Abstract
Metabolomics has been increasingly applied to discovering biomarkers and identifying perturbed pathways. Berberine has been shown to exhibit anti-inflammatory, antioxidant, and anticancer properties, but its mechanisms for treating nonbacterial prostatitis (NBP) remain unclear completely. We developed the untargeted metabolomics approach based on UPLC-Q-TOF-HDMS to profile the metabolite changes in urine samples in order to discover novel potential biomarkers to clarify mechanisms of berberine in treating a rat model of capsaicin-induced nonbacterial prostatitis (NBP). The changes in metabolic profiling were restored to their base-line values after berberine treatment according to the principal component analysis (PCA) score plots. Fourteen different potential biomarkers and five acutely perturbed metabolic pathways contributing to the treatment of NBP were discovered and identified. Specifically, the berberine-treated rats are located closer to the normal group, indicating that the NBP-induced disturbances to the metabolic profile were partially reversed by berberine treatment. After treatment with berberine, the relative contents of 12 potential biomarkers were effectively regulated, which suggested that the therapeutic effects of berberine on NBP may involve regulating disturbances to the metabolism. Our results show that the protective effect of berberine occurs in part through a reversal of the NBP-caused disturbances.
Introduction
Nonbacterial prostatitis (NBP) is the most common urological diagnosis in men under 50 years of age and is the third common urologic diagnosis in men over 50 years of age. An estimated 50% of all men experience prostatitis-like symptoms at some point during their lifetime (Zeng et al., 2014). A suitable preclinical animal model of prostatitis-induced pain or prostatodynia can help us to understand this clinical issue.
Capsaicin is a polysaccharide that is very commonly used to induce inflammation and subsequent pain in various inflammatory pain models (Chuang et al., 2008). Rat models of capsaicin-associated nonbacterial prostatitis (NBP) can be useful for elucidating the mechanisms of the pathogenesis of NBP (Chuang et al., 2007). Wistar rats spontaneously develop NBP, which makes them a good animal model for laboratory investigation of NBP (Hu et al., 2014). NBP has become an important clinical issue. However, effective therapies for treating NBP have yet to be found and this has contributed to an increased use by sufferers of natural products. Plant-derived compounds possess a wide range of pharmacological properties, and their action has been the subject of considerable interest in recent years.
To explain the action mechanism of drugs, metabolomics methodology has been widely used (Wang et al., 2013a; 2013b; Zhang et al., 2013; 2014; Zhao et al., 2013). Metabolomics is defined as the quantitative measurement of the time-related multiparametric metabolic responses of multicellular systems to pathophysiological stimuli or a genetic modification (Arakaki et al., 2008). The metabolomics approach has demonstrated potential in many fields, including disease diagnosis, investigations of toxicological mechanisms, plant metabolomics, determination of the mechanism of drug treatment, and assessing the effect of nutritional intervention (Derewacz et al., 2013; Sabidóet al., 2012; Suhre et al., 2014; Wu et al., 2012; Zheng et al., 2013). It has a great impact in the investigation of discovering biomarkers, and identifcation of perturbed pathways due to disease or drug treatment. Mass spectrometry (MS)-based metabolomics is well suited for reliably coping with high-throughput samples with respect to both technical accuracy and the identification and quantitation of low-molecular-weight metabolites (Sreekumar et al., 2009).
Limonoids are a group of highly oxygenated, modified terpenoids with a prototypical structure either containing or derived from a precursor with a 4,4,8-trimethyl-17-furanylsteroid skeleton (Roy et al., 2006). Members of this class of bioactive compounds have health-promoting and disease-preventing properties and represent secondary metabolites produced in plants (Manosroi et al., 2014). Berberine (Supplementary Fig. S1; supplementary material is available online at www.liebertpub.com/omi) is the major pharmacologically active protoberberine alkaloid in Phellodendri amurensis cortex and Coptidis rhizoma. Several studies have shown that berberine has antibacterial, antiviral, anti-nociceptive, anti-inflammatory, anti-carcinogenic in aflatoxin B1-induced hepatocellular carcinoma and inhibit p-glycoprotein (Mahmoud et al., 2014; Matsuda et al., 1998; Tundis et al., 2014). In vivo tests have shown that berberine inhibits carcinogen-induced tumor growth in various organs (Gong et al., 2010). In addition to anticancer properties, berberine shows anti-inflammatory activity by eliciting a suppressive effect on CD4+ T cells (Kim et al., 2009). Berberine can also be used as an antioxidant as revealed by the b-carotene bleaching assay and can inhibit HIV-1 protease activity in infected human mononuclear cells (Battinelli et al., 2003; Breksa et al., 2006).
Numerous studies have revealed that berberine exhibits anticancer activity, however, the effects of berberine against NBP are yet to be fully elucidated. In this study, we are the first to investigate the beneficial effects of berberine on NBP via a more holistic approach that uses metabolomics. An animal model for NBP in rats was developed with the use of intraprostatic injection of capsaicin, and the reliability and validity of this model was tested by changes in the extent of inflammatory changes of the prostate. We performed metabolomic approach by high-performance liquid chromatography combined with time-of-flight mass spectrometry (UPLC-Q-TOF-HDMS) with TransOmics program (integrated multivariate data analysis and pathway analysis) to investigate the protective effects of berberine on capsaicin-induced NBP in rats.
Experimental Methods
Materials and reagents
Acetonitrile, HPLC grade, was obtained from Merck (Darmstadt, Germany); methanol (HPLC grade) was purchased from Fisher Scientific Corporation (Loughborough, UK); water was purified by a Milli-Q water purification system (Millipore, Bedford, MA, USA) and used for the preparation of samples and mobile phase; leucine enkephalin was purchased from Sigma-Aldrich (St. Louis, MO, USA). Capsaicin (97%) was obtained from (Aladdin, USA); Tween 80 was obtained from Bodi Chemical Co., Ltd.(Tianjin, China). Standards of berberine were purchased from the Tianjin Chemical Reagent Co. (Tianjin, China). All other reagents were of analytical grade.
