Abstract
Ascidians present a striking dichotomy between conserved phenotypes and divergent genomes: embryonic cell lineages and gene expression patterns are conserved between distantly related species. Much research has focused on Ciona or Halocynthia spp. but development in other ascidians remains poorly characterized. In this study, we surveyed the multipotent myogenic B7.5 lineage in Molgula spp. Comparisons to the homologous lineage in Ciona revealed identical cell division and fate specification events that result in segregation of larval, cardiac, and pharyngeal muscle progenitors. Moreover, the expression patterns of key regulators are conserved, but cross-species transgenic assays uncovered incompatibility, or ‘unintelligibility’, of orthologous cis-regulatory sequences between Molgula and Ciona. These sequences drive identical expression patterns that are not recapitulated in cross-species assays. We show that this unintelligibility is likely due to changes in both cis- and trans-acting elements, hinting at widespread and frequent turnover of regulatory mechanisms underlying otherwise conserved aspects of ascidian embryogenesis.
DOI: http://dx.doi.org/10.7554/eLife.03728.001
Research organism: other
eLife digest
When two species have features that look similar, this may be because the features arise by the same processes during development. Other features may look similar yet develop by different mechanisms. ‘Developmental system drift’ refers to the process where a physical feature remains unaltered during evolution, but the underlying pathway that controls its development is changed. However, to date, there have been only a few experimental studies that support this idea.
Ascidians—also commonly known as sea squirts—are vase-like marine creatures, which start off as tadpole-like larvae that swim around until they find a place to settle down and attach themselves. Once attached, the sea squirts lose the ability to swim and start feeding, typically by filtering material out of the seawater. Sea squirts and their close relatives are the invertebrates (animals without backbones) that are most closely related to all vertebrates (animals with backbones), including humans. Furthermore, although different species of sea squirt have almost identical embryos, their genomes are very different.
Stolfi et al. have now studied whether developmental system drift may have occurred during the evolution of ascidians, by analyzing different species of sea squirt named Molgula and Ciona. Stolfi et al. compared the genomes of Molgula and Ciona and studied the expression of genes in the cells that give rise to the heart and the muscles of the head. As an embryo develops, specific genes are switched on or off, and these patterns of gene activation were broadly identical in the two species of sea squirt examined.
Enhancers are sequences of DNA that control when and how a gene is switched on. Given the similarities between the development of heart and head muscle cells in the different sea squirts, Stolfi et al. looked to see if the mechanisms of gene expression, and therefore the enhancers, were also conserved. Unexpectedly, this was not the case. When enhancers from Molgula were introduced into Ciona (and vice versa), these sequences were unable to switch on gene expression—thus enhancers from one sea squirt species could not function in the other.
Stolfi et al. conclude that the developmental systems may have drifted considerably during evolution of the sea squirts, in spite of their nearly identical embryos. This reinforces the view that different paths can lead to the formation of similar physical features.
Introduction
Transcriptional regulatory networks are indispensable for the generation of the diverse germ layers, anatomical structures, and cell types of multicellular organisms (Levine and Davidson, 2005). The impact of cis-regulatory DNA sequence changes on the evolution of development is undeniable but not yet fully understood (Wray, 2007; Wittkopp and Kalay 2011). Phenotypic differences between species have often been traced to differences in expression of orthologous genes. In turn, this differential expression can be attributed to changes in cis-regulatory modules (or ‘enhancers’), which may alter their binding by sequence-specific transcription factors (TFs) (Sucena and Stern, 2000; Gompel et al., 2005; Prud'homme et al., 2006; Miller et al., 2007; Shirangi et al., 2009).
In contrast, gene expression patterns may be identical in spite of differences in the cis-regulatory DNA sequences controlling their transcription. For instance, orthologous enhancers might be interchangeable between different species without conspicuous DNA sequence similarity (Maduro and Pilgrim, 1996; Ludwig et al., 1998; Piano et al., 1999; Romano and Wray, 2003; Oda-Ishii et al., 2005). This is often attributed to the conservation of gene regulatory networks (GRNs) and the flexibility of TF binding site distribution in a given enhancer, which contribute to conservation of enhancer function (Ludwig et al., 2000; Oda-Ishii et al., 2005; Hare et al., 2008; Weirauch and Hughes, 2010). This cryptic regulatory turnover is termed ‘developmental system/systems drift’ (DSD) (True and Haag, 2001). This term broadly applies to the divergence in the molecular or morphogenetic basis for the development of identical homologous characters.
However, gene expression patterns that appear conserved between species may also be the output of divergent regulatory networks (Ludwig et al., 2005). In such cases, one species' trans environment may be unable to interpret the other species' enhancer, even if the expression patterns controlled by the orthologous enhancers are identical between the two species. This particularly acute manifestation of DSD is thought to contribute to developmental defects in interspecies hybrids (Takano, 1998), contributing to reproductive isolation and speciation as cis/trans combinations become so incompatible as to be lethal (Porter and Johnson, 2002). There is ample evidence that DSD is pervasive in metazoan evolution (Kiontke et al., 2007; Verster et al., 2014), but it has been difficult to study due to its cryptic nature.
Sea squirts, or ascidians, are well suited for investigating the evolution of gene regulation in development (Satoh, 2013). Species on the opposite branches of the ascidian tree, estimated to have diverged ∼520 million years apart, have extremely conserved embryos that share identical stereotyped cell divisions up until the late gastrula stage and beyond (Swalla, 2006; Lemaire, 2009). This allows one to unequivocally infer homology between specific embryonic cells, gene expression patterns, and GRNs. This is in contrast to the striking lack of non-coding sequence conservation, revealed by bioinformatic alignment of genomic sequences from disparate ascidian species. This means that any two randomly selected ascidian species are likely to be more genetically divergent from one another than humans are from fish, which share several highly conserved non-coding sequences (Woolfe et al., 2004). This dichotomy between near-identical embryos and un-alignable cis-regulatory sequences represents a singular paradox in the study of evolution and developmental biology today (Lemaire, 2011).
Modern molecular tools for embryo manipulation and visualization are available for several ascidian species, including a protocol for transfection en masse by electroporation of plasmid DNA into fertilized eggs of the solitary ascidian Ciona intestinalis (Corbo et al., 1997; Christiaen et al., 2009b). This experimental tractability has allowed researchers to begin addressing the ascidian embryological paradox. Regulation of the Otx gene is a prime example, as enhancers upstream of the Otx genes from C. intestinalis and the distantly related Halocynthia roretzi do not show any DNA sequence similarity but are broadly functional in cross-species assays. This was shown to be due to conservation of cis-regulatory logic in spite of a reshuffling of functional TF binding sites (Oda-Ishii et al., 2005). On the other hand, comparisons of C. intestinalis and H. roretzi development have also revealed clear examples of acute DSD in regulation of the Brachyury gene (Takahashi et al., 1999), cell signaling upstream of pigment cell differentiation (Darras and Nishida, 2001; Abitua et al., 2012), secondary tail muscle specification (Kim and Nishida, 2001; Tokuoka et al., 2007; Hudson et al., 2007; Hudson and Yasuo, 2008), and embryonic marginal zone patterning (Takatori et al., 2010; Hudson et al., 2013).
These few examples suggest that considerable DSD may have occurred during the evolution of ascidians, even with their presupposed highly constrained mode of embryogenesis. To get a better idea of how DSD may have shaped ascidian evolution, it was necessary to identify species comparable to Ciona spp. in their experimental tractability but phylogenetically distant enough as to maximize the genetic differences between them.
Molgula spp. belong to the order Stolidobranchia like H. roretzi, but produce embryos that are more comparable in size and developmental rate to those of C. intestinalis, a member of the order Phlebobranchia (Figure 1A). The genus is also remarkable for containing several species that have independently evolved anural development (Berrill, 1931; Swalla and Jeffery, 1990; Hadfield et al., 1995; Jeffery et al., 1999; Huber et al., 2000). These anural, or ‘tail-less’, Molgula species produce immotile larvae lacking the differentiated structures that are required for swimming (Swalla and Jeffery, 1990; Kusakabe et al., 1996), and some species even appear to bypass the larval stage altogether (Tagawa et al., 1997; Maliska and Swalla, 2010). Thus, Molgula species comprise a unique group in which to study body plan and life cycle evolution.
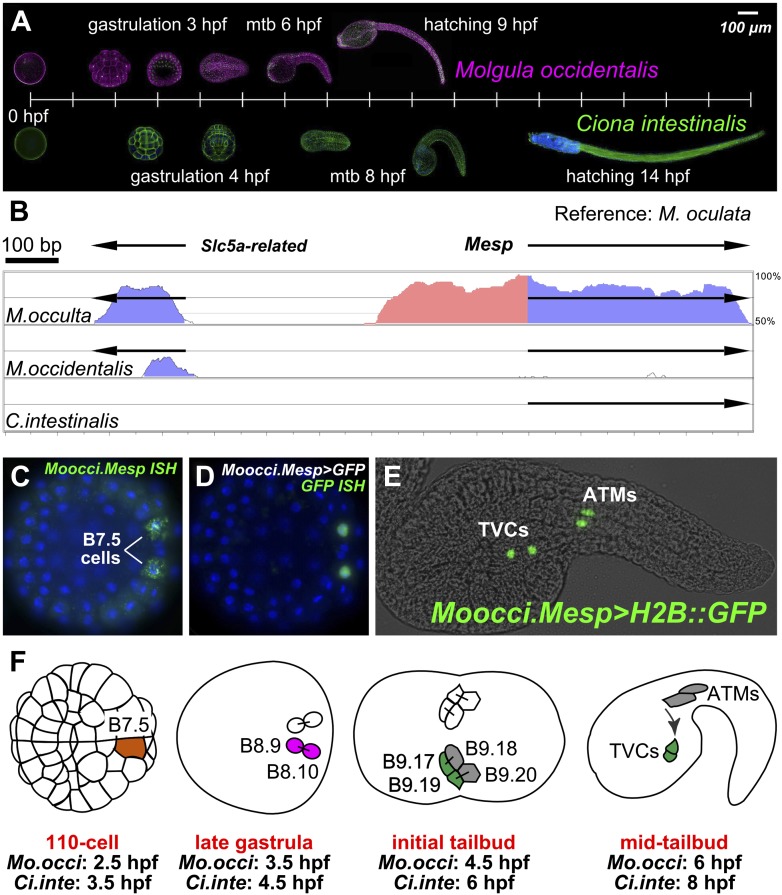
Figure 1. The B7.5 lineage in M. occidentalis.
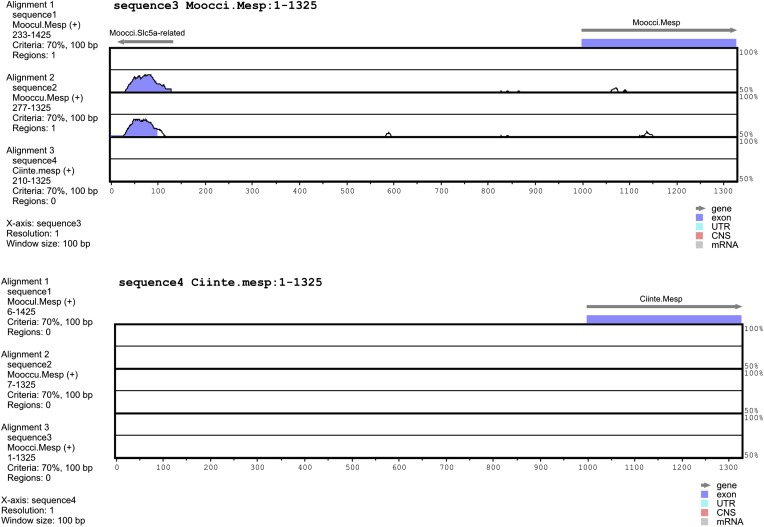
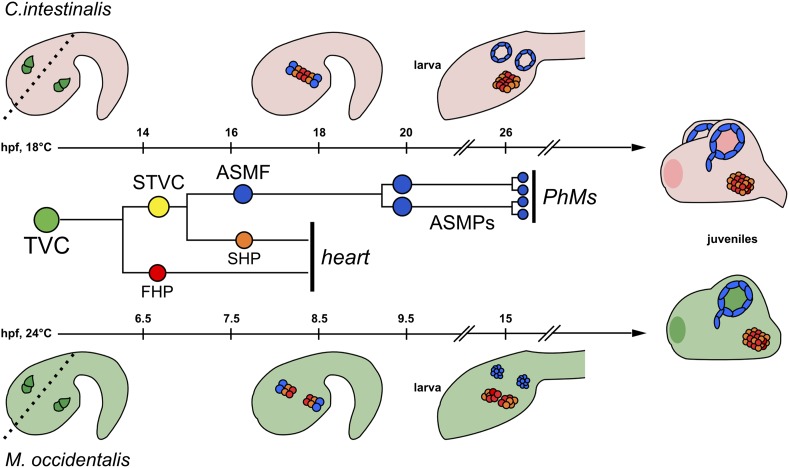
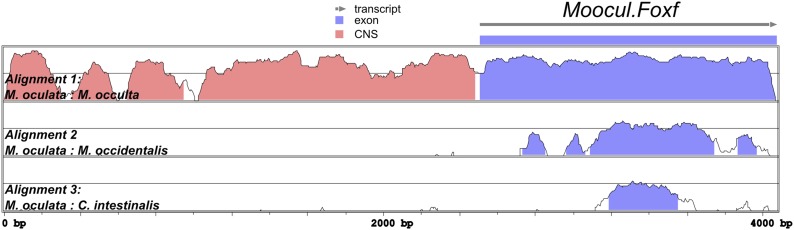
(A) Diagram comparing M. occidentalis (top) and C. intestinalis (bottom) embryogenesis at 24°C. Embryos were stained with Alexa Fluor dye-conjugated phalloidin to visualize cell outlines and DAPI to visualize cell nuclei. (B) Diagram of mVISTA (Frazer et al., 2004; genome.lbl.gov/vista/) alignment of M. oculata Mesp (Moocul.Mesp) locus to orthologs in M. occulta, M. occidentalis, and C. intestinalis. Shaded peaks indicate sequence conservation above 70% over 100-bp windows (blue = protein-coding, pink = non-coding). Arrows indicate direction of transcription of protein-coding genes. Non-coding sequences upstream of Mesp are only conserved between M. oculata and M. occulta. M. occidentalis and C. intestinalis show considerable divergence even in protein-coding sequences. Note that microsynteny with SLC5A-related gene supports the orthology of these sequences among the Molgulids. (C) In situ hybridization (ISH) for Moocci.Mesp in 110-cell stage embryo (vegetal view), showing mRNA detection (green) in B7.5 blastomeres. Nuclei were counterstained with DAPI (blue). Staging is given by hours post-fertilization (hpf). (D) Vegetal view of a 110-cell stage embryo electoporated with Moocci.Mesp>GFP reporter construct. Reporter gene expression was detected by ISH for GFP transcripts (green). Nuclei were stained with DAPI (blue). (E) Lateral view of a mid-tailbud stage embryo electroporated with Moocci.Mesp>Histone2B::GFP reporter construct. GFP fluorescence reveals B7.5 descendants on left side of embryo: two trunk ventral cells (TVCs) and two anterior tail muscles (ATMs). (F) Diagram of B7.5 lineage divisions from 110-cell stage to mid-tailbud stage, inferred from previous C. intestinalis studies. Cells are named according to Conklin's method (Conklin, 1905). The lineage is bilaterally symmetric, but only cells on the left side are indicated and named. Relative staging given for M. occidentalis (Mo.occi) and C. intestinalis (Ci.inte). 110-cell and late gastrula: vegetal view. Initial tailbud: dorsal view. Mid-tailbud: lateral view. Anterior pole is on the left in all images and illustrations.
Figure 1—figure supplement 1. Alignment of 5′ flanking sequences from Mesp orthologs.
Figure 1—figure supplement 2. The B7.5 lineage of the tail-less species M. occulta.
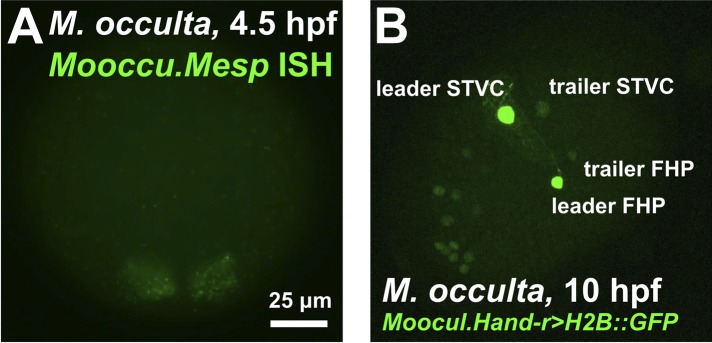
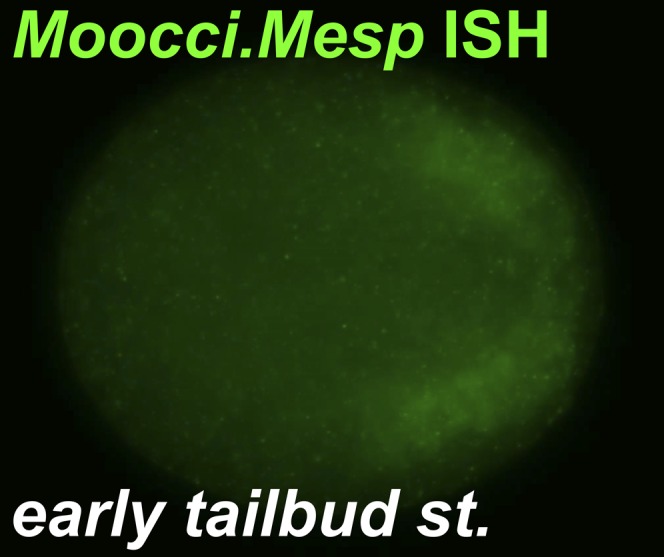
Figure 1—figure supplement 3. Moocci.Mesp in situ hybridization at tailbud stage.
