Abstract
Background
Epigenetics is tissue-specific and potentially even cell-specific, but little information is available from human reproductive studies about the concordance of DNA methylation patterns in cord blood and placenta, as well as within-placenta variations. We evaluated methylation levels at promoter regions of candidate genes in biological ageing pathways (SIRT1, TP53, PPARG, PPARGC1A, and TFAM), a subtelomeric region (D4Z4) and the mitochondrial genome (MT-RNR1, D-loop).
Methods
Ninety individuals were randomly chosen from the ENVIRONAGE birth cohort to evaluate methylation concordance between cord blood and placenta using highly quantitative bisulfite-PCR pyrosequencing. In a subset of nineteen individuals, a more extensive sampling scheme was performed to examine within-placenta variation.
Results
The DNA methylation levels of the subtelomeric region and mitochondrial genome showed concordance between cord blood and placenta with correlation coefficients ranging from r = 0.31 to 0.43, p ≤ 0.005, and also between the maternal and foetal sides of placental tissue (r = 0.53 to 0.72, p ≤ 0.05). For the majority of targets, an agreement in methylation levels between four foetal biopsies was found (with intra-class correlation coefficients ranging from 0.16 to 0.72), indicating small within-placenta variation.
Conclusions
The methylation levels of the subtelomeric region (D4Z4) and mitochondrial genome (MT-RNR1, D-loop) showed concordance between cord blood and placenta, suggesting a common epigenetic signature of these targets between tissues. Concordance was lacking between the other genes that were studied. In placental tissue, methylation patterns of most targets on the mitochondrial-telomere axis were not strongly influenced by sample location.
Keywords: ageing, cord blood, concordance, DNA methylation, epigenetics, placenta, within-placenta variation
1. Introduction
Epigenetic modifications represent a potential link between adverse insults and altered foetal development. Epigenetic changes, of which DNA methylation is the most commonly characterized, can occur throughout the course of life, but much of the epigenome is already established in germ cells and embryos as it appears to be particularly important for the regulation of embryonic growth and placental development [1]. Disruption of the epigenome has been demonstrated in human placenta-related pathologies such as intrauterine growth restriction [2] and preeclampsia [3, 4].
Although cord blood and placental tissue are the most frequently used specimens in human reproductive studies due to their functional significance, non-invasive collection and good accessibility, some challenges need to be taken into consideration. Gene methylation is tissue-specific and potentially even cell-specific [5–7]. In this regard, little information is available about DNA methylation patterns in cord blood and placental tissue, as well as within-placenta variations. Considerable variation in the DNA methylation of imprinted genes is observed in tissues derived from monozygotic and dizygotic twin pairs (such as cord blood-derived mononuclear cells and granulocytes, umbilical vein endothelial cells, buccal epithelial cells and placental tissue), with greater discordance in dizygotic twin pairs [7]. These data highlight that both genetic and intrauterine exposure factors contribute to the establishment of the neonatal epigenome of different tissues.
Rationale of target selection
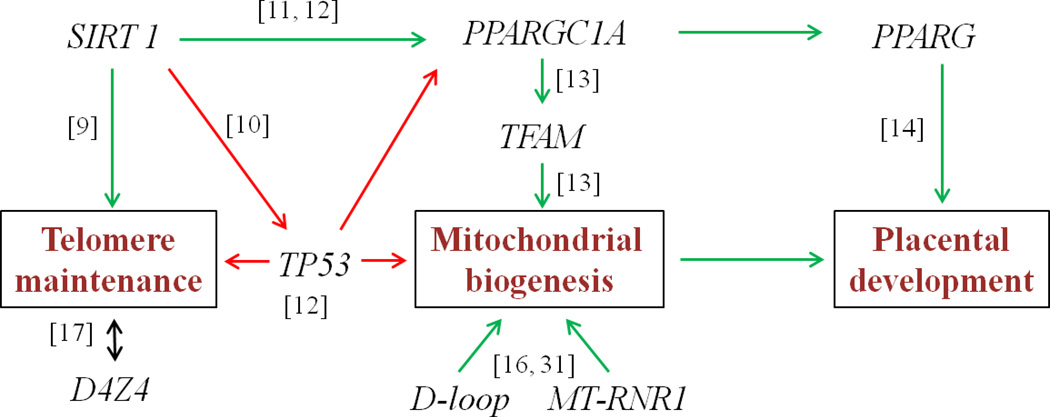
Due to the growing interest in biological ageing processes in utero, a better understanding of epigenetic alterations of age-and mitochondrial-related targets in cord blood and placenta is desired. Within the ENVIRONAGE project (ENVIRonmental influence ON AGEing in early life), we impart new dimensions to the present understanding of human ageing and its environmental influence from early life onwards. The integration of mitochondria into the ‘core axis of ageing’ is supported by the premature ageing conditions exemplified by telomere dysfunction as well as mutations or deficiencies in key regulators of mitochondrial biogenesis and function [8]. Our selected targets are partly based on the hypothesized ageing model outlined by Sahin et al. [8] proposing that telomere attrition activates the ‘guardian of the genome’, tumour protein 53 (TP53), resulting in a TP53-mediated mitochondrial dysfunction. We defined eight candidate targets that relate to the mitochondrial-telomere axis of ageing (Figure 1).