Animal handling
Male Wistar rats (weighting 280–300 g) were provided by the experimental animal centre of Heilongjiang University of Chinese Medicine. The care and handling of rats were in accordance with the standard of Specific Pathogen Free. The rats were housed in an animal room maintained at a constant temperature and humidity with a 12-hour light/dark cycle. The treatment protocols were approved by the Animal Care and Use Committee (HUCM-2014-03401) and handled according to NIH guidelines. All rats were randomly divided into three groups as follows: Sham group (n=12, control group), model group (surgery group, n=14), berberine group (pretreatment group, n=14). All animals were allowed to acclimatize in metabolism cages for 1 week prior to treatment. 2.10 g of berberine was added to 420 mL of H2O and then vortex mixed for 5 min. Rats were administrated an oral accurate volume of 1 mL/100g of each animal once daily for the following 7 days, rats in the normal control group were given saline alone under the same conditions.
For injection of capsaicin, rats were anesthetized with 0.25 mL/100g sodium pentobarbital (0.2%) and were fixed in a supine position. Then, the lower abdomen above the penis of rats was shaved and the skin in this area sterilized using applications of 10% povidone-iodine solution. A small midline incision was made in the sterile area, then the bladder and the prostate were carefully exposed. With a 30-gauge needle, 50 μL sterile suspension of 3% capsaicin (Aladdin, USA) was injected into both right and left ventral lobes of the prostate gland. For the control group, commensurable sterile normal saline was injected. After the injection, a 2% lidocaine solution was applied to the wound, and then the wound was closed in layers. Preparation of model requires histopathology, PCR testing, and metabolomics evaluation. For histological analysis, one part of the prostate was fixed in buffered 10% formaldehyde for 24 h, embedded in paraffin, cut with a microtome, and stained with hematoxylin-eosin. After 1 week of drug treatment, the prostatic proinflammatory cytokine TNF-α level and histological findings were analyzed.
Collection and preparation of biosamples
Urine samples for metabolomic study were collected. Urine was collected from metabolism cages at ambient temperature throughout the whole procedure and centrifuged at 13,000 rpm at 4°C for 10 min; the supernatants were stored frozen at −80°C until metabolomic analysis. The quality control sample was used to optimize the condition of UPLC-MS, as it contained most information of whole urine samples.
Metabolic profiling
Chromatography
Chromatography was performed using a Waters ACQUITY™ ultra performance liquid chromatography system (Waters Corporation, Milford, USA) controlled with Masslynx (V4.1, Waters Corporation). An aliquot of 5 μL of sample solution was injected onto an ACQUITY UPLC HSS T3 column (50 mm×2.1 mm i.d., 1.8 μm, Waters Corporation) at 45°C with a flow rate of 0.4 mL/min. The mobile phases were composed of 0.1% formic acid in acetonitrile (solvent A) and 0.1% formic acid in water (solvent B), the gradient was used as follows: a linear gradient of 1%–10% A over initial–2.0 min, 10%–20% A over 2–4 min, 20%–60% A over 4–7.5 min, 60%–99% A over 7.5–8.5 min, 99% A over 8.5–10.5 min, 99–1% A over 10.5–11.0 min, 1% A over 11–13 min. The eluant was introduced to the mass spectrometer directly. After every 10 samples injected, a pooled sample as the QC sample followed by a blank was injected in order to ensure the stability and repeatability of the LC-MS systems.
Mass spectrometry
The eluant was introduced into the high-definition mass spectrometer (Waters Corporation) analysis, and the optimal conditions of analysis were as follows: In positive ion mode, the capillary voltage was 3.0 kV, the sampling cone voltage was 30 V, and extraction cone voltage was 4.0 V, the source temperature was set at 110°C, desolvation gas temperature was 350°C, desolvation gas flow was 600 L/h; in negative ion mode, the capillary voltage was 2.6 kV, the sampling cone voltage was 30 V, and extraction cone voltage was 3.5 V, the source temperature was set at 120°C, desolvation gas temperature was 350°C, desolvation gas flow was 650 L/h. The data acquisition rate was set to 0.4 sec/scan, with a 0.1 sec inter scan delay. Data were collected in centroid mode and the mass range was set at m/z 50–1000 using extended dynamic range. For accurate mass acquisition, a lock-mass of leucine enkephalin at a concentration of 0.2 ng/mL was used via a lock spray interface at a flow rate of 100 μL·min-1 monitoring for positive ion mode ([M+H]+=556.2771) and negative ion mode ([M - H]−=554.2615) to ensure accuracy during the MS analysis.
TransOmics data processing and multivariate data analysis
All the LC–MS raw files were converted by a TransOmics program that enables peak identification and quantification using an in-house database. The detailed analysis workflow of TransOmics informatics for metabolomics data from large biological data sets was shown in Rf (Zhang et al., 2014). Subsequently the converted files were calculated for generation of alignment, peak picking, deconvolution, filter data, identification of compounds. The metabolites were exported in EXCEL format or exported to EZinfo software for compound statistics (principal component analysis (PCA) and orthogonal partial least square discriminant analysis (OPLS-DA)), and compound validation. PCA were carried out to visualize the metabolic alterations after mean centering. The variable importance for projection (VIP) plot derived from the OPLS-DA analysis was carried out to select distinct variables as potential markers. Besides the multivariate approaches, one univariate method, Student's t test, was selected to measure the significance of each metabolite in separating model from controls.
Biomarkers identification
The identification of potential biomarkers was determined by Q-TOF. The MS collision energy was 35 ev, and the data were obtained in the positive and negative ion mode, with MarkerLynx software used for data analysis. The identities of the specific metabolites were confirmed by elements information comparison of their mass spectra using the elemental composition information provided by the software. Furthermore, MassFragment conducts rapid putative ion identification and putative biologically relevant analysis via incorporation of four major small molecule databases: ChemSpider, KEGG, HMDB, and Lipid Maps.
