Abstract
Studies of hemoglobin S haplotypes in African subpopulations have potential implications for patient care and our understanding of genetic factors that have shaped the prevalence of sickle cell disease (SCD). We evaluated HBB gene cluster haplotypes in SCD patients from Cameroon, and reviewed the literature for a global distribution. We reviewed medical records to obtain pertinent socio-demographic and clinical features for 610 Cameroonian SCD patients, including hemoglobin electrophoresis and full blood counts. RFLP-PCR was used to determine the HBB gene haplotype on 1082 chromosomes.
A systematic review of the current literature was undertaken to catalogue HBB haplotype frequencies in SCD populations around the world. Benin (74%; n=799) and Cameroon (19%; n=207) were the most prevalent haplotypes observed among Cameroonian patients. There was no significant association between HBB haplotypes and clinical life events, anthropometric measures, hematological parameters, or fetal hemoglobin (HbF) levels. The literature review of the global haplotype distributions was consistent with known historical migrations of the people of Africa. Previously reported data from Sudan showed a distinctly unusual pattern; all four classical haplotypes were reported, with an exceptionally high proportion of the Senegal, Cameroon, and atypical haplotypes.
We did not observe any significant associations between HBB haplotype and SCD disease course in this cohort. Taken together, the data from Cameroon and from the wider literature suggest that a careful reassessment of African HBB haplotypes may shed further light on the evolutionary dynamics of the sickle allele, which could suggest a single origin of the sickle mutation.
Introduction
Sickle cell disease (SCD) is caused by a point mutation (A>T) in the sixth codon of the β-globin gene on chromosome 11, resulting in the substitution of the amino acid valine for glutamic acid.
SCD affects the structure of erythrocytes by altering the normal biconcave shape to a crescent. During this process the hemoglobin S (HbS) mutation leads to polymerization and precipitation of hemoglobin during deoxygenation or dehydration, resulting in sickling, abnormal adhesion of leukocytes and platelets, inflammation, hypercoagulation, hemolysis, and hypoxia, in addition to microvascular obstruction and ultimately organ damage (Bartolucci et al., 2012). SCD is most prevalent among populations in regions of the world where malaria has been endemic and it is estimated that 305,800 births with SCD occur annually, nearly two-third of which take place in Africa (Piel et al., 2013).
It is well accepted that the sickle mutation exists in Africa on diverse genetic haplotype backgrounds (Labie al., 1985). Five typical haplotypes have been described across the β-globin gene cluster based upon the pattern of specific RFLPs across the region. Four haplotypes are associated with HbS in Africa (Benin, Bantu/Central African Republic (CAR), Senegal, and Cameroon) and the fifth is thought to have arisen in India and/or the Arabian Peninsula (Arab/Hindu) (Elion et al., 1992; Pagnier et al., 1984). It has been suggested that these haplotypes also have an effect on the severity of the disease (Asultan et al., 2012; Steinberg, 2009) and possibly the clinical response to hydroxyurea (Friedrisch et al., 2008)—currently the only available treatment for SCD. Furthermore, these haplotypes have a well-defined geographic distribution in Africa, making it possible to establish the origin of African-descendant populations in America (Pagnier et al., 1984; Serjeant, 1989), but also the origin of SCD among Europeans from Mediterranean regions. Studies of SCD haplotypes in African subpopulations therefore have potential implications for the clinical care of patients and for our understanding of the dynamic population genetic factors that shape the SCD phenotype.
Cameroon is a sub-Saharan African country with a population of about 20 million inhabitants and an annual growth rate of 3%, with a carrier frequency of SCD ranging from 8% to 34% (Wheatherall et al., 2001). Universal medical insurance coverage does not exist, and care of SCD patients is therefore dependent on financial support and care-giving by family members (Wonkam et al., 2013a). Sadly, poverty in Cameroon affects more than 50% of the rural population and up to 30% of the urban population (World Bank, 2010), and the financial burden of medical care for SCD is often not met (Wonkam et al., 2013a, 2014a).
SCD patients in Cameroon frequently suffer exceptionally severe complications such as stroke (Njamnshi et al., 2006) and executive neurocognitive compromise (Ruffieux et al., 2013). Consanguinity, ascertained by pedigree analysis, is observed in 5.7% of the population, most of which is found in the northern Sahelian regions of Cameroon where the Arabo-Muslim influence is stronger (Wonkam et al. 2013b). Because of its central location on the continent, Cameroonians cultural, linguistic, and genetic diversity mimics that of various ethno-linguistic groups found in Africa (Tishkoff et al., 2009). It is therefore anticipated that genetic studies in this population might reveal important insights to other African populations. The aim of this study was to explore the frequency and influence of HBB haplotypes amongst a group of Cameroonian SCD patients and to compare our findings to published data from Africa and worldwide.
Materials And Methods
Ethics statement
Ethical approval was granted by the National Ethical Committee Ministry of Public Health, Republic of Cameroon (No 033/CNE/DNM/07), and the University of Cape Town, Faculty of Health Sciences Human Research Ethics Committee (HREC RE: 132/2010).
Patients and assessment of clinical events
The study was conducted at the Yaoundé and Douala, the two largest cities in Southern Cameroon. Socio-demographic and clinical data were collected by means of a structured questionnaire for parents/guardians and patients. Patients' medical records were reviewed to delineate their clinical features over the past 3 years. Specifically, the occurrences of vaso-occlusive painful crisis (VOC), hospital outpatient visits, hospitalizations, overt strokes, blood transfusions, and administration of hydroxyurea were recorded. Anthropomorphic variables (height, weight, body mass index (BMI)) and systolic and diastolic blood pressures (SBP and DBP) were measured in the outpatient setting. Detailed descriptions of sampling methods used in the Cameroonian have been previously reported (Wonkam et al, 2014c).
Hematological phenotypes
Hemoglobin electrophoresis and routine complete blood count were conducted on each patient upon arrival at the hospital. Two methods of fetal hemoglobin (HbF) detection were employed; the alkali denaturation test (ADT) in 55.5% (n=344) of the patients, and subsequent high performance liquid chromatography (HPLC) in the remainder. HbF levels measurements done in patients <5 years old were excluded from the analysis, because HbF levels are not yet stable at this age (Wonkam et al, 2014c).
Genotypes
Cameroonian patients
DNA samples were extracted from peripheral blood, following manufacturer's instructions (Puregene blood kit® (Qiagen®, USA) at the molecular diagnostic laboratory, Gyneco-Obstetric and Paediatric Hospital, Yaoundé, Cameroon. Genotype analyses were performed in the Division of Human Genetics, Faculty of Health Sciences, University of Cape Town.
