Abstract
PURPOSE
To investigate the presence of TBK1 copy number variations in a large, well-characterized Australian cohort of patients with glaucoma comprising both normal-tension glaucoma and high-tension glaucoma cases.
DESIGN
A retrospective cohort study.
METHODS
DNA samples from patients with normal-tension glaucoma and high-tension glaucoma and unaffected controls were screened for TBK1 copy number variations using real-time quantitative polymerase chain reaction. Samples with additional copies of the TBK1 gene were further tested using custom comparative genomic hybridization arrays.
RESULTS
Four out of 334 normal-tension glaucoma cases (1.2%) were found to carry TBK1 copy number variations using quantitative polymerase chain reaction. One extra dose of the TBK1 gene (duplication) was detected in 3 normal-tension glaucoma patients, while 2 extra doses of the gene (triplication) were detected in a fourth normal-tension glaucoma patient. The results were further confirmed by custom comparative genomic hybridization arrays. Further, the TBK1 copy number variation segregated with normal-tension glaucoma in the family members of the probands, showing an autosomal dominant pattern of inheritance. No TBK1 copy number variations were detected in 1045 Australian patients with high-tension glaucoma or in 254 unaffected controls.
CONCLUSION
We report the presence of TBK1 copy number variations in our Australian normal-tension glaucoma cohort, including the first example of more than 1 extra copy of this gene in glaucoma patients (gene triplication). These results confirm TBK1 to be an important cause of normal-tension glaucoma, but do not suggest common involvement in high-tension glaucoma.
Glaucomas are a group of eye diseases with a common feature of progressive irreversible degeneration of the optic nerve with corresponding loss of the peripheral visual field.1 Glaucomas are the leading cause of irreversible blindness worldwide, and primary open-angle glaucoma is the most prevalent subtype worldwide.2 The main risk factor for glaucoma is elevated intraocular pressure; however, approximately 20%–50% of all primary open-angle glaucoma cases present with normal intraocular pressure range (10–21 mm Hg) and are termed normal-tension glaucoma.3
The genetic contribution to primary open-angle glaucoma is well documented.4 Around half of all primary open-angle glaucoma patients have a positive family history,5 and first-degree relatives of primary open-angle glaucoma patients have an approximately 9-fold increased risk of developing glaucoma.6,7 The first gene identified to be associated with familial normal-tension glaucoma was Optineurin (OPTN) in the GLC1E region on chromosome 10p15-14.8 Subsequent studies reported that mutations in OPTN cause 1%–2% of primary open-angle glaucoma or normal-tension glaucoma.9-12 Despite several studies on the OPTN gene, its exact role in causing primary open-angle glaucoma remains elusive.3,13,14
Recently, a novel genetic locus (GLC1P) on chromosome 12q14 was reported to be linked to normal-tension glaucoma in an African-American pedigree.15 A duplication that spans the TANK binding kinase 1 (TBK1) gene was subsequently detected in this pedigree, as well as in 2 out of 153 unrelated normal-tension glaucoma subjects from Iowa (1.5%), 1 out of 252 unrelated Japanese normal-tension glaucoma patients (0.4%), and 1 out of 96 unrelated patients from New York (1.0%).15-17 These data suggest that abnormal TBK1 dosage (duplication) causes normal-tension glaucoma in these patients. The association between copy number variations of the TBK1 gene and normal-tension glaucoma is supported by several additional observations. First, copy number variations are known to be involved in influencing gene expression and are risk factors for primary open-angle glaucoma (GALC gene)17,18 and Axenfeld-Rieger syndrome (FOXC1 gene),19 as well as a number of diseases such as HIV dementia complex,20 autism,21 and Alzheimer disease.22 Second, TBK1 is specifically expressed in the ganglion cells and the nerve fiber layer of the human retina, which are involved in the pathogenesis of glaucoma.15,23 Third, OPTN binds the TBK1 protein, particularly in the presence of the recurrent severe glaucoma-causing mutation E50K in the OPTN gene.24 Interestingly, 3 known normal-tension glaucoma genes (TBK1, OPTN, and TLR4) each encode proteins that directly interact with each other in a biological pathway that activates autophagy,25,26 a process by which intracellular materials (eg, proteins, organelles, or pathogens) are degraded. Together these data further implicate the role of the TBK1 gene in the pathogenesis of normal-tension glaucoma.
