X-ray data were collected for two Dscam1 Ig7 isoforms and were processed to resolutions of 1.95 and 2.37 Å, respectively. Comparison of different Ig7 isoforms will provide insights into the mechanism of its homophilic binding specificity.
Keywords: Dscam1, RNA splicing, Ig7, homophilic dimer
Abstract
Drosophila Down syndrome cell adhesion molecule 1 (Dscam1) plays a critical role in neural development. It can potentially form 38 016 isoforms through alternative RNA splicing, and exhibits isoform-specific homophilic interaction through three variable Ig domains (Ig2, Ig3 and Ig7). The diversity and homophilic interaction are essential for its functions. Ig7 has 33 isoforms and is the most variable among the three variable Ig domains. However, only one isoform of Ig7 (isoform 30) has been structurally determined to date. Here, two isoforms of Dscam1 Ig7 (isoforms 5 and 9; Ig75 and Ig79) were produced and crystallized. Diffraction data from Ig75 and Ig79 crystals were processed to resolutions of 1.95 and 2.37 Å, respectively. Comparison of different Dscam1 Ig7 isoforms will provide insight into the mechanism of its binding specificity.
1. Introduction
The Down syndrome cell adhesion molecule (Dscam) was first identified in 1998; it is encoded by the DSCAM gene, which is located within human chromosome band 21q22, and it plays a critical role in Down syndrome and neural development (Yamakawa et al., 1998 ▶). Four Dscam paralogues are found in Drosophila. Accumulating evidence shows the founding member of the Drosophila Dscam family, Dscam1, is essential for neuronal wiring (Schmucker et al., 2000 ▶; Hattori et al., 2007 ▶), axon guidance (Wang et al., 2002 ▶; Ly et al., 2008 ▶), dendritic self-avoidance (Matthews et al., 2007 ▶; Hattori et al., 2009 ▶) and the innate immune system (Watson et al., 2005 ▶; Dong et al., 2006 ▶). Dscam1 belongs to the largest discovered immunoglobulin (Ig) superfamily of cell adhesion molecules (CAMs), which contain ten Ig domains and six fibronectin type III repeats, a single transmembrane domain and a cytoplasmic tail. Dscam1 can potentially form 38 016 isoforms through alternative RNA splicing (Schmucker et al., 2000 ▶), and exhibits isoform-specific homophilic interaction (Wojtowicz et al., 2004 ▶). Each isoform binds to itself but rarely binds to other isoforms. The homodimerization of Dscam1 relys on the three variable Ig domains (Ig2, Ig3 and Ig7) and directs the binding of matching Ig domains (Ig2/Ig2, Ig3/Ig3 and Ig7/Ig7; Wojtowicz et al., 2007 ▶; Sawaya et al., 2008 ▶).
Among the three variable Ig domains, Ig7 is the most variable. The whole domain of Ig7 is variable while only half of Ig2 or Ig3 is variable (Schmucker et al., 2000 ▶; Graveley et al., 2004 ▶; Watson et al., 2005 ▶; Brites et al., 2008 ▶). The expression pattern of Ig7 shows much greater changes both temporally and spatially during development than those of Ig2 or Ig3 (Neves et al., 2004 ▶). Ig7 has 33 isoforms. However, only one isoform of Ig7 has been structurally determined to date (Sawaya et al., 2008 ▶).
Here, we report the cloning, expression, purification, crystallization and preliminary X-ray analysis of two Dscam1 Ig7 isoforms (isoforms 5 and 9; Ig75 and Ig79). Structural analysis of these isoforms will make it possible to compare different Dscam1 Ig7 isoforms and will provide insight into the mechanism of its isoform-specific homophilic binding.
2. Materials and methods
2.1. Macromolecule production
The target DNA was amplified by PCR from a prepared Dscam1 cDNA library. The PCR products were inserted using BamHI and HindIII restriction-enzyme sites into a vector derived from pET-28a(+) (Novagen), which contains a TEV protease cleavage site after the N-terminal His6 tag. The primer details are shown in Table 1 ▶. All constructs were authenticated by DNA sequencing.
Table 1. Macromolecule-production information for Dscam1 Ig7.