In order to realize the full potential of Molgulid ascidians as model organisms for molecular and comparative developmental genetics on par with Ciona spp., we sequenced and assembled the genomes of three Molgula species: the tailed species M. occidentalis and M. oculata, and the tail-less M. occulta. Moreover, we adapted the electroporation protocol to M. occidentalis embryos. To illustrate the power of comparative approaches between Molgula and Ciona, we characterized the B7.5 lineage in M. occidentalis. In both C. intestinalis and H. roretzi, this lineage is descended from the B7.5 pair of blastomeres of the early gastrula embryo and gives rise to the anterior tail muscles of the larva, and to the heart and atrial siphon/pharyngeal muscles of the adult (Hirano and Nishida, 1997; Davidson and Levine, 2003; Stolfi et al., 2010). A growing body of research has focused on the molecular basis of B7.5 lineage development in C. intestinalis (reviewed in Tolkin and Christiaen, 2012; Cota et al., 2013), some of which have guided the discovery of vertebrate heart developmental mechanisms (Islas et al., 2012). Given its detailed characterization in C. intestinalis, we chose the B7.5 lineage for an in-depth comparative analysis. Our analysis revealed a remarkable degree of conservation of the clonal topology and gene expression patterns (together referred to as the ‘ontogenetic motif’) underlying ascidian cardiopharyngeal mesoderm development (Wang et al., 2013), but also uncovered divergent regulatory mechanisms underlying conserved gene expression profiles between Ciona and Molgula.
Results
De novo sequencing of Molgula spp. genomes
Genomes of three Molgula species (M. occidentalis, M. oculata, and M. occulta) were sequenced using next-generation sequencing technology and assembled. A common metric for judging the quality of a genome assembly is the contig N50 length, which is determined such that 50% of the assembly is contained in contigs of this length or greater. We used the contig N50 length to select the best assembly for each species given the varying ‘k’ parameter (length of k-mer overlap). A ‘k’ of 39 yields the best assembly for both M. occidentalis and M. occulta. The best ‘k’ for M. oculata was 61. M. occidentalis, M. occulta, and M. oculata N50 lengths were approximately 26.3 kb, 13 kb, and 34 kb, respectively (Table 1).
Table 1.
Genome assembly statistics
| Species | N50 | Mean contig length | Total | Total number of base pairs | CEGMA C1 | CEGMA P2 |
|---|---|---|---|---|---|---|
| M. occidentalis | 26,298 | 5072 | 51,761 | 262,547,660 | 81.45 | 96.77 |
| M. occulta | 13,011 | 3233 | 58,489 | 189,110,562 | 77.42 | 98.79 |
| M. oculata | 34,042 | 6270 | 25,497 | 159,886,716 | 89.92 | 99.19 |
The contig N50 length, mean contig length, total number of contigs, total number of base pairs and CEGMA scores were collected for each draft assembly. The CEGMA scores is a metric of completeness measured against highly Conserved eukaryotic genes. Alignments of 70% or greater of the protein length are called complete (C1) and all other statistically significant alignments are called partial (P2).
In addition to N50 lengths, we also used CEGMA (Core Eukaryotic Genes Mapping Approach) scores, in order to evaluate the assemblies' representative completeness (Parra et al., 2007). CEMGA reports scores for complete and partial alignments to a subset of core eukaryotic genes. An alignment is considered ‘complete’ if at least 70% of a given protein model aligns to a contig in the assembly, while a partial alignment indicates that a statistically significant portion of the protein model aligns. The partial alignment scores are ∼97% or higher for all assemblies. M. oculata has the best complete alignment score at ∼90%. M. occidentalis and M. occulta have complete alignment scores of 81% and 77% respectively (Table 1). These scores indicate that our assemblies contain at least partial sequences for the vast majority of protein-coding genes in the genomes of these species.
Various factors make it unreliable to predict genome size and gene density based on assembly metrics alone (Bradnam et al., 2013). Of the handful of sequences we isolated and analyzed, we found that the sizes of introns and upstream regulatory regions were roughly comparable to those from their Ciona orthologs. This suggests that the Molgula genomes may be as compact as the C. intestinalis genome (i.e., ∼150–170 Mb, ∼16,000 genes, Laird, 1971; Simmen et al., 1998; Satou et al., 2008).
Our sequencing efforts revealed extreme genetic divergence not only between Ciona and Molgula, as expected, but even within the Molgulids. For example, we used BLAST to identify the Molgula orthologs of C. intestinalis Mesp (Ciinte.Mesp, as per the proposed tunicate gene nomenclature rules, see Stolfi et al., 2014). Ciinte.Mesp is the sole ortholog of vertebrate genes coding for MesP and Mesogenin bHLH transcription factor family members (Satou et al., 2004). VISTA alignment shows high sequence similarity between sequences 5′ upstream of the Mesp genes from the closely related M. oculata and M. occulta (Figure 1B). However, there is no conservation of Mesp DNA sequences, coding or non-coding, between M. oculata/occulta and M. occidentalis, nor between C. intestinalis and any of the three Molgula species (Figure 1—figure supplement 1). In previous phylogenetic surveys, M. occidentalis has been placed as an early-branching Molgula species, often grouped together in a subfamily with species ascribed to the genera Eugyra and Bostrichobranchus instead (Hadfield et al., 1995; Huber et al., 2000; Tsagkogeorga et al., 2009). Our sequencing results support the view that M. occidentalis is highly diverged from other Molgula spp.
Expression of the M. occidentalis Mesp gene marks the B7.5 cells
Ciinte.Mesp specifies the B7.5 cells as the sole progenitors of the cardiopharyngeal lineage (Satou et al., 2004; Davidson et al., 2005; Hirano and Nishida, 1997; Stolfi et al., 2010). We performed RNA in situ hybridization (ISH) for M. occidentalis Mesp (Moocci.Mesp) and found that this gene is also expressed only in the B7.5 cells of M. occidentalis embryos (Figure 1C). These cells are unequivocally identified due to the perfect conservation of early embryonic cell cleavage patterns in all ascidians. ISH for the M. occulta Mesp gene (Mooccu.Mesp) also revealed conserved expression in this tail-less species (Figure 1—figure supplement 2A).
We successfully adapted the Ciona electroporation protocol for simultaneous transfection of reporter gene plasmids into hundreds of synchronized M. occidentalis embryos (Figure 1D,E). We were also able to electroporate M. occulta embryos (Figure 1—figure supplement 2B). However, only M. occidentalis was routinely available to us for in vivo studies, so we focused our experiments on this species. Development of M. occidentalis embryos was optimal at 24°C and faster than that of C. intestinalis (Figure 1A). Using electroporation-based transfection, we determined that an ∼1.1 kb genomic DNA fragment upstream of Moocci.Mesp is able to drive expression of fused reporter genes specifically in the B7.5 cells with no ‘leaky’ expression in other cells as is commonly observed in C. intestinalis (Figure 1D; Stolfi and Christiaen, 2012).
This faithful recapitulation of Moocci.Mesp expression and the persistence of GFP allows for visualization of the descendants of B7.5 long after endogenous Moocci.Mesp transcription has ceased (Figure 1—figure supplement 3; Davidson et al., 2005). At the tailbud stage, we find that each B7.5 blastomere gives rise to four grand-daughter cells (Figure 1E). The two anterior B7.5 grand-daughter cells on either side of the bilaterally symmetric embryo migrate anteriorly and are termed the trunk ventral cells (TVCs) due to their final position in the C. intestinalis and H. roretzi embryos (Nishida, 1987). Their posterior sister cells remain in the tail and become anterior tail muscles (ATMs). As far as we can tell, B7.5 lineage ontogeny is perfectly conserved between M. occidentalis and C. intestinalis (Figure 1F).
Cardiopharyngeal mesoderm gene expression
Our focus shifted to the TVCs, which in H. roretzi and C. intestinalis have been shown to be multipotent cardiopharyngeal progenitor cells that give rise to the heart and pharyngeal muscles of the atrial siphon (Hirano and Nishida, 1997; Stolfi et al., 2010). We performed a small ISH screen for orthologs of transcription factors that are expressed in C. intestinalis TVCs (Satou et al., 2004; Davidson et al., 2005; Davidson and Levine, 2003; Christiaen et al., 2008; Beh et al., 2007). These include the orthologs of conserved cardiac regulators Ets1/2, GATA4/5/6, and NK4/Nkx2.5/tinman (Cripps and Olson, 2002) (Figure 2).
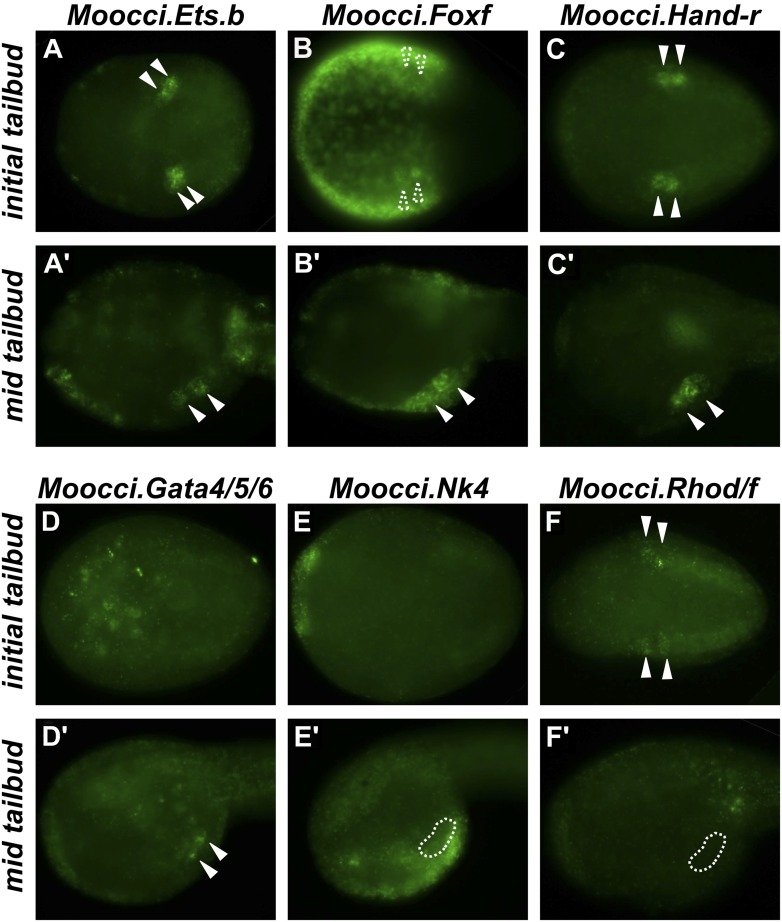
Figure 2. Expression of conserved TVC/heart markers in M. occidentalis embryos.
In situ hybridization (ISH) in M. occidentalis embryos for (A and A′) Moocci.Ets.b, (B and B′) Moocci.Foxf, (C and C′) Moocci.Hand-related (Moocci.Hand-r), (D and D′) Moocci.Gata4/5/6, (E and E′) Moocci.Nk4, (F and F′) Moocci.Rhod/f. ISH was performed on initial tailbud (A–F) and mid tailbud (A′–F′) stage embryos. Solid arrowheads indicate definitive expression in TVCs. Dotted arrowheads indicate potential expression of Moocci.Foxf in initial tailbud, obscured by strong epidermal expression. Dotted outline indicates probable position of TVCs, not visible due to lack of mRNA hybridization signal. Initial tailbud embryos were imaged ventrally or dorsally, while tailbud embryos were imaged laterally.
Figure 2—figure supplement 1. Ets.b expression in B7.5 of M. occidentalis.
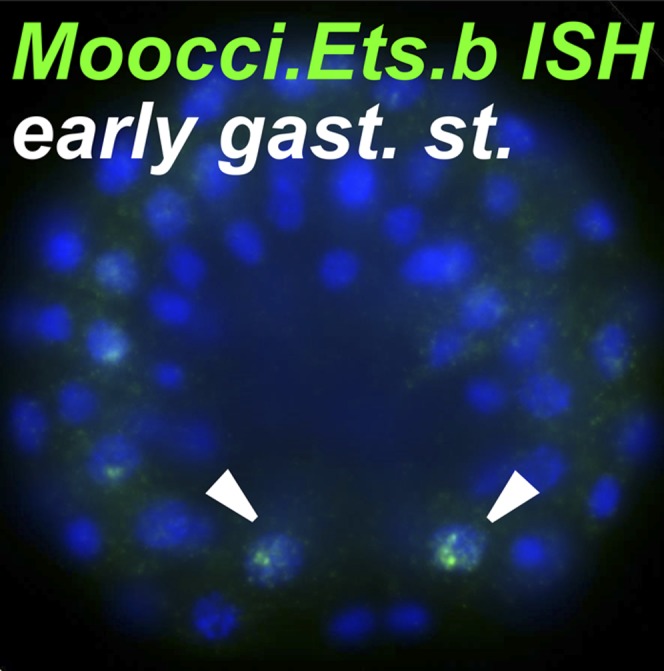
Figure 2—figure supplement 2. In situ hybridizations reveal TVC gene expression in M. occidentalis.
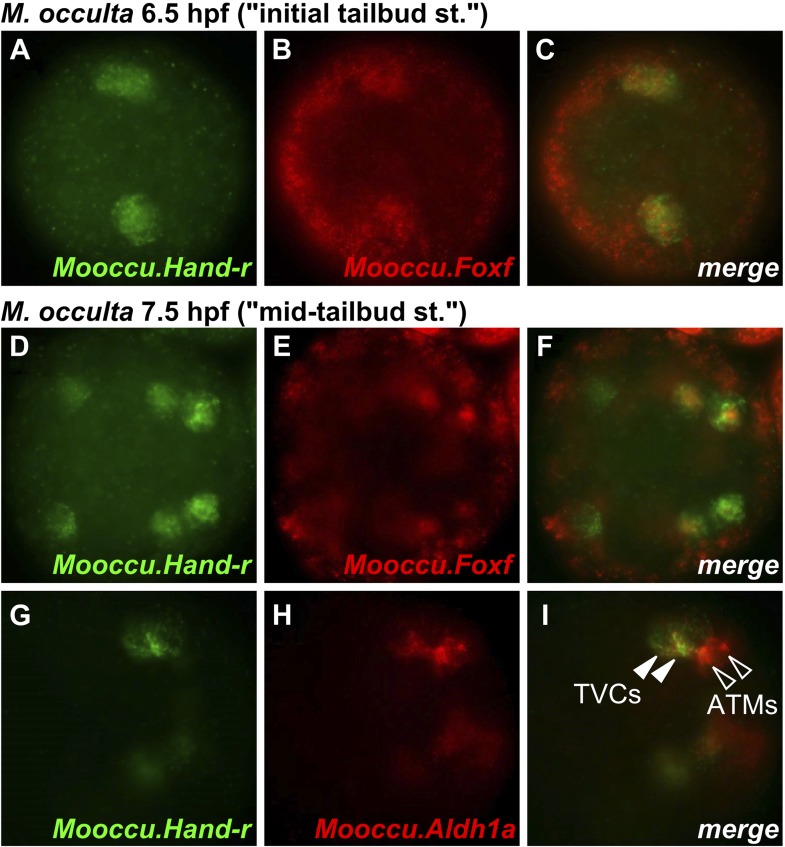
Figure 2—figure supplement 3. Hand-r and FoxF co-expression reveals TVCs of M. occulta.
Figure 2—figure supplement 4. Aldh1a expression in M. occidentalis embryo.