Metabolic pathway analysis
MetPA software was employed to all significant upregulated and downregulated metabolites and related biological pathways. The potential markers identified were compared with the accurate mass charge ratio in some databases, including HMDB, KEGG, METLIN, LIPID MAPS, and PUBCHEM, to discover related pathways. T-test and impact value >0.01 was considered to be criteria for statistically significance and would be selected.
Statistical analysis
The PCA was used to uncover unknown trends in the treated groups. Statistically significant differences in mean values were tested by using two-tailed, two-sample Student's t-test; p<0.05 was considered statistically significant. Prior to multivariate analysis, the resultant data matrices were mean-centered and paretoscaled.
Results
Histopathology
Extensive infiltration of inflammatory cells in the lumina, mononuclear cells in the stroma of the gland, and epithelial degeneration were observed in the NBP group. In microscopic findings from the berberine-treatment group, there were fewer inflammatory cells in the lumina and the epithelial cells of the gland and stroma showed more improvement compared to the NBP group. In the berberine-treated groups, the inflammatory cells in the lumina and the epithelial cells of the gland and stroma showed more improvement than the NBP group (Supplementary Fig. S2). These result showed that treatment with the berberine led to the decreased inflammation level from our NBP rat model. These effects are presumed to be the result of the anti-inflammatory effects of berberine.
Plasma protein extravasation and analysis of changes in pro-inflammatory cytokine level
As shown in Supplementary Table S1, after capsaicin injection, Evans blue extravasation was significantly increased in model group, which implied neurogenic plasma extravasation in the prostate. Administration with berberine could effectively ameliorate and recovered to control level (Supplementary Table S1). To investigate the effects of berberine on NBP, analyses of the inflammatory cytokines TNF-α was conducted. Rats in the capsaicin-induced NBP treatment group showed significant increases in pro-inflammatory cytokine TNF-α when compared to the control group (p<0.01) (Supplementary Table S1). Similar to the inflammatory cell accumulation, these changes implied inflammatory processes in prostate induced by capsaicin injection. Based on the above results, the NBP was considered to be successfully established. All these results indicated that the rats presented the typical pathological features of NBP. Of note, administration with berberine could effectively ameliorate these symptoms, as demonstrated by the pro-inflammatory cytokine level of TNF-α of the berberine group showed significant decreases when compared to the NBP group (p<0.01).
Method development and validation
For the method validation study, 5 mL of urine samples from each group were pooled to get a quality control (QC) specimen, and preparation of the QC specimen was the same as the samples. A number of consecutive injections of the QC sample were made to obtain a stable QTOF/MS system before experimental data acquisition, and then acquisition of data for the urine samples was started. QC specimens were analyzed every ten specimens throughout the whole analysis procedure. For the QC sample, 8 selected characteristic ions (ESI+:138.0914, 314.1236, 180.1032, 447.0922; ESI+:124.0068, 312.1077, 173.0810, 407.2794) were chosen to examine the drift of retention times, m/z, and peak areas (Supplementary Table S2). The results showed that variations in the retention times were less than 0.05 min, drift values of m/z were less than 5 ppm, and the relative standard deviation (RSD) of each peak area was below 8.5 %, demonstrating that the system had excellent stability and repeatability during the analysis procedure. According to the optimized conditions of urine analysis, principal component analysis of QCs urine samples from prostatitis rats was shown in Supplementary Figure S3, QC samples were gathered together to determine during the data collection and instrument was stable during collected sample data, demonstrating that the system had excellent stability during the analysis procedure.
Metabolic profile analysis
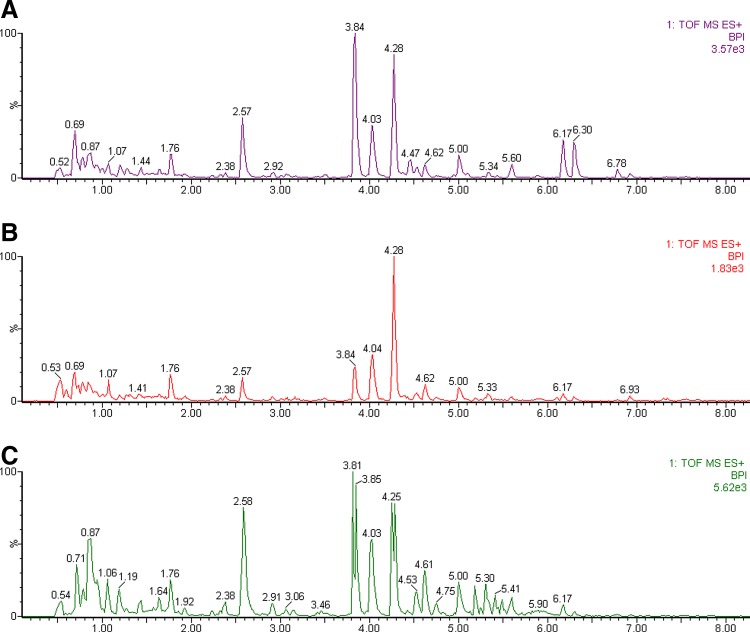
The representative basic ion chromatograms of urine samples derived from control, model, and dose groups in negative modes are presented in Figure 1 by using the optimal UPLC-Q-TOF-HDMS conditions described above. Low molecular mass metabolites could be separated well in the short time of 6 min. Visual comparison of each group at the 9th day revealed slightly different contours, some ingredients may be hidden differences in the peaks, and the visual comparison is not easy to distinguish thousands of signals of each group. In order to better visualize the subtle similarities and differences among these complex data sets, multiple pattern recognition methods were employed to phenotype the urine metabolome of rats.
FIG. 1.
Based Peak Intensity (BPI) chromatograms of urine samples based on UPLC-Q-TOF/HDMS. (A) control group; (B) model group; (C) berberine group.