In this study, we aimed to investigate the presence of copy number variations of the TBK1 gene in unrelated normal-tension glaucoma cases and unaffected controls recruited from the Australian population. We also explored the presence of the gene copy number variations in patients with high-tension glaucoma, thus attempting to define an overall contribution of TBK1 copy number variations to glaucoma blindness.
METHODS
Approval of this retrospective cohort study was obtained from the Southern Adelaide Clinical Human Research Ethics Committee. This study has been conducted in accordance with the Declaration of Helsinki and its subsequent revisions. The committee prospectively approved the recruitment of individuals and family members with primary open-angle glaucoma and its subtypes, the collection of blood or saliva samples for deoxyribonucleic acid extraction, the screening for genetic mutations, the data analysis, and the making of genotype and phenotype correlations. Written informed consent was obtained from each individual to participate in this study. Recruitment was conducted through the Australian and New Zealand Registry of Advanced Glaucoma.5 The unaffected control cohort was collected from retirement villages in Adelaide, South Australia, as previously described.27
Each participant was examined by his or her specialist and received a complete eye examination, including slitlamp examination of the anterior chamber, gonioscopy, measurement of central corneal thickness (CCT), visual acuity, intraocular pressure, fundus examination with special attention to optic disc health and size, and automated perimetry. The diagnosis of glaucoma followed the definition of the International Society of Geographical and Epidemiological Ophthalmology described by Foster and associates,28 with optic nerve damage and corresponding visual loss detected in at least 1 eye. Patients recruited in the study and identified as having normal-tension glaucoma followed the same criteria described by Fingert and associates15 (intraocular pressure less than or equal to 21 mm Hg in both eyes, unadjusted for CCT). High-tension glaucoma patients were diagnosed with intraocular pressure greater than 21 mm Hg in at least 1 eye, along with glaucomatous optic nerve and visual field damage. Patients diagnosed with advanced glaucoma presented with either fixation involving visual field loss (at least 2 of the 4 central fixation squares having a pattern standard deviation of less than 0.5% on a reliable Humphrey 24-2 field) or severe global field loss at baseline (mean deviation of less than _22 dB) in at least 1 eye.5 Family members of TBK1 copy number variation carriers were recruited when available. The controls had no evidence of glaucomatous optic nerve damage, intraocular pressure of less than or equal to 21 mm Hg, and no family history of glaucoma, and were slightly older than cases by design for this aging disease. The study was first conducted using a total of 334 unrelated patients with normal-tension glaucoma and 254 unaffected controls. Sixty-three percent of patients (n 1⁄4 212) had advanced normal-tension glaucoma, while the remainder (n 1⁄4 122) had less severe (nonadvanced) normal-tension glaucoma. A positive family history of glaucoma was present in 133 patients (40%).