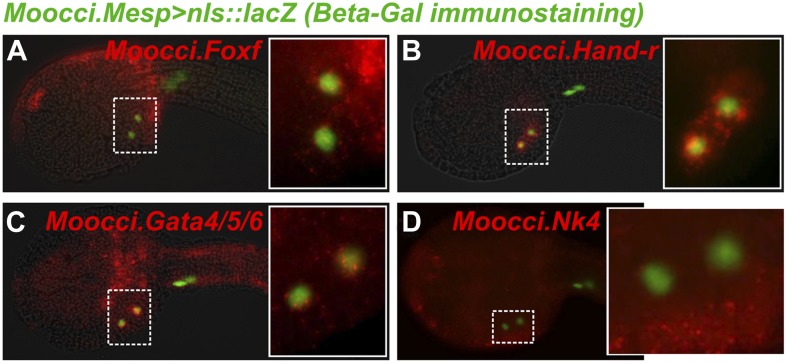
In M. occidentalis, Ets.b expression is initiated in B7.5 blastomeres (Figure 2—figure supplement 1), is maintained in their daughter cells the cardiopharyngeal founders and in the TVCs during their migration (Figure 2A). This profile is similar to the expression of C. intestinalis Ets.b (Ciinte.Ets.b), previously named Ets/pointed2 or Ets1/2 (see Supplementary file 2 for list of old and new gene name correspondences). In C. intestinalis, Ets.b mediates the FGF/MAPK-dependent induction of TVCs in part by the activation of key regulators such as Foxf, Hand-related (Hand-r, also known as Hand-like or NoTrlc), and Gata4/5/6 (also known as GATA-a) prior to the onset of TVC migration (Davidson et al., 2006; Beh et al., 2007). In M. occidentalis, orthologs of Foxf and Hand-r are also activated in the TVCs shortly before and throughout their migration away from the ATMs (Figure 2B,C). Moocci.Gata4/5/6 expression was detected in migrating TVCs but not before migration (Figure 2D). This is slightly different from Ciinte.Gata4/5/6, which is expressed in C. intestinalis TVCs prior to migration (Christiaen et al., 2010; Ragkousi et al., 2011). Expression of Moocci.Foxf and Moocci.Gata4/5/6 in surrounding epidermis and endoderm, respectively, is identical to the expression domains of their orthologs in C. intestinalis and sometimes obscured TVC expression. However, double ISH/immunohistochemical detection (ISH/IHC) of Moocci.Mesp promoter-driven reporter gene clearly shows transcripts in the migrating B7.5-derived TVCs (Figure 2—figure supplement 2A–C).
Foxf and Hand-r are also expressed in the TVCs of tail-less M. occulta embryos. Because these embryos do not extend a tail and lack functional larval tail muscles, it is unclear whether their TVCs move away from the posterior pole of the embryo (Figure 2—figure supplement 3A–F). However, their TVCs occupy the ventro-lateral regions of the trunk (Figure 1—figure supplement 2B), similar to what is observed in M. occidentalis, suggesting they might be migrating. We cannot draw any further conclusions about the TVCs of M. occulta, but have not found any evidence for fundamental molecular or morphological differences relative to M. occidentalis.
Notably, the ATMs appear to be specified in M. occulta, as revealed by expression of Aldh1a (also known as Raldh2, Figure 2—figure supplement 3G–I), which encodes the rate-limiting enzyme for retinoic acid (RA) synthesis. Therefore, although the larval muscles of M. occulta do not differentiate (Swalla and Jeffery, 1990; Kusakabe et al., 1996), their ATMs may still be required for RA-mediated embryonic patterning like in C. intestinalis (Nagatomo and Fujiwara, 2003) and probably also in M. occidentalis (Figure 2—figure supplement 4).
Unexpectedly, we could not detect unequivocal expression of the sole M. occidentalis ortholog of NK4/Nkx2.5/tinman (Moocci.Nk4) in the TVCs (Figure 2E). Expression in the ventral ectoderm mirrored that seen in C. intestinalis, indicating that the probe used was functional. It is possible that expression of Nk4 in the TVCs of M. occidentalis is below the threshold of reliable detection by ISH. Our ISH data are not entirely incompatible with Moocci.Nk4 being expressed in the TVCs at very low levels (Figure 2—figure supplement 2D).
Conserved TVC gene expression patterns were not limited to TF-coding genes. The small GTPase-coding Rhod/f gene was found to be transcriptionally upregulated in M. occidentalis TVCs just prior to migration (Figure 2F). In C. intestinalis, Rhod/f was identified as an effector of cytoskeleton dynamics in TVC migration and a direct transcriptional target of Foxf and activated Ets.b (Christiaen et al., 2008). This suggests that the conservation of B7.5 development in ascidians extends to the interface between transcriptional regulators and cellular effectors of cell migration.
Specification of heart and pharyngeal muscles from common progenitors
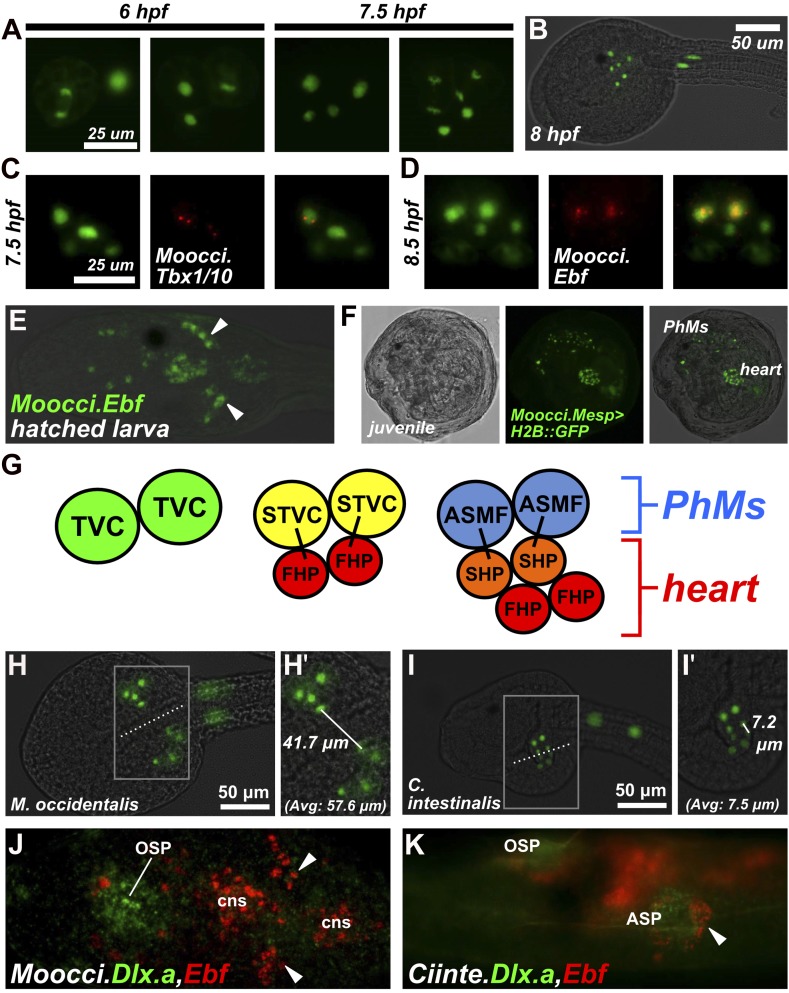
In C. intestinalis, heart and atrial siphon (pharyngeal) muscle precursors are derived from TVCs in a two-step process involving asymmetric cell divisions (Stolfi et al., 2010; Wang et al., 2013). First, each TVC undergoes an asymmetric cell division along the medial/lateral (M/L) axis to produce a medial First Heart Precursor (FHP) and a lateral Secondary TVC (STVC). Each STVC then also undergoes a M/L asymmetric division to give rise to a medial Second Heart Precursor (SHP) and a lateral atrial siphon muscle founder cell (ASMF). In C. intestinalis, this step-wise segregation of heart vs pharyngeal muscle fate is coordinated by (1) activation of Tbx1/10 in the STVCs (Wang et al., 2013) and (2) Tbx1/10-dependent activation of Collier/Olf/EBF (Ciinte.Ebf, previously named COE) in the ASMFs (Stolfi et al., 2010; Wang et al., 2013; Razy-Krajka et al., 2014). The shared clonality of heart and pharyngeal muscles and similarity of associated TF expression patterns point to a single evolutionary origin for this cardiopharyngeal ‘ontogenetic motif’ in the last common ancestor of tunicates and vertebrates (Wang et al., 2013).
In M. occidentalis, we found that the segregation of heart and pharyngeal muscle fates occurs in identical fashion to that of C. intestinalis, albeit on a faster timescale. At 6 hr post-fertilization (hpf) the first asymmetric division of the TVCs begins, with each TVC giving rise to a smaller, more ventral/medial cell and a larger, more dorsal/lateral cell. At 7.5 hpf, the resulting STVCs also undergo an asymmetric division along the same axis (Figure 3A). The end result is a cluster consisting of two larger cells (presumptive ASMFs) lateral to four smaller cells (presumptive heart precursors), on either side of the embryo (Figure 3B).
Figure 3. Specification of heart vs atrial siphon muscle precursors in M. occidentalis.
(A) First and second asymmetric division of TVCs in M. occidentalis embryos. B7.5 lineage was visualized by electroporation of Moocci.Mesp>H2B::GFP (green). The first division occurs at ∼6 hpf at 24°C, and the second division occurs at ∼7.5 hpf at 24°C. (B) Result of two asymmetric divisions of TVCs on left side of embryo electroporated with Moocci.Mesp>H2B::GFP (green). At 8 hpf at 24°C, a cluster of 6 cells derived from the TVCs is located in the ventro-lateral region of the trunk. From top to bottom: 2 atrial siphon muscle founder cells (ASMFs), 2 second heart precursors (SHPs), and 2 first heart precursors (FHPs). (C) In situ hybridization (ISH, red) for Moocci.Tbx1/10 + β-galactosidase immunodetection (IHC, green) in embryos electoporated with Moocci.Mesp>nls::lacZ. Moocci.Tbx1/10 nascent transcripts are detected as two dots in the nuclei of Secondary TVCs (STVCs), between the first and second asymmetric divisions. (D) ISH + IHC for Moocci.Ebf (red) in embryos electoporated with Moocci.Mesp>nls::lacZ (green), revealing Moocci.EBF expression in ASMFs after the second asymmetric division. (E) ISH for Moocci.Ebf in a swimming larva, viewed dorsally, revealing Moocci.Ebf+ migrating atrial siphon muscle precursors (ASMPs, arrowheads). (F) Lateral view of a M. occidentalis juvenile (>100 hpf) electroporated with Moocci.Mesp>H2B::GFP. GFP + nuclei reveal contributions of B7.5 lineage to atrial siphon muscle-derived pharyngeal muscles (PhMs) and heart. (G) Diagram of the TVC divisions giving rise to the pharyngeal muscles and heart of the adult. (H) Ventral view of a M. occidentalis embryo electroporated with Moocci.Mesp>H2B::GFP, just after the first asymmetric division of the TVCs. (H′) Inset from (H) is focused on the distance between FHPs on either side of the embryo (mean = 57.6 μm, n = 9). (I) Ventrolateral view of a C. intestinalis embryo electroporated with Ciinte.Mesp>H2B::GFP, right after the first asymmetric division. (I′) The distance between FHPs from either side is very small (mean = 7.5 μm, n = 14) since they contact each other at the midline to form a single cluster of cells. (J) Double ISH for Moocci.Dlx.a (green) and Moocci.Ebf (red) in a M. occidentalis larva, viewed dorsally. Moocci.Dlx.a marks the siphon primordia, while Moocci.Ebf marks siphon muscle precursors. Arrowheads point to atrial siphon muscle precursors (ASMPs), which have migrated dorsally but do not encounter an atrial siphon primordium around which to encircle. In contrast, oral siphon muscle precursors form a ring around the oral siphon primordium (OSP). Other Moocci.Ebf+ cells seen are neurons or neuronal precursors in the central nervous system (cns). (K) Double ISH for Ciinte.Dlx.a (green) and Ciinte.Ebf (red) in a C. intestinalis larva viewed dorsolaterally. Arrowhead indicates Ciinte.Ebf+ ASMPs on one side of the embryo encircling one of two bilaterally paired atrial siphon primordia (ASP). The same configuration of Ciinte.Ebf+ muscle precursors is seen encircling the OSP.
Figure 3—figure supplement 1. Mosaic transgene labeling of M. occidentalis suggests bilateral origin of juvenile heart.
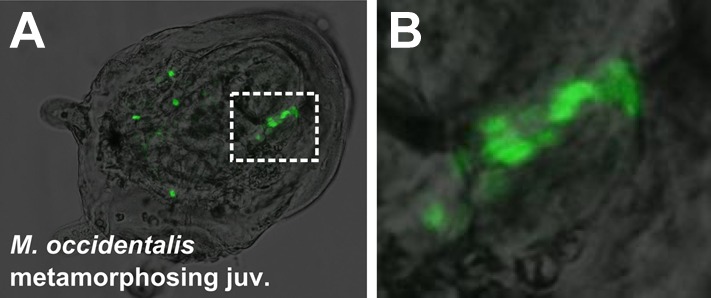
Figure 3—figure supplement 2. Delayed atrial siphon primordium specification in M. occidentalis.
Figure 3—figure supplement 3. Cardiopharyngeal development in C. intestinalis vs. M. occidentalis.
Gene expression during heart vs pharyngeal muscle segregation is also conserved. Namely, Moocci.Tbx1/10 is specifically activated in the STVCs immediately following the first asymmetric division (Figure 3C), while Moocci.Ebf is specifically activated in the ASMFs immediately following the second asymmetric division (Figure 3D). Moocci.Ebf expression is maintained in ASMFs and their daughter cells, the atrial siphon muscle precursors (ASMPs) during their migration to the dorsal regions of the now swimming larva (Figure 3E). This step-wise progression of gene expression, cell division, and migration are virtually identical to those observed in C. intestinalis, demonstrating deep evolutionary conservation of the cardiopharyngeal ontogenetic motif in ascidians.
We allowed embryos electroporated with Moocci.Mesp>H2B::GFP to settle and undergo metamorphosis from the larval to the juvenile stage. Visualization of H2B::GFP revealed contributions of the B7.5 lineage to the heart and pharyngeal muscles of juveniles (Figure 3F). Thus, the post-metamorphic tissues derived from B7.5 are identical to those previously reported for C. intestinalis and H. roretzi (Hirano and Nishida, 1997; Stolfi et al., 2010). A summary of the TVC divisions and their post-metamorphic fates is shown in Figure 3G. Based on the perfectly conserved parallels to C. intestinalis, we hypothesize that specification of ASM vs heart fate in Molgula occurs through a conserved Tbx1/10-Ebf-dependent regulatory cascade.
In summary, the cardiopharyngeal mesoderm of Molgula and Ciona display unequivocally homologous ontogenies, as revealed by nearly identical cell divisions, cell fate choices, and gene expression patterns. These data indicate that the key developmental features of the B7.5 lineage evolved at the base of the tunicate radiation.
Species-specific differences in TVC migration and heart primordium formation
Despite perfect conservation of B7.5 lineage cell divisions and cell fates between M. occidentalis and C. intestinalis, we noticed a striking species-specific difference in TVC behavior (summarized in Figure 3, Figure 3—figure supplement 3). Namely, in M. occidentalis, the TVCs appear to migrate along a more lateral path than their counterparts in C. intestinalis. As a result, the TVCs on one side of the embryo do not contact those on the other side, resulting in two discrete heart precursor clusters during the tailbud stage (Figure 3H). In C. intestinalis, the TVCs converge towards the ventral midline prior to dividing, such that the resulting FHPs and SHPs from the two sides contact each other as soon as they are born, forming a single cluster of heart precursors (Figure 3I; Davidson and Levine, 2003). However, observation of juveniles electroporated with Mocci.Mesp>H2B::GFP showed some individuals with hearts that were only half-labeled with H2B::GFP, likely due to left/right transgene mosaicism (Figure 3—figure supplement 1). This indicates that heart precursors from both sides of the embryo ultimately contribute to a single heart, as they do in C. intestinalis. Therefore, we assume this naturally occurring cardia bifida is only a temporary configuration of a single embryonic heart primordium. This odd configuration has not been previously described in the embryos of other ascidians and may represent a Molgula-specific feature.
Differences in ASMP behavior related to heterochrony of atrial siphon formation
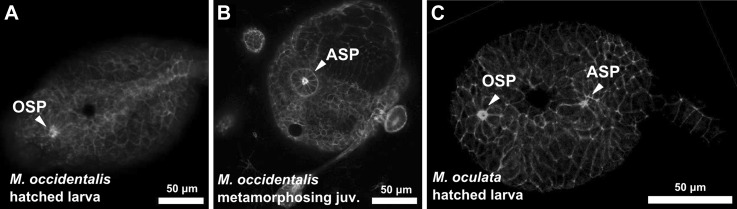
We also noticed a difference in ASM precursor behavior. In C. intestinalis, migrating ASMPs from either side of the embryo migrate dorsally, forming rings around a bilateral pair of atrial siphon primordia (ASPs) on the corresponding side (Stolfi et al., 2010). The ASPs are placode-like rosettes of molecularly and morphologically distinct ectodermal cells that arise through a retinoic acid/Hox1- and FGF/MAPK-dependent program and are proposed to be homologous to vertebrate otic placodes (Wada et al., 1998; Mazet et al., 2005; Kourakis and Smith, 2007; Sasakura et al., 2012). In juveniles of C. intestinalis and other phlebobranch ascidians, there are initially two atrial siphons, one on either side of the animal, that eventually fuse into a single one at the end of the ‘1st Ascidian’ stage (Chiba et al., 2004; Kourakis et al., 2010). In contrast, all stolidobranch ascidians initially specify a single ASP, bypassing the two-siphon fusion process (Manni et al., 2004; Grave, 1926; Figure 3—figure supplement 2A–C). Indeed, we observed the formation of a single ASP in M. occidentalis late in metamorphosis and in the larvae of M. oculata (Figure 3—figure supplement 2A,B). This flexibility in atrial siphon development in ascidians is an example of DSD at the morphogenetic level. It is believed that the dual primordium condition is ancestral and that the specification of a single primordium is a derived, Stolidobranchia-specific trait (Kourakis et al., 2010).