Metabolomic study of NBP
In order to identify the various different metabolic changes affecting the NBP rats, the MS spectrum were preprocessed in order to be able to carry out multivariate statistical analysis (PCA, PLS-DA, and OPLS-DA). The data files that were generated by TransOmics were imported into urine database and filtered by Metascope that is a flexible search engine which is designed to work with databases that can set thresholds for mass and retention time as required. TransOmics showed the detailed information for each individual feature including statistics, extracted ion chromatograms, spectrum details, and putative identifications (Supplementary Fig. S4). Raw data from UPLC/MS were analyzed by the TransOmics were imported into EZinfo 2.0 software for data analysis. Multivariate statistical analysis can give a comprehensive view of the clustering trend for the metabolomics data. Here, PCA were used to classify the metabolic phenotypes and identify the differing metabolites. In the PCA scores, each point represents an individual sample. The PCA results are displayed as score plots indicating the scatter of the samples, which indicate similar metabolomics compositions when clustered together and compositionally different metabolomes when dispersed. The PCA scores plot could divide the different urine samples into different blocks, respectively, suggesting the metabolic profiles have changed. NBP rats deviated from control rats in their metabolic profile, indicating that the metabolic networks of NBP model rat were disordered. The differential metabolites associated with NBP were found according to the loading plot of OPLS and the p value of Student's t-test.
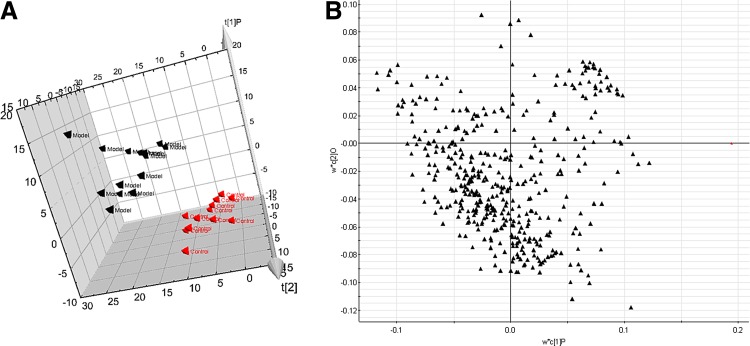
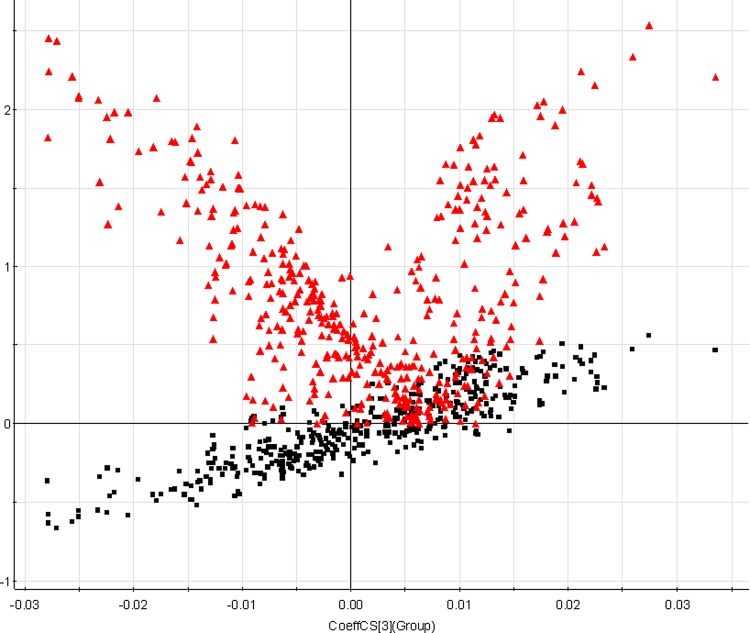
With regard to information analysis of PCA in our experiment shown in Figure 2A, the control and model groups were significantly divided into two classes, indicating that the model of NBP was successfully reproduced. We further used PLS-DA with coefficient plots to directly visualize the results of the loadings and correlation coefficients. Loading-plot of metabolomic profiling of urine samples is shown in Figure 2B. Additionally, to exhibit how each of these variables is responsible for the separation more intuitively, we created an S-plot that is combined with a VIP plot and a color coefficient scale bar. More subtle changes can be found by the pattern recognition approach-score plots of OPLS. An OPLS score plot base on all 407 compounds showed a obvious separation of the control group and the NBP model group (Fig. 3). For further analysis of feature ions, VIP-plot (Fig. 3) from the OPLS was carried out to select distinct variables as potential biomarkers for distinguishing NBP from controls. We generated VIP plots from the OPLS-DA with a threshold of 2 to identify the metabolites that significantly contribute to the clustering between groups. Following the criterions above, 14 metabolite ions were selected as potential biomarkers related with NBP rats and are listed in Supplementary Table S3.
FIG. 2.
Multivariate data analysis result for control and model group in positive ion mode. (A) Principal component analysis score plots of urine samples in control vs. NBP (model) groups at 9th day. Note: Each colored point represents a sample. (B) Loading plot of orthogonal partial least square discriminant analysis of NBP in positive ion mode.
FIG. 3.
The variable importance for projection (VIP)-plot of orthogonal partial least square discriminant analysis of NBP urine samples at 9th day.