Venous blood samples were obtained from the participants for the study. Genomic DNA was extracted from peripheral whole blood using the QiaAmp Blood Maxi Kit (Qiagen, Valencia, California, USA). DNA from each subject was tested for TBK1 duplications using TaqMan Copy Number Assays (Life Technologies, Carlsbad, California, USA). The segment of the TBK1 gene was amplified in 4 replicates for each DNA sample. The experiment was conducted using the StepOne Plus real-time polymerase chain reaction instrument, which quantitates the gene of interest, normalized to an endogenous reference gene (RNase P) known to be present in 2 copies in a diploid genome. Evaluation of the copy number of genomic DNA targets was performed using the CopyCaller 2.0 software (Life Technologies) with default settings. For detailed mapping of duplication events, patients with detected TBK1 duplications were analyzed using custom 8x60K SurePrint G3 Human custom comparative genomic hybridization microarrays (Agilent, Santa Clara, California, USA) that interrogated over 55 000 probes in the GLC1P locus that spans 9.5 Mbp between rs12227270 and rs7488555 on chromosome 12q14, using the manufacturer’s protocol.15
To further explore the relationship between 2 apparently unrelated individuals with an identical duplication, we analyzed the haplotypes surrounding the duplication region. The 3 carriers with primary open-angle glaucoma were also part of a previously reported genome-wide association scan (GWAS).29 Along with 590 other participants with primary open-angle glaucoma, they were genotyped on the Omni1 array (Illumina, San Diego, California, USA). The most likely haplotype pair across the duplication region (chr12:64173733–65613733, hg19) in each participant in the GWAS was estimated using Beagle 3.3.2 (http://faculty. washington.edu/browning/beagle/beagle.html).30 The haplotypes across the whole region were visually compared between patients AG624 and AG724. The haplotypes for AG604 with a different duplication were also compared.
Mutation screening of TBK1 was performed on 95 unrelated cases with high-tension glaucoma, 100 unrelated cases with normal-tension glaucoma, and 104 unaffected unrelated controls from Australia. Exome capture was performed using the SureSelect system (Agilent) and paired-end libraries were sequenced on an Illumina HiSeq 2000 by Macrogen Inc (Seoul, South Korea). Reads were mapped to the human reference genome (hg19) using BWA (http://bio-bwa.sourceforge.net/), and duplicates were marked and removed using picard. Variants were called using SAMtools and annotated with ANNOVAR (http://www.openbioinformatics.org/annovar/). Variants were described according to the recommendations of the Human Genome Variation Society (http://www.hgvs.org/) and referenced against the NHLBI Exome Variant Server (http://evs.gs.washington.edu/EVS/[July 2014]), 1000 Genomes,31 and dbSNP v138 databases (http://www.ncbi.nlm.nih.gov/snp).
RESULTS
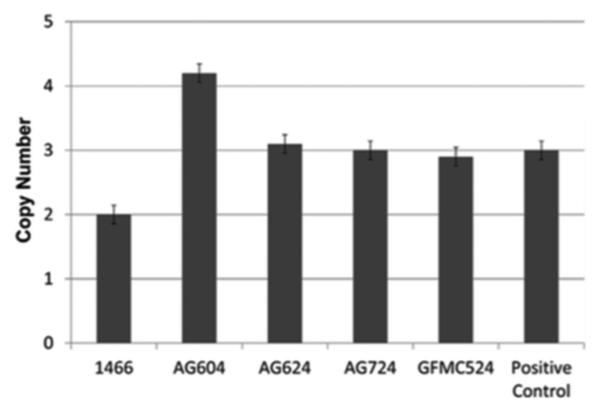
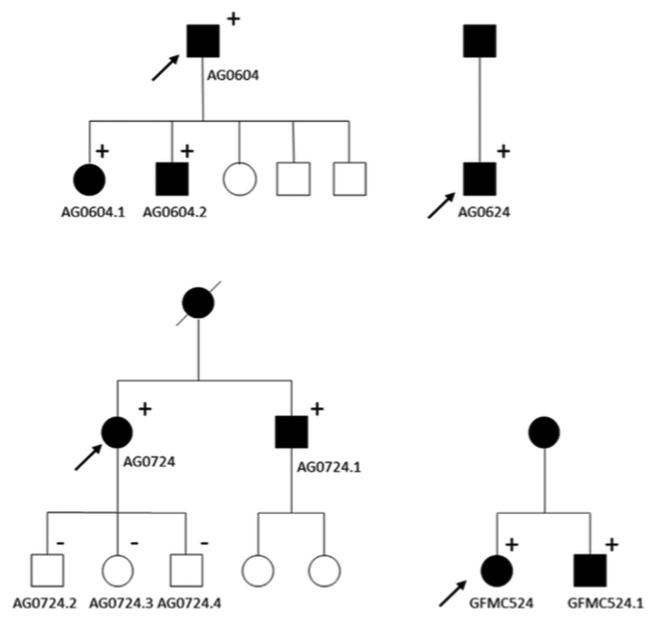
TBK1 copy number variations were detected in 4 of 334 Australian cases with normal-tension glaucoma (1.2%) using quantitative polymerase chain reaction assays (Figure 1). Three unrelated probands, GFMC524, AG604, and AG624, were found to have 3 copies of the gene (1 extra dose), while AG724 participant was found to carry 4 total copies of TBK1 (2 extra doses). No copy number variations were detected in any of the unaffected controls. This rate is similar to previously published data where overlapping copy number variations were found in 1.3% of white normal-tension glaucoma subjects from Iowa and in 1% of