Molecular diagnostic testing for sickle cell anemia (SCA; HbSS)
SCA diagnosis was carried out using 200 ng of DNA. A thermocycler (Biorad®, USA) was used to amplify a 770 bp segment of the β-globin gene, followed by Dde I (Gibco-BRL®, USA) restriction analysis of the PCR product, as previously reported (Saiki et al., 1985).
Haplotyping of β-globin gene cluster
Five restriction fragment length polymorphism (RFLP) regions in the β-gene cluster were amplified using published methods designed to analyze the XmnI (5'Gγ), HindIII (Gγ), HindIII (Aγ), HincII (3?'Ψβ), and HinfI (5'β) restriction sites (Steinberg et al., 1998). RFLP sites and the fragments were visualized by agarose gel electrophoresis, and β-globin gene haplotypes were defined by examining the combination of the restriction sites. The βS haplotypes were constructed from the absence (−) and presence (+) of each of the five restriction enzyme sites in 541 individuals with HbSS. Of the samples studied, sickle (βS) haplotypes were identified through a specific algorithm (Supplementary Table S1; supplementary material is available online at www.liebertonline.com/omi)). Haplotypes were classified as ‘atypical’ if they did not match the five known typical βS haplotypes.
Literature review
A review of the current literature on SCD haplotypes was conducted from June 2013 to July 2014 using Pubmed (National Library of Medicine), Medline, and Google Scholar. Key words included a combination of the following: “Haplotype,” “Sickle Cell,” “Africa,” and specific country names were also used. Prior knowledge of research groups working on sickle cell disease in Africa and globally further facilitated the identification and selection of research articles. Only available full-length articles, in English, with the use of at least five (5) RFLP to differentiate the five main types of haplotype, were selected. In cases where multiple studies were reported in the same population, the most recent with the largest sample size was included. The main search was conducted by a PhD student in Human Genetics and the search was reviewed by a Medical/Human Geneticist with expertise in SCD.
Statistical analysis
Descriptive statistics were obtained for all quantitative data using SPSS (IBM, USA version 21.0). Normality was confirmed by the Shapiro-Wilk Test followed by the use of parametric (Chi-squared test and t-test) or nonparametric tests (Mann-Whitney U test for two samples or the Kruskal-Wallis one-way ANOVA for more than two samples). To correct for the skewed HbF distribution, the data were log10-transformed and normalized to obtain the quantitative trait used in the association analysis (after correcting for age, gender, and electrophoresis technique and history of transfusion).
Results
Socio-demographic of Cameroonian SCD patients
The description of the study sample is presented in Table 1. Among the 610 patients; 50.3% were female with mean age of 17.3±10 years (range: 5–54 years) and the majority of patients were children aged between 5–10 years (32.2%; n=196) and adolescents 11–20 years (50%; n=305). Patients lived mostly in the urban areas of Yaoundé and Douala (93%; n=567), both located in the southern part of Cameroon. The median age at diagnosis of SCD, was 3 years (range: 1 month—29 years). Forty two percent (n=158) of fathers and 23% (n=108) of mothers were formally employed; and in 75%, the monthly direct incomes were <300 USD.
Table 1.
Description of SCD Cameroonian Patients
| Variables | N | Mean±SD | Minimum–Maximum |
|---|---|---|---|
| M/F | 303/307 | ||
| Age (Yrs) | 610 | 17.3±10 | 5–58 |
| RBC (1012/L) | 610 | 2.8±0.7 | 1.2–6.5 |
| Hb (g/dL) | 610 | 7.8±1.6 | 3.8–16.5 |
| MCV (fL) | 610 | 84.4±9.9 | 59–117 |
| MCHC (g/dL) | 610 | 34.0±3.8 | 21.5–54.3 |
| WBC (109/L) | 610 | 13.7±5.6 | 2.9–49.8 |
| Monocytes (109/L) | 610 | 1.5±1 | 0.1–11.9 |
| Pl atelets (109/L) | 610 | 374.6±123 | 97–837 |
| HbA2 (%) (HPLC)* | 244 | 3.3±1.3 | 0–9.7 |
| HbF (%) (HPLC)* | 244 | 7.5±4.8 | 0–22.7 |
| N VOC /Yr | 572 | 3±3 | 0–40 |
| N hospital attendance (per Yr) | 608 | 2.2±3.2 | 0–60 |
| N hospital admission (per Yr) | 606 | 2.3±3 | 0–30 |
| N patients with at least one overt stroke | 25/608 | 4.1% |
Hb electrophoresis was also obtained from 344 patients (55.5%) using alkali denaturation test (ADT), with a mean of 11.4±9.4 for HbF and 4.1±2.1 for HbA2 levels.
Haplotypes in the beta-globin gene cluster, clinical events, and HbF in Cameroonian patients
Following the full blood count and HbF electrophoresis and RFLP-PCR, the vast majority of patients were determined to have SCA (HbSS) (97.4%; n=594). In addition, 61 randomly selected patients were Sanger sequenced, and SCA diagnosis was confirmed in all of them. There was no evidence of the deletional form of HbS β-thalassemia.
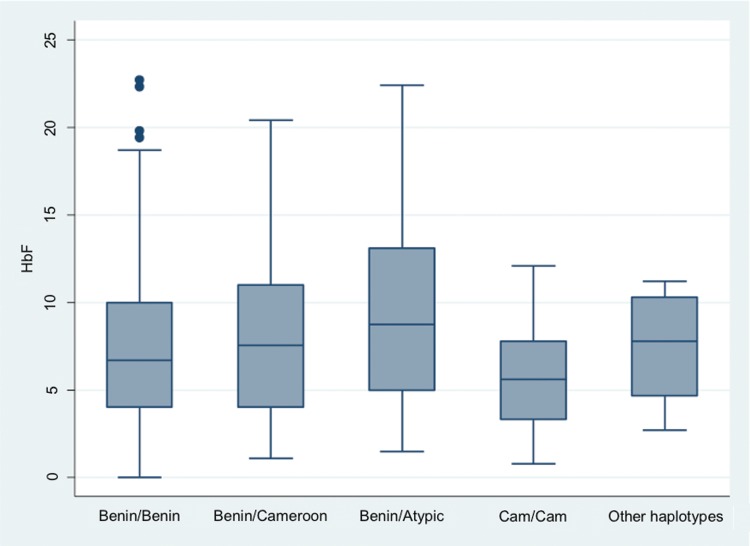
Based on analysis of 1082 chromosomes, the allele frequencies of various haplotypes showed that Benin (74%; n=799) and Cameroon (19%; n=207) were the most prevalent forms. Bantu/CAR (n=5), Senegal (n=3), and Indian-Arab (n=2) haplotype were very rare. Atypical haplotypes accounted for 6% (n=66). Homozygous Benin/Benin haplotypes represented 57% (n=307), Benin/Cameroon 25% (n=137), Benin/atypical 8% (n=45), and Cameroon/Cameroon 5% (n=26) of diploid haplotype frequencies. There were no significant associations between major haplotype combinations and hematological indices (Table 2), clinical events (Table 3), or HbF levels (Fig. 1).
