Abstract
Insults to nuclear DNA induce multiple response pathways to mitigate the deleterious effects of damage and mediate effective DNA repair. G-protein-coupled receptor kinase-interacting protein 2 (GIT2) regulates receptor internalization, focal adhesion dynamics, cell migration, and responses to oxidative stress. Here we demonstrate that GIT2 coordinates the levels of proteins in the DNA damage response (DDR). Cellular sensitivity to irradiation-induced DNA damage was highly associated with GIT2 expression levels. GIT2 is phosphorylated by ATM kinase and forms complexes with multiple DDR-associated factors in response to DNA damage. The targeting of GIT2 to DNA double-strand breaks was rapid and, in part, dependent upon the presence of H2AX, ATM, and MRE11 but was independent of MDC1 and RNF8. GIT2 likely promotes DNA repair through multiple mechanisms, including stabilization of BRCA1 in repair complexes; upregulation of repair proteins, including HMGN1 and RFC1; and regulation of poly(ADP-ribose) polymerase activity. Furthermore, GIT2-knockout mice demonstrated a greater susceptibility to DNA damage than their wild-type littermates. These results suggest that GIT2 plays an important role in MRE11/ATM/H2AX-mediated DNA damage responses.
INTRODUCTION
Maintaining genomic integrity through DNA repair is of fundamental importance for cellular processes and for the overall life span of an organism. Compromised genomic stability underlies human disorders, including developmental defects, immune deficiency, cancer, and neurological disease. The human central nervous system (CNS), comprising mostly postmitotic tissue, is profoundly affected by DNA repair deficiencies. Defective DNA repair in mature neural tissues is linked to premature aging (Werner's/Bloom syndrome) as well as to neurodegenerative diseases, such as Alzheimer's disease and amyotrophic lateral sclerosis (1, 2). One of the syndromes linking DNA damage and neurodegeneration first to be identified was ataxia telangiectasia (A-T). Patients with A-T have severe neurodegeneration and an extreme sensitivity to ionizing radiation (IR) (1, 3, 4). A-T established a compelling link between the failure of responses to DNA double-strand breaks (DSBs) and central neurodegenerative disorders. A-T was subsequently found to result from the mutation of a single gene, ataxia telangiectasia, mutated (ATM). The ATM gene encodes a 370-kDa protein that belongs to the phosphoinositide 3-kinase (PI3K) superfamily (5). The ATM kinase, however, phosphorylates proteins rather than lipids (6, 7) and is crucial for the initiation of signaling pathways in mammalian cells following exposure to IR and other agents that introduce DSBs into DNA. The ATM protein kinase is one of the key factors in DNA DSB repair. ATM typically exists as an inactive homodimer, and exposure to IR induces intermolecular autophosphorylation at serine-1981 (ATM-pS1981), causing homodimer dissociation into active monomers through the MRE11-RAD50-NBS1 (MRN) complex at DSB sites (marked by H2AX phosphorylation at serine-139 [γ-H2AX]) (8, 9). The subsequent DNA damage response (DDR) cascade transduces signals to downstream targets that initiate cell cycle arrest, DNA repair, or apoptosis. ATM forms just one component of DNA damage repair complexes, and more than 30 ATM substrates that maintain genome stability and reduce the risk of disease have been identified, including NBS1 (10, 11), p53 (2, 3), CHK1/CHK2 (12, 13), BRCA1 (14), SMC1 (15), BID (16), FANCD2 (17), and H2AX (18). The phosphorylation of these targets has been shown to be critical for their function in DDR cascades.
G-protein-coupled receptor kinase-interacting protein 2 (GIT2) is one of the members of the ADP-ribosylation factor (Arf) GTPase-activating protein (GAP) subfamily (19). GIT proteins are multidimensional molecular scaffolds that serve as regulators of G-protein-coupled receptor (GPCR) internalization (20, 21), cell migration (22, 23), and Cdc42-mediated focal adhesion turnover (24). In the immune system, GIT2 is necessary for directional chemotaxis, suppression of superoxide production in GPCR-stimulated neutrophils, and regulation of chemokine-mediated motility of double-positive thymocytes (25). GIT2 is necessary for the orientation of superoxide production toward chemoattractant sources, and the loss of GIT2 in vivo leads to an immunodeficient state (26). In neuronal tissue, an analog of GIT2, GIT1, localizes to both pre- and postsynaptic terminals in hippocampal neurons, and its downregulation/mislocalization results in aberrant dendritic spine morphogenesis and synapse formation (27, 28). Furthermore, GIT1 promotes α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor targeting in primary hippocampal neurons (29) and mediates ephrin-B signaling during spine formation (30). Currently, less is known about the neuronal functions of GIT2, despite the fact that both GIT2 and GIT1 are widely expressed and have predominantly overlapping expression patterns throughout the mouse brain (31). Recently, neuronal expression of GIT2 across multiple mammalian species has been shown to be modulated by oxidative stresses associated with the aging process (32). In addition, we have recently demonstrated that GIT2 acts as a functional keystone factor in aging-related physiological processes (33). As the aging process is closely linked to the stability/repair of DNA, we investigated the potential role of GIT2 in the DDR. We found that IR or cisplatin treatment of neuroblastoma cells increases GIT2 expression in the nucleus. Elevated GIT2 promotes the repair of damaged DNA, while GIT2 silencing by RNA interference (RNAi) diminishes repair. GIT2 associates and colocalizes with multiple DDR complex proteins, including γ-H2AX, MRE11, and ATM, to form nuclear foci in response to DNA damage. GIT2 is phosphorylated by ATM at T195 and S384 upon DNA damage. In addition, we found that GIT2 promotes DNA repair by facilitating the coordinated regulation and protein complex stabilization of repair proteins through a poly(ADP-ribose) (PADPR) polymerase (PARP)-linked process. Mice with a gene trap abolition of GIT2 expression demonstrated an age-related heightened susceptibility to DNA-damaging insults compared to that of age-matched wild-type (WT) mice. Our findings suggest that GIT2 plays important roles in the repair of damaged DNA and can trophically regulate the aging process.
MATERIALS AND METHODS
Cell culture.
Cells of the ATM+/+ GM0637 simian virus 40-transformed human fibroblast cell line and ATM−/− GM5849 cells were cultured in alpha minimum essential medium with 10% fetal bovine serum, 1% glutamine, 1% penicillin-streptomycin, 1% vitamin E, and 1% nonessential amino acids. H2AX+/+ and H2AX−/− immortalized mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, sodium pyruvate, and glutamine at 37°C with 5% CO2. Human neuroblastoma SH-SY5Y cells were grown in DMEM supplemented with 10% fetal bovine serum (Gibco), 2 mM l-glutamine, penicillin (20 units/ml), and streptomycin (20 mg/ml) at 37°C in a saturated humidity atmosphere containing 95% air and 5% CO2.
GIT2-KO mice.
GIT2 gene-trap-knockout (GIT2-KO) mice were genotyped by PCR using genomic DNA isolated from tail clips as previously described (34). Amplification was carried out by standard PCR protocols. Primers used to screen the mice for either WT or GIT2-KO status were forward primer 5′-TCTCCTGGAACTCAGGGATT and reverse primers 5′-CATTTCAGAGTCTGCTGCCTTA for wild-type (WT) mice and 5′-GGCTACCGGCTAAAACTTGA for GIT2-KO mice. Male wild-type (C57BL/6) and GIT2-KO mice were separately housed in groups in temperature-controlled (22°C) and humidity-controlled (45%) rooms with a 12-h light and 12-h dark cycle and were given food and water ad libitum. Four individual mice were employed for each specific experimental genotype group. Experiments were conducted during the light phase of the light-dark cycle, in accordance with NIH guidelines. Animal care and experimental procedures followed NIH guidelines and were approved by the National Institute on Aging Animal Care and Use Committee (protocol numbers 321-LMBI-2013, 432-LCI-2015, and 433-LCI-2015).
Plasmids and siRNA.
Flag-tagged plasmids pBK GIT2-His, GIT2, and GIT2 R39A (Arf GAP-dead mutant) have been previously described (20). Flag-tagged plasmid pcDNA3 ATM was provided by Didier Trono (National Center of Competence in Research, Switzerland). Small interfering RNA (siRNA) specific for human GIT2 (Santa Cruz) was a pool of three target-specific 19- to 25-nucleotide siRNAs. The control siRNA (siRNA-A) consisted of a scrambled sequence that did not lead to the specific degradation of any cellular mRNA. Control siRNA and siRNAs against H2AX, mediator of DNA damage checkpoint protein 1 (MDC1), and RNF8 were purchased from Dharmacon (Thermo Scientific). Cells were transfected with siRNA oligonucleotides using the Lipofectamine RNAiMAX reagent (Invitrogen, Life Technologies) according to the manufacturer's protocol. See the materials and methods in the supplemental material for the sequences.
IR treatment of cells or mice.
SH-SY5Y cells, human fibroblasts, and MEFs were irradiated with a 137Cs IR source. Cells were seeded at 70% confluence 24 h before irradiation. Cells were γ irradiated in the presence of medium with a 137Cs source emitting the dose (in Gy) indicated below at a fixed dose rate (irradiator model 0103; U.S. Nuclear Corp.). After irradiation, fresh medium was added and cells were incubated for the times indicated below. The WT and GIT2-KO mice were irradiated with the IR source and sacrificed, and then cortical brain tissue was collected for immunohistochemistry analysis 1 h after irradiation.
Laser-induced DSBs in multiple cell types.
Cells were seeded in a 35-mm glass-bottom culture dish (MatTek Corporation). Briefly, localized irradiation was conducted using a Nikon Eclipse TE2000 confocal microscope equipped with an SRS NL100 nitrogen laser-pumped dye laser (Photonics Instruments) that fires 3-ns pulses at a repetition rate of 10 Hz at 365 nm with a power of 0.7 nW, measured at the back aperture of the 60× objective. The laser was directed to a specified rectangular region of interest (ROI) within the nucleus of a cell visualized with a Plan Fluor 60× (numerical aperture, 1.25) oil objective. The laser beam was oriented by galvanometer-driven beam displacers and fired randomly throughout the ROI until the entire region was exposed. See the materials and methods in the supplemental material for details of the experiments.