normal-tension glaucoma patients from New York.15,17 Affected siblings of the probands AG724 and GFMC524 (AG724.1 and GFMC524.1, respectively) were also shown to carry TBK1 duplications using the quantitative polymerase chain reaction assay. The inheritance of TBK1 copy number variations and normal-tension glaucoma is shown in Figure 2 for these pedigrees. Interestingly, all of the members of 1 pedigree that were diagnosed with normal-tension glaucoma (AG604, AG604.1, and AG604.2) had 2 extra copies of TBK1 (triplication), while previously reported cases had 1 extra copy (Figure 1). All families display an autosomal dominant inheritance pattern of TBK1 copy number variations and normal-tension glaucoma, providing further evidence that these copy number variations are pathogenic. Moreover, these data also suggest that the extra copies of the TBK1 gene are tandem repeats on the same allele, that is, a gene duplication in pedigrees with 1 extra copy of TBK1 and a gene triplication in pedigrees with 2 extra copies.
FIGURE 1.
Assessment of TBK1 gene dosage by quantitative polymerase chain reaction in Australian patients with primary open-angle glaucoma. The x-axis shows the number of copies of the TBK1 gene that were detected in each subject. The normal dosage of 2 copies of TBK1 was detected in the control (Subject 1466). Three probands from unrelated pedigrees (AG624, AG724, and GFMC524) were found to have 1 extra copy of TBK1 (3 total copies) while the proband AG604 was found to have 2 extra copies of TBK1 (4 total copies). AG, advanced glaucoma; GFMC, nonadvanced glaucoma; the positive control was from a normal-tension glaucoma patient previously reported to carry a TBK1 gene duplication.16
FIGURE 2.
Pedigrees of the Australian probands with normal-tension glaucoma carrying the TBK1 copy number variations. Black symbols indicate individuals with normal-tension glaucoma. The proband is indicated by an arrow. Participants carrying a TBK1 duplication or triplication are indicated by a (D), and tested wild-type individuals are denoted with a (L).
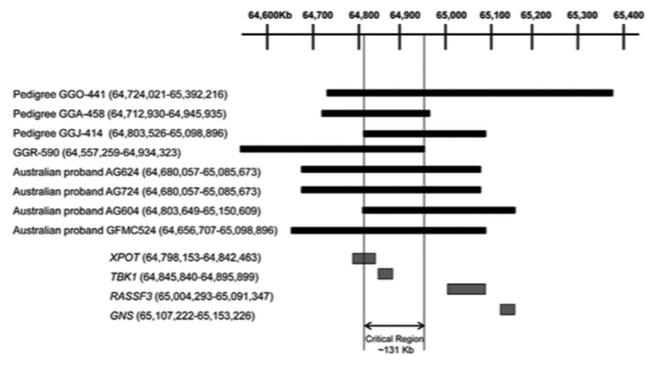
The borders of the copy number variations detected in normal-tension glaucoma probands GFMC524, AG604, AG624, and AG724 were assessed using comparative genomic hybridization (Figure 3). The copy number variations in these Australian normal-tension glaucoma patients are all novel and differ from previously reported copy number variations in the extent of chromosome 12q14 that is involved. Both probands AG624 and AG724 had a duplication, extending from approximately 64,68 Mbp to 65,09 Mbp on chromosome 12. These probands were not known to be related; however, detection of identical copy number variation borders suggested a founder effect. This hypothesis was investigated by comparing haplotypes spanning the TBK1 locus using genotypes obtained from a prior genome-wide association study. These 2 patients were found to share a common haplotype over a greater than 1.4 Mbp segment of chromosome 12q14 (between rs10506464 and rs1909340), which further supports a founder effect in these 2 individuals. A 300 kbp duplication was detected in normal-tension glaucoma proband AG604 that has similar borders as a previously reported copy number variation in a Japanese normal-tension glaucoma patient, GGJ-414 (Figure 3).15 Genotype data were not available to explore a possible founder effect between these 2 patients. When the copy number variations from the current report and those from prior reports were analyzed,15-17 the overlap defined a critical region (w131 kbp), which harbors the TBK1 gene and part of the XPOT gene (Figure 3).