Table 2.
Median Values of Hematological Indices Among Cameroonian SCD Patients Conditioned on HBB Haplotypes
| Hematological variables | Benin/Benin (n=307) | Benin/Cam (n=137) | Benin/Atypical (n=45) | Cameroon/Cameroon (n=26) | P values |
|---|---|---|---|---|---|
| RBC (1012/L) | 2.7 | 2.6 | 2.8 | 2.6 | 0.6 |
| Hb (g/dL) | 7.7 | 7.5 | 7.8 | 7.6 | 0.5 |
| MCV (fL) | 85 | 84 | 86 | 83.5 | 0.9 |
| MCHC (g/dL) | 33.4 | 34.1 | 32.6 | 34.3 | 0.5 |
| Platelets (109/L) | 395 | 374 | 350 | 382 | 0.4 |
| WBC (109/L) | 12.6 | 13.6 | 13.1 | 14.3 | 0.5 |
| Lymphocytes (109/L) | 5.2 | 5.6 | 5.4 | 6.5 | 0.1 |
| Monocytes (109/L) | 1.3 | 1.3 | 1.3 | 1.3 | 1 |
Table 3.
Clinical Event Among Cameroonian SCD Patients Conditioned on HBB Haplotypes
| Benin/Benin | Benin/Cameroon | Benin/Atypical | Cameroon/Cameroon | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Median (25th-75th centiles) | N | Median (25th-75th centiles) | N | Median (25th-75th centiles) | N | Median (25th-75th centiles) | P values | |
| BMI (kg/m2) | 251 | 18 (14.9–21) | 103 | 17.2 (15–20.1) | 39 | 17 (14.4–19.7) | 23 | 17.9 (15.1–18.5) | 0.12 |
| SBP mmHg | 259 | 108 (100–115) | 107 | 107 (98–115) | 40 | 109 (99–113.5) | 21 | 108 (99–115) | |
| DBP mmHg | 259 | 58 (54–65) | 107 | 58 (52–64) | 40 | 59 (53.5–64.5) | 21 | 56 (53–60) | 0.56 |
| No. of VOC/Year | |||||||||
| Stroke | |||||||||
| Yes | 13 | – | 6 | – | – | – | – | 0.8 | |
| No | 294 | – | 131 | – | 45 | – | 25 | – | |
| No. of consultation/Year | 301 | 1 (0–4) | 135 | 2 (0–4) | 45 | 1 (0–3) | 26 | 1 (0–4) | 0.66 |
| No. of hospitalization/Year | 301 | 1 (0–2) | 135 | 1 (0–2) | 45 | 1 (0–2) | 26 | 1 (1–2) | 0.54 |
FIG. 1.
Distribution of fetal hemoglobin levels conditioned on HBB haplotypes. Boxes have lines at the lower quartile, median, and upper quartile. The plot whiskers extend up and down from the median a distance 1.5 times the interquartile range of the boxes (truncated at zero where necessary). Outliers are the points outside the whiskers indicated as circles. There were not significant differences across various combination of haplotypes (p=0.54) and between them.
Global distribution of haplotypes
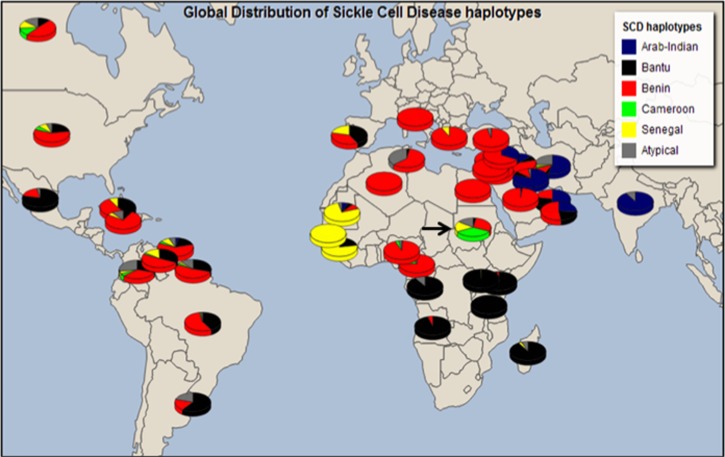
As demonstrated in Figure 2 and Table 4, the Benin haplotype was the most prevalent globally with its highest frequency in West Africa, the Mediterranean region, America, and Europe. The Bantu/CAR haplotype was most prevalent in central, southern, and East Africa, and in relatively high proportion in Central and South America and parts of Europe, as compared to North America. The Senegal haplotype was restricted to the far west regions of Africa, with the exception of Sudan and had a relatively low frequency in America; the Indian-Arab haplotype was almost completely restricted to part of India and the Middle East.
FIG. 2.
Global distribution of haplotypes among various world SCD populations. Previously reported data allow drawing a global haplotype distribution that was consistent with known historical migrations of the people of Africa. Among Indigenous African populations, published data from Sudan (arrow) showed a distinctly unusual pattern; all four classical haplotypes were reported, with an exceptionally high proportion of the Senegal and Cameroon haplotypes.
Table 4.