Immunoblotting and immunoprecipitation.
All immunoblot and immunoprecipitation experiments were performed as described previously (34). For details of the immunoblot and immunoprecipitation experiments, see the materials and methods in the supplemental material.
Immunofluorescence microscopy.
Cells were seeded into 4-well slide culture chambers (LabTek, Scotts Valley, CA). After 24 h, cells were treated with IR at 5 Gy or 4 μM cisplatin. Cells were fixed with 4% paraformaldehyde for 10 min at room temperature before being permeabilized in 0.5% Triton X-100. Slide culture chambers were washed with phosphate-buffered saline (PBS), blocked with PBS containing 2% bovine serum albumin, and incubated with antibodies specific for GIT2, ATM-pS1981, γ-H2AX, p53 binding protein 1 (53BP1), NBS1, and MDC1, followed by Alexa Fluor 568 (red)-conjugated anti-mouse and Alexa Fluor 488 (green)-conjugated anti-rabbit secondary antibodies (Molecular Probes). Cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole) for identification of the nucleus. Specific staining was visualized and images were captured with a Zeiss LSM 710 confocal microscope.
Mutation of potential GIT2 ATM phosphorylation sites.
Threonine-195 (Thr195) and serine-384 (Ser384) on human GIT2 were mutated to alanine by using a QuikChange site-directed mutagenesis kit from Agilent Technologies (La Jolla, CA) according to the protocol in the product manual. The primers used for mutation of Thr195 to alanine were forward primer GGAGCAGACCCAGGCGCACAGGATTCTAGTGG and reverse primer CCACTAGAATCCTGTGCGCCTGGGTCTGCTCC. Primers for mutation of Ser384 to alanine were forward primer ATAACCAGCACAGCGTTGAGGCTCAAGACAACGATCAGC and reverse primer GCTGATCGTTGTCTTGACTCTCAACGGCGTGCTGGTTAT.
Neutral comet assay single-cell electrophoresis.
SH-SY5Y cells were treated with 10 Gy IR or 4 μM cisplatin and subjected to a neutral comet assay (Trevigen, Gaithersburg, MD) to detect DNA damage and repair of double-strand breaks. According to the manufacturer's instructions, after treatment, cells were harvested and mixed with low-melting-temperature agarose. After lysis, electrophoresis was performed in neutral electrophoresis buffer at 1 V cm−1 and 15 mA for 40 min. Slides were stained with SYBR green dye for 10 min. One hundred randomly selected cells per sample were visually captured under a Zeiss fluorescence microscope, and digital fluorescent images were obtained using AxioVision software. The relative length and intensity of SYBR green-stained DNA streaks from head to tail were proportional to the amount of DNA damage present in the individual nuclei and were measured by determining olive tail movement using Comet Score software (TriTek, Sumerduck, VA).
Cell viability assays.
SH-SY5Y cells were transfected with 5 μg of Flag-tagged GIT2 or treated with siRNA specific for GIT2 (GIT2 siRNA; 200 nM) and then subjected to various short-term cellular insults (IR at 10 Gy for 10 min, 1 μM etoposide for 1 h, 10 μM hydrogen peroxide for 1 h), followed by removal of the stressor and refreshment of the growth medium before eventual assessment of the percentage of viable cells remaining at 0.5, 24, or 48 h postinsult using a standard trypan blue (Sigma-Aldrich, St. Louis, MO) exclusion assay.
Immunohistochemistry.
For immunohistochemistry, mice were sacrificed and immediately perfused with 4% paraformaldehyde (PFA) in PBS. Isolated brains were postfixed at 4°C for 24 h in 4% PFA–PBS and then for 48 h in 4% PFA–20% sucrose in PBS. Brain cortices were cut into 40-μm sections using a sliding microtome. Floating sections were pretreated with 1% H2O2 in PBS at room temperature for 20 min, blocked with 3% goat serum in PBS-Tween 20 for 1 h, and incubated with mouse anti-γ-H2AX antibody (1:300; Abcam, Cambridge, MA) in blocking solution overnight at 4°C. Staining was visualized using a Vectastain universal ABC kit and ImmPACT diaminobenzidine substrate (Vector Laboratories, Cambridgeshire, United Kingdom).
SILAC labeling and mass spectrometry.
Human SH-SY5Y neuroblastoma cells were maintained in DMEM (Invitrogen) supplemented with 10% dialyzed fetal bovine serum and 1% penicillin-streptomycin (Invitrogen). Customized arginine- and lysine-free DMEM was obtained from AthenaES (Baltimore, MD). l-Lysine-D4 (K4), [U-13C6-15N2]l-lysine (K8), [U-13C6]l-arginine (R6), and [U-13C6-15N4]l-arginine (R10) were purchased from Cambridge Isotope Laboratories (Andover, MA). For GIT2 siRNA or Flag-tagged GIT2 cDNA expression studies, cells were grown for 7 days (eight division cycles) in DMEM containing medium (M; K4, R6) or heavy (H; K8, R10) forms of arginine and lysine. Approximately 92% amino acid labeling was achieved using this protocol. To reduce labeling errors, experiments were performed as doublets (control versus GIT2 siRNA or Flag-tagged GIT2 cDNA) with random labeling by stable isotope labeling by amino acids in cell culture (SILAC), with the conditions with the M and H forms being compared to avoid nonlabeled peptide errors. Details are described in the materials and methods in the supplemental material.
Statistical analysis.
Measurement of statistical significance between any experimental group employed within the study was assessed using a nonpaired Student's t test within the basic statistical suite of GraphPad Prism (version 5.0) software. For data represented in any histogram, the values indicated are means ± standard errors of the means (SEMs) obtained from at least three independent experimental replicates.
RESULTS
The GIT2 protein constellation is linked to the DDR.
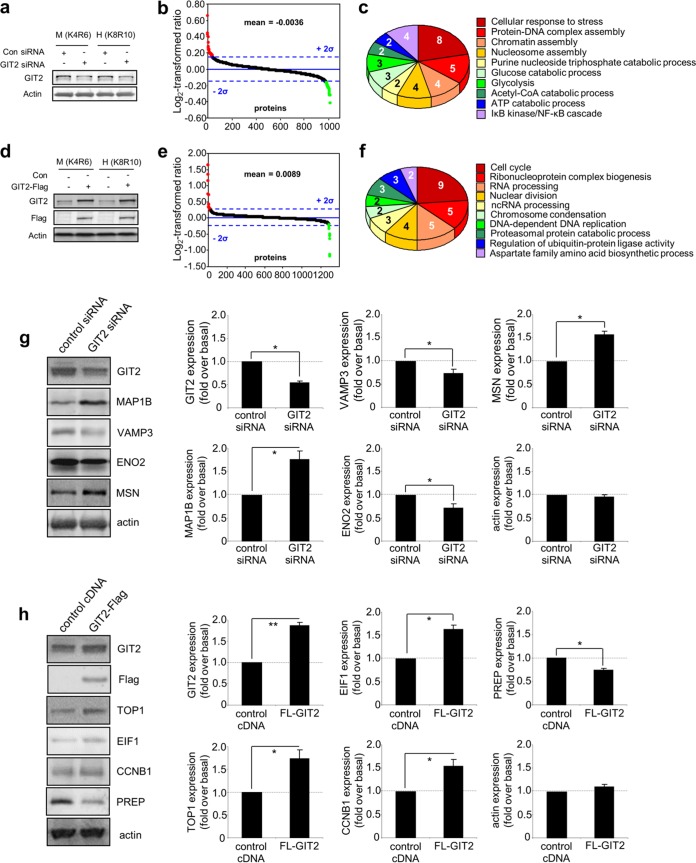
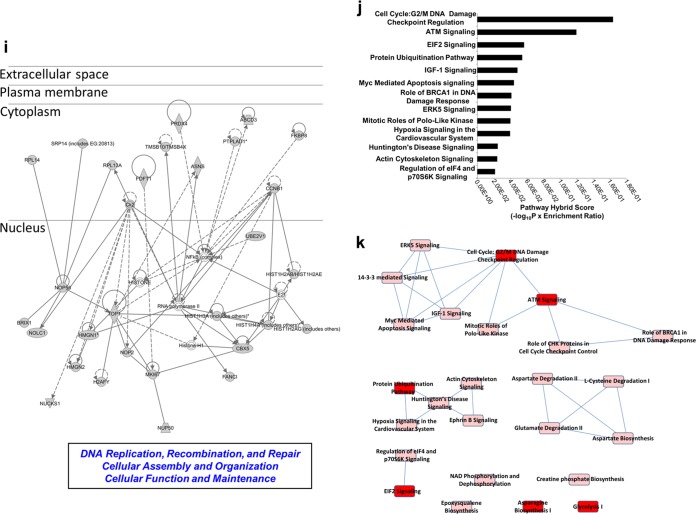
To investigate the constellation of proteins functionally associated with GIT2, we assessed the quantitative proteomic response in neuroblastoma cells to bidirectional modulation of GIT2 expression. To avoid nonspecific global proteomic alterations, which can distort the analysis (35), only moderate decreases (with GIT2 siRNA) or increases (with Flag-tagged GIT2) in the level of GIT2 expression were employed. Using label-swap variation of conditions with the labeled medium (K4R6) or heavy (K8R10) form, we observed the significant regulation of 69 proteins (36 upregulated, 33 downregulated) of 1,020 total identified/quantified proteins (Fig. 1a and b; see also Table S1 in the supplemental material) following a reduction in the level of endogenous GIT2 expression. Gene ontology (GO) annotation was performed with these 69 significantly regulated proteins, and a strong representation of proteins involved in cell stress and DNA damage management was evident (Fig. 1c; see also Table S2 in the supplemental material). With moderate increases in the level of GIT2 expression with ectopically tagged GIT2 introduction, we observed the significant regulation of 65 proteins (45 upregulated, 20 downregulated) of 1,300 total identified/quantified proteins (Fig. 1d and e; see also Table S3 in the supplemental material). GO term annotation of the proteins significantly affected by the overexpression of GIT2 again demonstrated a strong functional pattern of nucleic acid maintenance and cell cycle control (Fig. 1f; see also Table S4 in the supplemental material). Western blot validation of protein expression responses was performed for six proteins from each GIT2 modulatory polarity (Fig. 1g for GIT2 siRNA Fig. 1h for GIT2 overexpression). To appreciate the functional constellation of GIT2-associated proteins, we performed functional network analysis (Ingenuity Pathway Analysis) with all 134 significantly regulated proteins from both knockdown and ectopic expression experiments. The highest-scoring network was specifically associated with cellular assembly and organization, cellular function and maintenance, and DNA replication, recombination, and repair and was primarily focused toward the nucleus (Fig. 1i; see also Table S5 in the supplemental material). Signaling pathway analysis of the 134 significantly regulated proteins also reinforced the strong association of GIT2 with DNA damage checkpoint regulation and ATM signaling (Fig. 1j; see also Table S6 in the supplemental material). Investigating further this potential functional role of GIT2 in DNA repair, we found that the well-characterized DDR protein HMGN1 (36) was both elevated in expression in response to GIT2 overexpression and attenuated in its expression in response to siRNA-mediated GIT2 reduction (see Tables S1 and S3 in the supplemental material). The expression profile of other DDR-related proteins was also shown to be specifically sensitive to the GIT2 expression modulation polarity; i.e., a reduction in GIT2 siRNA resulted in a similar reduction of HMGB1, MDC1, and FANCI (see Table S1 in the supplemental material), while GIT2 overexpression resulted in a similar potentiation of TOP1, HMGN2, and RFC1 (see Table S3 in the supplemental material). Therefore, the bimodal expression profile of GIT2 is highly synergistic with the coherent expression of multiple DNA damage/repair-related factors. In addition, we noted that multiple pathways linked to DNA damage and repair (e.g., DNA damage checkpoint regulation, ATM signaling, the role of BRCA1 in the DNA damage response, and the role of CHK proteins in cell cycle checkpoint control) were significantly populated by GIT2 constellation proteins and overlapped each other (Fig. 1k), indicating a potential strong trophic role of GIT2 in DNA repair. As we have previously demonstrated that GIT2 expression is sensitive to both aging and reactive oxygen species exposure (32, 33), we postulated that GIT2 may also play a role in maintaining DNA stability during periods of stress.
