA processing in human mitochondria. Cell Cycle. 2011;10:2904–2916. doi: 10.4161/cc.10.17.17060. [DOI] [PubMed] [Google Scholar]
- Sass JO, Forstner R, Sperl W. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency: impaired catabolism of isoleucine presenting as neurodegenerative disease. Brain Dev. 2004;26:12–14. doi: 10.1016/s0387-7604(03)00071-8. [DOI] [PubMed] [Google Scholar]
- Smet J, Devreese B, Van Beeumen J, Van Coster R. Nondenaturing polyacrylamide gel electrophoresis as a method for studying protein interactions: applications in the analysis of mitochondrial OXPHOS complexes. Cell biology : a laboratory handbook. 2005;4:259–264. [Google Scholar]
- Smet J, Seneca S, De Paepe B, Meulemans A, Verhelst H, Leroy J, De Meirleir L, Lissens W, Van Coster R. Subcomplexes of mitochondrial complex V reveal mutations in mitochondrial DNA. Electrophoresis. 2009;30:3565–3572. doi: 10.1002/elps.200900213. [DOI] [PubMed] [Google Scholar]
- SMITH L. Spectrophotometric assay of cytochrome c oxidase. Methods Biochem Anal. 1955;2:427–434. doi: 10.1002/9780470110188.ch13. [DOI] [PubMed] [Google Scholar]
- Sriram G, Martinez JA, McCabe ERB, Liao JC, Dipple KM. Single-gene disorders: what role could moonlighting enzymes play? Am. J. Hum. Genet. 2005;76:911–924. doi: 10.1086/430799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs--like members of all families, similar but different. Biochim. Biophys. Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Coster R, Smet J, George E, De Meirleir L, Seneca S, Van Hove J, Sebire G, Verhelst H, De Bleecker J, Van Vlem B, Verloo P, Leroy J. Blue native polyacrylamide gel electrophoresis: a powerful tool in diagnosis of oxidative phosphorylation defects. Pediatr. Res. 2001;50:658–665. doi: 10.1203/00006450-200111000-00020. [DOI] [PubMed] [Google Scholar]
- Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase--extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–11593. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-Y, He X-Y, Schulz H. Multiple functions of type 10 17beta-hydroxysteroid dehydrogenase. Trends Endocrinol. Metab. 2005;16:167–175. doi: 10.1016/j.tem.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Zschocke J. HSD10 disease: clinical consequences of mutations in the HSD17B10 gene. J. Inherit. Metab. Dis. 2012;35:81–89. doi: 10.1007/s10545-011-9415-4. [DOI] [PubMed] [Google Scholar]
- Zschocke J, Ruiter JP, Brand J, Lindner M, Hoffmann GF, Wanders RJ, Mayatepek E. Progressive infantile neurodegeneration caused by 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency: a novel inborn error of branched-chain fatty acid and isoleucine metabolism. Pediatr. Res. 2000;48:852–855. doi: 10.1203/00006450-200012000-00025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.