Figure 1.
Sirtuin 1 (SIRT1) is an important determinant of longevity in humans that influences telomerase activity [9] and also inactivates TP53 [10]. On the other hand, SIRT1 activates peroxisome proliferator-activated receptor γ-coactivator1α (PPARGC1A) boosting mitochondrial biogenesis [11]. Indeed, SIRT1 activity has been found to decrease in aged tissues, and this might contribute to the increased TP53 activity and suppressed PPARGC1A activity seen in aged mouse and human tissues [12].
In addition to its close relationship with TP53, PPARGC1A is involved in mitochondrial function by acting as a transcriptional co-activator of several nuclear-encoded transcription factors, including mitochondrial transcription factor A (TFAM) which regulates mitochondrial biogenesis [13]. PPARGC1A is also a co-activator of peroxisome proliferator-activated receptor γ (PPARG), regulating trophoblast invasion and early placental development [14].
Recently, mitochondrial DNA methylation has been shown to control mitochondrial gene transcripts [15] and therefore identifies a novel series of targets for in utero changes. The displacement loop (D-loop) is particularly important in this regard because it contains promoters for mitochondrial DNA transcription and nearly the entire mitochondrial genome transcribes from this region. In addition, 12S ribosomal RNA (MT-RNR1) encodes for a protein that facilitates the formation of RNA secondary structures, assembly of the mitochondrial ribosome, and mitochondrial translation [16].
Finally, we studied the non-satellite tandem repeat D4Z4 in the subtelomeric region. In contrast to mammalian telomeric repeats (TTAGGG), the subtelomeric region has a high density of CpG sequences. In vitro and in vivo mouse studies indicate a conserved link between telomere length and the epigenetic status of subtelomeres [17–19]. Evidence from human studies shows an inverse correlation between DNA methylation of the subtelomeric D4Z4 repeat and average telomere length in a panel of cancer cell lines [20], while a positive correlation with telomere length is observed in patients with dyskeratosis congenital [21]. These data thereby demonstrate that the epigenetic status of the telomeric region is affected by disease conditions. Variations in telomere length among adults may already be established in utero and might be linked to epigenetic changes induced by environmental factors, making early life an important time of susceptibility to change [22, 23].
Focus of the study
Few studies have investigated within-placenta variation of (global) DNA methylation levels and these only focus upon a specific set of genes [5, 24–26]. Since the placenta shows a remarkable amount of normal variability in size and structure, these results cannot be extrapolated to other genes. To provide a representative snapshot of placental biomarkers, it requires multiple samples to be taken from a single placenta. However, it is not always feasible given the relatively large number of samples or subjects under investigation in an epidemiological context and the related costs for the epigenetic measurements. Therefore, the number of study participants versus the numbers of sampling sites or biological replicates is an important consideration in molecular epidemiological study design. A solution to minimize the impact of regional differences in methylation or gene expression patterns within a mother’s placenta is to standardize the sampling method by selecting one specific site from which biopsies are taken [27]. In this study, we evaluated the concordance of DNA methylation patterns in cord blood and placental tissue, as well as within-placenta variations, at specific regions of the aforementioned eight candidate-target regions that were selected due to their relevance to ageing.