FIGURE 3.
Relative positions of copy number variations detected in Australian cases (AG604, AG624, AG724, and GFMC524) with normal-tension glaucoma in the current report and copy number variations in pedigrees GGO-441, GGA-458, GGJ-414, and GGR-590, which were previously reported.16-18 The extent of each copy number variation in base pairs is in parentheses (hg19 build) and is also depicted by black boxes; the genes encompassed by duplications are depicted as gray boxes.
Table 1 shows the clinical features of patients carrying the TBK1 copy number variations. All of them presented with a family history of glaucoma, large cup-to-disc ratio (ranges from 0.80 to 0.95), and intraocular pressure in the normal range (the maximum recorded untreated intraocular pressure ranged from 12 mm Hg to 17 mm Hg). However, the central corneal thickness varied between the affected probands. GFMC524 had thin CCT (496 mm OD, 505 mm OS), and AG624 had thick CCT (622 mm OD, 621 mm OS). Most of the patients who carry the TBK1 copy number variations were diagnosed at a relatively young age, except for Patient AG604, who was diagnosed at age 60. However, the onset of the disease in Patient AG604 is likely to have been much earlier, given the advanced visual field loss that was observed at the time of diagnosis.
TABLE 1.
Clinical Features of Normal-Tension Glaucoma Patients Carrying TBK1 Copy Number Variations. AG = advanced glaucoma; CCT = central corneal thickness; CDR = cup-to-disc ratio; GFMC = nonadvanced glaucoma; IOP = intraocular pressure; N/A = not available.
| Patient ID | Age of Diagnosis (y) |
Highest IOP OD (mm Hg) |
Highest IOP OS (mm Hg) |
CCT OD (μm) |
CCT OS (μm) |
CDR OD |
CDR OS |
|---|---|---|---|---|---|---|---|
| GFMC524 | 32 | 13 | 13 | 496 | 505 | 0.85 | 0.85 |
| AG604 | 60 | 12 | 12 | N/A | N/A | 0.95 | 0.95 |
| AG624 | 44 | 17 | 17 | 622 | 621 | 0.95 | 0.90 |
| AG724 | 43 | 14 | 14 | 560 | 550 | 0.80 | 0.90 |
To further explore the role of TBK1 copy number variations in primary open-angle glaucoma in general, 1045 patients with high-tension glaucoma were screened by quantitative polymerase chain reaction. No TBK1 copy number variations were identified, indicating that in our dataset TBK1 duplications were found only in normal-tension glaucoma cases. The demographic features and clinical differences between the 2 subtypes of primary open-angle glaucoma and normal controls are illustrated in Table 2.
TABLE 2.