FIG 1.
The GIT2 proteomic constellation is strongly associated with cellular DNA management. Statistically significant protein expression responses to experimental modulation of GIT2 neuroblastoma expression were measured using SILAC labeling. (a) Reduction of GIT2 expression induced by GIT2 siRNA. Con, control. (b) GIT2 attenuation induces the significant alteration of proteins (red, upregulated proteins; green, downregulated proteins) 48 h after siRNA introduction. (c) Gene ontology annotation of the proteins significantly affected by attenuation of GIT2 expression demonstrates a strong relationship of these proteins to the cell stress response and nuclear DNA protective behavior. Acetyl-CoA, acetyl coenzyme A. (d) cDNA clone-mediated elevation of GIT2 expression. (e) GIT2 potentiation induces the significant alteration of proteins 48 h after cDNA introduction. (f) Gene ontology annotation of the proteins significantly affected by GIT2 overexpression demonstrates a strong relationship of these proteins to cell cycle control, chromosome stability, and DNA replication processes. ncRNA, noncoding RNA. (g and h) Expression validations were performed for the GIT2 attenuation (g) and potentiation (h) experiments. FL-GIT2, Flag-tagged GIT2. (i) Protein interaction network analysis was performed on the protein identities from the combined data sets of the GIT2-knockdown and overexpression experiments. The most significantly populated network (83% network coverage by input data set) was associated with DNA replication, recombination, and repair. (j) Canonical signaling pathway analysis of this combined GIT2-responsive data set revealed that these correlated proteins are strongly associated with DNA damage response and ATM-dependent signaling activity. (k) Interaction and overlap between predicted signaling pathway functions for the GIT2 protein constellation. An increased red intensity of the pathway indicates a greater probability of pathway enrichment. EIF2, eukaryotic initiation factor 2; IGF-1, insulin-like growth factor 1; ERK5, extracellular signal-regulated kinase 5; eIF4, eukaryotic initiation factor 4. *, P < 0.05; **, P < 0.01.
GIT2 modulates DNA repair and associates with classical DDR complexes.
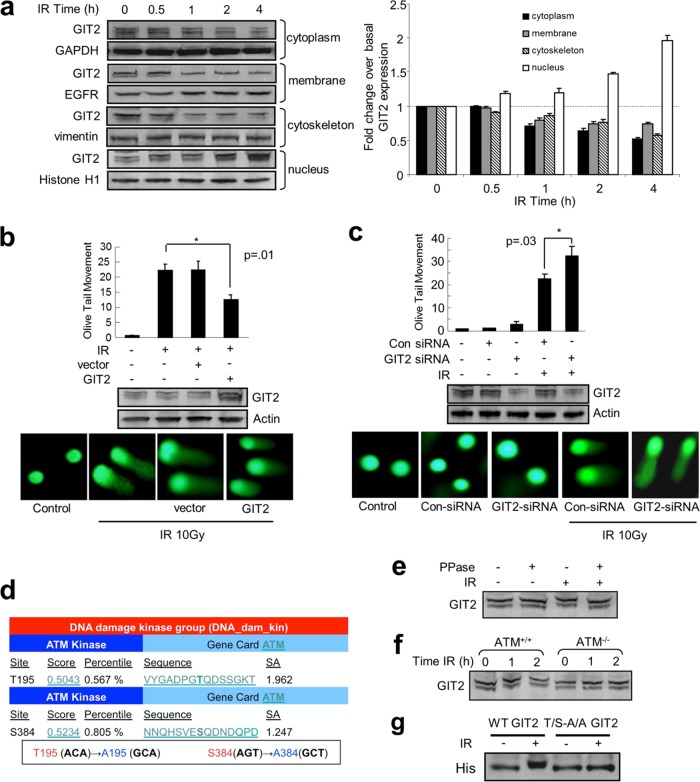
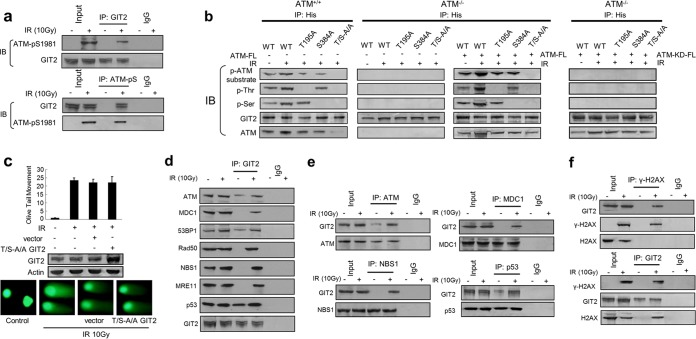
We examined the cellular expression and disposition of GIT2 in response to DNA damage. GIT2 was identified in all major cell compartments under control cell conditions (Fig. 2a). Exposure of SH-SY5Y cells to the DNA damage-inducing agents IR (Fig. 2a) or cisplatin (cis-diamminedichloroplatinum II [CDDP]) (see Fig. S1a in the supplemental material) enhanced the GIT2 nuclear expression level, mediated in part by a transition from the other cellular compartments. To investigate the potential role(s) of GIT2 in the DDR, we assessed the effects of GIT2 expression modulation on DNA repair. The neutral comet assay was used to specifically measure DNA double-strand breaks (DSBs) after exposure to the DNA-damaging agents (IR, CDDP). Ectopically increasing GIT2 expression enhanced the repair of DNA damage caused by either IR (Fig. 2b) or CDDP (see Fig. S1b in the supplemental material) exposure, whereas the siRNA-mediated reduction of GIT2 expression reduced the repair of DNA damage induced by IR (Fig. 2c) or CDDP (see Fig. S1c in the supplemental material). Our GIT2 constellation analysis (Fig. 1) strongly suggested a role for GIT2 in ATM-mediated pathways. ATM is a member of the phosphatidylinositol 3-kinase-related kinase (PIKK) family, which has the following common phosphorylation sites: Ser/Thr followed by Gln, a consensus motif commonly termed SQ-TQ (37). We screened the human GIT2 protein sequence and found two such motifs (Thr195Gln and Ser384Gln) (http://scansite3.mit.edu/) (Fig. 2d). Protein phosphorylation of DDR proteins, e.g., via ATM activity, can reduce their electrochemical migration rate in acrylamide gels (38). GIT2 protein from IR-stimulated (Fig. 2e) or CDDP-stimulated (see Fig. S1d in the supplemental material) cells demonstrated this characteristic retarded gel migration, whereas GIT2 protein from nontreated cells did not. Cell extracts treated with λ-protein phosphatase prevented the slower migration of the GIT2 protein observed in the presence of IR (Fig. 2e) or CDDP (see Fig. S1d in the supplemental material), suggesting that GIT2 is likely phosphorylated upon DNA damage. In order to determine whether the GIT2 phosphorylation in response to DNA damage involved the ATM kinase, we employed ATM wild-type (ATM+/+; GM0637) and ATM-null (ATM−/−; GM5849) human fibroblasts exposed to either IR or CDDP. GIT2 gel retardation caused by the DNA damage induced by IR (Fig. 2f) and CDDP (see Fig. S1e in the supplemental material) was observed in ATM+/+ cells but not in the ATM−/− cells. We generated a double mutant form of GIT2, termed the GIT2 T/S-A/A mutant, in which we disrupted both SQ/TQ motifs (Thr195 → Ala195 and Ser384 → Ala384). In a manner similar to that for GIT2 in ATM−/− cells, the GIT2 T/S-A/A mutant failed to demonstrate retarded gel migration upon IR exposure (Fig. 2g). These findings indicate that ATM kinase is required for the phosphorylation of GIT2 in response to DNA damage. As IR generates a cleaner, more definitive DNA DSB than CDDP, we chose to further focus upon the relationship between GIT2 and IR exposure. Using reciprocal coimmunoprecipitation, we found that GIT2 associated with activated ATM kinase (ATM-pS1981) upon IR stimulation (Fig. 3a), further supporting the identification of GIT2 as a substrate of ATM kinase. We next investigated the role of the predicted ATM phosphorylation sites in GIT2 on DDR activity. In ATM−/− or ATM+/+ cells, we expressed epitope-tagged (His) WT, single mutant (Thr195 → Ala195 or Ser384 → Ala384), and double mutant (Thr195 → Ala195/Ser384 → Ala384) GIT2 to investigate the physical interaction mechanics of GIT2 and ATM. Using antibodies raised against phospho-ATM/ATR substrates, as well as against generic phosphothreonine or phosphoserine residues, we found that IR enhanced the physical association of WT GIT2 with the ATM kinase in ATM+/+ cells (Fig. 3b). Mutation of either Thr195 or Ser384 to alanine attenuated the IR-induced phosphorylation of these GIT2 mutants and ATM in the ATM+/+ cells. The double GIT2 T/S-A/A mutant failed to demonstrate any significant ATM site-associated phosphorylation upon IR exposure and demonstrated a marked reduction in its ability to coprecipitate with the endogenous ATM. Unsurprisingly, there was no observed association of GIT2 with ATM in ATM−/− cells transfected with any of the GIT2 constructs. Reintroduction of ATM into the ATM−/− cells recovered the IR-mediated association of WT GIT2 with the ectopically expressed ATM kinase (Fig. 3b). These findings further imply that GIT2 is phosphorylated by ATM kinase on the predicted ATM consensus sites Thr195Gln and Ser384Gln. In contrast to the results presented in Fig. 2b, we found that overexpression of the double T/S-A/A GIT2 mutant failed to reduce the levels of DNA damage induced by either IR (Fig. 3c) or CDDP (see Fig. S1f in the supplemental material), suggesting that phosphorylation of GIT2 by ATM is important for its role in the repair of DNA damage.