2. Material and Methods
2.1 Study population
We randomly selected cord blood and placental tissue from the ongoing ENVIRONAGE birth cohort. In this cohort, we recruit mother-newborn pairs from the East-Limburg Hospital in Genk, Belgium of which extensive medical and lifestyle data are available as described previously [28, 29]. All subjects provided informed consent, and all procedures were approved by the Ethical Committee of Hasselt University and East-Limburg Hospital in Genk. We used a subset of ninety placenta-cord blood samples (sample set A) to examine the relationship between DNA methylation in those two tissues. In a subset of nineteen placentas (sample set B), a more extensive sampling scheme was performed to examine within-placenta variation in DNA methylation.
2.2 Cord blood collection and placental sampling
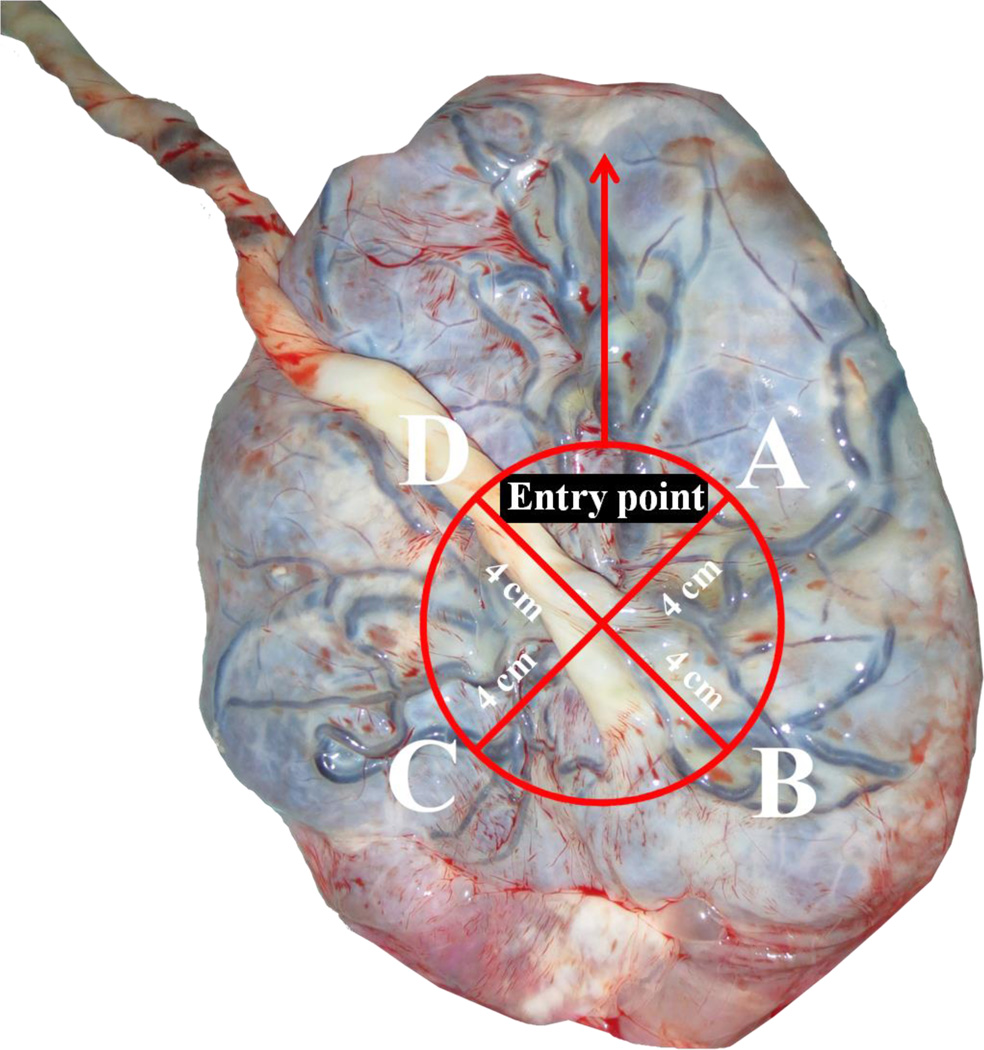
Umbilical cord blood was collected immediately after delivery in Vacutainer® Plus Plastic K2EDTA Tubes (BD, Franklin Lakes, NJ, USA). Samples were centrifuged at 3,200 rpm for 15 min to retrieve buffy coats and instantly frozen, first at −20°C and later at −80°C. Placentas were deep-frozen within ten minutes of delivery. Afterwards, we took biopsy samples of approximately 1 to 2 cm3 for DNA extraction using a standardized protocol as described by Adibi et al. [30]. Four distinct sites from nineteen placentas were sampled from the foetal side across the middle region of the placenta approximately four cm away from the umbilical cord and 1–1.5 cm below the chorio-amniotic membrane (Figure 2). An extra biopsy was taken from the maternal side. Chorio-amniotic membrane contamination was avoided by careful visual examination and dissection. Via histological examination (hematoxylin & eosin staining) we compared four fetal biopsies from four placentas. This confirmed that biopsies were taken from chorionic villous tissue with normal architecture composed of trophoblasts. In the fetal biopsies we could identify terminal and intermediate villi with cytotrophoblasts and syncytiotrophoblasts, vessels, fibrin and the intervillous space. We did not observe any consistent differences in the histology or cell type composition between the fetal samples, nor between the four placentas.