Demographic and Clinical Characteristics of the Australian Cohort Including Normal-Tension Glaucoma Patients, High-Tension Glaucoma Patients, and Normal Unaffected Controls. CCT = central corneal thickness; CDR = cup-to-disc ratio; HTG = high-tension glaucoma patients; IOP = intraocular pressure; NTG = normal-tension glaucoma patients.
| Variables |
|||||
|---|---|---|---|---|---|
| Cohort | Mean Age, Years (SD) | Sex (% Female) | Mean IOP. mm Hg (SD) | Mean CCT. μ (SD) | Mean COR (SD) |
| NTG (n = 334) | 62.4 (11.4) | 61% | 16.9 (2.4) | 510.7 (40.2) | 0.8 (0.1) |
| HTG (n = 1045) | 53.0 (14.6) | 50% | 25.8 (8.9) | 519.8 (43.3) | 0.8 (0.2) |
| Normal controls (n = 254) | 75.9 (8.9) | 58% | 12.8 (2.3) | 544.7 (7.2) | 0.2 (0.12) |
A cohort of 195 Australian cases with primary-open angle glaucoma (including 100 normal-tension glaucoma cases) and 104 unrelated unaffected controls were screened for disease-causing variants in the coding sequence of TBK1. A total of 3 single nucleotide variants were identified. Two synonymous variants were detected in 87 unaffected controls (p.N22N, p.I326I). One previously published,15 nonsynonymous variant (p.V464A) was found in 3 normal-tension and 5 high-tension glaucoma cases and 7 unaffected controls. None of these variants is likely to account for disease.
DISCUSSION
Primary open-angle glaucoma is known to be a genetically heterogeneous disease. Recently, Fingert and associates identified a large duplication within a novel locus (GLC1P) to be associated with primary open-angle glaucoma and its subtype, normal-tension glaucoma, located on chromosome 12q14.15 Although the overlapping duplication encompassed 4 genes (TBK1, XPOT, RASSF3, and GNS), TBK1 was considered the strongest candidate gene for normal-tension glaucoma by virtue of its biology and the critical region defined by duplications in multiple patients. TBK1 is expressed in cells affected by glaucoma (human retina),15 with a clearly documented direct interaction with OPTN, another gene known to cause normal-tension glaucoma.24,32 TBK1 encodes a protein kinase that participates in both autophagy and NF-kB signaling pathways.25,26 The specific mechanism by which TBK1 duplication causes normal-tension glaucoma is still undetermined; however, there is a plausible hypothesis that copy number variations of TBK1 cause a dysregulation either of autophagy or of NF-kB signaling pathways that ultimately leads to apoptosis of retinal ganglion cells and the development of normal-tension glaucoma.7
In addition to confirming the association of the TBK1 gene copy number variations with normal-tension glaucoma, we also provide the first report of a TBK1 gene triplication in a family with normal-tension glaucoma. After making this discovery, we retested our American pedigrees that were previously reported to have TBK1 gene duplications, and we found compelling evidence that 1 of these pedigrees (GGA-458)15 in fact has a TBK1 triplication (data not shown). It is tempting to hypothesize that patients with 2 extra doses of the TBK1 gene may have a more severe phenotype than those patients with 1 extra dose (ie, earlier onset of disease). Moreover, such a genotype-phenotype relationship might be mediated by increased TBK1 gene expression. A previous study reported a 1.60-fold increase in the expression level of TBK1 in patients carrying the duplication than in controls.15 It would be interesting to examine the expression level in our patients with a TBK1 triplication.
The absence of TBK1 duplication in the Australian high-tension glaucoma cohort provides confirmation that TBK1 duplications appear to occur specifically associated with the normal-tension glaucoma subtype. Nonetheless, it is interesting to speculate that the likely phenotype if a TBK1 copy number variation carrier had elevated intraocular pressure by chance could be significantly more severe. The Australian cohort shows a similar rate of mutation in normal-tension glaucoma as other studies of white subjects. TBK1 copy number variations are responsible for 0.4%– 1.3% of normal-tension glaucoma cases in different populations.15-17 As such, it is a rare but easily detectable marker for significant disease, which appears to be highly penetrant within families. Analysis of coding variants in our Australian cohort did not show any mutations likely to cause disease. Considering these data in conjunction with the previously published data by Fingert and associates15 indicates that coding variants in TBK1 are not a common cause of normal-tension glaucoma.