We found that M. occidentalis ASMPs, marked by Ebf expression, migrate dorsally but remain as a cluster, unlike the characteristic rings of cells stretched around the invaginations of the ASPs, as observed in C. intestinalis (Figure 3J,K). On the other hand, oral siphon muscle precursors form a ring of Moocci.Ebf+ cells around the presumptive oral siphon primordium (Figure 3J). ISH for the siphon primordium marker Dlx.a reveals that only the oral siphon primordium is specified at this stage in M. occidentalis larvae. In C. intestinalis, both oral and atrial siphon primordia expressing Dlx.a are specified by the larval stage and are encircled by Ebf+ siphon muscle precursors (Figure 3K). Thus, we conclude that this difference in ASMP ring formation is related to the heterochrony of atrial siphon formation between M. occidentalis and C. intestinalis.
Divergence of Mesp cis-regulatory sequence function between M. occidentalis and C. intestinalis
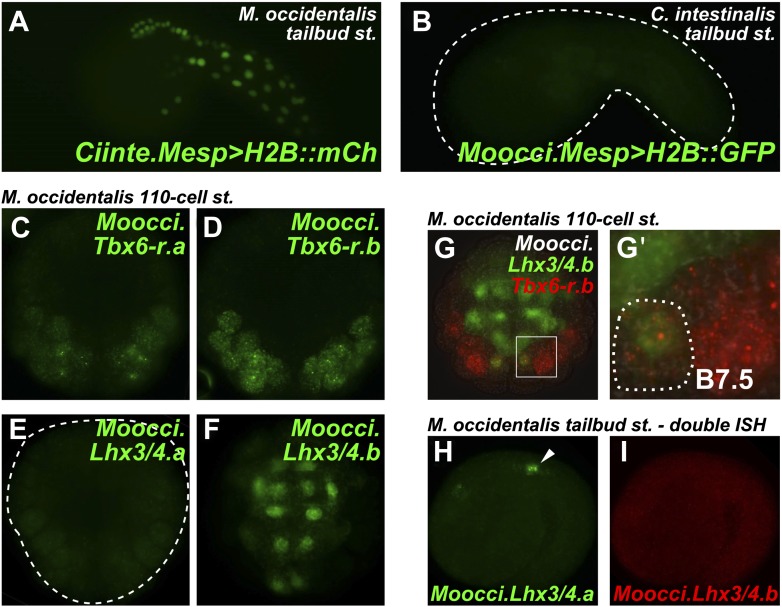
Given the obvious parallels between C. intestinalis and M. occidentalis cardiopharyngeal development, we expected transcriptional regulatory mechanisms to also be highly conserved between the two species. We tested this assumption by electroporating C. intestinalis reporter constructs into M. occidentalis embryos, and vice-versa. We observed that a Ciinte.Mesp reporter construct (Davidson et al., 2005), when electroporated into M. occidentalis embryos, drives relatively weak reporter gene expression in B7.5 with substantial leaky expression in other tissues (Figure 4A, Figure 4—figure supplement 1). Conversely, the Moocci.Mesp enhancer fails to drive any reporter gene expression when electroporated into C. intestinalis embryos (Figure 4B), despite recapitulating robust B7.5-specific expression in M. occidentalis embryos (Figure 1D,E).
Figure 4. Developmental system drift of Mesp regulation between C. intestinalis and M. occidentalis.
(A) M. occidentalis tailbud embryo electroporated with a Ciinte.Mesp>H2B::mCherry reporter construct. Weak reporter gene expression was observed in the B7.5 lineage and occasionally in other territories including B-line mesenchyme and tail muscle cells, and A-line neural plate derivatives. (B) C. intestinalis tailbud embryo electroporated with Moocci.Mesp>H2B::GFP reporter. No fluorescence was seen in any cells, indicating complete lack of activity of Moocci.Mesp enhancer in wild-type C. intestinalis embryos. (C) In situ hybridization (ISH) for Moocci.Tbx6-r.a, (D) Moocci.Tbx6-r.b, (E) Moocci.Lhx3/4.a, and (F) Moocci.Lhx3/4.b in 110-cell stage embryos. (G) Double ISH in 110-cell stage embryo reveals co-expression of Moocci.Lhx3/4b (green) and Moocci.Tbx6-r.b (red) exactly in the B7.5 cells of M. occidentalis. (G′) Magnified view of inset in (G). (H) Double ISH for Moocci.Lhx3/4.a (green) and (I) Moocci.Lhx3/4.b (red) in a mid-tailbud embryo. Moocci.Lhx3/4.a but not Moocci.Lhx3/4.b is expressed in motor ganglion neurons (arrowhead).
Figure 4—figure supplement 1. Weak and leaky expression of Ciinte.Mesp reporter in M. occidentalis embryos.
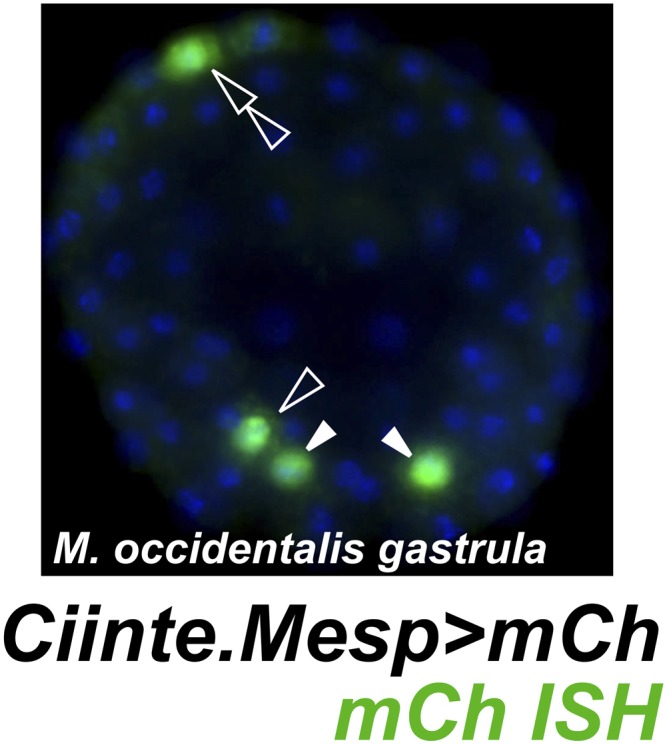
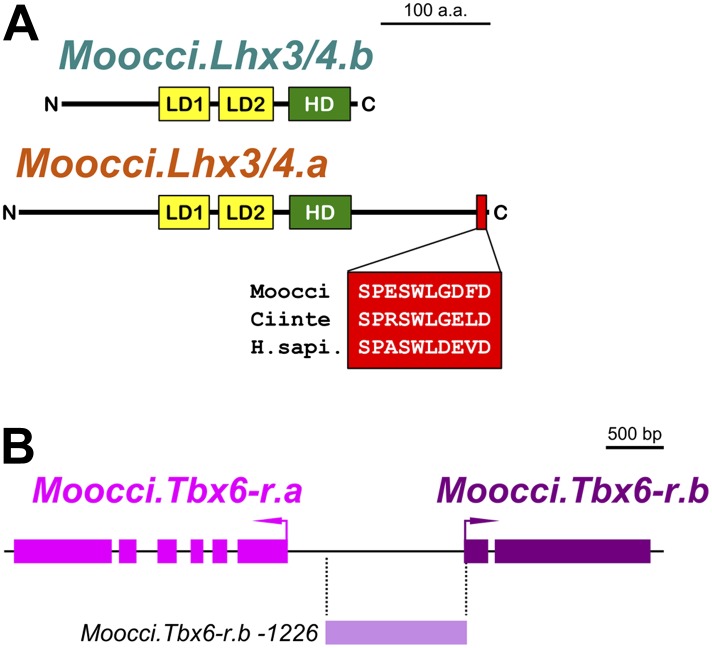
Figure 4—figure supplement 2. Configuration of Lhx3/4 protein domains and Tbx6-r locus in M. occidentalis.
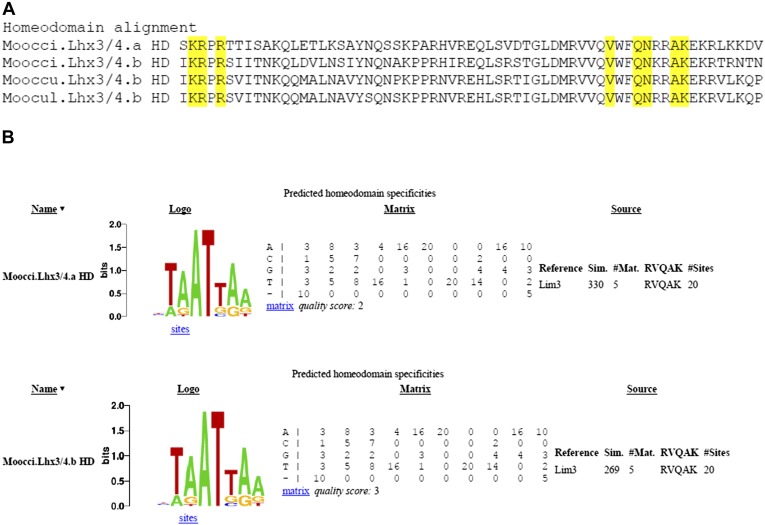
Figure 4—figure supplement 3. Divergent Molgula Lhx3/4.b homeodomains are not predicted to have altered DNA binding specificities.
These data suggest acute DSD of transcriptional regulatory mechanisms underlying otherwise identical Mesp expression patterns. More specifically, the trans-regulatory environment of the B7.5 blastomeres has diverged between Molgula and Ciona, and compensatory changes in the respective Mesp cis-regulatory sequences must have rendered these unable to function adequately outside of that milieu.
Tbx6-related and Lhx3/4 gene expression patterns in M. occidentalis
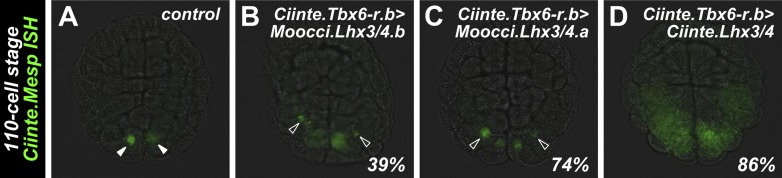
The drift of Mesp regulation between M. occidentalis and C. intestinalis prompted us to investigate potential species-specific differences in upstream trans-acting regulators. In C. intestinalis, Mesp is transcriptionally activated downstream of two zygotically expressed TFs: Tbx6-related.b (Ciinte.Tbx6-r.b) and Lhx3/4 (Ciinte.Lhx3/4) (Satou et al., 2004; Davidson et al., 2005; Christiaen et al. 2009a). Ciinte.Tbx6-r.b is directly activated in the posterior mesodermal lineages by the maternal determinant Macho-1/Zic-related.a (Ciinte.Zic-r.a) (Yagi et al., 2004), while Ciinte.Lhx3/4 is activated in the vegetal pole in response to stabilized maternal Beta-catenin (Satou et al., 2001). Since the expression domains of Ciinte.Tbx6-r.b and Ciinte.Lhx3/4 overlap only in B7.5, activation of Ciinte.Mesp is achieved through the synergistic interaction between Ciinte.Tbx6-r.b and Ciinte.Lhx3/4 proteins at the Ciinte.Mesp promoter in these cells (Christiaen et al., 2009a).
In each of the three Molgula genome assemblies, we identified two Tbx6-r genes of uncertain orthology relationships to the four Tbx6-r genes in the C. intestinalis genome (Stolfi et al., 2014). In all three Molgula genomes, the two Tbx6-r genes are arranged in head-to-head configuration with ∼2 kb separating the transcript start sites (Figure 4—figure supplement 2B). Despite the uncertain phylogeny of tunicate Tbx6-r genes, we named these genes Tbx6-r.a and Tbx6-r.b in Molgula. In M. occidentalis, both Tbx6-r.a and Tbx6-r.b are expressed in a broad posterior swath of the pre-gastrula embryo in a manner similar to the expression pattern of Tbx6-r genes in C. intestinalis (Figure 4C,D; Yagi et al., 2004).
We also identified two Lhx3/4 genes in each Molgula genome. This was unexpected because no Lhx3/4 duplications have been identified in any ascidian species (Christiaen et al., 2009a; Kobayashi et al., 2010). We named these genes Lhx3/4.a and Lhx3/4.b. In all three Molgula species, Lhx3/4.a is the more conserved paralog, while Lhx3/4.b is more divergent, lacking a well-conserved C-terminal motif (Figure 4—figure supplement 2A). However, the amino acid sequence changes are not predicted to change the DNA-binding preference of the homeodomain (Figure 4—figure supplement 3; Noyes et al., 2008). In each of the three Molgula species’ genome assemblies, the two Lhx3/4 genes were found to be located on separate contigs.
In M. occidentalis, Lhx3/4.b is expressed in vegetal cells of the early embryo, overlapping with Tbx6-r genes only in the B7.5 cells (Figure 4F,G). In contrast, Moocci.Lhx3/4.a is not zygotically expressed in the early embryo, but is expressed in larval motor neurons (Figure 4E,H), where Moocci.Lhx3/4.b is not transcriptionally active (Figure 4I). In both C. intestinalis and H. roretzi, transcription of a single Lhx3/4 gene from separate proximal and distal promoters results in two distinct transcript variants. Early vegetal cells express one transcript variant while motor neurons express the other transcript variant (Wada et al., 1995; Satou et al., 2001; Christiaen et al., 2009a; Kobayashi et al., 2010). Since this pleiotropic expression profile appears to be strictly partitioned between the two Lhx3/4 paralogs in M. occidentalis, this constitutes a clear example of regulatory sub-functionalization following gene duplication (Ohno, 1970; Hahn, 2009).
In summary, M. occidentalis embryos express Tbx6-r and Lhx3/4 genes in early B7.5 blastomeres, where they could potentially synergize to activate Moocci.Mesp following a trans-acting logic shared with C. intestinalis. The next set of experiments sought to test this potentially conserved logic.
Moocci.Tbx6-r.b and Moocci.Lhx3/4 proteins are conserved enough to activate Ciinte.Mesp
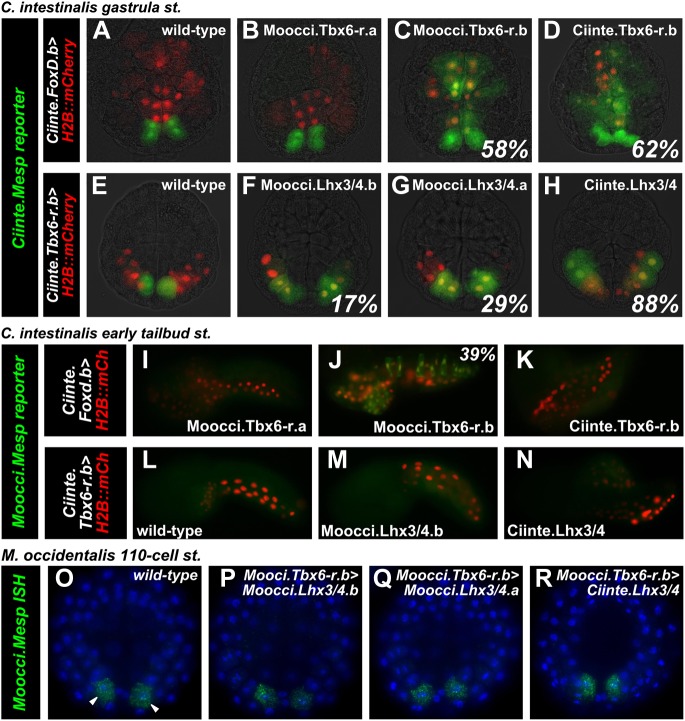
We asked whether cis/trans co-evolution between Mesp and Tbx6-r and Lhx3/4 could account for incompatibility of Mesp regulatory sequences between M. occidentalis and C. intestinalis. We previously demonstrated that in C. intestinalis, overexpression of Tbx6-r.b in the vegetal pole cells of the embryo, using the cis-regulatory sequences from the Foxd.b gene (Shi and Levine, 2008), results in ectopic Mesp activation in these cells due to expanded overlap of Tbx6-r.b and Lhx3/4 (Christiaen et al., 2009a). Similarly, overexpression of Lhx3/4 in the posterior pole of the embryo, using the cis-regulatory sequences from Tbx6-r.b, is sufficient to cause ectopic Mesp expression throughout this territory. We used these gain-of-function assays in C. intestinalis with the orthologous M. occidentalis proteins to test potential conservation of function in synergistic activation of Mesp expression.
Overexpression of Moocci.Tbx6-r.b but not Moocci.Tbx6-r.a was sufficient to activate ectopic Ciinte.Mesp reporter construct expression in vegetal pole cells (Figure 5A–C). In this regard, Moocci.Tbx6-r.b function seemed comparable to that of Ciinte.Tbx6-r.b (Figure 5D). Conversely, overexpression of either Lhx3/4.b or Lhx3/4.a from M. occidentalis in the posterior C. intestinalis embryo using the Ciinte.Tbx6-r.b promoter caused ectopic expression of both endogenous Ciinte.Mesp (Figure 5—figure supplement 1) and Ciinte.Mesp reporter construct (Figure 5E–G). These experiments revealed that both M. occidentalis Lhx3/4 proteins are conserved enough to activate low levels of Ciinte.Mesp expression (presumably by synergizing with endogenous Ciinte.Tbx6-r.b). However, neither was as effective as Ciinte.Lhx3/4 in activating Ciinte.Mesp (Figure 5H). These data suggested that cis/trans or trans/trans co-evolution involving the highly divergent Moocci.Lhx3/4.b paralog may only partially account for the incompatibility of Mesp cis-regulatory logic between the two species.