The BamHI and HindIII restriction sites are underlined.
| Isoform | 5 | 9 |
|---|---|---|
| Source organism | D. melanogaster | D. melanogaster |
| DNA source | cDNA | cDNA |
| Forward primer | 5-CGGGATCCGTTCCACCGTCGATAGC-3 | 5-CGGGATCCGTGCCGCCCCAGGTTT-3 |
| Reverse primer | 5-CCCAAGCTTTCAGTTGACTTTCAGCTCCGA-3 | 5-CCCAAGCTTTCAATTAACTTGTAATTCA-3 |
| Expression vector | pET-28a(+) | pET-28a(+) |
| Expression host | E. coli strain Rosetta(DE3) | E. coli strain SHuffle |
Ig75 and Ig79 were overexpressed in Escherichia coli strains Rosetta(DE3) (Novagen) and SHuffle (NEB), respectively. An overnight culture (grown at 310 K) of freshly transformed cells in 20 ml Luria–Bertani (LB) medium was transferred to 1000 ml LB medium. The culture was incubated at 310 K and shaken at 250 rev min−1 until an OD600 of 0.6–0.8 was reached. The expression of recombinant protein was then induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 0.5 mM at 300 K for 6 h (Ig75) or 291 K for 18 h (Ig79). The cells were harvested by centrifugation at 5000 rev min−1 at 277 K and the cell pellet was resuspended in binding buffer (20 mM Tris–HCl pH 7.4, 250 mM NaCl). The cells were lysed by a ultrahigh-pressure homogenizer (JNBIO) and the lysate was centrifuged at 15 000 rev min−1 and 277 K for 30 min. The supernatant was collected and loaded onto an Ni–NTA column (Roche) pre-equilibrated with binding buffer. The Ni–NTA column was washed with binding buffer supplemented with 5 mM imidazole to remove nonspecifically binding proteins. The target protein was eluted using binding buffer supplemented with 250 mM imidazole.
The eluted target protein was collected and the His6 tag was removed by TEV protease (1:100) at 277 K overnight. The completeness of the protein digestion was checked by SDS–PAGE. The digested proteins were then concentrated to 1.5 ml using Amicon Ultra centrifugal filters (Merck Millipore) and passed through a desalting column to remove imidazole and then an Ni–NTA column to remove free His6 tag, uncleaved protein and TEV protease. The flowthrough was collected and was further purified via gel filtration (Superdex 75 10/300 GL, GE Healthcare) in a buffer consisting of 20 mM HEPES, 100 mM NaCl pH 7.5 on an ÄKTA system (GE Healthcare). The purified proteins were concentrated to 15 mg ml−1 and stored at 193 K until use.
2.2. Crystallization
Crystals (Fig. 1 ▶) were grown using the hanging-drop vapour-diffusion method. Details of the crystallization conditions are summarized in Table 2 ▶.
Figure 1.
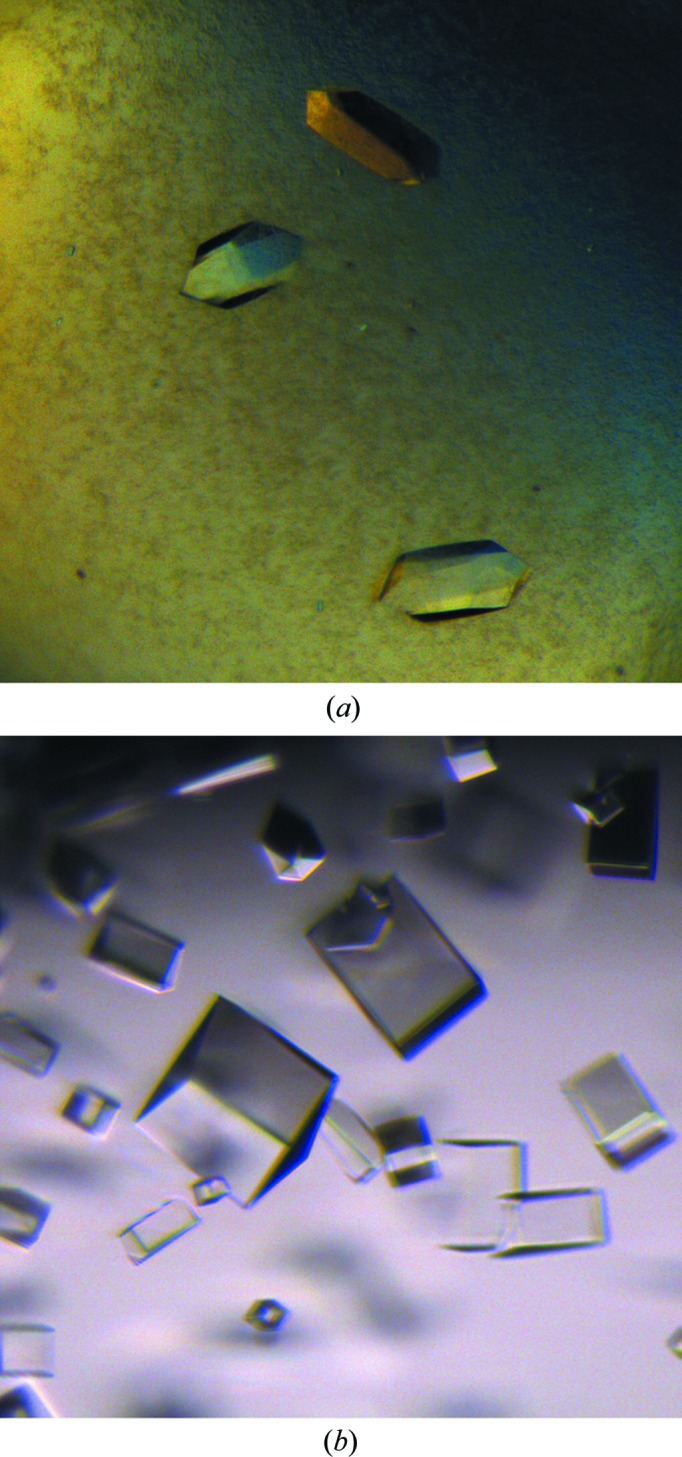
Crystals of Dscam1 Ig7 isoforms 5 (a) and 9 (b).
Table 2. Crystallization of the two Dscam1 Ig7 isoforms.
| Isoform | 5 | 9 |
|---|---|---|
| Method | Hanging-drop vapour diffusion | Hanging-drop vapour diffusion |