Figure 2.
2.3 DNA methylation analyses
Genomic DNA was isolated from buffy coat of cord blood and from placental tissue using the QIAamp DNA mini kit (Qiagen, Inc., Venlo, the Netherlands). We performed DNA methylation analysis by highly quantitative bisulfite-PCR pyrosequencing. Bisulfite conversions were performed using 1 µg of extracted genomic DNA with the EZ-96 DNA methylation Gold kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions. The final elution volume was 40 µl, using M-elution buffer. We interrogated CpG sites within promoter regions (SIRT1, TP53, PPARG, PPARGC1A, TFAM) or other specific regions (subtelomere: D4Z4; mitochondrial genome: MT-RNR1, D-loop) of targets on the mitochondrial-telomere axis. We combined data from the literature and data derived from the UCSC Genome Browser (http://genome.ucsc.edu,assemblyGRCh37/hg19), such as transcription factor binding sites, DNase I hypersensitive sites and histone modifications to identify regions with potentially methylated CpG sites. The MethPrimer program (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) was used for PCR primer design. The assays for D4Z4, MT-RNR1 and D-loop have been previously described [31, 32]. Detailed information regarding primer sequences and genomic regions is given in Supplementary file 1, Table 1. Methylation levels of the mitochondrial regions MT-RNR1 and D-loop showed a strong correlation in placental tissue (n = 81, r = 0.85, p < 0.0001) and cord blood (n = 82, r = 0.68, p < 0.0001).
Table 1. Characteristics of the two sample sets.
Sample set A: Ninety individuals for the correlation between cord blood and placental tissue. Sample set B: Nineteen individuals for within-placenta variation analysis. Data are presented as mean ± SD or range and number (%).
| Variables | Sample set A (n = 90) | Sample set B (n = 19) |
|---|---|---|
| Maternal age, y | 29.0 (19–40) | 27.8 (18–38) |
| Pre-gestational BMI, kg/m2 | 25.5 ± 5.1 | 26.0 ± 6.6 |
| Smoking | ||
| Never-smoker | 64 (71.1%) | 10 (52.7%) |
| Past-smoker | 11 (12.2%) | 7 (36.8%) |
| Smoker | 15 (16.7%) | 2 (10.5%) |
| Alcohol | ||
| No | 69 (76.6%) | 12 (63.2%) |
| Yes | 21 (23.3%) | 7 (36.8%) |
| Parity | ||
| 1 | 44 (48.9%) | 11 (57.9%) |
| 2 | 36 (40%) | 6 (31.6%) |
| ≥ 3 | 10 (11.1%) | 2 (10.5%) |
| Newborn’s gender | ||
| Male | 47 (52.2%) | 7 (36.8%) |
| Female | 43 (47.8%) | 12 (63.2%) |
| Gestational age, w | 39.2 (35–42) | 38.9 (36–41) |
| Apgar score after 5 min | ||
| 6 | 1 (1.1%) | - |
| 7 | 2 (2.2%) | - |
| 8 | 2 (2.2%) | 1 (5.3%) |
| 9 | 20 (22.2%) | 7 (36.8%) |
| 10 | 65 (72.2%) | 11 (57.9%) |
| Birth weight, g | 3462 ± 461 | 3515 ± 381 |
| Birth length, cm | 50.4 ± 1.8 | 50.2 ± 2.0 |
| Caesarean section | 5 (5.6%) | 4 (21.0%) |
| Epidural anaesthesia | 64 (71.1%) | 13 (68.4%) |
PCR amplification of regions of interest prior to pyrosequencing was performed in a total reaction volume of 30 µl, using 15 µl GoTaq Hot Start Green Master Mix (Promega, Madison, WI, USA), 10 pmol forward primer, 10 pmol reverse primer, 1 µl bisulfite-treated genomic DNA and water. One primer was biotin-labelled in order to purify PCR products in conjunction with Sepharose beads. PCR products were bound to Streptavidin Sepharose High Performance beads (Amersham Biosciences, Uppsala, Sweden) via the biotin label, and immobilized on the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Inc., Westborough, MA, USA) before being purified, washed, denatured using 0.2 mol/L NaOH solution, and washed again according to the manufacturer’s protocol. 