FIG 2.
GIT2 enhances repair of DNA damage induced by IR. (a) SH-SY5Y cells were irradiated with 5 Gy IR for the indicated times, followed by subcellular fractionation of proteins and subsequent immunoblotting analysis. The histogram depicts the relative quantitation of GIT2 expression changes in diverse cellular compartments in response to IR. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; EGFR, epidermal growth factor receptor. (b) Overexpression of GIT2 promotes repair of DNA double-strand breaks. The neutral comet assay was performed 1 h after irradiation (100 nuclei were counted for each group; data are expressed as means ± SEMs). (c) Knockdown of GIT2 exacerbates IR-mediated DNA damage. (d) Schematic representation (http://scansite3.mit.edu/) of potential ATM kinase phosphorylation sites in the GIT2 protein and sites of engineered alanine mutations. SA, surface accessibility. (e) GIT2 is phosphorylated in response to DNA damage in an ATM-dependent manner. IR-treated SH-SY5Y cells were harvested at 1 h after irradiation, and cell extracts were incubated with or without λ-protein phosphatase (PPase) and subjected to immunoblot gel migration analysis using GIT2 antibody. λ-Protein phosphatase incubation abrogated IR-induced GIT2 gel retardation. (f) GIT2 gel retardation is dependent upon ATM. IR-treated ATM+/+ (GM0637) and ATM−/− (GM5849) cells were harvested at the indicated time point, and cell extracts were subjected to immunoblot gel migration analysis using GIT2 antibody. IR-mediated GIT2 gel retardation was lost in ATM−/− cells. (g) The GIT2 T/S-A/A mutant fails to display the IR-mediated gel retardation observed with WT GIT2.
FIG 3.
GIT2 interacts with ATM kinase and multiple classical DDR proteins in response to DNA damage. (a) GIT2 coprecipitates with activated ATM in response to IR. The input represents 1% of the total protein used in the immunoprecipitation (IP) assays. Employment of either GIT2 (top) or active ATM (bottom) as the antigenic target resulted in the coprecipitation of active ATM (ATM-pS1981) or GIT2, respectively. IB, immunoblotting. (b) ATM+/+ (GM0637) and ATM−/− (GM5849) cells were transfected with His-tagged wild-type GIT2, GIT2-T195A, GIT2-S384A, or GIT2-T195A/S384A (T/S-A/A) constructs or cotransfected with Flag-tagged (FL) WT or kinase-dead (KD) ATM for 24 h; cells were harvested 1 h after IR stimulation. Coimmunoprecipitation and immunoblot assays were performed using the antibodies indicated. Generic serine or threonine phosphorylation of immunoprecipitated GIT2 was assessed using antiphosphoserine (p-Ser) or antiphosphothreonine (p-Thr) immunoblotting assays. Reintroduction of WT ATM into ATM−/− cells reinstated the IR-induced GIT2 phosphorylation. (c) The DNA-protective role of GIT2 is dependent on ATM phosphorylation sites. SH-SY5Y cells were transfected with the GIT2 T/S-A/A mutant for 24 h before IR treatment. The neutral comet assay was performed 1 h after irradiation (100 nuclei were counted for each group). Overexpression of the GIT2 T/S-A/A mutant failed to confer protection against IR-mediated DNA damage. (d) Proteins associating with immunoprecipitated GIT2 in an IR-dependent manner were identified by specific immunoblot assays. (e) Reciprocal IR-dependent coimmunoprecipitations were performed between ATM, MDC1, p53, NBS1, and GIT2. (f) GIT2 dynamically coprecipitates with phosphorylated histone H2AX (γ-H2AX) in an IR-dependent manner.
The efficient repair of DNA damage requires the coordinated recruitment and assembly of multiple DDR proteins. We therefore investigated whether GIT2 could bind to the components of the MRE11-RAD50-NBS1 (MRN) complex and other known DDR proteins. GIT2 coprecipitated with ATM, as well as with multiple components of the MRN complex, mediator of DNA damage checkpoint protein 1 (MDC1), p53 binding protein 1 (53BP1), and the p53 tumor suppressor, upon exposure to IR (Fig. 3d) or CDDP (see Fig. S1g in the supplemental material). In the reciprocal coimmunoprecipitation experiments, GIT2 was recovered with ATM, MDC1, NBS1, and p53 (Fig. 3e). It was evident that, in addition to the increase caused by DNA damage, there were also various levels of constitutive association of GIT2 with some DNA damage response proteins, i.e., ATM, 53BP1, and p53. The GIT2 T/S-A/A double mutant associated with p53 but failed to demonstrate any profound interaction with the other DDR proteins tested in the presence of IR (see Fig. S2a in the supplemental material) or CDDP (see Fig. S2b in the supplemental material).
It is therefore likely that GIT2 forms part of a contextually sensitive multiprotein complex consisting of DNA damage regulatory and repair proteins. The phosphorylated histone H2AX (Ser139–γ-H2AX) is a marker for sites of DNA DSBs and is critical for the recruitment and concentration of DNA repair proteins (39, 40). We therefore performed reciprocal coimmunoprecipitation experiments with GIT2 and γ-H2AX. We found a strong IR-dependent association of GIT2 with γ-H2AX (Fig. 3f).
As GIT2 clearly interacts with multiple DDR proteins, we next investigated the ability of GIT2 to engender repair and cell survival following DNA damage. Overexpression of GIT2 resulted in the significant dose-dependent elevation of the expression of multiple DNA repair proteins, e.g., HMGN1 (36) and RFC1 (41), as well as proteins involved in nuclear responses to cytotoxic DNA damage (NOP2 [42]) or involved in aging/neurodevelopment (SPEN [43]) (see Fig. S3a and b in the supplemental material). Overexpression of GIT2 also attenuated the degree of residual DNA damage (24 to 48 h after irradiation-induced DSBs), suggesting an ability of GIT2 to facilitate DNA repair (see Fig. S3c in the supplemental material). Not only was DNA repair enhanced, but also after various acute cell stressors (IR, 10 Gy; etoposide, 1 μM; peroxide, 10 μM), long-term cell survival (24 and 48 h postinsult) was potentiated by GIT2 overexpression (see Fig. S3d to f in the supplemental material) and attenuated by GIT2 silencing (see Fig. S3g to i in the supplemental material).
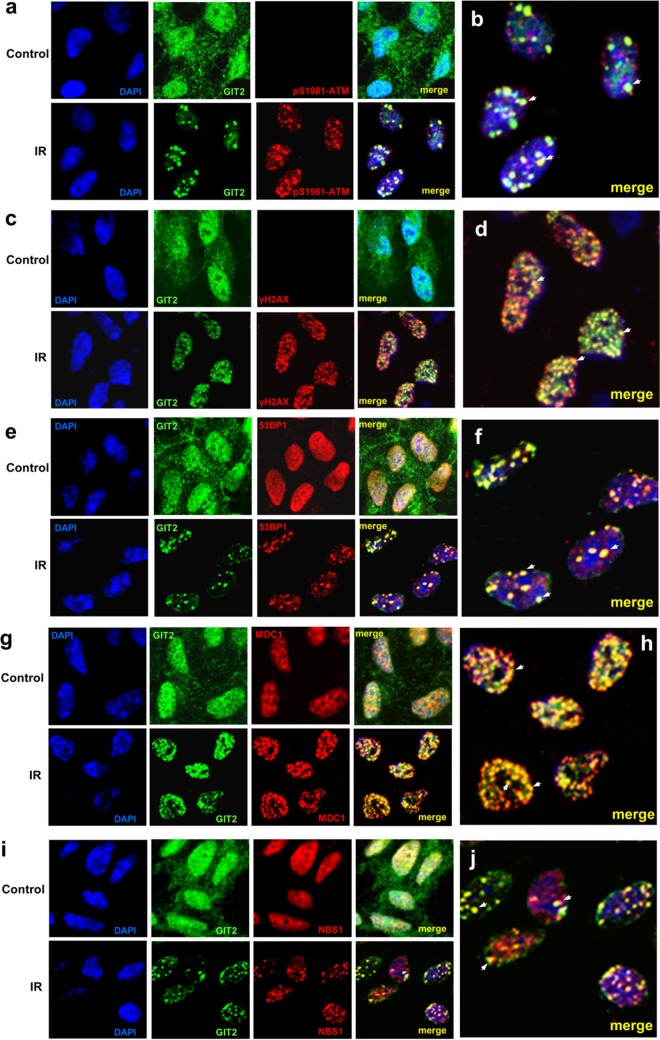
GIT2 forms nuclear foci in response to DNA damage.