Figure 5. Divergence of Mesp regulation due to changes in cis and trans.
(A) Wild-type C. intestinalis gastrula embryo showing expression of Foxd.b reporter (red, Shi and Levine, 2008) in vegetal pole cells and Ciinte.Mesp reporter (green) in the B7.5 cells. (B) Electroporation of Ciinte.Foxd.b>Moocci.Tbx6-r.a has no effect on Ciinte.Mesp reporter expression. (C) Electroporation of Ciinte.Foxd.b>Moocci.Tbx6-r.b results in ectopic Ciinte.Mesp reporter expression in the vegetal pole in 58% of embryos (n = 100). (D) This is indistinguishable from the effect of Ciinte.Foxd.b>Ciinte.Tbx6-r.b, which results in ectopic Ciinte.Mesp reporter activation in 62% of embryos (n = 100). (E) Wild-type C. intestinalis gastrula embryo showing expression of Tbx6-r.b reporter (Christiaen et al., 2009a, red) in all B-line cells and Ciinte.Mesp reporter (green) in the B7.5 cells. (F) Electroporation of Ciinte.Tbx6-r.b>Moocci.Lhx3/4.b results in ectopic Ciinte.Mesp reporter expression in other B-line cells in 17% of embryos (n = 100) (G) Ciinte.Tbx6-r.b>Moocci.Lhx3/4.a results in ectopic Ciinte.Mesp reporter expression in 29% of embryos (n = 100). (H) Ciinte.Tbx6-r.b>Ciinte.Lhx3/4 induces ectopic Ciinte.Mesp reporter expression in 88% of embryos (n = 100). (I) C. intestinalis tailbud embryo showing no expression of Moocci.Mesp reporter, as expected. (J) Moocci.Mesp reporter expression is not rescued by electroporation of Ciinte.Foxd.b>Moocci.Tbx6-r.a but (K) electroporation of Ciinte.Foxd.b>Moocci.Tbx6-r.b is sufficient to activate Moocci.Mesp reporter in 39% of embryos (n = 100). (K) Moocci.Mesp reporter is not transactivated upon electroporation of Foxd.b>Ciinte.Tbx6-r.b. (L) Wild-type C. intestinalis tailbud embryo, showing no expression of electroporated Moocci.Mesp reporter. (M) Electroporation of Ciinte.Tbx6-r.b>Moocci.Lhx3/4.b or (N) Ciinte.Tbx6-r.b>Ciinte.Lhx3/4 is not sufficient to activate co-electroporated Moocci.Mesp reporter expression. (O–R) ISH showing Moocci.Mesp expression (green, arrowheads) in 110-cell stage M. occidentalis embryos. Ectopic Moocci.Mesp was not observed upon overexpression of any Lhx3/4 orthologs in the posterior embryo using the Moocci.Tbx6-r.b driver (P–R). All gastrula stage embryos are viewed vegetally, with anterior to the top of the image. All tailbud embryos are viewed laterally, anterior to the left. M. occidentalis embryo nuclei were visualized by staining with DAPI (blue).
Figure 5—figure supplement 1. Lhx3/4 proteins from M. occidentalis are sufficient to activate ectopic expression of endogenous Mesp in C. intestinalis.
Moocci.Tbx6-r.b but not Moocci.Lhx3/4 can activate Moocci.Mesp reporter expression in C. intestinalis embryos
Having established that M. occidentalis proteins can activate ectopic Ciinte.Mesp expression, we next asked whether these could be sufficient to activate Moocci.Mesp reporter expression in C. intestinalis embryos. Indeed, Moocci.Mesp reporter activation in the vegetal pole was observed upon overexpression of Moocci.Tbx6-r.b (Figure 5J), but not Moocci.Tbx6-r.a (Figure 5I), consistent with their effects on Ciinte.Mesp activation. Surprisingly, overexpression of Ciinte.Tbx6-r.b was not sufficient to activate Moocci.Mesp reporter (Figure 5K). These results hint at cis/trans co-evolution between Moocci.Mesp cis-regulatory sequences and Moocci.Tbx6-r.b, even though the latter retains enough functional information to activate Ciinte.Mesp.
In stark contrast to the rescue by Moocci.Tbx6-r.b, Moocci.Mesp reporter expression was never observed in C. intestinalis embryos upon overexpression of Moocci.Lhx3/4.b or Ciinte.Lhx3/4 in the posterior pole (Figure 5L–N). This indicates that a functional modification extending beyond cis/trans co-evolution accounts for the lack of Moocci.Mesp reporter activity in C. intestinalis embryos.
Moocci.Lhx3/4.b overexpression is not sufficient to activate ectopic Moocci.Mesp in M. occidentalis embryos
The heterologous experiments in C. intestinalis suggested that Moocci.Mesp regulation could be independent of Moocci.Lhx3/4.b. To test this hypothesis, we recapitulated the experiments in M. occidentalis embryos. We isolated cis-regulatory sequences from the Moocci.Tbx6-r.b gene (Figure 4—figure supplement 2) and used this driver to overexpress Lhx3/4 coding sequences in the posterior pole of M. occidentalis embryos. All Lhx3/4 proteins tested failed to induce ectopic Moocci.Mesp activation (Figure 5O–R). These data suggest that Moocci.Mesp is not responsive to Moocci.Lhx3/4.b, even in permissive Moocci.Tbx6-r.b-expressing cells. This differs markedly from the results of equivalent experiments in C. intestinalis, indicating major changes to the cis-regulatory logic governing identical Mesp spatiotemporal expression patterns in Ciona and Molgula.
Conserved requirement for MAPK activation in specification of migratory TVCs
Given the cis-regulatory re-wiring of Mesp regulation, we extended our analysis to search for other instances of acute DSD between Molgula and Ciona. We shifted our focus to the next major cell fate decision in the B7.5 lineage, namely the fate choice between TVCs and ATMs. In C. intestinalis, the FGF/MAPK pathway, mediated in part by MEK-dependent phosphorylation and activation of Ets.b, is both necessary and sufficient for TVC specification. Inhibition of the FGF/MAPK/Ets pathway converts TVCs to ATMs, inhibiting cell migration and expression of TVC markers (Davidson et al., 2006; Christiaen et al., 2008; Woznica et al., 2012). Conversely, constitutive activation of the pathway induces ectopic TVCs at the expense of ATMs.
We ought to test the requirement of the MAPK pathway for TVC induction in M. occidentalis. We treated embryos with the MEK small molecule inhibitor U0126 prior to the estimated time window of TVC specification (∼5 hpf at 22°C). U0126-treated embryos displayed inhibited TVC migration relative to DMSO-treated control embryos (Figure 6A–D). Moreover, expression of Moocci.Foxf and Moocci.Hand-r in the TVCs was severely downregulated by U0126 treatment relative to controls (Figure 6A–D). These data suggest a conserved requirement for MAPK pathway activity in TVC induction and expression of important TVC regulators in M. occidentalis.
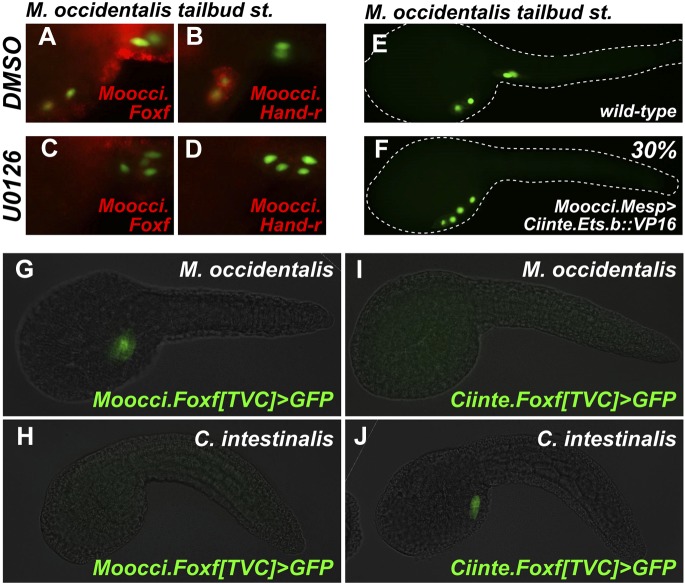
Figure 6. Mutual unintelligibility of orthologous Foxf enhancers in spite of shared requirement for the MAPK pathway in TVC fate specification.
In situ hybridization (ISH, red) + β-galactosidase immunodetection (IHC, green) in embryos electoporated with Moocci.Mesp>nls::lacZ and treated with DMSO (control treatment), showing normal TVC induction in 85% of embryos (n = 49) and TVC-localized expression of (A) Moocci.Foxf and (B) Moocci.Hand-r in 96% (n = 25) and 100% (n = 23) of embryos, respectively. Treatment with MEK inhibitor U0126 inhibited TVC induction and migration in 66% of embryos (n = 128). Treatment with U0126 also abolished TVC-specific (C) Moocci.Foxf expression in 91% of embryos (n = 64) and (D) Moocci.Hand-r expression in 91% of embryos (n = 64). (E) Wild-type M. occidentalis tailbud embryo with B7.5 lineage labeled by electroporation of Moocci.Mesp>H2B::GFP. Note two TVCs and two ATMs. (F) TVC induction and migration was enhanced upon electroporation with Moocci.Mesp>Ciinte.Ets.b::VP16, which converted ATMs into TVC-like cells in 30% of electroporated embryos (n = 200). (G) A fragment from −1517 to −425 upstream of the Moocci.Foxf start codon (Moocci.Foxf[TVC]), fused to the minimal promoter (−230/+21) from Moocci.Foxf and GFP (bpMoFF>GFP), recapitulates expression in the TVCs of M. occidentalis. (H) This same exact construct is silent in C. intestinalis embryos. (I) Conversely, a published TVC enhancer from the Ciinte.Foxf gene (Beh et al., 2007) fused to bpMoFF>GFP is silent in M. occidentalis embryos. (J) This same construct is strongly expressed in the TVCs of C. intestinalis. All panels represent lateral views.
Figure 6—figure supplement 1. Alignment of Foxf coding and non-coding sequences.
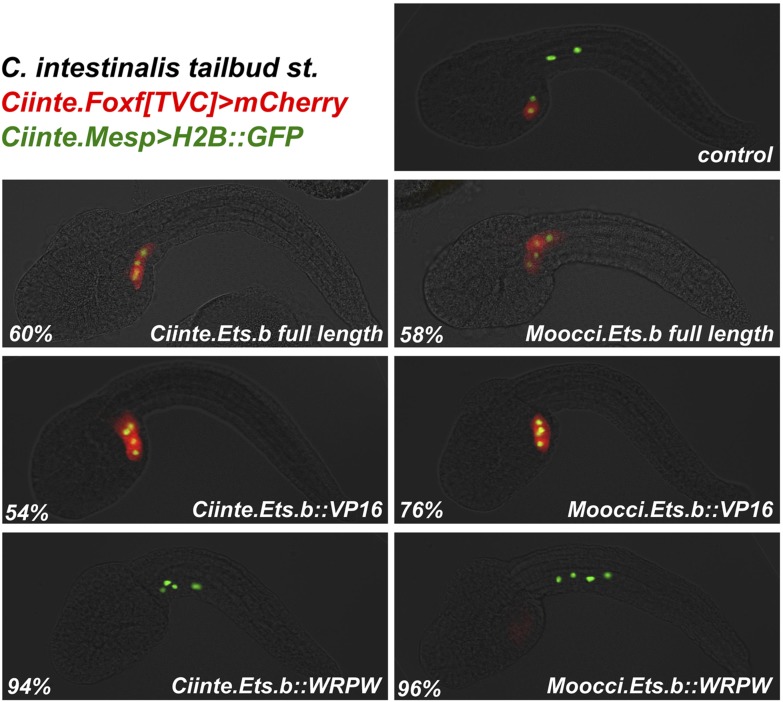
Figure 6—figure supplement 2. Moocci.Ets.b and Ciinte.Ets.b are functionally very similar.
Figure 6—figure supplement 3. Ets.b proteins are not sufficient to transactivate Moocci.Foxf reporter construct activation in C. intestinalis embryos.
We also tested the potential involvement of Ets factors in mediating the MAPK-dependent activation of the TVC program. To do so, we expressed a constitutively active fusion of the Ciinte.Ets.b DNA-binding domain to a VP16 transactivation domain (Ciinte.Ets.b::VP16, Davidson et al., 2006), in M. occidentalis B7.5 cells using the Moocci.Mesp driver. Electroporation of Moocci.Mesp>Ciinte.Ets.b::VP16 induced ectopic TVCs at the expense of ATMs, in 31% of transfected embryos (Figure 6E,F). In sum, our results are consistent with a conserved function for MAPK-activated Ets.b upstream of the cardiopharyngeal gene regulatory network in M. occidentalis.
Foxf TVC enhancers are mutually unintelligible in cross-species assays
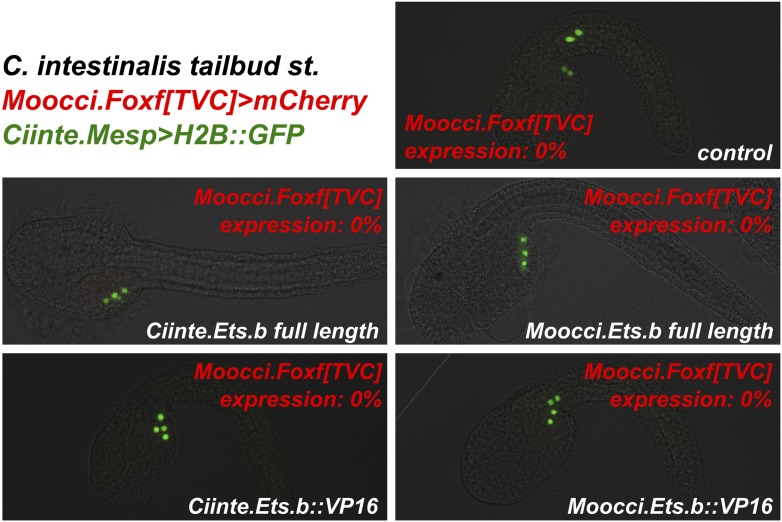
Given the conservation of the MAPK/Ets pathway in ascidian TVC specification, we reasoned that Foxf cis-regulatory sequences should be compatible in cross-species assays. We identified a sequence upstream of the Moocci.Foxf gene that is sufficient to drive TVC-specific expression in M. occidentalis (Figure 6G, Figure 6—figure supplement 1). However, this sequence is completely non-functional when tested in C. intestinalis (Figure 6H). Likewise, a previously identified Ciinte.Foxf TVC enhancer (Figure 6J; Beh et al., 2007) is incapable of driving TVC-specific expression in M. occidentalis embryos (Figure 6I).
Thus, to borrow a term from the field of linguistics, these enhancers are said to be ‘mutually unintelligible’, since the enhancer from M. occidentalis cannot be interpreted by embryos of C. intestinalis, and the enhancer from C. intestinalis cannot be interpreted by M. occidentalis. This mutual unintelligibility is due to differences between the enhancers and not the promoters, since we used the same Moocci.Foxf basal promoter in both constructs. Although the Ciinte.Foxf enhancer is able to interact with and potentiate transcription from Moocci.Foxf promoter, nucleotide sequence is poorly conserved around Foxf proximal promoter regions, even between M. occidentalis and M. occulta/oculata (Figure 6—figure supplement 1).
We asked whether the observed unintelligibility was indicative of changes in Ets.b protein properties between C. intestinalis and M. occidentalis embryo. At first glance, differences in DNA binding domain amino acid sequences between the Ets.b orthologs do not predict a change in binding site preference (Donaldson et al., 1996). Indeed, full-length Moocci.Ets.b and Moocci.Ets.b::VP16 were just as effective as their C. intestinalis counterparts to induce ectopic TVCs when expressed in the C. intestinalis B7.5 lineage (Figure 6—figure supplement 2). Moreover, C. intestinalis TVC induction was abolished by expression of the constitutive repressor form (Moocci.Ets.b::WRPW). However, neither full-length Moocci.Ets.b nor Moocci.Ets.b::VP16 was sufficient to induce activation of Moocci.Foxf reporter in C. intestinalis embryos (Figure 6—figure supplement 3). These surprising results suggest that, even though TVC induction and TVC-specific Foxf expression depends upon Ets.b-mediated MAPK activity in M. occidentalis embryos, there are profound species-specific differences in the regulatory mechanisms acting in parallel to MAPK/Ets.b upstream of Foxf, reflected in the mutual unintelligibility of the orthologous enhancers.
Extensive cis-regulatory unintelligibility between C. intestinalis and M. occidentalis
Since investigation into both Mesp and Foxf regulation revealed divergent cis-regulatory logic underlying identical gene expression patterns, we asked whether these examples constituted a general trend between C. intestinalis and M. occidentalis. Testing several other candidate enhancers, we identified other examples of cis-regulatory unintelligibility. For instance, Moocci.Tbx6-r.b and Mooci.Hand-r upstream sequences recapitulated their endogenous activity in M. occidentalis embryos (Figure 7B,E), but these sequences were conspicuously silent in the B7.5 lineage in C. intestinalis embryos (Figure 7C,F). Notably, activity in other tissues seemed normal: Moocci.Tbx6-r.b reporter expression was robust and precise in C. intestinalis primary tail muscle cells (Figure 7A,C) and Moocci.Hand-r reporter recapitulated expression in the C. intestinalis A7.6 and anterior endoderm lineages (Figure 7D,F). In the case of Hand-r, this partial intelligibility is due to the modular organization of its cis-regulatory sequences, which in C. intestinalis is comprised of discrete enhancers controlling expression in each of the distinct domains (Davidson and Levine, 2003; Woznica et al., 2012).