Metabolic pathway analysis
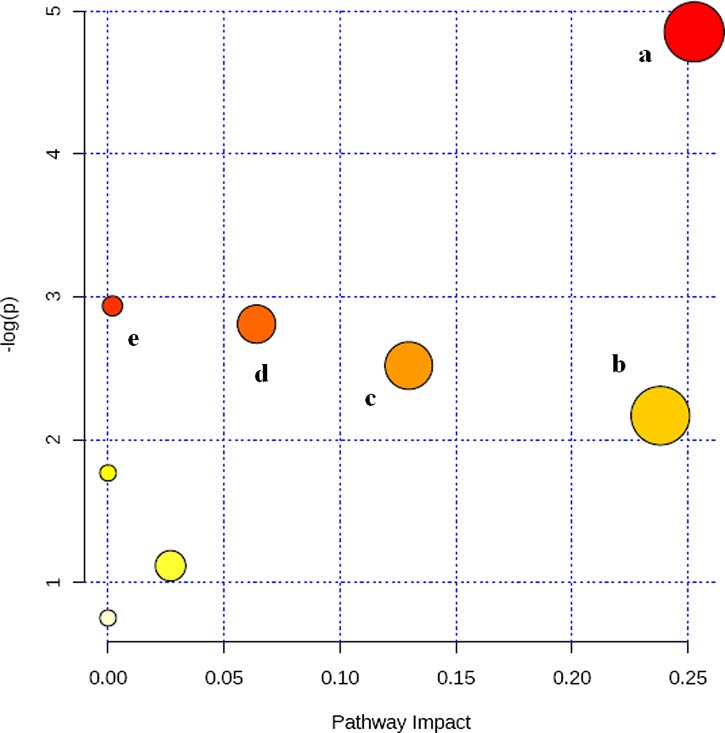
To gain insight into the metabolic mechanism of NBP, metabolic pathways of the significantly altered metabolites were analyzed using the “pathway analysis” module within the MetaboAnalyst software. The related pathway of biomarker was identified by searching KEGG PATHWAY Database. We identified a total of five distinct metabolic pathways (Supplementary Table S4) that were significantly altered in the urine samples from the model group. The detailed analysis of the most relevant pathways of NBP was performed by MetaboAnalyst's tool. A total of feature compounds in five pathways that were identified together are important for the host response to NBP. The predominant hits were pathways involved in histidine metabolism, nicotinate and nicotinamide metabolism, phenylalanine metabolism, arginine and proline metabolism, and tyrosine metabolis. The detailed construction of the metabolism pathways with higher score is shown in Figure 4.
FIG. 4.
Summary of pathway analysis with MetPA tool. (a) histidine metabolism; (b) nicotinate and nicotinamide metabolism; (c) phenylalanine metabolism; (d) arginine and proline metabolism; (e) tyrosine metabolism.
Protective effects of berberine against NBP
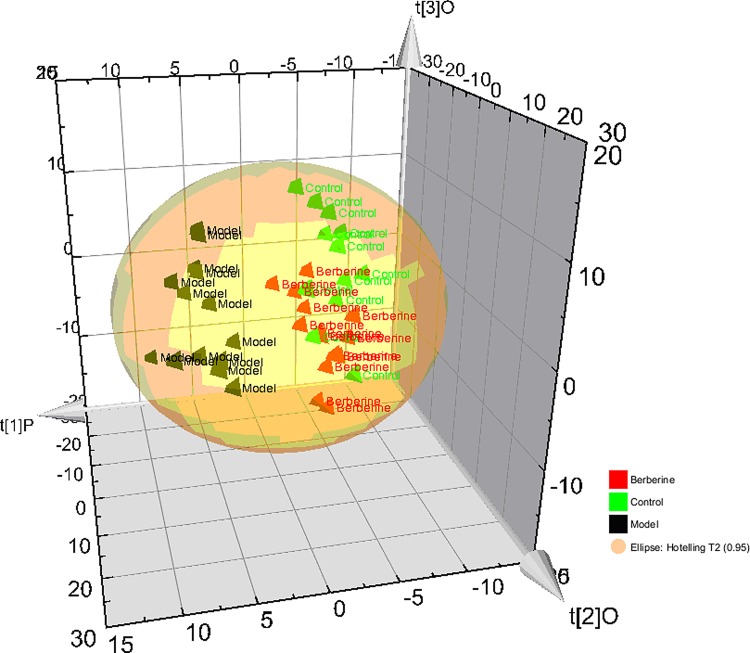
The results of differential metabolite identification suggest that the development of NBP involves serious disorders of the metabolism of histidine metabolism, nicotinate and nicotinamide metabolism, phenylalanine metabolism, arginine and proline metabolism, and tyrosine metabolism. The dysfunction of the immune system in NBP rats may be caused by metabolic disorders. Using the presented UPLC-MS method, the urinary metabolic profiles of the berberine-treated group were obtained. As the 14 potential biomarkers have been found, it is reasonable to take them as the potential drug targets for further investigating the intervening mechanisms of berberine to NBP. Subtle changes could be found using a pattern recognition approach, such as PCA and PLS-DA. Therefore, the levels of the biomarkers from UPLC-MS on the 9th day were introduced as variables to PCA, respectively, performed on control, NBP, and berberine-treated groups. Interestingly, it is important to note that the disturbed metabolic pathways are able to be partially reversed by berberine treatment. Using the 3D of PCA score plot (Fig. 5), the NBP group is clearly separated from the normal group, which implies that the metabolic characteristics of the various small molecules are distinctly different. The score plot of the PCA shows that the control, NBP, and berberine treatment groups were separated clearly (Fig. 5), and the berberine treatment group was closer to the control group than the NBP group, which suggested that berberine could reverse the pathological process of NBP. After treatment with berberine, the score plot of OPLS (Fig. 5) shows that there was a significant difference in the metabolic profile of the NBP model group, berberine-treated group, and control group. This difference confirmed that berberine and and its main components can play a role in regulating the abnormal metabolic network of NBP rat.
FIG. 5.
Protective effects of berberine against NBP based on metabolite profile.