| Plate type | 24-well | 24-well |
| Temperature (K) | 289 | 289 |
| Protein concentration (mgml1) | 15 | 15 |
| Buffer composition of protein solution | 20mM HEPES pH 7.5, 100mM NaCl | 20mM HEPES pH 7.5, 100mM NaCl |
| Composition of reservoir solution | 0.1M bis-tris pH 6.5, 50mM calcium chloride, 30%(w/v) polyethylene glycol monomethyl ether 550 | 0.1M HEPES pH 7.5, 0.7M sodium phosphate monobasic, 0.7M potassium phosphate monobasic |
| Volume and ratio of drop | 1:1 | 1:1 |
| Volume of reservoir (l) | 500 | 500 |
2.3. Data collection and processing
The crystals of Ig75 were cryoprotected in reservoir solution supplemented with 20% glycerol. The crystals of Ig79 were cryoprotected in reservoir solution supplemented with 25% sucrose. After soaking in the cryosolution for a few seconds, the crystals were picked up in a nylon loop and flash-cooled in liqid nitrogen. Diffraction data were collected on beamline 3W1A at Beijing Synchrotron Radiation Facility (BSRF) and beamline BL17U1 at Shanghai Synchrotron Radiation Facility (SSRF). The data collected were processed by XDS (Kabsch, 2010 ▶) or the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶). Details of the data-collection and processing statistics are summarized in Table 3 ▶.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Isoform | 5 | 9 |
|---|---|---|
| Diffraction source | BSRF | SSRF |
| Wavelength () | 1.0 | 1.0 |
| Temperature (K) | 100 | 100 |
| Rotation range per image () | 1 | 1 |
| Space group | P32 | P222 |
| a, b, c () | 74.0, 74.0, 67.2 | 45.5, 117.4, 117.3 |
| , , () | 90, 90, 120 | 90, 90, 90 |
| Mosaicity () | 0.312 | 0.248 |
| Resolution range () | 501.95 (2.021.95) | 502.37 (2.522.37) |
| No. of unique reflections | 30014 (2913) | 26120 (4120) |
| Completeness (%) | 99.8 (98.3) | 99.5 (98.8) |
| Multiplicity | 3.8 (3.1) | 7.1 (7.2) |
| I/(I) | 17.7 (2.3) | 21.8 (6.2) |
| R meas (%) | 6.6 (36.6) | 6.2 (28.2) |
| Solvent content (%) | 53.6 | 68.5 |
3. Results and discussion
Drosophila Ig7 has 33 isoforms. We have attempted to express all of the nonglycosylated isoforms (isoforms 5, 6, 8, 9, 10 and 13) in E. coli. The E. coli host strain Rosetta(DE3) was used for our studies since it contains codons that are rarely used in E. coli to enhance the expression of eukaryotic proteins. Only isoform 5 was expressed solubly in a high yield; the other isoforms were mostly expressed as inclusion bodies. We then tried another E. coli strain, SHuffle, which may assist the oxidative folding of disulfide-bonded proteins. As a result, the solubility of the expressed Ig79 was greatly improved. The final yields were about 2 mg per litre for Ig75 and 1 mg per litre for Ig79, with at least 95% purity.