As we have demonstrated a strong association between GIT2 and ATM, we assessed the DNA damage-dependent physical nuclear colocalization of these two proteins. We found that GIT2 exhibited diffuse nuclear staining in undamaged quiescent cells. In response to IR, GIT2 and active ATM (ATM-pS1981) colocalized to sharp nuclear foci (Fig. 4a and b). We also assessed whether GIT2 foci contained other DDR factors, i.e., γ-H2AX, 53BP1, MDC1, and NBS1. Distinct GIT2 puncta induced by IR colocalized with γ-H2AX (Fig. 4c and d), 53BP1 (Fig. 4e and f), MDC1 (Fig. 4g and h), and NBS1 (Fig. 4i and j) foci. We also performed a similar analysis of GIT2 nuclear colocalization with the same DDR proteins (γ-H2AX, ATM-pS1981, 53BP1, MDC1, and NBS1) after CDDP exposure (see Fig. S4a to j in the supplemental material). As with IR, CDDP induced the generation of multiple GIT2-positive foci that demonstrated a strong colocalization with each respective DDR protein. These data indicate that GIT2 colocalizes at the subcellular level with key DDR proteins at sites of DNA DSBs. Furthermore, in contrast to WT GIT2, the double mutant GIT2 T/S-A/A failed to form nuclear foci at DNA damage sites upon IR (see Fig. S2c in the supplemental material) or CDDP (see Fig. S2d in the supplemental material) treatment.
FIG 4.
DNA damage induces GIT2 nuclear focus formation with classical DNA damage response factors. SH-SY5Y cells were treated with IR, left untreated for 1 h, and fixed; and then the subcellular localization and colocalization of endogenous GIT2 with active ATM-pS1981 (a and b [merge enlargement]), γ-H2AX (c and d [merge enlargement]), 53BP1 (e and f [merge enlargement]), MDC1 (g and h [merge enlargement]), and NBS1 (i and j [merge enlargement]) were assessed. Colocalizations are indicated with arrows. GIT2 or the DNA damage complex proteins were detected using specific primary antibodies followed by an Alexa Fluor 488 (green)- or Alexa Fluor 568 (red)-conjugated secondary antibody. Nuclear DAPI stain was employed to visualize cellular nuclei. Magnifications, ×63.
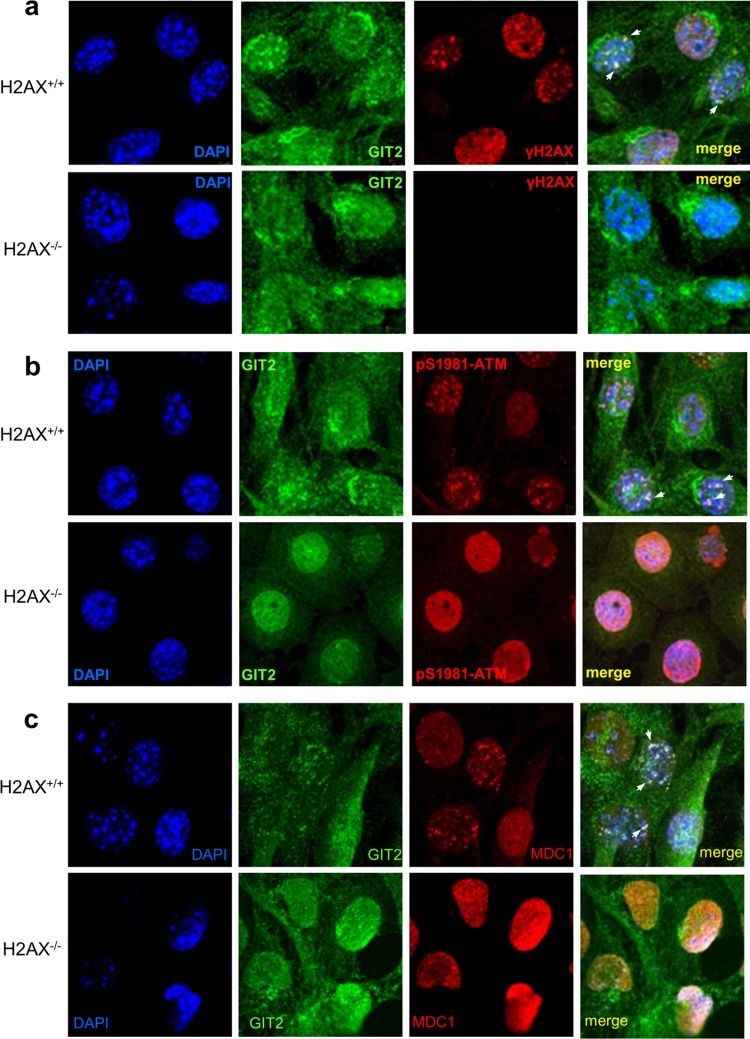
DNA damage-dependent GIT2 nuclear localization is dependent on multiple DDR proteins.
We have demonstrated that the GIT2 association with γ-H2AX is strongly promoted by IR (Fig. 3f). γ-H2AX is associated with chromatin domains flanking the sites of DNA DSBs and plays a key role in recruiting DNA repair proteins, such as BRCA1 (14, 15), NBS1 (10–12), 53BP1 (44), and MDC1 (45), to nuclear damage foci. Thus, we examined whether H2AX is required for nuclear GIT2 focus formation in response to DNA damage. We examined DNA damage-induced nuclear GIT2-containing focus formation in both H2AX+/+ and H2AX−/− MEFs. GIT2 colocalized with γ-H2AX-containing foci in H2AX+/+ cells exposed to IR (Fig. 5a) or CDDP (see Fig. S5a in the supplemental material). In contrast, no GIT2 foci were observed in IR-treated (Fig. 5a) or CDDP-treated (see Fig. S5a in the supplemental material) H2AX−/− cells. GIT2 foci colocalized with ATM-pS1981 (Fig. 5b) or MDC1 (Fig. 5c) foci in an H2AX-dependent manner after exposure to IR or CDDP (see Fig. S5b in the supplemental material for the results for ATM and Fig. S4c in the supplemental material for the results for MDC1), indicating that DNA damage-induced GIT2 focus formation is dependent on the presence of H2AX.
FIG 5.
DNA damage fails to generate GIT2-positive nuclear foci in H2AX−/− cells. GIT2 forms nuclear foci (arrows) that colocalize with γ-H2AX (a), active ATM (b), or MDC1 (c) in IR-treated H2AX+/+ cells but not H2AX−/− cells. Immunocytochemical foci were detected using antibodies against GIT2, γ-H2AX, ATM-pS1981, and MDC1, followed by an Alexa Fluor 488 (green)- or Alexa Fluor 568 (red)-conjugated secondary antibody. Nuclear DAPI stain was employed to visualize cellular nuclei. Magnifications, ×63.
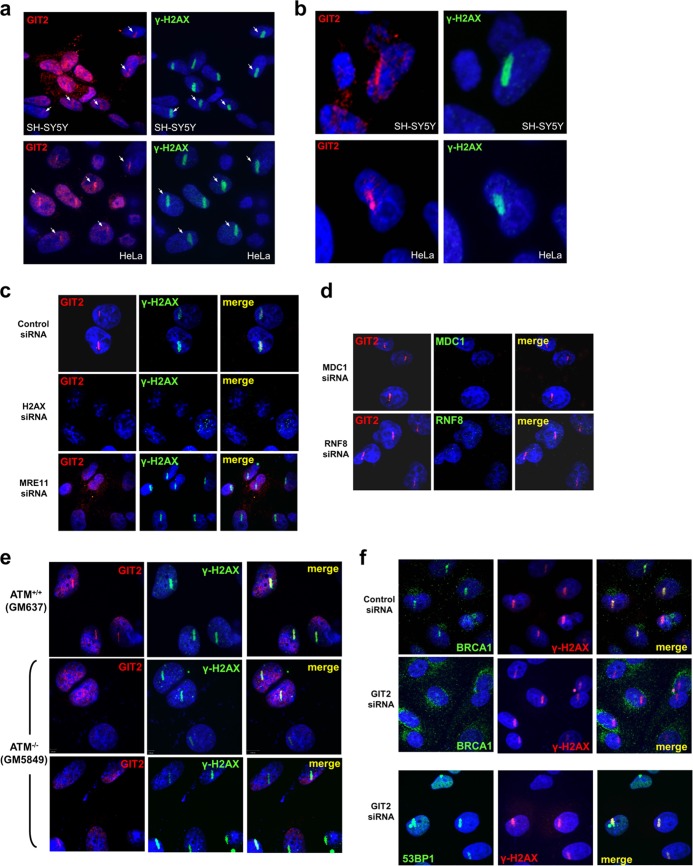
Supplementing our use of IR-induced DSBs, we also utilized a laser-induced DSB system. Using an SRS NL100 nitrogen laser-pumped dye laser, 3-ns pulses were aimed at specific regions in the cell nuclei to induce DSBs (Fig. 6). Rapidly after laser etching (2 min), the colocalization of GIT2 with γ-H2AX was observable in both SH-SY5Y cells and other human cell models, e.g., HeLa cells (Fig. 6a and b), demonstrating a rapid, cell type-independent role in the DDR for GIT2. As HeLa cells represent a good general cell model that is more permissive to genetic manipulation than neuronal tissue, we further investigated the recruitment of GIT2 to DSBs in these cells. siRNA-mediated H2AX suppression demonstrated that accumulation of GIT2 at damage sites was H2AX dependent (Fig. 6c), indicating that it participates in the DDR in multiple cellular contexts.