Figure 7. Cross-species reporter construct assays reveal multiple cases of cis-regulatory unintelligibility.
(A) C. intestinalis embryo electroporated with Ciinte.Tbx6-r.b>GFP reporter construct (Christiaen et al., 2009a), which drives GFP expression in tail muscles and the B7.5 lineage cells (arrowheads), recapitulating endogenous Ciinte.Tbx6-r.b expression. (B) M. occidentalis embryo electroporated with Moocci.Tbx6-r.b>GFP reporter construct, which recapitulates expression in tail muscle cells including B7.5 lineage cells (arrowheads). (C) C. intestinalis embryo electroporated with Moocci.Tbx6-r.b>GFP, which drives expression in B-line tail muscle and mesenchyme cells but is excluded from the B7.5 lineage. (D) C. intestinalis embryo electroporated with Ciinte.Hand-r>H2B::GFP reporter (Davidson and Levine, 2003), which drives H2B::GFP expression in anterior endoderm, A7.6 lineage, and TVCs (arrowheads), recapitulating endogenous Ciinte.Hand-r expression. (E) M. occidentalis embryo electroporated with Moccci.Hand-r>H2B::GFP construct, which recapitulates the same expression pattern. (F) C. intestinalis embryo electroporated with Moocci.Hand-r>H2B::GFP, which drives expression in endoderm and A7.6 lineage, but not in B7.5. (G) M. occidentalis embryo electroporated with Ciinte.Ebf neuron-specific YFP (green) and H2B::mCherry (red) reporter constructs electroporated (Stolfi and Levine, 2011), which drive very weak expression in a limited subset of motor ganglion neurons (green and red). (H) C. intestinalis embryo electroporated with a Moocci.Ebf>YFP reporter, which drives robust reporter gene expression in several brain, motor ganglion, and nerve cord neurons. (I) C. intestinalis embryos electroporated with Ciinte.Sox1/2/3 (left) and Moocci.Sox1/2/3 (right) H2B::mCherry reporter constructs, both of which recapitulate Sox1/2/3 expression in ectoderm. Panels A–F are lateral views of tailbud embryos, panels G is a dorsal view of a tailbud embryo, panel H is a dorso-lateral view of a hatched larva, and panel I shows vegetal views of mid-gastrula stage embryos. Anterior is to the right, except in Panel I, in which anterior is to the top.
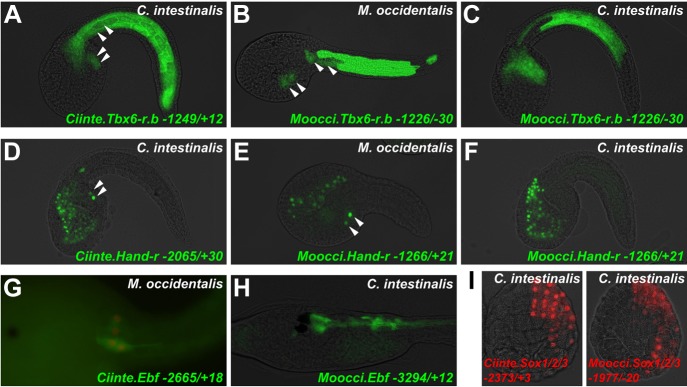
Figure 7—figure supplement 1. Asymmetric intelligibility between Molgula and Ciona Hand-r TVC enhancers.
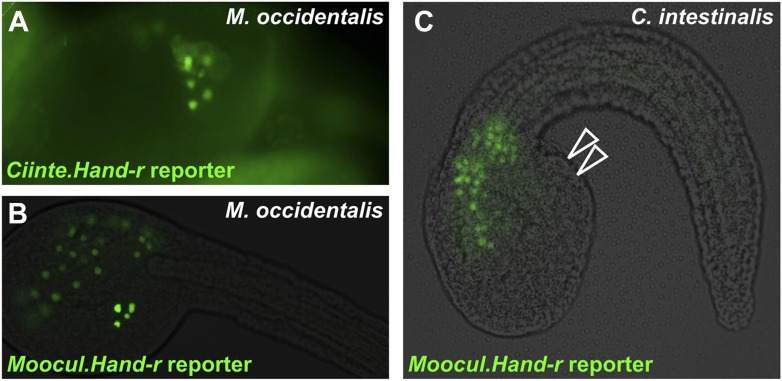
Curiously, we found that the Ciinte.Hand-r reporter can drive reporter gene expression in M. occidentalis TVCs (Figure 7—figure supplement 1). Thus, unlike the case of Foxf, there is an asymmetric intelligibility of Hand-r TVC enhancers between M. occidentalis and C. intestinalis. Moreover, a M. oculata Hand-r TVC enhancer is functional in M. occidentalis but not in C. intestinalis (Figure 7—figure supplement 1). Taken together, these data suggest that differences in enhancer logic may have accumulated over the course of the deep evolutionary history between Molgula and Ciona but not between M. occidentalis and M. oculata, and that some enhancers may have evolved asymmetrically in the two branches, retaining greater pan-ascidian ‘intelligibility’ in one or the other.
Finally, we found that acute DSD was not restricted to cis-regulatory sequences controlling gene expression in the B7.5 lineage. For instance, a Ciinte.Ebf driver that has been shown to recapitulate neuronal Ebf expression (Stolfi and Levine, 2011) was only very weakly active in a subset of M. occidentalis motor ganglion neurons (Figure 7G). However, the orthologous sequence from M. occidentalis recapitulated robust reporter gene expression in several C. intestinalis Ebf+ neurons throughout the central nervous system (Figure 7H). In contrast, expression of a Moocci.Sox1/2/3 reporter construct in the ectoderm of C. intestinalis embryos was found to be indistinguishable from that of the C. intestinalis ortholog (Figure 7I), suggesting that some cis-regulatory sequences may retain mutual intelligibility between Molgula and Ciona without sequence conservation, as is the case for various Otx enhancers between C. intestinalis and H. roretzi (Oda-Ishii et al., 2005). In sum, our preliminary cross-species enhancer activity screen suggests that DSD was rampant during ascidian evolution, resulting in mutual unintelligibility of cis-regulatory mechanisms underlying otherwise identical expression patterns of orthologous genes.
Discussion
Molgula spp. as model organisms for molecular developmental genetics
By sequencing the genomes of three Molgula species and adapting the electroporation protocol for routine transfection of M. occidentalis embryos, we have further expanded the reach of ascidian molecular genetics. We have demonstrated the usefulness of M. occidentalis as a species amenable to medium-throughput molecular perturbations and assays. While Ciona/Halocynthia comparisons have yielded fundamental insights into chordate development, the lack of a reliable electroporation protocol for transfection of Halocynthia eggs has hampered comparative studies on a larger scale. On the other hand, eggs from species such as Phallusia mammilata can be electroporated, but this species is much more closely related to Ciona. Thus, Molgula species may significantly contribute to understanding the extreme conservation of ascidian embryogenesis in spite of vast genetic differences and deep evolutionary histories.
We have shown that de novo sequencing and assembly of Molgulid genomes using next-generation technologies is relatively efficient and cost-effective, largely due to their compact nature. The sequence information obtained can be used to quickly generate a comprehensive set of molecular tools. We predict that a large number of ascidian genomes will be as compact and efficiently sequenced and assembled as these, unlocking the experimental potential of many ascidian species representing several different families (Tsagkogeorga et al., 2009).
Similarities and differences between Molgula and Ciona cardiopharyngeal regulatory mechanisms
In our survey of B7.5 development, we observed that the cell division events and cell fate choices in the lineage are perfectly conserved between Ciona and Molgula, demonstrating the deep evolutionary origins of the cardiopharyngeal ontogenetic motif in tunicates. Regulatory gene expression patterns in the lineage are also nearly identical, the only potential difference being Nk4, which was not confidently detected in M. occidentalis TVCs.
NK4 proteins are intimately linked to cardiac development throughout bilateria (Akazawa and Komuro, 2005). Moreover, Nk4 is expressed in C. intestinalis TVCs where it functions as a regulator of heart precursor fate (Davidson and Levine, 2003; Wang et al., 2013). However, there are distinct early and late phases of Nk4 expression in C. intestinalis heart development. Nk4 is expressed in the TVCs during the mid-tailbud stage, downregulated during the late tailbud and larval stages, and only re-activated in the differentiating heart of metamorphosing juveniles (Davidson and Levine, 2003). This points to distinct early (pre-metamorphic) and late (post-metamorphic) roles for Nk4 in C. intestinalis.
In C. intestinalis, early Nk4 expression does not appear to be sufficient for heart fate, as it is expressed in the TVCs prior to the segregation of heart and pharyngeal muscle fates (Davidson and Levine, 2003; Beh et al., 2007; Christiaen et al., 2008, 2010; Ragkousi et al., 2011; Wang et al., 2013). Second, this early expression may not be strictly required for heart fate either, as the first heart precursors (FHPs) do not adopt a pharyngeal muscle fate upon dominant-negative Nk4 overexpression like the second heart precursor (SHPs) do (Wang et al., 2013). Therefore, Nk4 acts as partially redundant mechanism to specify heart precursors in C. intestinalis, complementing parallel mechanisms of heart/pharyngeal muscle cell fate choice (e.g., activation of Tbx1/10 in the STVCs and Ebf in the ASMFs).
It is possible that Moocci.Nk4 is only activated during metamorphosis, an early role for it being dispensable in M. occidentalis TVCs. This might be the case if the complementary mechanism for pharyngeal muscle fate choice is more robust than in Ciona. In this way, the divergence of parallel regulatory mechanisms may have contributed to the loss of a key regulator like Nk4 from the early cardiopharyngeal mesoderm GRN of Molgula. A much more detailed investigation will be required to determine a possible role for Nk4 in the Molgula heart development.
Species-specific differences in B7.5 lineage morphogenetic events
M. occidentalis and C. intestinalis also display differences in certain morphogenetic behaviors of B7.5 derivatives (summarized in Figure 3, Figure 3—figure supplement 3). First, M. occidentalis TVCs migrate more laterally and do not meet at the ventral midline, resulting in a temporary condition of cardia bifida. A similar condition can be induced in C. intestinalis embryos by perturbing endoderm development (Ragkousi et al., 2011). Since M. occidentalis and C. intestinalis tailbud embryos look quite different in their shape, it will be interesting to assess endoderm formation and its relationship with TVC migration trajectory in Molgula.
Secondly, the ASM precursors in M. occidentalis migrate away from the heart progenitors but do not encounter atrial siphon primordia to encircle in the dorsal part of the embryo. This process is uncoupled in C. intestinalis Hox1 mutants, which fail to specify the ASPs. In these mutant larvae, the ASMPs still appear to migrate dorsally but do not form rings of muscle (Sasakura et al., 2012). It remains to be determined how the M. occidentalis ASMPs migrate to the general vicinity of the future atrial siphon opening and remain localized there until the later specification and differentiation of the siphon.
These two major morphogenetic differences prompted us to rethink our assumptions regarding the interdependence of certain processes in ascidian cardiopharyngeal development, as identified in C. intestinalis. For instance, one might assume that the convergence of Ciona TVCs at the ventral midline to be important for the asymmetric cell divisions and fate choices segregating heart from ASM precursors. Similarly, one might assume the atrial siphon primordia to be the source of a cue required for the dorsal migration of ASMPs. In M. occidentalis, these processes have been naturally uncoupled. These observations serve to emphasize that sampling diverse taxa provides insights into the modularity of specific developmental processes.
Developmental system drift hidden beneath the surface of highly conserved ascidian embryos
From our initial survey of a handful of enhancers from C. intestinalis, M. occidentalis, and M. oculata, we encountered several instances of either mutual unintelligibility, or asymmetric intelligibility of enhancers. Qualitative cross-species assays of M. occidentalis and C. intestinalis cis-regulatory sequences indicated that specific orthologous pairs of enhancers may be mutually intelligible (e.g., Sox1/2/3 ectodermal enhancer) while others are mutually unintelligible (e.g., Foxf TVC enhancer). Some orthologous gene pairs appear to be regulated by a mix of intelligible and unintelligible tissue-specific enhancers (e.g., Hand-r). The observation that cross-species intelligibility due to DSD occurs in a tissue-specific manner is consistent with the widely documented modularity of cis-regulatory elements controlling complex gene expression (Arnone and Davidson, 1997; Levine, 2010).
These results add to the mounting evidence suggesting that acute and pervasive DSD may have occurred over the course of ascidian evolution, obfuscated by the identical cell lineages and highly conserved gene expression patterns of ascidian embryos (Swalla, 2004). The multiple examples of cis-regulatory unintelligibility we identified were rather unexpected given (a) the extremely conserved pattern of expression of orthologous genes from Molgula and Ciona (Takada et al., 2002) and (b) previous observations of mutual intelligibility of enhancers between C. intestinalis and H. roretzi (e.g., Otx), and between C. intestinalis and the more closely related C. savignyi (Johnson et al., 2004; Brown et al., 2007). Large-scale, quantitative cross-species assays and detailed GRN studies will illuminate factors that may contribute to conservation or divergence in regulatory mechanisms.
Evolution of different activating inputs for Mesp expression in ascidians
Cis-regulatory unintelligibility can arise due to different underlying factors. We have presented evidence that, in the case of Mesp, this results in part from a change in an upstream activating input. In C. intestinalis, Lhx3/4 is an activator of Mesp, while in M. occidentalis, Mesp activation appears to be independent of Lhx3/4.b, despite the conserved expression pattern of this gene in vegetal pole cells. Regulation of Mesp in other ascidian species will need to be investigated in order to establish whether Lhx3/4-dependent activation of Mesp in ascidians is ancestral or derived. On the other hand, both C. intestinalis and M. occidentalis have retained a conserved role for Tbx6-r in regulating Mesp, a regulatory connection that may have arisen in the last common ancestor of tunicates and vertebrates (Yasuhiko et al., 2006; Davidson et al., 2005). In hindsight, it appears the weak expression of Ciinte.Mesp reporter constructs observed in the B7.5 cells of M. occidentalis embryos is not indicative of GRN conservation. This expression instead appears to be an artifact attributed in part to the conserved, overlapping expression profiles of Moocci.Tbx6-r.b and Moocci.Lhx3/4.b, even if this overlap is not instructive for endogenous Moocci.Mesp activation.
We can only speculate what is the connection, if any, between the re-wiring of Mesp regulation and the Molgula-specific duplication and sub-functionalization of Lhx3/4. Interestingly, in C. intestinalis and H. roretzi, the early and late functions of Lhx3/4 are partitioned between two transcript variants of the same locus, not between different loci. This substitution of alternate isoforms with gene duplicates has been documented for a few genes in teleost fish (Altschmied et al., 2002; Yu et al., 2003). Although allowing for some degree of sub-functionalization of protein function, alternative transcription/splicing does not completely relax the constraints imposed by pleiotropy. The Lhx3/4 variants in C. intestinalis and H. roretzi still share the bulk of their amino acid sequence, differing only at the portion of the N-terminus encoded by the alternate first exons (Christiaen et al., 2009a; Kobayashi et al., 2010). In Molgula spp. on the other hand, the two paralogs are considerably diverged from one another throughout their coding sequences, even though the divergence does not appear to alter DNA sequence-binding specificity. Pleiotropy has been hypothesized to both promote and suppress evolution, more specifically DSD (Hansen, 2003). Investigating the full breadth of Lhx3/4 functions in multiple ascidian species will be required in order to determine the relationship between Lhx3/4 duplication and re-wiring of Mesp regulation.
Multiple cases of cardiopharyngeal mesoderm enhancer unintelligibility point to differences in B7.5/TVC trans environments
The mutual unintelligibility of orthologous Foxf enhancers was unexpected, given that both M. occidentalis and C. intestinalis depend upon a shared MAPK/Ets-based switch for TVC fate induction. This discrepancy could be due to species-specific requirements for additional activating inputs, and/or silencing by species-specific repressors. Our observation that cis-trans complementation with Moocci.Ets.b::VP16 does not rescue the activity of the Moocci.Foxf enhancer in C. intestinalis embryos suggests that the latter possibility may be the case. It is thought that co-evolution between an enhancer and its trans environment is critical in order for it to faithfully direct a conserved pattern of gene expression, even if the overarching GRN is broadly conserved (Landry et al., 2005). Therefore, the incompatibility of B7.5 lineage-specific enhancers between M. occidentalis and C. intestinalis may reflect key differences in the trans-regulatory milieus of this lineage in the two species. Future comparative expression profiling of M. occidentalis and C. intestinalis B7.5 lineage cells will shed light on the nature of these differences.