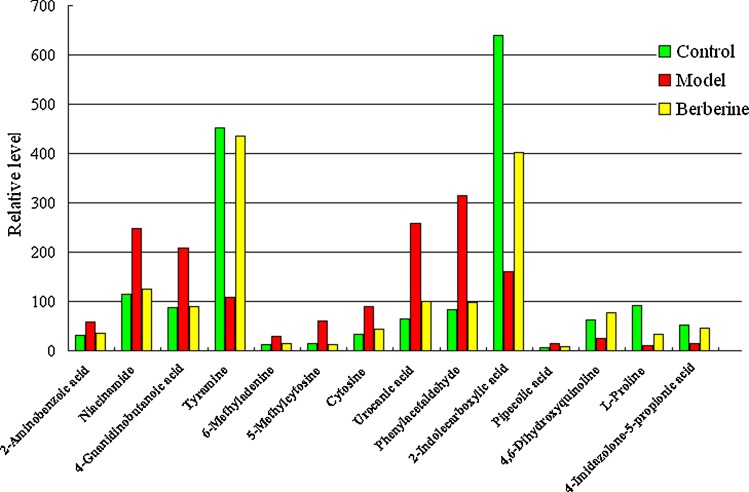
The relative mean height intensity of different metabolites are graphed in Figure 6. By comparing the identified biomarker level in the NBP group, 14 of the identified biomarkers were completely reversed by berberine. Interestingly, according to the parametric t-test, we found that the relative concentration of these metabolites could be reversed after berberine treatment. Compared with the alterations of NBP-related metabolites, most of them were reset to a normal state after berberine administration. Berberine, which exerts multi-pathway and multi-target roles in the body, effectively regulated the metabolic disorders of rat with NBP. The relative contents of 12 biomarkers in the limonin-treated group were prominently regulated, and the content of these differential metabolites trended to the level of the control group (Fig. 6). Berberine effectively repaired the metabolic network, thus alleviating the protective effects of differential metabolites on immune response and inflammation, ameliorating the dysfunction of the immune system and the multiorgan inflammatory response during the development of NBP, thereby inhibiting the progression of NBP. Metabolomics represents an emerging and powerful discipline that provides an accurate and dynamic picture of the phenotype of biosystems through the study of potential biomarkers of NBP that could be used for therapeutic targets and discovery of new drugs.
FIG. 6.
Changes in the relative intensity levels (mean) of target metabolites identified by UPLC-Q-TOF/HDMS.
Discussion
Metabolomics represents a comprehensive method for comprehensive assessment of endogenous metabolites in a biological system and may provide additional insight into the molecular mechanisms (Sun et al., 2014). It enables the parallel assessment of the levels of a broad range of endogenous and exogenous metabolites and has been shown to have a great impact in investigation of physiological status, discovering biomarkers, and identifying perturbed pathways. Small-molecule metabolites have an important role in biological systems to understand disease phenotypes, and help us in understanding a detailed analysis of complex reaction pathways and uncovering drug targets (Zhang et al., 2014). Metabolomics has recently demonstrated significant potential in many fields, including disease diagnosis, investigations of toxicological mechanisms, and determination of the mechanism of drug intervention. To explain the action mechanism of drugs, metabolomics methodology has been widely used (Wang et al., 2012a, b).
NBP is the most common urological diagnosis in men under 50 years of age and is the third common urologic diagnosis in men over 50 years of age. It has become an important clinical issue, but the causes and treatment of NBP are largely unknown. Thus, it is imperative to study the mechanisms underlying NBP to develop effective therapeutic treatments. Natural products possess a wide range of pharmacological properties and their action has been of interest in recent years. Berberine is the major pharmacologically active protoberberine alkaloids in Phellodendri amurensis cortex and Coptidis rhizoma. Previous studies showed that berberine has antibacterial, antiviral, antinociceptive, anti-inflammatory, and anti-carcinogenic effects (Mahmoud et al., 2014; Matsuda et al., 1998; Tundis et al., 2014). There is still little information concerning its anti-NBP mechanism. We are the first to investigate the beneficial effects of berberine on NBP by metabolomics analysis.
In the present study, using a UPLC-Q-TOF/MS based metabolomic study, we investigated the metabolic changes of molecular mechanism by which berberine conferred a protective effect on NBP. Interestingly, berberine exhibited a protective role and kept animals in the normal situation. Histopathology tests showed that treatment with berberine led to fewer inflammatory cells in the lumina and the epithelial cells of the gland, and stroma showed more improvement compared to the NBP group. Of note, administration with berberine could effectively ameliorate these symptoms, as demonstrated by the pro-inflammatory cytokine level of TNF-α of the berberine group showed significant decreases when compared to the NBP group. In order to more clearly characterize treatment effects of berberine, multiple pattern recognition methods were employed to phenotype the urine metabolome. PCA scores plot showed the control and model groups were significantly divided into two classes, indicating that the model of NBP was successfully reproduced. More subtle changes can be found by the pattern recognition approach-score plots of OPLS. For further analysis of feature ions, VIP-plot from the OPLS was carried out to select distinct variables as potential biomarkers for distinguishing NBP from controls.
As a result, 14 differential metabolites in the NBP model were identified and these metabolites demonstrated that abnormal metabolism occurred in the model animals and metabolic analysis of NBP was inferred from changes in the intermediates during substance metabolism. The related pathway of biomarker was identified by searching the KEGG PATHWAY Database. We identified a total of five distinct metabolic pathways that were identified together and are important for the host response to NBP. The predominant hits were pathways involved in metabolism of histidine, nicotinate and nicotinamide, phenylalanine, arginine and proline, and tyrosine.
A discussion of the newly identified biomarkers and implicated biochemical pathways is also presented. 2-Aminobenzoic acid is a substrate of enzyme anthranilate hydroxylase in benzoate degradation via a hydroxylation pathway. Niacinamide is an important compound functioning as a component of the coenzyme NAD. 4-Guanidinobutanoate is a normal metabolite present in low concentrations. Tyramine, a monoamine compound derived from the amino acid tyrosine, is metabolized by the enzyme monoamine oxidase. Tyramine acts as a neurotransmitter via a G protein-coupled receptor with high affinity for tyramine called TA1. The TA1 receptor is found in the brain as well as peripheral tissues including the kidney.
6-Methyladenine is a methylated adenine residue. The presence of 6-methyladenine residues increases the in vitro translation efficiency of dihydrofolate reductase; an inhibition of 6-methyladenine residues in dihydrofolate reductase transcripts significantly alters their rate of translation. 5-Methylcytosine is a methylated nucleotide base found in eukaryotic DNA. In animals, the DNA methylation of cytosine to form 5-methylcytosine is found primarily in the palindromic sequence. Cytosine is a pyrimidine base that is a fundamental unit of nucleic acids. The deamination of cytosine alone is apparent and the nucleotide of cytosine is the prime mutagenic nucleotide in diseases.