FIG 6.
GIT2 localization to laser-induced DNA double-strand breaks is partially dependent on H2AX, MRE11, and ATM. GIT2 is rapidly (within 2 min) recruited to laser-induced DNA DSB damage in SH-SY5Y cells as well as in HeLa cells (a and b [increased magnification]). Sites of laser-induced DNA damage demonstrate a strong and consistent colocalization of GIT2 with γ-H2AX. Arrows indicate GIT2–γ-H2AX colocalization at laser damage sites. (c) GIT2 recruitment to DSBs in HeLa cells is highly sensitive to the siRNA-mediated attenuation of H2AX and MRE11 expression. (d) GIT2 recruitment to DSBs is independent of MDC1 or RNF8 siRNA-mediated attenuation. (e) Loss of ATM expression (ATM−/− cells) partially inhibits GIT2 but not γ-H2AX recruitment to laser-induced DSBs. (f) Recruitment of the BRCA1 but not 53BP1 to laser-induced DSBs is strongly inhibited by the introduction of GIT2 siRNA.
In addition to H2AX dependence, we found that MRE11 was required for the recruitment of GIT2 to DSBs but not for the damage-induced formation of γ-H2AX (Fig. 6c). When we investigated other classical DDR components and their relationship to the dynamic recruitment of GIT2 to DSBs, we found that knockdown of MDC1 or RNF8 did not affect the recruitment of GIT2 to DSBs (Fig. 6d). We also assessed the contribution of ATM. In response to laser-induced DSBs, both GIT2 and γ-H2AX accumulated at sites of DSBs in ATM+/+ cells (Fig. 6e). In ATM−/− (GM5849) cells, γ-H2AX accumulation at DSBs was unaffected; however, the recruitment of GIT2 to DSBs was considerably decreased (25 to 30%) but not completely blocked, suggesting that other kinases might contribute to the phosphorylation of sites necessary for recruitment to DSBs (Fig. 6e). The genomic deletion of ATM did not greatly affect the IR-induced GIT2 cellular protein redistribution (see Fig. S6a in the supplemental material) or the formation of γ-H2AX-positive foci (see Fig. S6b in the supplemental material). However, in accordance with the results of the laser-induced damage, the presence of GIT2 containing nuclear foci upon IR exposure was largely suppressed in ATM−/− cells (see Fig. S6c in the supplemental material).
Our constellation expression experiment (Fig. 1) and DNA damage assays (Fig. 2 and 3) demonstrated that GIT2 is associated with DDR and DNA repair proteins. Our unbiased informatics analysis of the GIT2 functional constellation (Fig. 1j) suggested a potential link between GIT2 and ATM (which we subsequently validated), as well as between GIT2 and BRCA1. Consequently, we asked if GIT2 knockdown would influence the recruitment of key members of the DDR identified by our informatics analyses. The breast cancer susceptibility gene BRCA1 and the 53BP1 protein are important regulators of pathway choices in DSB repair (44). siRNA-mediated knockdown of GIT2 prevented the proper recruitment of BRCA1 to DSBs. In contrast, the appearance of both γ-H2AX and 53BP1 at DSBs was unaffected (Fig. 6f). This selective disruption of BRCA1 recruitment indicates a relatively nuanced role of GIT2 in orchestrating DDR processes. Therefore, it seems that GIT2 DSB recruitment can be a rapid process, is found in multiple cellular lineages, and is strongly dependent on the presence of H2AX, MRE11, and ATM but is independent of MDC1 and RNF8. GIT2 also appears to control specific subsets of DDR complexes and does not universally regulate all DDR processes.
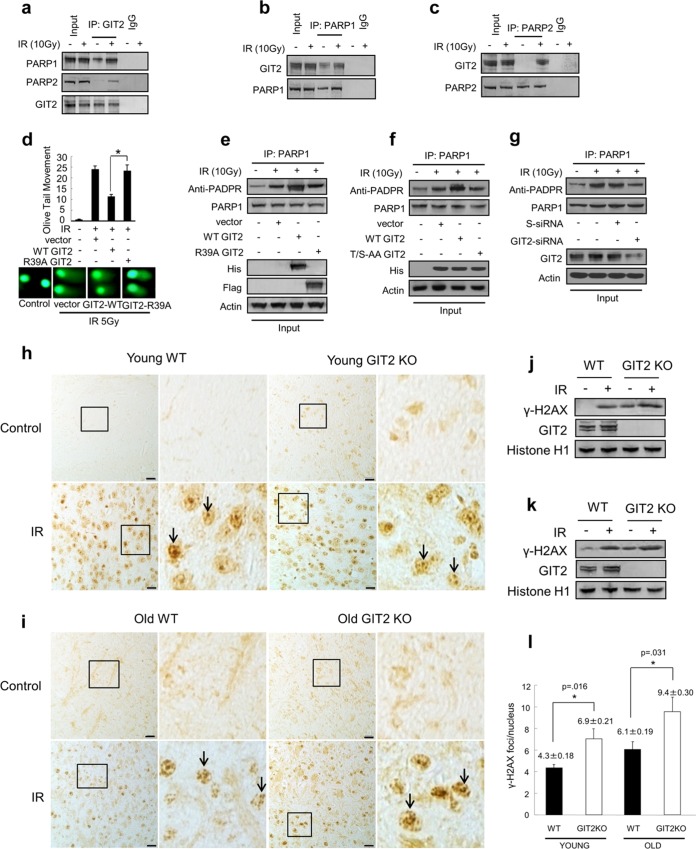
GIT2 promotes the activity of PARPs.
GIT2 is a member of the family of ADP-ribosylation factor (Arf) GTPase-activating proteins (GAPs), and the GIT2 GAP domain is present at the proximal amino terminus (19). Poly(ADP-ribose) (PADPR) polymerases (PARPs) are involved in the repair of damaged DNA. PARPs recognize and are activated by both single- and double-strand DNA breaks. PARP1 interacts physically and functionally with various proteins involved in DNA repair and recruits the repair proteins to sites of DNA damage (46, 47). We therefore asked if the Arf GAP activity of GIT2 plays a role in its DDR function. In human neuroblastoma cells, endogenous GIT2 coprecipitated with PARP1 and PARP2 in an IR-dependent manner (Fig. 7a). In reciprocal coimmunoprecipitation experiments, both PARP1 (Fig. 7b) and PARP2 (Fig. 7C) associated with GIT2 after IR treatment. Introduction of an Arg39-Ala (R39A) mutation into GIT proteins abolishes their GAP activity. While overexpression of WT GIT2 stimulated the repair of IR-induced DNA damage, ectopic expression of GIT2-R39A failed to reduce the olive tail movement (Fig. 7d), suggesting that Arf GAP activity is required for GIT2-mediated DNA repair. PADPR is a polymer synthesized by both PARP1 and PARP2. Under basal, nonstressed conditions, cells display low basal levels of PADPR polymers. Under conditions of cellular stress, e.g., DNA damage, PADPR levels increase dramatically. Ectopic expression of WT GIT2 promoted the synthesis of PADPR in IR-exposed cells (Fig. 7e and f) or CDDP-exposed cells (see Fig. S7a and b in the supplemental material), while overexpression of GIT2 R39A (Fig. 7e; see also Fig. S7a in the supplemental material) or GIT2 T/S-A/A (Fig. 7f; see also Fig. S7b in the supplemental material) failed to enhance IR-induced PADPR synthesis. Furthermore, siRNA-mediated depletion of GIT2 attenuated the synthesis of PARP1-associated PADPR in cells stimulated by IR (Fig. 7g) or CDDP (see Fig. S7c in the supplemental material). These data demonstrate that GIT2 may promote DNA repair by facilitating the activities of poly(ADP-ribose) polymerases which catalyze the synthesis of PADPR.
FIG 7.
GIT2 associates with PARP1 and PARP2, and its genomic deletion increases murine sensitivity to DNA damage. (a) GIT2 is associated with PARP1 and PARP2 in SH-SY5Y cells after IR exposure. Coimmunoprecipitations (IPs) were performed with an antibody specific to GIT2 or a nonspecific, species-matched IgG control, followed by immunoblotting (IB) analysis with antibodies against PARP1 or PARP2. Reciprocal coimmunoprecipitations of GIT2 with PARP1 (b) and PARP2 (c) were also performed in the presence of IR. (d) Overexpression of a GIT2 mutant lacking GAP activity (Flag-GIT2-R39A) does not confer the DNA repair activity mediated by the overexpression of wild-type GIT2 (GIT2-WT) assessed by comet assay 1 h after IR exposure (100 nuclei were counted for each group). Data are expressed as means ± SEMs. *, P < 0.05. (e) Overexpression of WT GIT2 but not R39A-GIT2 potentiates the increased synthesis of PARP1-associated PADPR in response to IR. (f) WT GIT2 overexpression but not GIT2 T/S-A/A overexpression enhances PARP1-associated PADPR generation in response to IR. (g) siRNA-mediated reduction of GIT2 attenuates PARP1-associated PADPR generation in response to IR. S-siRNA, scrambled siRNA. (h and i) Immunohistochemical staining of γ-H2AX in cortical brain tissue sections prepared from 4-month-old (young) (h) and 18-month-old (old) (i) WT or GIT2-KO mice. Arrows, nuclear foci containing γ-H2AX. Bars = 40 μm. (j and k) Detection of γ-H2AX expression levels in cortical tissues from 4-month-old (j) and 18-month-old (k) WT or GIT2-KO mice with or without IR treatment. Histone H1 was used as a loading control. (l) Quantification of γ-H2AX foci in each nucleus in young (age, 4 months) and old (age, 18 months) WT or GIT2-knockout mouse brain sections. At least 100 nuclei were counted for each group of animals. Data are expressed as means ± SEMs.