As no mutations have been reported in unaffected controls, this assay may be an important predictor of normal-tension glaucoma risk in select patient populations (ie, strongly familial normal-tension glaucoma, or in relatives of patients with known TBK1 copy number variations), leading to regular clinical screening of carriers of TBK1 copy number variants. Identifying the genetic risk(s) will facilitate early diagnosis and treatment of any complications arising from this condition and prevent the advanced vision loss seen in 3 of our 4 TBK1-associated normal-tension glaucoma cases.
ACKNOWLEDGMENTS
All authors have completed and submitted the icmje form for disclosure of potential conflicts of interest and the following were reported. Stuart Graham received research grants from AllerGen (Sydney, Australia), Biogen (Sydney, Australia), and Novartis (Sydney, Australia), licensed to VisionSearch for electrophysiology testing, and received a travel grant from Alcon (Sydney, Australia). Kathryn Burdon, Emmanuelle Souzeau, Mona Awadalla, and Jamie Craig also receive funding from the Ophthalmic Research Institute of Australia and the Channel Seven Foundation. David Mackey received an unrelated grant from the National Health and Medical Research Council. The remaining authors have no commercial financial support or financial conflict of interests to disclose. The sponsors or funding organizations had no role in the design or conduct of this research. Kathryn Burdon is funded in part by a National Health and Medical Research Council (NHMRC) of Australia Research Fellowship and Jamie Craig is an NHMRC Practitioner Fellow. This work was funded by a grant from the Australian National Health and Medical Research Council Centres of Research Excellence Grant 1023911 (2012–2016). Funding was also provided in part by the National Eye Institute (NEI EY018825 and EY023512), The Polakoff Foundation, and Robert and Sharon Wilson.
REFERENCES
- 1.Quigley HA, Broman AT. The number of people with glaucoma world wide in 2010 and 2020. Br J Ophthalmol. 2006;90(3):262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwon YH, Fingert JH, Kuehn MH, Alward WL. Primary open-angle glaucoma. N Engl J Med. 2009;360(11):1113–1124. doi: 10.1056/NEJMra0804630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quigley HA. Glaucoma. Lancet. 2011;377(9774):1367–1377. doi: 10.1016/S0140-6736(10)61423-7. [DOI] [PubMed] [Google Scholar]
- 4.Hewitt AW, Craig JE, Mackey DA. Complex genetics of complex traits: the case of primary open-angle glaucoma. Clin Experiment Ophthalmol. 2006;34(5):472–484. doi: 10.1111/j.1442-9071.2006.01268.x. [DOI] [PubMed] [Google Scholar]
- 5.Souzeau E, Goldberg I, Healey PR, et al. Australian and New Zealand Registry of Advanced Glaucoma: methodology and recruitment. Clin Experiment Ophthalmol. 2012;40(6):569–575. doi: 10.1111/j.1442-9071.2011.02742.x. [DOI] [PubMed] [Google Scholar]
- 6.Wolfs RC, Klaver CC, Ramrattan RS, van Duijn CM, Hofman A, de Jong PT. Genetic risk of primary open-angle glaucoma. Population-based familial aggregation study. Arch Ophthalmol. 1998;116(12):1640–1645. doi: 10.1001/archopht.116.12.1640. [DOI] [PubMed] [Google Scholar]
- 7.Fingert JH. Primary open-angle glaucoma genes. Eye (Lond) 2011;25(5):587–595. doi: 10.1038/eye.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rezaie T, Child A, Hitchings R, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295(5557):1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 9.Forsman E, Lemmela S, Varilo T, et al. TheroleofTIGRand OPTN in Finnish glaucoma families: a clinical and molecular genetic study. Mol Vis. 2003;9:217–222. [PubMed] [Google Scholar]
- 10.Tang S, Toda Y, Kashiwagi K, et al. The association between Japanese primary open-angle glaucoma and normal tension glaucoma patients and the optineurin gene. Hum Genet. 2003;113(3):276–279. doi: 10.1007/s00439-003-0964-y. [DOI] [PubMed] [Google Scholar]