0.3 µmol/L of pyrosequencing primer was annealed to the purified single-stranded PCR product, and pyrosequencing was performed using the PyroMark Q96 MD Pyrosequencing System (Qiagen, Inc., Germantown, MD, USA). We used 0% (PSQ-T oligo: 5′-TTGCGATACAACGGGAACAAACGTTGAATTC-3′) and 100% (PSQ-C oligo: 5′-TTGCGATACGACGGGAACAAACGTTGAATTC-3′) DNA methylation control oligos. The sequencing primer for the control oligo is: 5′-AACGTTTGTTCCCGT-3′. We mixed the PSQ-C oligo (or PSQ-T oligo) with the sequencing oligo in PyroMark Annealing Buffer (Qiagen, Inc., Valencia, CA, USA) and performed pyrosequencing with the sequencing entry C/TGTAT [31]. The degree of methylation was expressed as the percentage of methylated cytosines over the sum of methylated and unmethylated cytosines. The efficiency of the bisulfite-conversion process was assessed using non-CpG cytosine residues within the sequence.
2.4 Statistical analysis
Multiple CpGs within each region of interest were interrogated and we calculated the average across all the CpGs for the analysis. Spearman correlation coefficients were applied to assess the association between methylation levels in cord blood and placental tissue of ninety participants (sample set A). The Wilcoxon signed rank test was performed to compare methylation levels between the two different tissues.
In nineteen individuals (sample set B), we measured DNA methylation of the eight targets in the four quadrants of the placenta. We computed the intra-class correlation coefficient (ICC) from a variance components mixed model to evaluate within- versus between-placenta variability. Mixed models take into account regional differences of each individual placenta and calculate the proportion of variation that is explained by the variance between individuals. Additionally, we tested for differences between placental biopsies using the Friedman’s test and conducted post-hoc analysis with the Wilcoxon signed rank test. The Bonferroni method was applied to correct for multiple pairwise comparisons. All statistical analysis were performed using the SAS software program (version 9.2; SAS Institute Inc., Cary, NC, USA).
3. Results
The study included two sample sets; sample set A (n = 90) for the correlation of methylation levels between cord blood and placental tissue and sample set B (n = 19) for the assessment of within-placenta variation. Characteristics of the two samples are reported in Table 1.
3.1 Concordance between DNA methylation in cord blood and placenta
Low methylation levels, ranging from 1.7 % to 11.7%, were observed in both tissues for all targets except D4Z4, which had mean methylation levels of 65.2% in cord blood and 45.1% in placenta (Table 2). Mean cord blood methylation levels at the interrogated regions of D4Z4, SIRT1, PPARG, MT-RNR1 and D-loop were significantly higher than those observed in placenta (p ≤ 0.01). PPARGC1A showed significantly lower methylation levels (p < 0.0001) in cord blood compared with placental tissue. The examined CpGs in the promoter region of PPARGC1A in placental tissue showed strong correlation (r ≥ 0.60) but not in cord blood. No significant differences in mean methylation levels at the promoter regions of TP53 and TFAM were observed between tissues.
Table 2.
Summary of the correlation between cord blood and placental tissue methylation (sample set A).
| Target | n | Mean (%) ± SD cord blood |
Mean (%) ± SD placenta |
Spearman correlation |
|---|---|---|---|---|
| D4Z4 | 88 | 65.1 ± 8.4 | 45.1 ± 7.9a | 0.43b |
| SIRT1 | 78 | 2.7 ± 2.0 | 2.0 ± 1.1a | −0.08 |
| TP53 | 89 | 2.6 ± 0.4 | 2.6 ± 0.5 | 0.02 |
| PPARG | 88 | 3.2 ± 3.8 | 2.8 ± 4.2a | −0.13 |
| PPARGC1A | 89 | 2.4 ± 0.6 | 8.2 ± 6.7a | 0.06 |
| TFAM | 87 | 1.8 ± 0.3 | 1.7 ± 0.24 | 0.08 |
| MT-RNR1 | 86 | 11.7 ± 0.6 | 9.5 ± 0.4a | 0.36b |
| D-loop | 82 | 4.0 ± 1.1 | 3.7 ± 1.3a | 0.31b |
Statistically significant difference in methylation between cord blood and placental tissue (Wilcoxon signed rank test; p-value ≤ 0.05).