Global β-S Haplotype Distribution from Previously Reported Data
| β-S Haplotypes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Continents | Countries | Arab-Indian N* (%) | Bantu/Car N (%) | Benin N (%) | Cameroon N (%) | Senegal N (%) | Atypical N (%) | References |
| Africa | Algeria | 0 (0.0) | 0 (0.0) | 20 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | Pagnier et al., 1984 |
| Angola | 0 (0.0) | 42 (95.5) | 2 (4.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | Lavinha et al,. 1992 | |
| Cameroon | 3 (0.3) | 5 (0.5) | 799 (73.8) | 207 (19.1) | 2 (0.2) | 66 (6.1) | The present data | |
| Congo | 0 (0.0) | 211 (91.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 21 (9.1) | Mouele et al., 1999 | |
| Egypt | 0 (0.0) | 0 (0.0) | 28 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | El-Hazemi et al., 1999 | |
| Guinea | 0 (0.0) | 9 (22.5) | 0 (0.0) | 31 (77.5) | 0 (0.0) | 0 (0.0) | Sow et al., 1995 | |
| Kenya | 0 (0.0) | 109 (98.2) | 2 (1.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | Ojwang et al., 1987 | |
| Madagascar | 0 (0.0) | 32 (91.4) | 0 (0.0) | 0 (0.0) | 1 (2.9) | 2 (5.7) | Hewitt et al., 1996 | |
| Mauritania | 5 (5.6) | 4 (4.4) | 8 (8.9) | 0 (0.0) | 70 (77.8) | 3 (3.3) | Veten et al., 2012 | |
| Nigeria | 0 (0.0) | 6 (1.0) | 624 (93.2) | 23 (3.4) | 0 (0.0) | 16 (2.4) | Adekile et al., 1992 | |
| Senegal | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 90 (100.0) | 0 (0.0) | Currat et al,. 2002 | |
| Sudan | 0 (0.0) | 4 (2.8) | 42 (29.4) | 50 (35.0) | 26 (18.2) | 21 (14.6) | Elderdery et al., 2012 | |
| Tanzania | 0 (0.0) | 41 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | Oner et al., 1992 | |
| Tunisia | 0 (.0) | 9 (2.7) | 201 (60.5) | 0 (0.0) | 0 (0.0) | 122 (36.7) | Imen et al,. 2011 | |
| Uganda | 0 (0.0) | 207 (99.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | Ndugwa et al., 2012 | |
| Asia | India | 128 (91.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 12 (8.6) | Mukherjee et al., 2004 |
| Bahrain | 33 (89.2) | 2 (5.4) | 1 (2.7) | 0 (0.0) | 0 (0.0) | 1 (2.7) | Al-Arrayed,1995 | |
| Iran | 87 (53.7) | 5 (3.1) | 19 (11.7) | 4 (2.5) | 6 (3.7) | 41 (25.3) | Rahimi et al., 2003 | |
| Iraq | 16 (12.5) | 10 (7.8) | 89 (69.5) | 0 (0.0) | 0 (0.0) | 13 (10.2) | Al-Allawi et al., 2012 | |
| Jordan | 4 (20.0) | 0 (0.0) | 16 (80.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | El-Hazemi et al., 1999 | |
| Kuwait | 101 (80.8) | 7 (5.6) | 14 (11.2) | 0 (0.0) | 0 (0.0) | 3 (2.4) | Adekile, 2001 | |
| Lebanon | 10 (10.0) | 15 (15.0) | 73 (73.0) | 0 (0.0) | 0 (0.0) | 2 (2.0) | Inati et al., 2003 | |
| Oman | 31 (26.5) | 25 (21.4) | 61 (52.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | Daar et al,. 2000 | |
| Palestine (wb)# | 0 (0.0) | 6 (5.1) | 104 (88.1) | 0 (0.0) | 0 (0.0) | 8 (6.8) | Samarah et al., 2009 | |
| Saudi-arabia (se)^ | 2 (1.6) | 0 (0.0) | 122 (98.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | El-Hazemi et al., 1999 | |
| Syria | 6 (33.3) | 0 (0.0) | 12 (66.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | El-Hazemi et al., 1999 | |
| United Arab Emirates | 49 (52.0) | 24 (26.0) | 21 (22.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | El-Kalla & Baysal, 1998 | |
| North America | Canada | 0 (0.0) | 7 (11.5) | 30 (49.2) | 8 (13.1) | 8 (13.1) | 8 (13.1) | Oner et al., 1992 |
| Cuba | 0 (0.0) | 81 (40.9) | 101 (51.0) | 0 (0.0) | 16 (8.1) | 0 (0.0) | Muniz et al., 1995 | |
| Jamaica | 0 (0.0) | 37 (8.3) | 339(76.0) | 0 (0.0) | 23 (5.2) | 47 (10.5) | Ndugwa et al., 2012 | |
| Mexico | 0 (0.0) | 26 (78.8) | 6 (18.2) | 0 (0.0) | 0 (0.0) | 1 (3.0) | Magana et al., 2002 | |
| Trinidad | 9 (3.2) | 49 (17.3) | 175 (61.8) | 10 (3.5) | 24 (8.5) | 16 (5.7) | Jones-Lecointe et al., 2008 | |
| USA$ | 12 (0.8) | 129 (21.6) | 503 (58.9) | 38 (6.1) | 76 (8.0) | 48 (4.6) | Crawford et al., 2002 | |
| South America | Uruguay | 0 (0.0) | 6 (60.0) | 2 (20.0) | 0 (0.0) | 0 (0.0) | 2 (20.0) | Luz et al., 2006 |
| Brazil | 1 (0.4) | 104 (41.6) | 138 (55.2) | 3 (1.2) | 1 (0.4) | 3 (1.2) | Adorno et al., 2008 | |
| Colombia | 1 (0.4) | 68 (29.7) | 76 (33.2) | 10 (4.4) | 10 (4.4) | 64 (27.9) | Fong et al., 2013 | |
| Suriname | 0 (0.0) | 23 (29.9) | 41 (53.2) | 2 (2.6) | 2 (2.6) | 9 (11.7) | Oner et al, 1992 | |
| Venezuela | 0 (0.0) | 57 (32.2) | 90 (50.8) | 4 (2.3) | 25 (14.1) | 1 (0.6) | Arends et al., 2000 | |
| Europe | Portugal | 0 (0.0) | 14 (42.4) | 12 (36.4) | 0 (0.0) | 7 (21.2) | 0 (0.0) | Lavinha et al., 1992 |
| Italy | 0 (0.0) | 0 (0.0) | 64 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | Schiliro et al., 1992 | |
| Turkey | 1 (0.5) | 0 (0.0) | 206 (96.3) | 0 (0.0) | 0 (0.0) | 7 (3.3) | Oner et al., 1992 | |
| Greece | 0 (0.0) | 0 (0.0) | 13 (92.9) | 0 (0.0) | 1 (7.1) | 0 (0.0) | Oner et al., 1992 | |
N, number of chromosomes; #WB, West Bank; ^SE, South Eastern; $USA, combined African-American and Hispanic data.
Exceptional HBB haplotype distribution in Sudan?