Genomic ablation of GIT2 in mice potentiates susceptibility to DNA damage.
Our previous findings demonstrated that GIT2 expression is elevated in the brains of multiple mammalian species with natural aging (32, 33). As DNA damage can specifically accumulate in the central nervous system with aging, we next examined the effect of GIT2 genomic suppression upon DNA damage in the brains of elderly mice. We examined the presence of extant γ-H2AX nuclear foci in the brain (cortex) from young (4 months) and old (18 months) WT or GIT2-knockout (KO) mice after acute nonlethal IR exposure. Immunohistochemical staining of cortical sections from young mice demonstrated the presence of significantly more γ-H2AX foci in the cellular nuclei of GIT2-KO mice than young WT mice after IR exposure (Fig. 7 h, i). In accordance with a potential regulatory role of GIT2 in the aging process, considerably larger amounts of γ-H2AX foci were observed in the nuclei from the cortices of aged GIT2-KO mice than in those from the cortices of WT mice (Fig. 7j and k). The levels of γ-H2AX-positive foci in the young GIT2-KO mice were commensurate with those seen in aged WT mice, indicating that an advanced aging event potentially due to reduced DNA repair was occurring (Fig. 7l). Taken together, these findings demonstrate that the loss of GIT2 expression potentiates the sensitivity of these mice to DNA damage events in an age-related manner and may thus contribute to the multiple mechanisms in which GIT2 deficiency is associated with aging and cellular deterioration.
DISCUSSION
We have demonstrated that GIT2 is associated both functionally (Fig. 1) and physically (Fig. 2 and 3) with multiple protein components of the DDR process. To date, more than 30 ATM-dependent substrates that maintain genome stability and reduce the risk of disease have been identified (48). As has been observed for other DDR proteins that are ATM kinase substrates, such as BRCA1 and NBS1 (38), GIT2 exhibited a retarded gel migration velocity in response to DNA damage that was reversed by phosphatase incubation or prevented by the genomic deletion of the ATM kinase (Fig. 2). In addition, GIT2 was able to physically associate with the autophosphorylated (active) form of ATM in a DNA damage-dependent manner (Fig. 3). Using an unbiased informatics analysis, we identified two candidate consensus ATM phosphorylation motifs in the GIT2 protein sequence, i.e., Thr195Gln and Ser384Gln. Disrupting the ability of these sites to be phosphorylated by ATM (by introduction of an alanine mutation) attenuated the ability of DNA-damaging agents to induce protein phosphorylation of GIT2 (Fig. 3). In addition, overexpression of the double alanine mutant form of GIT2 (Ala195-Ala384) failed to demonstrate an ability to promote DNA repair in neuroblastoma cells, in contrast to the enhanced repair resulting from overexpression of the wild-type form (Fig. 3). These predicted ATM phosphorylation sites are also conserved in GIT2 from multiple species, both mammalian (Mus musculus, Thr195Gln and Ser385Gln; Macaca mulatta, Thr195Gln and Ser384Gln; Canis lupus familiaris, Thr195Gln and Ser386Gln; Bos taurus, Thr195Gln and Ser384Gln) and nonmammalian (Gallus gallus, Thr195Gln and Ser384Gln). Furthermore, we demonstrated that GIT2 physically associates and colocalizes at the cellular level, in a DNA damage-dependent manner, with multiple DDR proteins, including ATM, NBS1, 53BP1, MDC1, and γ-H2AX (Fig. 3). Using cellular systems with genetic ablation of H2AX, we found that GIT2 nuclear DSB focus formation is H2AX, ATM, and MRE11 dependent (Fig. 5 and 6), which is consistent with the findings for other DNA repair proteins, such as BRCA1, NBS1, 53BP1, and MDC1. In addition, we demonstrated that the loss of the putative ATM phosphorylation sites in GIT2 abrogated its physical association and subcellular colocalization with endogenously expressed DDR proteins, with the interesting exception of p53, in response to DNA damage (see Fig. S2 in the supplemental material). Given the evident important role of ATM-phosphorylated GIT2 in DNA repair, in future studies we will investigate the effects of this GIT2 posttranslational modification upon specific domain interaction regions of DDR proteins, such as MDC1 or MRE11. Taken together, these findings indicate that GIT2 is associated with DNA repair complexes at the sites of DNA DSBs and phosphorylation by ATM kinase is vital for the promotion of repair by GIT2.
With respect to the specific role of GIT2 in DNA repair mechanisms, we found that a relatively subtle elevation of GIT2 expression potentiated the cellular expression of multiple other proteins associated with DNA repair, including HMGN1 and RFC1. In addition, overexpression of GIT2 increased the expression of SPEN and NOP2, proteins that are involved in stress-responsive polyadenylated RNA metabolism (49), nuclear responses to cytotoxic DNA damage (42, 50), as well as aging and neurodevelopmental processes (43) (see Fig. S3 in the supplemental material). In the converse scenario, i.e., forced downregulation of GIT2 expression, the expression of multiple DNA repair proteins, e.g., HMGB1, FANCI, HMGN1, and UBE2V1, was significantly diminished (51–54). The poly(ADP-ribose) (PADPR) polymerases (PARPs) play pivotal roles in DNA damage detection and repair. Both single- and double-strand DNA breaks are recognized by PARPs, and the recruitment of PARPs to the damage sites triggers their enzymatic activity. Activated PARPs generate PADPR by cleavage of the nicotinamide-ribose bond of NAD+ and subsequent polymerization of the ADP-ribose units. PADPR formation is an important step in DNA base excision and strand break repair pathways (46, 47). GIT2 possesses ADP-ribosylation factor (Arf) GTPase-activating protein activity within its N-terminal GAP domain (19). We found that GIT2 associates with both PARP1 and PARP2 in response to DNA damage and promotes the activities of both PARPs. In addition, a GIT2 Arf GAP activity-null mutant (R39A) protein and the GIT2 double ATM phosphorylation site mutant (the GIT2 T/S-A/A mutant) are not able to enhance PARP1 or PARP2 activity and thus fail to promote the repair of damaged DNA by WT GIT2 (Fig. 7; see also Fig. S7 in the supplemental material). In association with these multiple findings concerning the role of GIT2 in DNA repair, we found that the levels of GIT2 expression were positively associated with long-term (48 h postinsult) prosurvival activity in the face of multiple cellular stressors (see Fig. S3 in the supplemental material). Therefore, it appears that GIT2 possesses a beneficial and multidimensional role in DNA damage mitigation and long-term survival.
Previously, GIT2-KO mice have been shown to display anxiety-like behaviors in the zero-maze and light-dark emergence tests, indicating that GIT2 has neurobehavioral functions (35). Here, we demonstrated that cortical neurons from GIT2-KO mice have more DNA damage than those from age-matched WT mice (Fig. 7), which suggests that GIT2 plays an important role in DNA repair in vivo. In line with our previous demonstration of an age-related functionality of GIT2 in the CNS (32, 33), we found that young GIT2-KO mice possess an impairment of DNA damage repair capacity that was even higher than that possessed by old WT mice. Therefore, the ability of the animal to maintain functional GIT2 activity may be a factor which dictates the rate of aging in CNS tissues.
In conclusion, the results of our study demonstrate that upon exposure to diverse forms of DNA-damaging agents, GIT2 is rapidly recruited to DSBs in a manner closely regulated by γ-H2AX, ATM, and MRE11. It is likely that H2AX and MRE11 control the recruitment of GIT2, while ATM controls GIT2 phosphorylation, required for the activity of GIT2 in response to DSBs. At these sites of damage, GIT2 promotes the repair of DNA damage (Fig. 2) via interaction with PARPs (Fig. 7), stabilization of repair factors, such as BRCA1, in the repair complex (Fig. 6), and the regulation of expression of multiple DNA repair factors (see Fig. S3 in the supplemental material). Based on our findings, we postulate a novel role for GIT2 in DNA damage repair and long-term genome stability. Disruption of GIT2 expression or functionality may lead to compromised DNA repair in the CNS, eventually leading to the premature induction of neuronal aging-related pathogenic mechanisms. Advanced neuronal aging and accumulated damage may underpin several well-characterized neurodegenerative disorders, such as Alzheimer's or Parkinson's disease (55). Therefore, further elucidation of the role(s) of GIT2, including the potential contribution of naturally occurring GIT2 SQ-TQ motif mutations, in maintaining DNA integrity in these processes would be of interest.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the National Institute on Aging, NIH.
We thank Fred Indig (National Institute on Aging) for his assistance with our confocal laser microscopy. We also thank Vilhelm Bohr (National Institute on Aging) for his advice concerning the preparation of the experimental work flow and manuscript preparation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01432-14.