The crystals of Ig75 and Ig79 diffracted to 1.95 and 2.37 Å resolution, respectively. Initially, the data for Ig79 were processed in space group P422. Although the density map appeared to be good, the R free was above 30% and was difficult to reduce. We then lowered the symmetry to P222 and the R free could easily be reduced to below 26%.
Acknowledgments
Financial support for this work was provided by Sichuan University Research Start-up Funds YJ201318 and YJ201327.
References
- Brites, D., McTaggart, S., Morris, K., Anderson, J., Thomas, K., Colson, I., Fabbro, T., Little, T. J., Ebert, D. & Du Pasquier, L. (2008). Mol. Biol. Evol. 25, 1429–1439. [DOI] [PubMed]
- Dong, Y., Taylor, H. E. & Dimopoulos, G. (2006). PLoS Biol. 4, e229. [DOI] [PMC free article] [PubMed]
- Graveley, B. R., Kaur, A., Gunning, D., Zipursky, S. L., Rowen, L. & Clemens, J. C. (2004). RNA, 10, 1499–1506. [DOI] [PMC free article] [PubMed]
- Hattori, D., Chen, Y., Matthews, B. J., Salwinski, L., Sabatti, C., Grueber, W. B. & Zipursky, S. L. (2009). Nature (London), 461, 644–648. [DOI] [PMC free article] [PubMed]
- Hattori, D., Demir, E., Kim, H. W., Viragh, E., Zipursky, S. L. & Dickson, B. J. (2007). Nature (London), 449, 223–227. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Ly, A., Nikolaev, A., Suresh, G., Zheng, Y., Tessier-Lavigne, M. & Stein, E. (2008). Cell, 133, 1241–1254. [DOI] [PMC free article] [PubMed]
- Matthews, B. J., Kim, M. E., Flanagan, J. J., Hattori, D., Clemens, J. C., Zipursky, S. L. & Grueber, W. B. (2007). Cell, 129, 593–604. [DOI] [PubMed]
- Neves, G., Zucker, J., Daly, M. & Chess, A. (2004). Nature Genet. 36, 240–246. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Sawaya, M. R., Wojtowicz, W. M., Andre, I., Qian, B., Wu, W., Baker, D., Eisenberg, D. & Zipursky, S. L. (2008). Cell, 134, 1007–1018. [DOI] [PMC free article] [PubMed]
- Schmucker, D., Clemens, J. C., Shu, H., Worby, C. A., Xiao, J., Muda, M., Dixon, J. E. & Zipursky, S. L. (2000). Cell, 101, 671–684. [DOI] [PubMed]
- Wang, J., Zugates, C. T., Liang, I. H., Lee, C.-H. J. & Lee, T. (2002). Neuron, 33, 559–571. [DOI] [PubMed]
- Watson, F. L., Püttmann-Holgado, R., Thomas, F., Lamar, D. L., Hughes, M., Kondo, M., Rebel, V. I. & Schmucker, D. (2005). Science, 309, 1874–1878. [DOI] [PubMed]
- Wojtowicz, W. M., Flanagan, J. J., Millard, S. S., Zipursky, S. L. & Clemens, J. C. (2004). Cell, 118, 619–633. [DOI] [PMC free article] [PubMed]
- Wojtowicz, W. M., Wu, W., Andre, I., Qian, B., Baker, D. & Zipursky, S. L. (2007). Cell, 130, 1134–1145. [DOI] [PMC free article] [PubMed]
- Yamakawa, K., Huot, Y.-K., Haendelt, M. A., Hubert, R., Chen, X.-N., Lyons, G. E. & Korenberg, J. R. (1998). Hum. Mol. Genet. 7, 227–237. [DOI] [PubMed]