p-value ≤ 0.05
The correlation between methylation levels in cord blood and placental tissue was significant for three of the eight targets (D4Z4: r = 0.43, p < 0.0001; MT-RNR1: r = 0.36, p = 0.0006; D-loop: r = 0.31, p = 0.005). SIRT1, TP53, PPARG, PPARGC1A, and TFAM showed no significant correlation between cord blood and placental tissue (Table 2). The correlation between tissues did not alter after adjustment for maternal age, gender, gestational age or parity.
3.2 Within-placenta variation in DNA methylation
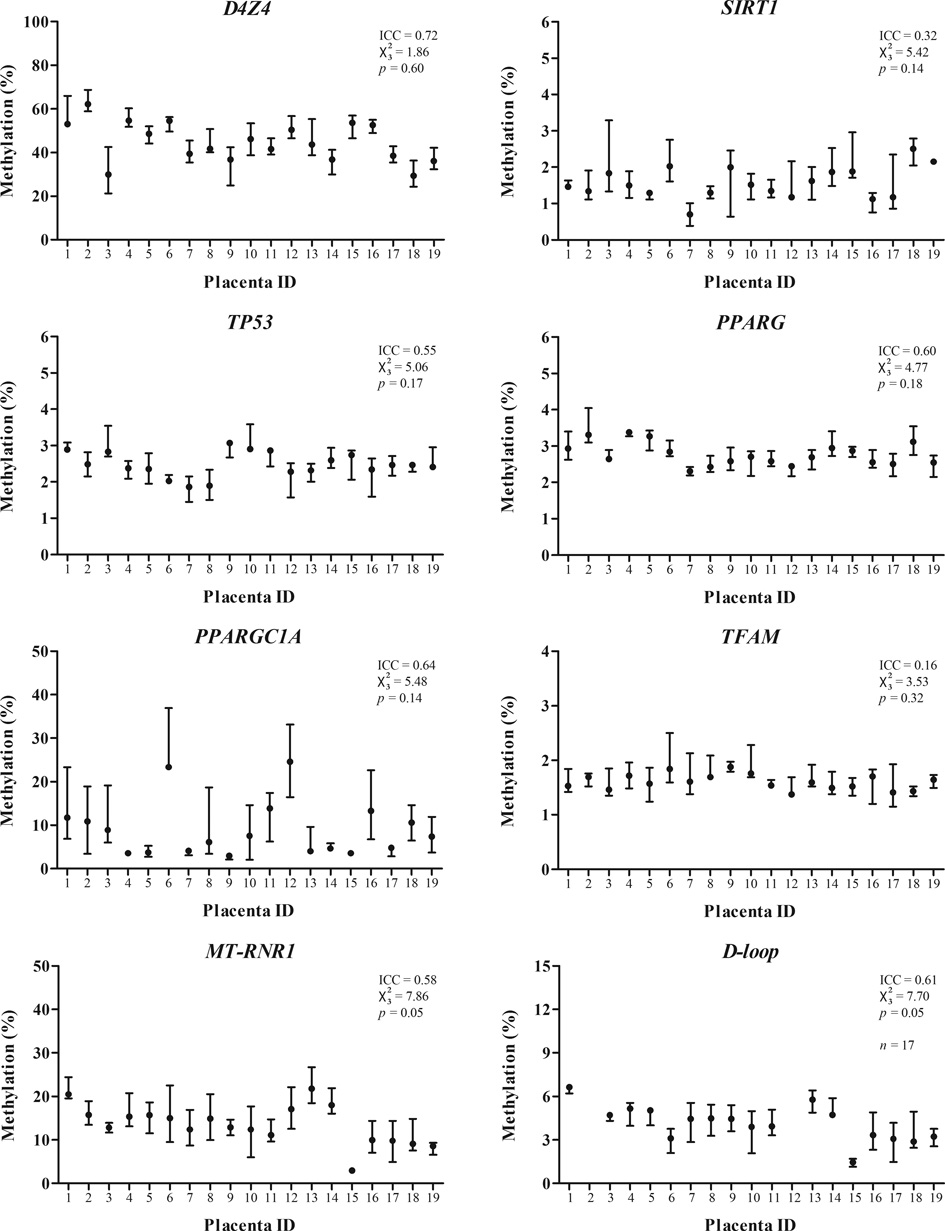
We measured DNA methylation of the eight targets in four quadrants of nineteen placentas and evaluated the within-placenta variation in DNA methylation using the ICC and Friedman test (). Between-placenta variability, exemplified by the ICC, was higher than within-placenta variability for D4Z4 (72% vs. 28%, p = 0.003), TP53 (55% vs. 45%, p = 0.04), PPARG (60% vs. 40%, p = 0.006), PPARGC1A (64% vs. 37%, p = 0.005), MT-RNR1 (58% vs. 42%, p = 0.009), and D-loop (61% vs. 39%, p = 0.01), but not for SIRT1 (32% vs. 68%, p = 0.16) and TFAM (16% vs. 84%, p = 0.42) (Figure 3). For PPARGC1A, two placentas showed much higher methylation levels (>20%) compared to the other placentas. For the D-loop assay, all four biopsies from two placentas failed pyrosequencing twice and were excluded from analysis.
Figure 3.
The methylation levels between different biopsies of each individual, exemplified by the statistic, showed no significant difference for D4Z4 (p = 0.60), SIRT1 (p = 0.14), TP53 (p = 0.17), PPARG (p = 0.18), PPARGC1A (p = 0.14), and TFAM (p = 0.32). Only for MT-RNR1 and D-loop there was a borderline significant difference between biopsies observed (both p = 0.05). Pairwise comparisons showed no significant differences between biopsies after Bonferroni adjustment. Mean methylation levels of each biopsy averaged by all individuals are shown in Supplementary file, Figure 1.
Additionally, we compared the methylation levels of all the targets between biopsy A, taken from the foetal side, and the corresponding biopsy A2, taken from the maternal side. No significant difference in methylation levels was observed between the maternal and foetal side for any of the targets (p ≥ 0.1). However, the correlation between the foetal and maternal sides was only significant for the same targets that also correlated between cord blood and placental tissue (i.e. D4Z4: r = 0.72, p = 0.005; MT-RNR1: r = 0.53, p = 0.05; D-loop: r = 0.60, p = 0.04).