When data within Africa was compared, the literature review revealed four major features in Sudanese SCD patients: 1) all four classical SCD haplotypes were reported in significant proportions; 2) Cameroon haplotype was reported in a much higher proportion in Sudan than in Cameroon; 3) surprisingly, there was a higher proportion of Senegal haplotypes in Sudan when compared to the rest of East Central and parts of West Africa, and lastly 4) there was a relatively high proportion of atypical haplotypes (Elderdery et al., 2012) (Table 4, Fig. 2).
Discussion
To the best of our knowledge, the present study represents the largest study of HBB haplotypes in a single African country, and is the most comprehensive review of the global distribution of HBB SCD haplotypes thus far. The data presented confirm the origin and flow of people from Africa through the slave trade and/or migration, and emphasize differences in settlement on the American coasts dependent on whether slaves were brought from the Gulf of Guinea or from Angola (Fong et al., 2013). Similarly, trade and migration routes to the Mediterranean areas and the Middle East from West Africa explain the high prevalence of Benin haplotypes and general pattern of haplotype distribution in those parts of the world (Fig. 2).
The present study also confirms that the majority of the chromosomes with the β(S) globin gene are consistent with one of the five common RFLP haplotypes, designated as Benin, Bantu/CAR, Senegal, Cameroon, and Arab-Indian haplotypes. In most settings, 5%–10% of the chromosomes are less commonly observed haplotypes, usually referred to as “atypical” haplotypes (Table 4). However, on the African continent and in two independent studies, atypical haplotypes appear in relatively high frequency in Sudan, in concert with all the other types of African haplotypes, particularly a relatively high proportion of Senegal and Cameroon haplotypes (Elderdery et al., 2012; Mohammed et al., 2006). The relatively high frequency of the Cameroon haplotype in Sudan versus in Cameroon (Elderdery et al., 2012) is not completely anomalous, but rather the reflection that the geographical nomenclature of haplotypes (named according to the first region of observation) does not necessarily reflect their place of origin. The refinement of the molecular structure (i.e., haplotype) in HBB and migration patterns is still urgently needed, specifically in Africa.
Two decades ago, the observation of a specific RFLP pattern of the Cameroon haplotype strongly argued for yet another independent origin of the sickle cell mutation in Africa (Lapoumeroulie et al., 1992). The Cameroon haplotype was then said to be restricted to a single ethnic group, the Eton of Central Cameroon (Lapoumeroulie et al., 1992), which is contrary to current data from Sudan (Elderdery et al., 2012; Mohammed et al., 2006). More recently, a specific atypical haplotype in Tunisia was considered by authors as specific to the Tunisian chromosome β(S) (Imen et al., 2011). In the case of the Cameroon and Tunisia, these specific haplotypes were likely generated by a variety of genetic mechanisms including (a) isolated nucleotide changes in one of the polymorphic restriction sites, (b) simple and double crossovers between two typical β(S) haplotypes, or much more frequently between a typical β(S) haplotype and a different β(A)-associated haplotype that was present in the population, and (c) gene conversions (Zago et al., 2000)—all of which have been observed at the beta-globin locus (Borg et al., 2009; Holloway et al., 2006; Liu et al., 2009; Neumann et al., 2010; Patrinos et al., 2005; Sankaran et al., 2011).
Thus, a relatively high frequency of what was originally an atypical β(S) haplotype might subsequently be labeled a “new” haplotype supposedly (albeit unlikely) bearing a new independent β(S) mutation, as was postulated by the authors of the original globin haplotype report (Wainscoat et al., 1983). To date, the existence of at least five different geographic centers of origin has been postulated on the basis of the predominance of major haplotypes associated with the β(S) mutation. The multiple and independent origins of the β(S) mutation (Chakravarti et al., 1984; Nagel and Fleming, 1992) also fails to account for the absence of β(S) mutation from malarial regions in Southeast Asia and Oceania (Flint et al., 1993), although many other hemoglobin gene mutations also confer malaria protection, including α- and β-thalassemia traits and other structural variants of hemoglobin that are common in Southeast Asia.
In addition, the hypothesis of multiple independent origins raises the question of how several identical mutations could have occurred in a short period of time in Africa after the appearance of malaria, despite the low mutation rate of nuclear DNA (Currat et al., 2002). It is also possible that gene conversion has played a role in moving the β(S) mutation to different haplotype backgrounds in sub-Saharan Africa. Resolution of RFLP haplotypes with recently described long-range sickle haplotypes (Ghansah et al., 2012; Hanchard et al., 2007), together with next-generation sequencing, should provide definitive answers to this long-standing question.
It is then perhaps provocative and ambitious, but not unreasonable, to hypothesize that, there could be a single origin of the sickle cell mutation, in the region of East Africa/Sudan, with additional haplotypes generated by diverse structural mechanisms, and the spread to the rest of the continent through traditional migratory patterns. Recently, both phylogenetic and network analysis indicate that east Africans possess more ancestral mitochondrial lineages in comparison to various continental populations placing them at the foot of the human evolutionary tree. Interestingly, the two most ancestral mitochondrial sequences in the NJ tree figure refer to Nubian individuals in Sudan (Elhassan et al., 2014). Moreover, a compound associated with a lithic Middle Stone Age industry was discovered in Dhofar Oman and regarded as evidence of human migration out of Africa through an Arabian route (Rose et al., 2011).
Therefore, the shared rs7482144 SNP in both the Senegal and Indian-Arab haplotypes invites the question of whether the Indian-Arab haplotype evolved from an east African chromosome with Senegal haplotype. In fact, a specific study of the Senegal haplotype concluded that migration from the east of Senegal was implicated in the spread of this mutation across west African populations and the exact geographic origin of the Senegal β(S) mutation was not located (Currat et al., 2002). It could well be even further east of Senegal, perhaps as far east as Sudan/East Africa region. It was suggested more than 2 decades ago, that although β(S) is attributed to recent independent mutations occurring approximately 2000 years ago (Currat et al., 2002), the separation and geographical distribution of African populations allows for the possibility of an ancient origin of β(S) mutation. The authors even suggested that β(S) could have protected selected populations from malaria in rain forest refuges during the most recent Ice Age (Stine et al., 2011).
The data reviewed here provides evidence that the detailed geographic distribution of known sickle haplotypes is still not well established, as many African countries do not have reported data. Were this available, it is conceivable that a gradient of haplotype frequencies from East Africa to other parts of the continent could have emerged differently. Further genetic studies in well-defined populations from across Africa are urgently needed, to allow determination of the exact extent of the spread of various HBB haplotypes, as this would help to identify the mutation's geographical origin precisely, and to explore the recent increase in frequency of the β(S) mutation, irrespective of its haplotypic background. The data reported here emphasize the need to re-explore the origin of sickle cell mutation as well as past and recent evolutionary dynamics of African human populations, as this could yield important information about the evolutionary history of SCD, which of course hold significant clinical relevance (Adorno et al., 2008).