- 11.Weisschuh N, Neumann D, Wolf C, Wissinger B, Gramer E. Prevalence of myocilin and optineurin sequence variants in German normal tension glaucoma patients. Mol Vis. 2005;11:284–287. [PubMed] [Google Scholar]
- 12.Alward WL, Kwon YH, Kawase K, et al. Evaluation of optineurin sequence variations in 1,048 patients with open-angle glaucoma. Am J Ophthalmol. 2003;136(5):904–910. doi: 10.1016/s0002-9394(03)00577-4. [DOI] [PubMed] [Google Scholar]
- 13.Abu-Amero KK, Azad TA, Spaeth GL, et al. Absence of altered expression of optineurin in primary open angle glaucoma patients. Mol Vis. 2012;18:1421–1427. [PMC free article] [PubMed] [Google Scholar]
- 14.Chi ZL, Akahori M, Obazawa M, et al. Overexpression of optineurin E50K disrupts Rab8 interaction and leads to a progressive retinal degeneration in mice. Hum Mol Genet. 2010;19(13):2606–2615. doi: 10.1093/hmg/ddq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fingert JH, Robin AL, Stone JL, et al. Copy number variations on chromosome 12q14 in patients with normal tension glaucoma. Hum Mol Genet. 2011;20(12):2482–2494. doi: 10.1093/hmg/ddr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawase K, Allingham RR, Meguro A, et al. Confirmation of TBK1 duplication in normal tension glaucoma. Exp Eye Res. 2012;96(1):178–180. doi: 10.1016/j.exer.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritch R, Darbro B, Menon G, et al. TBK1 gene duplication and normal-tension glaucoma. JAMA Ophthalmol. 2014;132(5):544–548. doi: 10.1001/jamaophthalmol.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Gibson J, Wheeler J, et al. GALC deletions increase the risk of primary open-angle glaucoma: the role of Mendelian variants in complex disease. PLoS One. 2011;6(11):e27134. doi: 10.1371/journal.pone.0027134. http://dx.doi.org/10.1371/journal.pone.0027134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strungaru MH, Dinu I, Walter MA. Genotype-phenotype correlations in Axenfeld-Rieger malformation and glaucoma patients with FOXC1 and PITX2 mutations. Invest Ophthalmol Vis Sci. 2007;48(1):228–237. doi: 10.1167/iovs.06-0472. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV1/AIDS susceptibility. Science. 2005;307(5714):1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 21.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman JL, Perry GH, Feuk L, et al. Copy number variation: new insights in genome diversity. Genome Res. 2006;16(8):949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 23.Fingert JH, Darbro BW, Qian Q, et al. TBK1 and flanking genes in human retina. Ophthalmic Genet. 2014;35(1):35–40. doi: 10.3109/13816810.2013.768674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 2008;582(6):997–1002. doi: 10.1016/j.febslet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 25.Wild P, Farhan H, McEwan DG, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333(6039):228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galluzzi L, Kepp O, Kroemer G. Autophagy and innate immunity ally against bacterial invasion. EMBO J. 2011;30(16):3213–3214. doi: 10.1038/emboj.2011.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimasi DP, Burdon KP, Hewitt AW, et al. Genetic investigation into the endophenotypic status of central corneal thickness and optic disc parameters in relation to open-angle glaucoma. Am J Ophthalmol. 2012;154(5):833–842. doi: 10.1016/j.ajo.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 28.Foster PJ, Buhrmann R, Quigley HA, Johnson GJ. The definition and classification of glaucoma in prevalence surveys. Br J Ophthalmol. 2002;86(2):238–242. doi: 10.1136/bjo.86.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011;43(6):574–578. doi: 10.1038/ng.824. [DOI] [PubMed] [Google Scholar]
- 30.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minegishi Y, Iejima D, Kobayashi H, et al. Enhanced optineurin E50K-TBK1 interaction evokes protein insolubility and initiates familial primary open-angle glaucoma. Hum Mol Genet. 2013;22(17):3559–3567. doi: 10.1093/hmg/ddt210. [DOI] [PubMed] [Google Scholar]