Concluding remarks
Even though the word ‘drift’ implies a greater role for chance over natural selection, adaptive processes are very likely to play a role in DSD. True and Haag hypothesized as much in their seminal introduction to the concept of DSD, predicting that cis/trans co-evolution and other compensatory changes should result in enhancers resembling species-specific ‘Rube Goldberg machines’ that give superficially similar outputs but are functionally non-interchangeable (True and Haag, 2001). Models incorporating selection, pleiotropy, and compensation (SPC) have refined this idea by ascribing ‘drift’ in one biological process to adaptation in another (Landry et al., 2005; Johnson and Porter, 2007; Pavlicev and Wagner, 2012; Martinez et al., 2014). For instance, if the same pleiotropic gene is involved in both processes, any mutations in that gene that are adaptive for the process under selection will require some compensatory changes simply to maintain the same output of the other process. This may occur with or without deleterious intermediate states, with the latter occurring through a process termed ‘pseudocompensation’ (Haag, 2007).
We speculate that a high frequency of compensatory changes, required for the rapidly evolving ascidians to accommodate the constraints imposed by their invariant embryonic cell lineages and highly compact genomes, has given rise to a preponderance of cross-species cis-regulatory unintelligibility, following the DSD/SPC model. This perfect storm of intrinsic factors may be the key to explaining the dichotomy observed between highly conserved embryos and divergent cis-regulatory structure/function in ascidians.
Materials and methods
Genomic DNA library preparation and sequencing
Genomic DNA was phenol/chloroform extracted from dissected gonads of Molgula occulta (Kupffer) and Molgula oculata (Forbes) adults from Roscoff, France, and a Molgula occidentalis (Traustedt) adult from Panacea, Florida, USA (Gulf Specimen Marine Lab). Genomic DNA was sheared using an M220 Focused-ultrasonicator (Covaris, Woburn, MA). Sequencing libraries were prepared using KAPA HiFi Library Preparation Kit (KAPA Biosystems, Wilmington, MA) indexed with DNA barcoded adapters (BioO, Austin, TX). Size selection was performed using Agencourt (Beckman–Coulter, Brea, CA) AMPure XP purification beads (300–400 bp fragments), or Sage Science (Beverly, MA) Pippin Prep (650–750 bp and 875–975 bp fragments). For M. occulta and M. occidentalis libraries, 6 PCR cycles were used. For M. oculata libraries, 8 cycles were used for the 300–400 bp library, and 10 cycles were used for the 650–750 and 875–975 bp libraries. Libraries of different species but same insert size ranges were multiplexed for sequencing in three 100 × 100 PE lanes on a HiSeq 2000 sequencing system (Illumina, San Diego, CA) at the Genomics Sequencing Core Facility, Center for Genomics and Systems Biology at New York University (New York, NY). Thus, each lane was dedicated to a mix of species, specifically barcoded libraries of a given insert size range. Raw sequencing reads were deposited as a BioProject at NCBI under the ID# PRJNA253689.
Genome sequence assembly
All genomes were assembled on Michigan State University High Performance Computing Cluster (http://contact.icer.msu.edu). Prior to assembly, read quality was examined using FastQC v0.10.1 (Babraham Bioinformatics, 2014). Reads were then quality trimmed on both the 5′ and 3′ end using seqtk trimfq (https://github.com/lh3/seqtk) which uses Phred algorithm to determine the quality of a given base pair. Seqtk trimfq only trims bases, so no reads were discarded. Each library per species was then abundance filtered using 3-pass digital normalization to remove repetitive and erroneous reads (Brown et al., 2012; Schwarz et al., 2013; Howe et al., 2014). Genome assembly was done using Velvet v1.2.08 (Zerbino and Birney, 2008) with k-mer overlap length (‘k’) ranging from 19 to 69 and scaffolding was done by Velvet, by default. Velvet does not produce separate files for contigs and scaffolds; because Velvet scaffolded conservatively, contigs dominated the assemblies so we refer to both contigs and scaffolds as contigs. CEGMA scores were then computed to evaluate genome completeness (Parra et al., 2007).
The latest versions of three species' genome assemblies have been deposited on the ANISEED (Ascidian Network for In Situ Expression and Embryological Data) database for browsing and BLAST searching at http://www.aniseed.cnrs.fr/ (Tassy et al., 2010). Scripts for genome assembly and CEGMA analysis can be found in the following github repository: https://github.com/elijahlowe/molgula_genome_assemblies.git.
Molecular cloning of Molgula sequences
Putative orthologs of Ciona intestinalis (Linnaeus) and Halocynthia roretzi (Drasche) protein-coding genes were initially identified by TBLASTN. Identified sequences were aligned to each other to support orthology relationships within Molgula, and then queried by BLASTP against the NCBI non-redundant protein sequence database to further support orthology relationships to known tunicate and vertebrate proteins. cDNAs were cloned by RT-PCR, or by 5′ and/or 3′ RACE (SMARTer RACE kit, Clontech, Mountain View, CA). The template cDNA libraries were prepared from total RNA extracted from embryos of various stages. Upstream regulatory sequences were cloned by PCR from genomic DNA. For non-RACE PCRs, we used Phusion high-fidelity polymerase (New England Biolabs, Ipswich, MA). Refer to Supplementary file 1 for detailed sequence information. Genes are named according to the proposed unified nomenclature system for tunicate genetic elements (Stolfi et al., 2014). Refer to Supplementary file 2 for table with previously used gene names and the corresponding new proposed gene symbols and names.
Electroporation of plasmid DNA into embryos
M. occidentalis gravid adults were obtained from Gulf Specimen Marine Lab between March and December. Gravid adults were rare between the winter months from December to March. Animals were kept in a small aquarium with circulating artificial sea water at 24°C until needed. All subsequent manipulations using eggs and embryos were performed between 20°C and 24°C, though 24°C appeared to be the optimal temperature for both dechorionation and embryonic development.
Individual gonads were dissected in filtered TAPS-buffered artificial sea water (T-ASW) to release eggs and sperm. Eggs were allowed to undergo germinal vesicle breakdown for 10 min and then pooled along with sperm to fertilize for 5 min. Eggs were rinsed thoroughly and dechorionated in solution (168 μl 10N NaOH, 1.2 ml 2.5% pronase E, 0.3 g sodium thioglycolate in 40 ml T-ASW) with constant pipetting for 8–10 min (modified after Christiaen et al., 2009c). Viewing under a high-powered dissecting microscope was often necessary to verify removal of the thin, translucent chorion that becomes wrapped tightly around the egg upon immersion in dechorionation solution. After confirming chorion removal, eggs were quickly rinsed by the passage through multiple dishes of T-ASW.
Electroporation conditions and procedures were identical to those previously described for C. intestinalis (Christiaen et al., 2009b). Unless otherwise specifically noted, all fluorescent reporter genes were fused at the N-terminus to unc-76 tags (Dynes and Ngai, 1998), to ensure distribution throughout the whole cytosol, electroporated typically at ∼50 μg to 90 μg per 700 μl electroporation solution, while nuclei were visualized with histone2B::fluorescent protein constructs electroporated at ∼5 μg to 25 μg per electroporation unless otherwise specifically noted. Perturbation constructs were electroporated at ∼35 μg to 100 μg per electroporation. To image juveniles, larvae were allowed to settle and metamorphose in plastic Petri dishes or on glass coverslips in Petri dishes for several days with frequent changes of T-ASW.
Fertilization of M. occulta eggs was performed as previously described (Swalla and Jeffery, 1990). Embryos were dechorionated and electroporated as described for M. occidentalis embryos, using a custom-built electroporation machine (Zeller et al., 2006). C. intestinalis (Type A ‘robusta’) adults were collected in San Diego, CA (M-Rep).
MEK1/2 inhibitor U0126 treatment
Embryos were treated in 10 μM U0126 (Cell Signaling Technology, Danvers, MA) in T-ASW at least 30 min prior to the targeted MAPK signaling event.
In situ hybridization
Fluorescent in situ hybridization assays were performed as previously described, with modifications (Beh et al., 2007; Ikuta and Saiga, 2007; Christiaen et al., 2009d). Embryos were fixed in MEM-3.2% to 4% paraformaldehyde buffers for at least 2 hr. Pre-hybridization proteinase K concentrations ranged from 0.25 μg/ml (110-cell stage) to 1 μg/ml (tailbud) and 5 μg/ml (larvae) for Molgula spp. embryos. The Ciinte.Ebf probe was synthesized from template plasmid from the C. intestinalis Gene Collection Release 1 (Satou et al., 2002). See Supplementary file 1 for more detailed probe template sequence information.
Imaging
Embryos were counterstained with DAPI and/or Phalloidin conjugates (LifeTechnologies/Thermo Fisher Scientific, Waltham, MA). Images were taken using a combination of different Leica Microsystems (Wetzlar, Germany) microscopes: TCS SP8 X confocal microscope, TCS SP5 confocal microscope, and DM2500 epifluorescence microscope.
Acknowledgements
The authors are greatly indebted to Paul Scheid, members of the NYU CGSB Genomics Sequencing Core facility, Rodoniki Athanasiadou and the Gresham Lab for help in planning and performing library preparation and sequencing. We thank ASSEMBLE MARINE for funding the MoEvoDevo project in 2013. We thank the Station Biologique faculty and staff, especially Stéphane Hourdez, Sophie Booker, and Xavier Bailly for their help in carrying out our studies at Roscoff. We would also like to thank Nadine Peyriéras and her research group for sharing of reagents, equipment, and expertise for M. occulta and M. oculata imaging. We are grateful to Wei Wang for subcloning the full-length Ciinte.Ets.b sequence. Work in the laboratory of LC is supported by grants from the National Institute of General Medical Sciences (R01GM096032), the American Heart Association (10SDG4310061), and by the New York Cardiac Center and New York University College of Arts and Sciences. AS is supported by the National Science Foundation Postdoctoral Research Fellowship in Biology (under grant NSF-1161835). EL, BJS, and CTB are supported by the National Science Foundation under Cooperative Agreement No. DBI-0939454. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. This project was supported by Agriculture and Food Research Initiative Competitive Grant 2010-65205-20361 to CTB.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
National Institute of General Medical Sciences R01GM096032 to Lionel Christiaen.
National Science Foundation NSF-1161835 to Alberto Stolfi.
Agriculture and Food Research Initiative 2010-65205-20361 to C Titus Brown.
ASSEMBLE MARINE to Billie J Swalla.
American Heart Association 10SDG4310061 to Lionel Christiaen.
New York Cardiac Center to Lionel Christiaen.
National Science Foundation DBI-0939454 to Elijah K Lowe, C Titus Brown, Billie J Swalla.
New York University College of Arts and Science to Lionel Christiaen.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
AS, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
EKL, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
BJS, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
CR, Conception and design, Acquisition of data, Contributed unpublished essential data or reagents.
FR, Conception and design, Contributed unpublished essential data or reagents.
CTB, Conception and design, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
LC, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
Additional files
DNA sequences of probes, enhancers, promoters, protein-coding cDNAs, in situ hybridization probe templates, primers, etc used in this study.
Table of newly proposed tunicate gene names and symbols (Stolfi et al., 2014), and their aliases and synonyms.
Major dataset
The following dataset was generated:
Stolfi A, Lowe EK, Racioppi C, Ristoratore F, Brown CT, Swalla BJ, Christiaen LC, 2014, Molgula occidentalis, M. occulta, and M. oculata genome samples, http://www.ncbi.nlm.nih.gov/bioproject/PRJNA253689, BioProject - NCBI.
References
- Abitua P, Wagner E, Navarrete IA, Levine M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature. 2012;492:104–107. doi: 10.1038/nature11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacology & Therapeutics. 2005;107:252–268. doi: 10.1016/j.pharmthera.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Altschmied J, Delfgaauw J, Wilde B, Duschl J, Bouneau L, Volff JN, Schartl M. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics. 2002;161:259–267. doi: 10.1093/genetics/161.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnone M, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- Babraham Bioinformatics Quality control tool for high throughput sequence data. 2014 Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134:3297–3305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- Berrill NJ. Studies in tunicate development. Part II. Abbreviation of development in the Molgulidae. Philosophical Transactions of the Royal Society of London Series B, Containing Papers of a Biological Character. 1931:281–346. [Google Scholar]
- Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, Chitsaz H, Chou WC, Corbeil J, Del Fabbro C, Docking TR, Durbin R, Earl D, Emrich S, Fedotov P, Fonseca NA, Ganapathy G, Gibbs RA, Gnerre S, Godzaridis E, Goldstein S, Haimel M, Hall G, Haussler D, Hiatt JB, Ho IY, Howard J, Hunt M, Jackman SD, Jaffe DB, Jarvis ED, Jiang H, Kazakov S, Kersey PJ, Kitzman JO, Knight JR, Koren S, Lam TW, Lavenier D, Laviolette F, Li Y, Li Z, Liu B, Liu Y, Luo R, Maccallum I, Macmanes MD, Maillet N, Melnikov S, Naquin D, Ning Z, Otto TD, Paten B, Paulo OS, Phillippy AM, Pina-Martins F, Place M, Przybylski D, Qin X, Qu C, Ribeiro FJ, Richards S, Rokhsar DS, Ruby JG, Scalabrin S, Schatz MC, Schwartz DC, Sergushichev A, Sharpe T, Shaw TI, Shendure J, Shi Y, Simpson JT, Song H, Tsarev F, Vezzi F, Vicedomini R, Vieira BM, Wang J, Worley KC, Yin S, Yiu SM, Yuan J, Zhang G, Zhang H, Zhou S, Korf IF. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience. 2013;2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CD, Johnson DS, Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–1560. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH. A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv preprint arXiv. 2012;1203 [Google Scholar]
- Chiba S, Sasaki A, Nakayama A, Takamura K, Satoh N. Development of Ciona intestinalis juveniles (through 2nd ascidian stage) Zoological Science. 2004;21:285–298. doi: 10.2108/zsj.21.285. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–1352. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Stolfi A, Davidson B, Levine M. Spatio-temporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Developmental Biology. 2009a;328:552–560. doi: 10.1016/j.ydbio.2009.01.033. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Stolfi A, Levine M. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Developmental Biology. 2010;340:179–187. doi: 10.1016/j.ydbio.2009.11.006. Ciona savignyi, whole genome shotgun sequencing project - Nucleotide - NCBI. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Wagner E, Shi W, Levine M. Electroporation of transgenic DNAs in the sea squirt Ciona. Cold Spring Harbor Protocols. 2009b;2009 doi: 10.1101/pdb.prot5345. pdb-prot5345. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Wagner E, Shi W, Levine M. Isolation of sea squirt (Ciona) gametes, fertilization, dechorionation, and development. Cold Spring Harbor Protocols. 2009c;2009 doi: 10.1101/pdb.prot5344. pdb-prot5344. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Wagner E, Shi W, Levine M. Whole-mount in situ hybridization on sea squirt (Ciona intestinalis) embryos. Cold Spring Harbor Protocols. 2009d;2009 doi: 10.1101/pdb.prot5348. pdb-prot5348. [DOI] [PubMed] [Google Scholar]
- Conklin EG. The organization and cell-lineage of the ascidian egg. 1905;13 [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Cota CD, Segade F, Davidson B. Heart genetics in a small package, exploiting the condensed genome of Ciona intestinalis. Briefings in Functional Genomics. 2013;13:3–14. doi: 10.1093/bfgp/elt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Developmental Biology. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- Darras S, Nishida H. The BMP/CHORDIN antagonism controls sensory pigment cell specification and differentiation in the ascidian embryo. Developmental Biology. 2001;236:271–288. doi: 10.1006/dbio.2001.0339. [DOI] [PubMed] [Google Scholar]
- Davidson B, Levine M. Evolutionary origins of the vertebrate heart: specification of the cardiac lineage in Ciona intestinalis. Proceedings of the National Academy of Sciences of USA. 2003;100:11469–11473. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes & Development. 2006;20:2728–2738. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Shi W, Levine M. Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development. 2005;132:4811–4818. doi: 10.1242/dev.02051. [DOI] [PubMed] [Google Scholar]
- Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Solution structure of the ETS domain from murine Ets-1: a winged helix-turn-helix DNA binding motif. The EMBO Journal. 1996;15:125–134. [PMC free article] [PubMed] [Google Scholar]
- Dynes JL, Ngai J. Pathfinding of olfactory neuron axons to stereotyped glomerular targets revealed by dynamic imaging in living zebrafish embryos. Neuron. 1998;20:1081–1091. doi: 10.1016/S0896-6273(00)80490-0. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Research. 2004;32(suppl 2):W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Grave C. Molgula citrina (Alder and Hancock). Activities and structure of the free‐swimming larva. Journal of Morphology. 1926;42:453–471. doi: 10.1002/jmor.1050420205. [DOI] [Google Scholar]
- Haag ES. Compensatory vs. pseudocompensatory evolution in molecular and developmental interactions. Genetica. 2007;129:45–55. doi: 10.1007/s10709-006-0032-3. [DOI] [PubMed] [Google Scholar]
- Hadfield KA, Swalla BJ, Jeffery WR. Multiple origins of anural development in ascidians inferred from rDNA sequences. Journal of Molecular Evolution. 1995;40:413–427. doi: 10.1007/BF00164028. [DOI] [PubMed] [Google Scholar]
- Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. Journal of Heredity. 2009;100:605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- Hansen TF. Is modularity necessary for evolvability? Remarks on the relationship between pleiotropy and evolvability. Bio Systems. 2003;69:83–94. doi: 10.1016/S0303-2647(02)00132-6. [DOI] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLOS Genetics. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Nishida H. Developmental fates of larval tissues after metamorphosis in ascidian halocynthia roretzi I. Origin of mesodermal tissues of the juvenile. Developmental Biology. 1997;192:199–210. doi: 10.1006/dbio.1997.8772. [DOI] [PubMed] [Google Scholar]
- Howe AC, Jansson JK, Malfatti SA, Tringe SG, Tiedje JM, Brown CT. Tackling soil diversity with the assembly of large, complex metagenomes. Proceedings of the National Academy of Sciences of USA. 2014;111:4904–4909. doi: 10.1073/pnas.1402564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JL, Burke da Silva K, Bates WR, Swalla BJ. Seminars in cell & developmental biology. Vol 11. Academic Press; 2000. The evolution of anural larvae in molgulid ascidians. [DOI] [PubMed] [Google Scholar]
- Hudson C, Kawai N, Negishi T, Yasuo H. β-Catenin-driven binary fate specification segregates germ layers in ascidian embryos. Current Biology. 2013;23:491–495. doi: 10.1016/j.cub.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Hudson C, Lotito S, Yasuo H. Sequential and combinatorial inputs from Nodal, Delta2/Notch and FGF/MEK/ERK signalling pathways establish a grid-like organisation of distinct cell identities in the ascidian neural plate. Development. 2007;134:3527–3537. doi: 10.1242/dev.002352. [DOI] [PubMed] [Google Scholar]
- Hudson C, Yasuo H. Similarity and diversity in mechanisms of muscle fate induction between ascidian species. Biology of the Cell. 2008;100:265–277. doi: 10.1042/BC20070144. [DOI] [PubMed] [Google Scholar]
- Ikuta T, Saiga H. Dynamic change in the expression of developmental genes in the ascidian central nervous system: revisit to the tripartite model and the origin of the midbrain–hindbrain boundary region. Developmental Biology. 2007;312:631–643. doi: 10.1016/j.ydbio.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, Mercola M, Oshima RG, Willerson JT, Potaman VN, Schwartz RJ. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proceedings of the National Academy of Sciences of USA. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR, Swalla BJ, Ewing N, Kusakabe T. Evolution of the ascidian anural larva: evidence from embryos and molecules. Molecular Biology and Evolution. 1999;16:646–654. doi: 10.1093/oxfordjournals.molbev.a026147. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Davidson B, Brown CD, Smith WC, Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Research. 2004;14:2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NA, Porter AH. Evolution of branched regulatory genetic pathways: directional selection on pleiotropic loci accelerates developmental system drift. Genetica. 2007;129:57–70. doi: 10.1007/s10709-006-0033-2. [DOI] [PubMed] [Google Scholar]
- Kim GJ, Nishida H. Role of the FGF and MEK signaling pathway in the ascidian embryo. Development, Growth & Differentiation. 2001;43:521–533. doi: 10.1046/j.1440-169X.2001.00594.x. [DOI] [PubMed] [Google Scholar]
- Kiontke K, Barrière A, Kolotuev I, Podbilewicz B, Sommer R, Fitch DH, Félix MA. Trends, stasis, and drift in the evolution of nematode vulva development. Current Biology. 2007;17:1925–1937. doi: 10.1016/j.cub.2007.10.061. [DOI] [PubMed] [Google Scholar]
- Kobayashi Mi, Takatori N, Nakajima Y, Kumano G, Nishida H, Saiga H. Spatial and temporal expression of two transcriptional isoforms of Lhx3, a LIM class homeobox gene, during embryogenesis of two phylogenetically remote ascidians, Halocynthia roretzi and Ciona intestinalis. Gene Expression Patterns. 2010;10:98–104. doi: 10.1016/j.gep.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Kourakis MJ, Newman-Smith E, Smith WC. Key steps in the morphogenesis of a cranial placode in an invertebrate chordate, the tunicate Ciona savignyi. Developmental Biology. 2010;340:134–144. doi: 10.1016/j.ydbio.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourakis MJ, Smith WC. A conserved role for FGF signaling in chordate otic/atrial placode formation. Developmental Biology. 2007;312:245–257. doi: 10.1016/j.ydbio.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe T, Swalla BJ, Satoh N, Jeffery WR. Mechanism of an evolutionary change in muscle cell differentiation in ascidians with different modes of development. Developmental Biology. 1996;174:379–392. doi: 10.1006/dbio.1996.0082. [DOI] [PubMed] [Google Scholar]
- Laird CD. Chromatid structure: relationship between DNA content and nucleotide sequence diversity. Chromosoma. 1971;32:378–406. doi: 10.1007/BF00285251. [DOI] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire P. Evolutionary crossroads in developmental biology: the tunicates. Development. 2011;138:2143–2152. doi: 10.1242/dev.048975. [DOI] [PubMed] [Google Scholar]
- Lemaire P. Unfolding a chordate developmental program, one cell at a time: invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Developmental Biology. 2009;332:48–60. doi: 10.1016/j.ydbio.2009.05.540. [DOI] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Current Biology. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Davidson EH. Gene regulatory networks for development. Proceedings of the National Academy of Sciences of USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLOS Biology. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Patel NH, Kreitman M. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development. 1998;125:949–958. doi: 10.1242/dev.125.5.949. [DOI] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Conservation of function and expression of unc-119 from two Caenorhabditis species despite divergence of non-coding DNA. Gene. 1996;183:77–85. doi: 10.1016/S0378-1119(96)00491-X. [DOI] [PubMed] [Google Scholar]
- Maliska ME, Swalla BJ. Molgula pugetiensis is a Pacific tailless ascidian within the Roscovita Clade of molgulids. The Biological Bulletin. 2010;219:277–282. doi: 10.1086/BBLv219n3p277. [DOI] [PubMed] [Google Scholar]
- Manni L, Lane NJ, Joly JS, Gasparini F, Tiozzo S, Caicci F, Zaniolo G, Burighel P. Neurogenic and non‐neurogenic placodes in ascidians. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2004;302:483–504. doi: 10.1002/jez.b.21013. [DOI] [PubMed] [Google Scholar]
- Martinez C, Rest JS, Kim AR, Ludwig M, Kreitman M, White K, Reinitz J. Ancestral resurrection of the Drosophila S2E enhancer reveals accessible evolutionary paths through compensatory change. Molecular Biology and Evolution. 2014;31:903–916. doi: 10.1093/molbev/msu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet F, Hutt JA, Milloz J, Millard J, Graham A, Shimeld SM. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Developmental Biology. 2005;282:494–508. doi: 10.1016/j.ydbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. cis-Regulatory changes in kit Ligand expression and parallel evolution of Pigmentation in Sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatomo Ki, Fujiwara S. Expression of Raldh2, Cyp26 and Hox-1 in normal and retinoic acid-treated Ciona intestinalis embryos. Gene Expression Patterns. 2003;3:273–277. doi: 10.1016/S1567-133X(03)00051-6. [DOI] [PubMed] [Google Scholar]
- Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme: III. Up to the tissue restricted stage. Developmental Biology. 1987;121:526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda-Ishii I, Bertrand V, Matsuo I, Lemaire P, Saiga H. Making very similar embryos with divergent genomes: conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development. 2005;132:1663–1674. doi: 10.1242/dev.01707. [DOI] [PubMed] [Google Scholar]
- Ohno S. London/Berlin, Heidelberg and New York: George Alien & Unwin Ltd/Springer-Verlag; 1970. Evolution by gene duplication. [Google Scholar]
- Pavlicev M, Wagner GP. A model of developmental evolution: selection, pleiotropy and compensation. Trends in Ecology & Evolution. 2012;27:316–322. doi: 10.1016/j.tree.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Piano F, Parisi MJ, Karess R, Kambysellis MP. Evidence for redundancy but not trans factor-cis element coevolution in the regulation of Drosophila Yp genes. Genetics. 1999;152:605–616. doi: 10.1093/genetics/152.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter AH, Johnson NA. Speciation despite gene flow when developmental pathways evolve. Evolution. 2002;56:2103–2111. doi: 10.1111/j.0014-3820.2002.tb00136.x. [DOI] [PubMed] [Google Scholar]
- Prud'Homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Ragkousi K, Beh J, Sweeney S, Starobinska E, Davidson B. A single GATA factor plays discrete, lineage specific roles in ascidian heart development. Developmental Biology. 2011;352:154–163. doi: 10.1016/j.ydbio.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razy-Krajka F, Lam K, Wang W, Stolfi A, Joly M, Bonneau R, Christiaen L. Collier/OLF/EBF-Dependent transcriptional dynamics control pharyngeal muscle specification from Primed cardiopharyngeal progenitors. Developmental Cell. 2014;29:263–276. doi: 10.1016/j.devcel.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano LA, Wray GA. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development. 2003;130:4187–4199. doi: 10.1242/dev.00611. [DOI] [PubMed] [Google Scholar]
- Sasakura Y, Kanda M, Ikeda T, Horie T, Kawai N, Ogura Y, Yoshida R, Hozumi A, Satoh N, Fujiwara S. Retinoic acid-driven Hox1 is required in the epidermis for forming the otic/atrial placodes during ascidian metamorphosis. Development. 2012;139:2156–2160. doi: 10.1242/dev.080234. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. Early embryonic expression of a LIM-homeobox gene Cs-lhx3 is downstream of β-catenin and responsible for the endoderm differentiation in Ciona savignyi embryos. Development. 2001;128:3559–3570. doi: 10.1242/dev.128.18.3559. [DOI] [PubMed] [Google Scholar]
- Satoh N. Developmental genomics of ascidians. John Wiley & Sons; 2013. [Google Scholar]
- Satou Y, Imai KS, Satoh N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131:2533–2541. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, Wiley GB, Macmil SL, Roe BA, Zeller RW, Hastings KE, Lemaire P, Lindquist E, Endo T, Hotta K, Inaba K. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biology. 2008;9:R152. doi: 10.1186/gb-2008-9-10-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Yamada L, Mochizuki Y, Takatori N, Kawashima T, Sasaki A, Hamaguchi M, Awazu S, Yagi K, Sasakura Y, Nakayama A, Ishikawa H, Inaba K, Satoh N. A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 2002;33:153–154. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- Schwarz EM, Korhonen PK, Campbell BE, Young ND, Jex AR, Jabbar A, Hall RS, Mondal A, Howe AC, Pell J, Hofmann A, Boag PR, Zhu XQ, Gregory T, Loukas A, Williams BA, Antoshechkin I, Brown C, Sternberg PW, Gasser RB. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biology. 2013;14:R89. doi: 10.1186/gb-2013-14-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Levine M. Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development. 2008;135:931–940. doi: 10.1242/dev.011940. [DOI] [PubMed] [Google Scholar]
- Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLOS Biology. 2009;7:e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen MW, Leitgeb S, Clark VH, Jones SJ, Bird A. Gene number in an invertebrate chordate, Ciona intestinalis. Proceedings of the National Academy of Sciences of USA. 1998;95:4437–4440. doi: 10.1073/pnas.95.8.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi A, Christiaen L. Genetic and genomic toolbox of the chordate Ciona intestinalis. Genetics. 2012;192:55–66. doi: 10.1534/genetics.112.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi A, Gainous TB, Young JJ, Mori A, Levine M, Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565–568. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi A, Levine M. Neuronal subtype specification in the spinal cord of a protovertebrate. Development. 2011;138:995–1004. doi: 10.1242/dev.061507. [DOI] [PubMed] [Google Scholar]
- Stolfi A, Sasakura Y, Chalopin D, Satou Y, Christiaen L, Dantec C, Endo T, Naville M, Nishida H, Swalla BJ, Volff JN, Voskoboynik A, Dauga D, Lemaire P. Guidelines for the nomenclature of genetic elements in tunicate genomes. Genesis. 2014 doi: 10.1002/dvg.22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena É, Stern DL. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proceedings of the National Academy of Sciences of USA. 2000;97:4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swalla BJ. Gastrulation. Vol. 49 Cold Spring Harbor: Cold Spring Harbor Press; 2004. Protochordate gastrulation: lancelets and ascidians; p. 139. [Google Scholar]
- Swalla BJ. Building divergent body plans with similar genetic pathways. Heredity. 2006;97:235–243. doi: 10.1038/sj.hdy.6800872. [DOI] [PubMed] [Google Scholar]
- Swalla BJ, Jeffery WR. Interspecific hybridization between an anural and urodele ascidian: differential expression of urodele features suggests multiple mechanisms control anural development. Developmental Biology. 1990;142:319–334. doi: 10.1016/0012-1606(90)90353-K. [DOI] [PubMed] [Google Scholar]
- Tagawa K, Jeffery WR, Satoh N. The recently-described ascidian species Molgula tectiformis is a direct developer. Zoological Science. 1997;14:297–303. doi: 10.2108/zsj.14.297. [DOI] [PubMed] [Google Scholar]
- Takada N, Satoh N, Swalla BJ. Expression of Tbx6, a muscle lineage T-box gene, in the tailless embryo of the ascidian Molgula tectiformis. Development Genes and Evolution. 2002;212:354–356. doi: 10.1007/s00427-002-0247-y. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Mitani Y, Satoh G, Satoh N. Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development. 1999;126:3725–3734. doi: 10.1242/dev.126.17.3725. [DOI] [PubMed] [Google Scholar]
- Takano TS. Loss of notum macrochaetae as an interspecific hybrid anomaly between Drosophila melanogaster and D. simulans. Genetics. 1998;149:1435–1450. doi: 10.1093/genetics/149.3.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatori N, Kumano G, Saiga H, Nishida H. Segregation of germ layer fates by Nuclear migration-dependent Localization of not mRNA. Developmental Cell. 2010;19:589–598. doi: 10.1016/j.devcel.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Tassy O, Dauga D, Daian F, Sobral D, Robin F, Khoueiry P, Salgado D, Fox V, Caillol D, Schiappa R, Laporte B, Rios A, Luxardi G, Kusakabe T, Joly JS, Darras S, Christiaen L, Contensin M, Auger H, Lamy C, Hudson C, Rothbächer U, Gilchrist MJ, Makabe KW, Hotta K, Fujiwara S, Satoh N, Satou Y, Lemaire P. The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Research. 2010;20:1459–1468. doi: 10.1101/gr.108175.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuoka M, Kumano G, Nishida H. FGF9/16/20 and Wnt-5α signals are involved in specification of secondary muscle fate in embryos of the ascidian, Halocynthia roretzi. Development Genes and Evolution. 2007;217:515–527. doi: 10.1007/s00427-007-0160-5. [DOI] [PubMed] [Google Scholar]
- Tolkin T, Christiaen L. Development and evolution of the ascidian cardiogenic mesoderm. Current Topics in Developmental Biology. 2012;100:107–142. doi: 10.1016/B978-0-12-387786-4.00011-7. [DOI] [PubMed] [Google Scholar]
- True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evolution & Development. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- Tsagkogeorga G, Turon X, Hopcroft RR, Tilak MK, Feldstein T, Shenkar N, Loya Y, Huchon D, Douzery EJ, Delsuc F. An updated 18S rRNA phylogeny of tunicates based on mixture and secondary structure models. BMC Evolutionary Biology. 2009;9:187. doi: 10.1186/1471-2148-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster AJ, Ramani AK, McKay SJ, Fraser AG. Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of Developmental System Drift on gene function. PLOS Genetics. 2014;10:e1004077. doi: 10.1371/journal.pgen.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Katsuyama Y, Yasugi S, Saiga H. Spatially and temporally regulated expression of the LIM class homeobox gene Hrlim suggests multiple distinct functions in development of the ascidian, Halocynthia roretzi. Mechanisms of Development. 1995;51:115–126. doi: 10.1016/0925-4773(95)00359-9. [DOI] [PubMed] [Google Scholar]
- Wada H, Saiga H, Satoh N, Holland PW. Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8, Hox and Otx genes. Development. 1998;125:1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- Wang W, Razy-Krajka F, Siu E, Ketcham A, Christiaen L. NK4 antagonizes Tbx1/10 to promote cardiac versus pharyngeal muscle fate in the ascidian second heart field. PLOS Biology. 2013;11:e1001725. doi: 10.1371/journal.pbio.1001725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends in Genetics. 2010;26:66–74. doi: 10.1016/j.tig.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nature Reviews Genetics. 2011;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLOS Biology. 2004;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woznica A, Haeussler M, Starobinska E, Jemmett J, Li Y, Mount D, Davidson B. Initial deployment of the cardiogenic gene regulatory network in the basal chordate, Ciona intestinalis. Developmental Biology. 2012;368:127–139. doi: 10.1016/j.ydbio.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yagi K, Satoh N, Satou Y. Identification of downstream genes of the ascidian muscle determinant gene Ci-macho1. Developmental Biology. 2004;274:478–489. doi: 10.1016/j.ydbio.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Yasuhiko Y, Haraguchi S, Kitajima S, Takahashi Y, Kanno J, Saga Y. Tbx6-mediated Notch signaling controls somite-specific Mesp2 expression. Proceedings of the National Academy of Sciences of USA. 2006;103:3651–3656. doi: 10.1073/pnas.0508238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WP, Brenner S, Venkatesh B. Duplication, degeneration and subfunctionalization of the nested synapsin–Timp genes in Fugu. Trends in Genetics. 2003;19:180–183. doi: 10.1016/S0168-9525(03)00048-9. [DOI] [PubMed] [Google Scholar]
- Zeller RW, Virata MJ, Cone AC. Predictable mosaic transgene expression in ascidian embryos produced with a simple electroporation device. Developmental Dynamics. 2006;235:1921–1932. doi: 10.1002/dvdy.20815. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]