HbF is the most powerful biological modifier of SCD phenotype and influenced by genomic variations. In a recent study, we confirmed in this group of Cameroonian SCD patients, robust genetic associations between HbF levels and BCL11A and HBS1L-MYB intergenic locus (Wonkam et al., 2014c), as previously reported in African Americans and Afro- Brazilians (Lettre et al., 2008) and Tanzanians (Makani et al., 2011). However, due to the virtual absence of the Senegal and Indian-Arab haplotypes, which contain the XmnI variant (rs7482144), we lacked power to replicate the association of rs7482144 in HBG2 with HbF levels in Cameroonian; this SNP explains 2.2% of the variation in HbF levels in African American SCD patients (Labie et al., 1985; Lettre et al., 2008). Indeed, there was no significant difference in HbF levels and various haplotypes combinations among this group of Cameroonian SCD patients (Fig. 1). Contrary to our findings, a recent report argues that the S haplotype itself (beyond HbF regulation) correlates with disease severity (Bean et al., 2013).
The Cameroon haplotype has previously been associated with poor clinical outcome and increased stoke episodes in SCD (Adorno et al., 2008; Steinberg et al., 1998), whereas the Benin haplotype is thought to confer a relatively favorable clinical outcome (Steinberg, 2009). In a previous report, we anticipated that the relatively high prevalence of stroke in SCD patients in Cameroon could be attributed to the high proportion of Cameroonian haplotype (Njamnshi et al., 2006). However, the present data do not confirm this hypothesis, since the most prevalent haplotype in Cameroon is Benin. The high prevalence of Benin haplotypes rendered the study underpowered to reveal a significant difference in phenotypic outcome with specific haplotype combinations. An alternative explanation could be the influence of environmental factors, specifically the limited access to the already inadequate health care and poverty, illustrated here by the late age at diagnosis, the low rate of formal employment, as well as modest direct monthly direct incomes among parents. In other studies, socioeconomic factors and poor healthcare infrastructure have been shown to be a major factor in the high psychosocial burden of SCD among Cameroonian adult SCD patients (Wonkam et al., 2014a,d) and parents of children with SCD (Wonkam et al., 2013).
Limitations and perspectives
In Cameroon, the data mostly originated from patients living in the southern part of the countries and are thus not fully representative of the general SCD population in Cameroon. Specifically, data from the northern Sahelian part will be needed to complete the picture and possibly provide additional clues of recent population migration from Sudan that brought the ‘Cameroon’ haplotype to Cameroon. Another limitation of this study was the self-report nature of some clinical variables such as VOC episodes, which can lack precision. In addition, pain tolerance and financial factors could have been limiting factors for hospital attendance (Wonkam et al., 2014c).
Despite the above limitations, this study represents an important step toward the understanding of the molecular structure of SCD in Cameroon, in sub-Saharan African patients and globally. This study also has a capacity-building dimension as it was fully performed from design, funding, molecular analysis, and reporting on the African continent and could in turn create major collaborative research opportunities at the regional and international levels.
Conclusion
This study has 1) confirmed the high frequency of Benin haplotype and lack of significant association between β(S) haplotypes and common SCD clinical outcomes in Cameroonian SCD patients; 2) highlighted previous data demonstrating the almost equivalent representation of the four major HBB haplotypes in Sudan with a much higher frequency of Cameroon haplotype; 3) provided a comprehensive review of published data illustrating the distribution of β(S) haplotypes in world populations, tracking their migration pattern within and outside of Africa. This preliminary report proposes extending the studies on HBB haplotype distribution in Africa and advances the opportunity to re-address the question of the origin(s) of the Sickle Cell mutation.
Supplementary Material
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Adekile AD, and Haider MZ. (1996). Morbidity, βS haplotype and α-globin gene patterns among sickle cell anemia patients in Kuwait. Acta Haematol 96, 150–154 [DOI] [PubMed] [Google Scholar]
- Adorno EV, Zanette Â, Lyra I, et al. (2008). Clinical and molecular characteristics of sickle cell anemia in the northeast of Brazil. Genet Mol Biol 31, 621–625 [Google Scholar]
- Al-Allawi NA, Jalal SD, Nerwey FF, et al. (2012). Sickle cell disease in the Kurdish population of northern Iraq. Hemoglobin 36, 333–342 [DOI] [PubMed] [Google Scholar]
- Alsultan A, Aleem A, Ghabbour H, et al. (2012). Sickle cell disease subphenotypes in patients from Southwestern Province of Saudi Arabia. J Pediatr Hematol Oncol 34, 79–84 [DOI] [PubMed] [Google Scholar]
- Arends A, Alvarez M, Velázquez D, et al. (2000). Determination of β-globin gene cluster haplotypes and prevalence of α-thalassemia in sickle cell anemia patients in Venezuela. Am J Hematol 64, 87–90 [DOI] [PubMed] [Google Scholar]
- Arrayed SSA, and Haltes N. (1995). Features of sickle-cell disease in Bahrain. East Mediterr Health J 1, 112–118 [Google Scholar]
- Bartolucci P, and Galactéros F. (2012) Clinical management of adult sickle-cell disease. Curr Opin Hematol 19,149–155 [DOI] [PubMed] [Google Scholar]
- Bean CJ, Boulet SL, Yang G, et al. (2013). Acute chest syndrome is associated with single nucleotide polymorphism-defined beta globin cluster haplotype in children with sickle cell anaemia. Br J Haematol 163, 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg J, Georgitsi M, Aleporou-Marinou V, Kollia P, and Patrinos GP. (2009). Genetic recombination as a major cause of mutagenesis in the human globin gene clusters. Clin Biochem 42, 1839–1850 [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Buetow KH, Antonarakis SE, et al. (1984). Nonuniform recombination within the human beta-globin gene cluster. Am J Hum Genet 36, 1239. [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Caggana M, Harris KB, et al. (2002). Characterization of the β-globin haplotypes using blood spots from a population-based cohort of newborns with homozygous HbS. Genet Med 4, 328–335 [DOI] [PubMed] [Google Scholar]