Urocanic acid, a breakdown product of histidine, is an intermediate in the conversion of histidine to glutamic acid, whereas in the epidermis, it accumulates and may be an immunoregulator. Phenylacetaldehyde, one important oxidation-related aldehyde, is readily oxidized to phenylacetic acid. Therefore it will eventually be hydrolyzed and oxidized to yield phenylacetic acid that will be excreted primarily in the urine in conjugated form. 2-Indolecarboxylic acid is a strong inhibitor of lipid peroxidation, similar to melatonin and some structurally related indole compounds.
Pipecolic acid is a metabolite of lysine found in human physiological fluids. It is known that pipecolic acid levels are also elevated in patients with chronic liver diseases. Pipecolic acid might therefore be a possible biochemical marker for selecting candidates for NBP therapy. 4,6-Dihydroxyquinoline is the product of the conversion of 5-hydroxykynurenamine by the enzyme monoamine oxidase, both metabolites from the 5-hydroxytryptophan metabolism. L-Proline is one of the twenty amino acids used in living organisms as the building blocks of proteins. It is an essential component of collagen and is important for proper functioning of joints and tendons.
4-Imidazolone-5-propanoate is a metabolite of histidine metabolism. It is produced from urocanic acid by the enzyme urocanate hydratase. 4-Imidazolone-5-propionic acid can spontaneously decay to 4-oxoglutaramate or formylisoglutamine. It is also converted to N-forminimo-L-glutamate by the enzyme imidazolonepropionase.
In the present study, we explored the metabolic profiling and potential metabolic markers to investigate the anti-NBP of berberine using well-established research strategy. It is noteworthy that all the changes of these metabolites could be reversed by berberine treatment, suggesting the possible pharmacological mechanisms of antipyretic effects of berberine. Most of them were reset to a normal state after berberine administration. It indicated that these metabolites may be the targets that were related to the action mechanism of berberine. Results of differential metabolite identification suggest that the development of NBP involves serious disorders of the metabolism of histidine, nicotinate and nicotinamide, phenylalanine, arginine and proline, and tyrosine. Interestingly, it is important to note that the disturbed metabolic pathways are able to be partially reversed by berberine treatment. In other words, berberine might contribute to repair of these metabolites involved pathways. In summary, we discovered a significant perturbance of metabolic profile in the urine of NBP rats, which could be obviously reversed by berberine treatment. Based on metabolic pathway analysis, we concluded that the anti-NBP mechanism of berberine lay on correcting perturbed metabolism according to a variety of metabolic pathways. These findings highlight the pharmacological characteristics of berberine. Further experiments should take into account the dose- and time-dependent manner of berberine.
The limitation of this study is that we only focused on the pharmacological mechanisms without the adverse drug reaction profiles and safety evaluation of berberine. Furthermore, our study also highlights the importance of metabolomics as a potential tool for us to increase research productivity toward metabolomics drug discovery.
Conclusion
In this study, for the first time, we report a comprehensive analysis of metabolic patterns of the treatment of NBP with berberine. UPLC-Q-TOF/MS-based urine metabolomics analysis, combined with multivariate statistical analysis, was applied to evaluate the pathogenesis of NBP and the mechanism of action of berberine in NBP model rats. As a result, 14 differential metabolites in the NBP model were identified by the MS and MS/MS information. Compared with the alterations of NBP-related metabolites, most were reset to a normal level after berberine administration. Our findings also show that berberine exhibited preventive efficacy against NBP by adjusting these multiple metabolic pathways to their normal states. Particularly, berberine can effectively regulate the metabolism pathways of histidine, nicotinate and nicotinamide, phenylalanine, arginine and proline, and tyrosine, and can exert a good therapeutic effect on NBP. These results provide a better understanding of the pathogenesis of NBP and the mechanism of action of berberine in NBP. In addition, this study of potential metabolites could be used to achieve multiple targets for treatment of NBP, lay foundation for finding therapeutic targets and discovering new multi-target drugs. Our results also proved that metabolomics is a powerful technology platform for researching complex disease mechanisms and evaluating the mechanism of action of natural products. The potential application of metabonomics in medicine is infinite and will have a significant impact on natural products and drug development.
Supplementary Material
Acknowledgments
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant No. 81430093, 81173500, 81373930, 81302905, 81102556, 81202639), National Key Technology Research and Development Program of the Ministry of Science and Technology of China (Grant No. 2011BAI03B03, 2011BAI03B06, 2011BAI03B08), National Key Subject of Drug Innovation (Grant No. 2009ZX09502-005).
Author Disclosure Statement
The authors declare no competing financial interests exist.