REFERENCES
- 1.Frappart PO, McKinnon PJ. 2006. Ataxia-telangiectasia and related diseases. Neuromolecular Med 8:495–511. doi: 10.1385/NMM:8:4:495. [DOI] [PubMed] [Google Scholar]
- 2.Madabhushi R, Pan L, Tsai LH. 2014. DNA damage and its links to neurodegeneration. Neuron 83:266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinnon PJ. 2004. ATM and ataxia telangiectasia. EMBO Rep 5:772–776. doi: 10.1038/sj.embor.7400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perlman S, Becker-Catania S, Gatti RA. 2003. Ataxia-telangiectasia: diagnosis and treatment. Semin Pediatr Neurol 10:173–182. doi: 10.1016/S1071-9091(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 5.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali SR, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NG, Taylor AM, Arlett CF, Miki T, Weissman SM, Lovett M, Collins FS, Shiloh Y. 1995. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 6.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 7.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 8.Falck J, Coates J, Jackson SP. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 9.Lee JH, Paull TT. 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 10.Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Ranganathan V, Weisman DS, Heine WF, Ciccone DN, O'Neill TB, Crick KE, Pierce KA, Lane WS, Rathbun G, Livingston DM, Weaver DT. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 12.Gatei M, Sloper K, Sorensen C, Syljuasen R, Falck J, Hobson K, Savage K, Lukas J, Zhou BB, Bartek J, Khanna KK. 2003. Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J Biol Chem 278:14806–14811. doi: 10.1074/jbc.M210862200. [DOI] [PubMed] [Google Scholar]
- 13.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A 97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu B, O'Donnell AH, Kim ST, Kastan MB. 2002. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res 62:4588–4591. [PubMed] [Google Scholar]
- 15.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. 2004. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev 18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamer I, Sarig R, Zaltsman Y, Niv H, Oberkovitz G, Regev L, Haimovich G, Lerenthal Y, Marcellus RC, Gross A. 2005. Proapoptotic BID is an ATM effector in the DNA-damage response. Cell 122:593–603. doi: 10.1016/j.cell.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Sobeck A, Stone S, Landais I, de Graaf B, Hoatlin ME. 2009. The Fanconi anemia protein FANCM is controlled by FANCD2 and the ATR/ATM pathways. J Biol Chem 284:25560–25568. doi: 10.1074/jbc.M109.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Wang M, Bocker W, Iliakis G. 2005. Complex H2AX phosphorylation patterns by multiple kinases including ATM and DNA-PK in human cells exposed to ionizing radiation and treated with kinase inhibitors. J Cell Physiol 202:492–502. doi: 10.1002/jcp.20141. [DOI] [PubMed] [Google Scholar]
- 19.Hoefen RJ, Berk BC. 2006. The multifunctional GIT family of proteins. J Cell Sci 119:1469–1475. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- 20.Premont RT, Claing A, Vitale N, Freeman JL, Pitcher JA, Patton WA, Moss J, Vaughan M, Lefkowitz RJ. 1998. β2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc Natl Acad Sci U S A 95:14082–14087. doi: 10.1073/pnas.95.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Premont RT, Claing A, Vitale N, Perry SJ, Lefkowitz RJ. 2000. The GIT family of ADP-ribosylation factor GTPase-activating proteins. Functional diversity of GIT2 through alternative splicing. J Biol Chem 275:22373–22380. doi: 10.1074/jbc.275.29.22373. [DOI] [PubMed] [Google Scholar]
- 22.Manabe R, Kovalenko M, Webb DJ, Horwitz AR. 2002. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J Cell Sci 115:1497–1510. [DOI] [PubMed] [Google Scholar]
- 23.Zhao ZS, Manser E, Loo TH, Lim L. 2000. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol Cell Biol 20:6354–6363. doi: 10.1128/MCB.20.17.6354-6363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank SR, Adelstein MR, Hansen SH. 2006. GIT2 represses Crk- and Rac1-regulated cell spreading and Cdc42-mediated focal adhesion turnover. EMBO J 25:1848–1859. doi: 10.1038/sj.emboj.7601092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phee H, Dzhagalov I, Mollenauer M, Wang Y, Irvine DJ, Robey E, Weiss A. 2010. Regulation of thymocyte positive selection and motility by GIT2. Nat Immunol 11:503–511. doi: 10.1038/ni.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazaki Y, Hashimoto S, Tsujimura T, Morishige M, Hashimoto A, Aritake K, Yamada A, Nam JM, Kiyonari H, Nakao K, Sabe H. 2006. Neutrophil direction sensing and superoxide production linked by the GTPase-activating protein GIT2. Nat Immunol 7:724–731. doi: 10.1038/ni1349. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Webb DJ, Asmussen H, Horwitz AF. 2003. Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol 161:131–142. doi: 10.1083/jcb.200211002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. 2005. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci 25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko J, Kim S, Valtschanoff JG, Shin H, Lee JR, Sheng M, Premont RT, Weinberg RJ, Kim E. 2003. Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting. J Neurosci 23:1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segura I, Essmann CL, Weinges S, Acker-Palmer A. 2007. Grb4 and GIT1 transduce ephrinB reverse signals modulating spine morphogenesis and synapse formation. Nat Neurosci 10:301–310. doi: 10.1038/nn1858. [DOI] [PubMed] [Google Scholar]
- 31.Schmalzigaug R, Phee H, Davidson CE, Weiss A, Premont RT. 2007. Differential expression of the ARF GAP genes GIT1 and GIT2 in mouse tissues. J Histochem Cytochem 55:1039–1048. doi: 10.1369/jhc.7A7207.2007. [DOI] [PubMed] [Google Scholar]
- 32.Chadwick W, Zhou Y, Park SS, Wang L, Mitchell N, Stone MD, Becker KG, Martin B, Maudsley S. 2010. Minimal peroxide exposure of neuronal cells induces multifaceted adaptive responses. PLoS One 5:e14352. doi: 10.1371/journal.pone.0014352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chadwick W, Martin B, Chapter MC, Park SS, Wang L, Daimon CM, Brenneman R, Maudsley S. 2012. GIT2 acts as a potential keystone protein in functional hypothalamic networks associated with age-related phenotypic changes in rats. PLoS One 7:e36975. doi: 10.1371/journal.pone.0036975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chadwick W, Mitchell N, Caroll J, Zhou Y, Park SS, Wang L, Becker KG, Zhang Y, Lehrmann E, Wood WH III, Martin B, Maudsley S. 2011. Amitriptyline-mediated cognitive enhancement in aged 3×Tg Alzheimer's disease mice is associated with neurogenesis and neurotrophic activity. PLoS One 6:e21660. doi: 10.1371/journal.pone.0021660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caffrey DR, Zhao J, Song Z, Schaffer ME, Haney SA, Subramanian RR, Seymour AB, Hughes JD. 2011. siRNA off-target effects can be reduced at concentrations that match their individual potency. PLoS One 6:e21503. doi: 10.1371/journal.pone.0021503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerlitz G. 2010. HMGNs, DNA repair and cancer. Biochim Biophys Acta 1799:80–85. doi: 10.1016/j.bbagrm.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim ST, Lim DS, Canman CE, Kastan MB. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 38.Kitagawa R, Kastan MB. 2005. The ATM-dependent DNA damage signaling pathway. Cold Spring Harbor Symp Quant Biol 70:99–109. doi: 10.1101/sqb.2005.70.002. [DOI] [PubMed] [Google Scholar]
- 39.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 40.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895. doi: 10.1016/S0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 41.Peng Z, Liao Z, Dziegielewska B, Matsumoto Y, Thomas S, Wan Y, Yang A, Tomkinson AE. 2012. Phosphorylation of serine 51 regulates the interaction of human DNA ligase I with replication factor C and its participation in DNA replication and repair. J Biol Chem 287:36711–36719. doi: 10.1074/jbc.M112.383570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blagosklonny MV, Iglesias A, Zhan Z, Fojo T. 1998. Like p53, the proliferation-associated protein p120 accumulates in human cancer cells following exposure to anticancer drugs. Biochem Biophys Res Commun 244:368–373. doi: 10.1006/bbrc.1998.8278. [DOI] [PubMed] [Google Scholar]
- 43.Yabe D, Fukuda H, Aoki M, Yamada S, Takebayashi S, Shinkura R, Yamamoto N, Honjo T. 2007. Generation of a conditional knockout allele for mammalian Spen protein Mint/SHARP. Genesis 45:300–306. doi: 10.1002/dvg.20296. [DOI] [PubMed] [Google Scholar]
- 44.Daley JM, Sung P. 2014. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol 34:1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 46.Hassa PO, Haenni SS, Elser M, Hottiger MO. 2006. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. 2005. Specific binding of poly(ADP-ribose) polymerase-1 to cruciform hairpins. J Mol Biol 348:609–615. doi: 10.1016/j.jmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Shiloh Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 49.Hornyik C, Terzi LC, Simpson GG. 2010. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18:203–213. doi: 10.1016/j.devcel.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 50.Fonagy A, Swiderski C, Freeman JW. 1995. Nucleolar p120 is expressed as a delayed early response gene and is inducible by DNA-damaging agents. J Cell Physiol 164:634–643. doi: 10.1002/jcp.1041640322. [DOI] [PubMed] [Google Scholar]
- 51.Elder RM, Jayaraman A. 2012. Sequence-specific recognition of cancer drug-DNA adducts by HMGB1a repair protein. Biophys J 102:2331–2338. doi: 10.1016/j.bpj.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sareen A, Chaudhury I, Adams N, Sobeck A. 2012. Fanconi anemia proteins FANCD2 and FANCI exhibit different DNA damage responses during S-phase. Nucleic Acids Res 40:8425–8439. doi: 10.1093/nar/gks638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masaoka A, Gassman NR, Kedar PS, Prasad R, Hou EW, Horton JK, Bustin M, Wilson SH. 2012. HMGN1 protein regulates poly(ADP-ribose) polymerase-1 (PARP-1) self-PARylation in mouse fibroblasts. J Biol Chem 287:27648–27658. doi: 10.1074/jbc.M112.370759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franko J, Ashley C, Xiao W. 2001. Molecular cloning and functional characterization of two murine cDNAs which encode Ubc variants involved in DNA repair and mutagenesis. Biochim Biophys Acta 1519:70–77. doi: 10.1016/S0167-4781(01)00223-8. [DOI] [PubMed] [Google Scholar]
- 55.Janssens J, Etienne H, Idriss S, Azmi A, Martin B, Maudsley S. 2014. Systems-level G protein-coupled receptor therapy across a neurodegenerative continuum by the GLP-1 receptor system. Front Endocrinol 5:142. doi: 10.3389/fendo.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.