- Currat M, Trabuchet G, Rees D, et al. (2002). Molecular analysis of the β-globin gene cluster in the Niokholo Mandenka population reveals a recent origin of the β-S Senegal mutation. Am J Hum Genet 70, 207–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar S, Hussain HM, Gravell D, Nagel RL, and Krishnamoorthy R. (2000). Genetic epidemiology of HbS in Oman: Multicentric origin for the βS gene. Am J Hematol 64, 39–46 [DOI] [PubMed] [Google Scholar]
- Elderdery AY, Mills J, Mohamed BA, et al. (2012). Molecular analysis of the B-globin gene cluster haplotypes in a Sudanese population with sickle cell anemia. Int J Lab Hematol 34, 262–266 [DOI] [PubMed] [Google Scholar]
- Elhassan N, Gebremeskel EI, Elnour MA, et al. (2014). The episode of genetic drift defining the migration of humans out of Africa is derived from a large East African population size. PloS One 9, e97674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hazmi MAF, Warsy AS, Bashir N, et al. (1999). Haplotypes of the ß-globin gene as prognostic factors in sickle-cell disease. East Mediterr Health J 5,1154–1158 [PubMed] [Google Scholar]
- Elion J, Berg PE, Lapoumeroulie C, et al. (1992). DNA sequence variation in a negative control region 5' to the beta-globin gene correlates with the phenotypic expression of the beta s mutation. Blood 79, 787–792 [PubMed] [Google Scholar]
- El-Kalla S, and Baysal E. (1998). Genotype-phenotype correlation of sickle cell disease in the United Arab Emirates. J Pediatr Hematol Oncol 15, 237–242 [DOI] [PubMed] [Google Scholar]
- Flint J, Harding RM, Boyce AJ, and Clegg JB. (1993). The population genetics of the haemoglobinopathies. Baillieres Best Pract Res Clin Haematol 6, 215–262 [DOI] [PubMed] [Google Scholar]
- Fong C, Lizarralde-Iragorri MA, Rojas-Gallardo D, and Barreto G. (2013). Frequency and origin of haplotypes associated with the beta-globin gene cluster in individuals with trait and sickle cell anemia in the Atlantic and Pacific coastal regions of Colombia. Genet Mol Biol 36, 494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrisch JR, Prá D, Maluf SW, et al. (2008). DNA damage in blood leukocytes of individuals with sickle cell disease treated with hydroxyurea. Mutat Res Genet Toxicol Environ Mutagen 649, 213–220 [DOI] [PubMed] [Google Scholar]
- Ghansah A, Rockett KA, Clark TG, et al. (2012). Haplotype analyses of haemoglobin C and haemoglobin S and the dynamics of the evolutionary response to malaria in Kassena-Nankana District of Ghana. PloS one 7, e34565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchard N, Elzein A, Trafford C, et al. (2007). Classical sickle beta-globin haplotypes exhibit a high degree of long range haplotype similarity in African and Afro-Caribbean populations. BMC Genet 8, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt R, Krause A, Goldman A, Campbell G, and Jenkins T. (1996). Beta-globin haplotype analysis suggests that a major source of Malagasy ancestry is derived from Bantu-speaking Negroids. Am J Hum Genet 58, 1303. [PMC free article] [PubMed] [Google Scholar]
- Holloway K, Lawson VE, and Jeffreys AJ. (2006). Allelic recombination and de novo deletions in sperm in the human β-globin gene region. Hum Mol Genet 15, 1099–1111 [DOI] [PubMed] [Google Scholar]
- Imen M, Ikbel BMM, Leila C, et al. (2011). Restriction mapping of βS locus among Tunisian sickle cell patients. Am J Hum Biol 23, 815–819 [DOI] [PubMed] [Google Scholar]
- Inati A, Taher A, Bou Alawi WW, et al. (2003). β-Globin gene cluster haplotypes and HbF levels are not the only modulators of sickle cell disease in Lebanon. Eur J Haematol 70, 79–83 [DOI] [PubMed] [Google Scholar]
- Jones-Lecointe A, Smith E, Romana M, et al. (2008). β-Globin gene cluster haplotypes and α-thalassemia in sickle cell disease patients from Trinidad. Am J Hum Biol 20, 342–344 [DOI] [PubMed] [Google Scholar]
- Labie D, Pagnier J, Lapoumeroulie C, et al. (1985). Common haplotype dependency of high G gamma-globin gene expression and high HbF levels in beta-thalassemia and sickle cell anemia patients. Proc Natl Acad Sci USA 82, 2111–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouméroulie C, Dunda O, Ducrocq R, et al. (1992). A novel sickle cell mutation of yet another origin in Africa: the Cameroon type. Hum Genet 89, 333–337 [DOI] [PubMed] [Google Scholar]
- Lavinha J, Goncalves J, Faustino P, et al. (1992). Importation route of the sickle cell trait into Portugal: Contribution of molecular epidemiology. Hum Biol 64, 891–901 [PubMed] [Google Scholar]
- Lettre G, Sankaran VG, Bezerra MA, et al. (2008). DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci USA 105, 11869–11874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Muralidhar S, Singh M, et al. (2009). High-density SNP genotyping to define β-globin locus haplotypes. Blood Cells Mol Dis 42, 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz JAD, Sans M, Kimura EM, et al. (2006). Alpha-thalassemia, HbS, and beta-globin gene cluster haplotypes in two Afro-Uruguayan sub-populations from northern and southern Uruguay. Genet Mol Biol 29, 595–600 [Google Scholar]
- Magaña MT, Ongay Z, Tagle J, et al. (2002). Analysis of β-S and β-A genes in a Mexican population with African roots. Blood Cells Mol Dis 28, 121–126 [DOI] [PubMed] [Google Scholar]
- Makani J, Menzel S, Nkya S, et al. (2011). Genetics of fetal hemoglobin in Tanzanian and British patients with sickle cell anemia. Blood 117, 1390–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed AO, Attalla B, Bashir FM, et al. (2006). Relationship of the sickle cell gene to the ethnic and geographic groups populating the Sudan. Community Genet 9, 113–120 [DOI] [PubMed] [Google Scholar]
- Mouélé R, Pambou O, Feingold J, and Galactéros F. (1999). α-Thalassemia in Bantu population from Congo-Brazzaville: Its interaction with sickle cell anemia. Hum Hered 50, 118–125 [DOI] [PubMed] [Google Scholar]