References
- Arakaki AK, Skolnick J, and McDonald JF. (2008). Marker metabolites can be therapeutic targets as well. Nature 456, 443. [DOI] [PubMed] [Google Scholar]
- Battinelli L, Mengoni F, Lichtner M, Mazzanti G, Saija A, Mastroianni CM, and Vullo V. (2003). Effect of limonin and nomilin on HIV-1 replication on infected human mononuclear cells. Planta Med 69, 910–913 [DOI] [PubMed] [Google Scholar]
- Breksa AP, 3rd, and Manners GD. (2006). Evaluation of the antioxidant capacity of limonin, nomilin, and limonin glucoside. J Agric Food Chem 54, 3827–3831 [DOI] [PubMed] [Google Scholar]
- Chuang YC, Yoshimura N, Wu M, Huang CC, Chiang PH, Tyagi P, and Chancellor MB. (2007). Intraprostatic capsaicin injection as a novel model for nonbacterial prostatitis and effects of botulinum toxin A. Eur Urol 51, 1119–1127 [DOI] [PubMed] [Google Scholar]
- Chuang YC, Yoshimura N, Huang CC, Wu M, Chiang PH, and Chancellor MB. (2008). Intraprostatic botulinum toxin a injection inhibits cyclooxygenase-2 expression and suppresses prostatic pain on capsaicin induced prostatitis model in rat. J Urol 180, 742–748 [DOI] [PubMed] [Google Scholar]
- Derewacz DK, Goodwin CR, McNees CR, McLean JA, and Bachmann BO. (2013). Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proc Natl Acad Sci USA 110, 2336–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Shen Y, Zhang Q, Sun Y, Tang J, Tao F, and Xu Q. (2010). Obaculactone suppresses Th1 effector cell function through down-regulation of T-bet and prolongs skin graft survival in mice. Biochem Pharmacol 80, 218–225 [DOI] [PubMed] [Google Scholar]
- Kim W, Fan YY, Smith R, Patil B, Jayaprakasha GK, McMurray DN, and Chapkin RS. (2009). Dietary curcumin and limonin suppress CD4+ T-cell proliferation and interleukin-2 production in mice. J Nutr 139, 1042–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Xiong L, Huang W, Cai H, Luo Y, Zhang Y, and Lu B. (2014). Anti-inflammatory effect and prostate gene expression profiling of steryl ferulate on experimental rats with non-bacterial prostatitis. Food Funct 5, 1150–1159 [DOI] [PubMed] [Google Scholar]
- Mahmoud MF, Gamal S, and El-Fayoumi HM. (2014). Limonin attenuates hepatocellular injury following liver ischemia and reperfusion in rats via toll-like receptor dependent pathway. Eur J Pharmacol 740, 676–682 [DOI] [PubMed] [Google Scholar]
- Manosroi A, Kitdamrongtham W, Ishii K, et al. (2014). Limonoids from Azadirachta indica var. siamensis extracts and their cytotoxic and melanogenesis-inhibitory activities. Chem Biodivers 11, 505–531 [DOI] [PubMed] [Google Scholar]
- Matsuda H, Yoshikawa M, Iinuma M, and Kubo M. (1998). Antinociceptive and anti-inflammatory activities of limonin isolated from the fruits of Evodia rutaecarpa var. bodinieri. Planta Med 64, 339–342 [DOI] [PubMed] [Google Scholar]
- Roy A, and Saraf S. (2006). Limonoids: Overview of significant bioactive triterpenes distributed in plants kingdom. Biol Pharm Bull 29, 191–201 [DOI] [PubMed] [Google Scholar]
- Sabidó E, Quehenberger O, Shen Q, et al. (2012). Targeted proteomics of the eicosanoid biosynthetic pathway completes an integrated genomics- proteomics-metabolomics picture of cellular metabolism. Mol Cell Proteomics 11, M111.014746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, et al. (2009). Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Suhre K, Shin SY, Petersen AK, et al. (2011). Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Zhang S, Zhang A, Yan G, Wu X, Han Y, and Wang X. (2014). Metabolomic analysis of diet-induced type 2 diabetes using UPLC/MS integrated with pattern recognition approach. PLoS One 9, e93384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tundis R, Loizzo MR, and Menichini F. (2014). An overview on chemical aspects and potential health benefits of limonoids and their derivatives. Crit Rev Food Sci Nutr 54, 225–250 [DOI] [PubMed] [Google Scholar]
- Wang X, Yang B, Zhang A, Sun H, and Yan G. (2012a). Potential drug targets on insomnia and intervention effects of Jujuboside A through metabolic pathway analysis as revealed by UPLC/ESI-SYNAPT-HDMS coupled with pattern recognition approach. J Proteomics 75, 1411–1427 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang A, and Sun H. (2012b). Future perspectives of Chinese medical formulae: Chinmedomics as an effector. OMICS 16, 414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang A, Yan G, Sun W, Han Y, and Sun H. (2013a). Metabolomics and proteomics annotate therapeutic properties of geniposide: Targeting and regulating multiple perturbed pathways. PLoS One 8, e71403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang A, Wang P, et al. (2013b). Metabolomics coupled with proteomics advancing drug discovery toward more agile development of targeted combination therapies. Mol Cell Proteomics 12, 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wu Z, Wang X, et al. (2012). Metabolomic analysis reveals that carnitines are key regulatory metabolites in phase transition of the locusts. Proc Natl Acad Sci USA 109, 3259–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Chen H, Yang J, et al. (2014) Development and validation of an animal model of prostate inflammation-induced chronic pelvic pain: Evaluating from inflammation of the prostate to pain behavioral modifications. PLoS One 9, e96824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Sun H, Han Y, et al. (2012). Exploratory urinary metabolic biomarkers and pathways using UPLC-Q-TOF-HDMS coupled with pattern recognition approach. Analyst 137, 4200–4208 [DOI] [PubMed] [Google Scholar]
- Zhang A, Sun H, Dou S, Sun W, Wu X, Wang P, and Wang X. (2013). Metabolomics study on the hepatoprotective effect of scoparone using ultra-performance liquid chromatography/electrospray ionization quadruple time-of-flight mass spectrometry. Analyst 138, 353–361 [DOI] [PubMed] [Google Scholar]
- Zhang A, Zhou X, Zhao H, et al. (2014). Rapidly improved determination of metabolites from biological data sets using the high-efficient TransOmics tool. Mol Biosyst 10, 2160–2165 [DOI] [PubMed] [Google Scholar]
- Zhang AH, Sun H, Han Y, et al. (2013). Ultraperformance liquid chromatography-mass spectrometry based comprehensive metabolomics combined with pattern recognition and network analysis methods for characterization of metabolites and metabolic pathways from biological data sets. Anal Chem 85, 7606–7612 [DOI] [PubMed] [Google Scholar]
- Zhao YY, Lei P, Chen DQ, Feng YL, and Bai X. (2013). Renal metabolic profiling of early renal injury and renoprotective effects of Poria cocos epidermis using UPLC Q-TOF/HSMS/MSE. J Pharm Biomed Anal 81–82, 202–209 [DOI] [PubMed] [Google Scholar]
- Zheng P, Wang Y, Chen L, et al. (2013). Identification and validation of urinary metabolite biomarkers for major depressive disorder. Mol Cell Proteomics 12, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.