- Mpalampa L, Ndugwa CM, Ddungu H, and Idro R. (2012). Foetal haemoglobin and disease severity in sickle cell anaemia patients in Kampala, Uganda. BMC Hematol 12, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee MB, Surve RR, Gangakhedkar RR, et al. (2004). β-Globin gene cluster haplotypes linked to the βS gene in western India. Hemoglobin 28, 157–161 [DOI] [PubMed] [Google Scholar]
- Muniz A, Corral L, Alaez C, et al. (1995). Sickle cell anemia and β-gene cluster haplotypes in Cuba. Am J Hematol 49, 163–164 [DOI] [PubMed] [Google Scholar]
- Nagel RL, and Fleming AF. (1992). Genetic epidemiology of the beta S gene. Baillieres Best Pract Res Clin Haematol 5, 331–365 [DOI] [PubMed] [Google Scholar]
- Neumann R, Lawson VE, and Jeffreys AJ. (2010). Dynamics and processes of copy number instability in human γ-globin genes. Proc Natl Acad Sci USA 107, 8304–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njamnshi AK, Mbong EN, Wonkam A, et al. (2006) The epidemiology of stroke in sickle cell patients in Yaounde, Cameroon. J Neurol Sci 250, 79–84 [DOI] [PubMed] [Google Scholar]
- Ojwang PJ, Ogada T, Beris P, et al. (1987). Haplotypes and α globin gene analyses in sickle cell anaemia patients from Kenya. Br J Haematol 65, 211–215 [DOI] [PubMed] [Google Scholar]
- Öner C, Dimovski AJ, Olivieri NF, et al. (1992). βS haplotypes in various world populations. Hum Genet 89, 99–104 [DOI] [PubMed] [Google Scholar]
- Pagnier J, Mears JG, Dunda-Belkhodja O, et al. (1984). Evidence for the multicentric origin of the sickle cell hemoglobin gene in Africa. Proc Natl Acad Sci USA 81, 1771–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinos GP, Kollia P, and Papadakis MN. (2005). Molecular diagnosis of inherited disorders: Lessons from hemoglobinopathies. Hum Mutat 26, 399–412 [DOI] [PubMed] [Google Scholar]
- Piel FB, Patil AP, Howes RE, et al. (2013). Global epidemiology of sickle haemoglobin in neonates: A contemporary geostatistical model-based map and population estimates. Lancet 381, 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi Z, Karimi M, Haghshenass M, and Merat A. (2003). β-Globin gene cluster haplotypes in sickle cell patients from southwest Iran. Am J Hematol 74, 156–160 [DOI] [PubMed] [Google Scholar]
- Rose JI, Usik VI, Marks AE, et al. (2011). The Nubian complex of Dhofar, Oman: An African Middle Stone Age industry in Southern Arabia. PLoS One 6, e28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffieux N, Njamnshi AK, Wonkam A, et al. (2013). Association between biological markers of sickle cell disease and cognitive functioning amongst Cameroonian children. Child Neuropsychol 19, 143–160 [DOI] [PubMed] [Google Scholar]
- Saiki RK, Scharf S, Faloona F, et al. (1985). Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230, 1350–1354 [DOI] [PubMed] [Google Scholar]
- Samarah F, Ayesh S, Athanasiou M, Christakis J, and Vavatsi N. (2009). βS-Globin gene cluster haplotypes in the West Bank of Palestine. Hemoglobin 33, 143–149 [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Byron R, et al. (2011). A functional element necessary for fetal hemoglobin silencing. N Engl J Med 365, 807–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiliro G, Samperi P, Consalvo C, et al. (1992). Clinical, hematological, and molecular features in Sicilians with sickle cell disease. Hemoglobin 16, 469–480 [DOI] [PubMed] [Google Scholar]
- Serjeant GR. (1989). Geography and the clinical picture of sickle cell disease. An overview. Ann N Y Acad Sci 565, 109–119 [DOI] [PubMed] [Google Scholar]
- Sow A, Peterson E, Josifovska O, et al. (1995). Linkage disequilibrium of the Senegal haplotype with the βs gene in the Republic of Guinea. Am J Hematol 50, 301–303 [DOI] [PubMed] [Google Scholar]
- Steinberg MH. (2009). Genetic etiologies for phenotypic diversity in sickle cell anemia. Sci World J 9, 46–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg MH, Lu ZH, Nagel RL, et al. (1998). Hematological effects of atypical and Cameroon beta-globin gene haplotypes in adult sickle cell anemia. Am J Hematol 59, 121–126 [DOI] [PubMed] [Google Scholar]
- Stine OC, Dover GJ, Zhu D, and Smith KD. (1992). The evolution of two West African populations. J Mol Evol 34, 336–344 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, et al. (2009). The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veten FM, Abdelhamid IO, Meiloud GM, et al. (2012). Hb S [β6 (A3) Glu→Val, GAG> GTG] and β-globin gene cluster haplotype distribution in Mauritania. Hemoglobin 36, 311–315 [DOI] [PubMed] [Google Scholar]
- Wainscoat JS, Bell JI, Thein SL, et al. (1983). Multiple origins of the sickle mutation: Evidence from beta S globin gene cluster polymorphisms. Mol Biol Med 1, 191–197 [PubMed] [Google Scholar]
- Weatherall DJ, and Clegg JB. (2001). Inherited haemoglobin disorders: An increasing global health problem. Bull World Health Organ 79, 704–712 [PMC free article] [PubMed] [Google Scholar]
- Wonkam A, Bitoungui VJN, Vorster AA, et al. (2014c). Association of variants at BCL11A and HBS1L-MYB with hemoglobin F and hospitalization rates among sickle cell patients in Cameroon. PloS one 9, e92506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonkam A, de Vries J, Royal CD, Ramesar R, and Angwafo FF. (2014d). Would you terminate a pregnancy affected by sickle cell disease? Analysis of views of patients in Cameroon. J Med Ethics 40, 615–620 [DOI] [PubMed] [Google Scholar]
- Wonkam A, Mba CZ, Mbanya D, et al. (2014a). Psychosocial stressors of sickle cell disease on adult patients in Cameroon. J Genet Couns 23, 948–956 [DOI] [PubMed] [Google Scholar]
- Wonkam A, Mba CZ, Mbanya D, et al. (2013). Psychosocial burden of sickle cell disease on parents with an affected child in Cameroon. J Genet Couns 23, 192–201 [DOI] [PubMed] [Google Scholar]
- Wonkam A, Noubiap JJ, Djomou F, Fieggen K, Njock R, and Toure GB. (2013b) Aetiology of childhood hearing loss in Cameroon (sub-Saharan Africa). Eur J Med Genet 56, 20–25 [DOI] [PubMed] [Google Scholar]
- World Bank. (2010). Education for all-fast track initiative: Support to the education sector. Report No. 48373-CM, 1–2
- Zago MA, Silva WA, Dalle B, et al. (2000). Atypical beta-S haplotypes are generated by diverse genetic mechanisms. Am J Hematol 63, 79–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.