4. Discussion
As suggested by Non and colleagues [24], researchers interested in epigenetic or gene expression analysis should evaluate each gene of interest across placental sampling sites, as each gene may display site-specific differences. Within the ENVIRONAGE birth cohort, integration of targets that reflect or determine the biological ageing process, including mitochondrial biogenesis, telomere length and candidate genes in placental development, will be investigated to understand the environmental influence on the ‘core axis of ageing’ from early life onwards. In this study, we found that: 1/ methylation levels of the subtelomeric region (D4Z4) and mitochondrial genome (MT-RNR1 and D-loop) showed concordance between umbilical cord blood and placental tissue, while promoter regions of candidate genes on the mitochondrial-telomere axis showed tissue-specific methylation patterns; 2/ most of our selected candidate targets, which play a potential role in biological ageing, did not display major differences in methylation levels across several biopsies in placental tissue.
4.1 Concordance between DNA methylation in cord blood and placenta
The mean methylation levels of some targets in our study differed between cord blood and placental tissue (i.e. D4Z4, SIRT1, PPARG, PPARGC1A, MT-RNR1, and D-loop). Similar results have been reported by other studies on imprinted genes and global methylation levels of the cord blood and placenta [6, 33]. It is noteworthy to mention that the promoter region of PPARGC1A was largely unmethylated in cord blood but not in placental tissue. In addition, much higher methylation levels in all four biopsies were observed in two out of nineteen placentas from sample set B. This is of particular interest because our interrogated region is a putative binding site for CREB transcription factors. The transcriptional activity of CREB is critical for the establishment and maintenance of energy homeostasis in mice neonates [34] and appears to be involved in the regulation of PPARGC1A, which is a key regulator of mitochondrial genes and energy metabolism. Methylation of the PPARGC1A promoter region may therefore present one mechanism by which the genes could be differentially regulated between cord blood and placental tissue.
Most of our studied genes had low methylation levels in both cord blood and placental tissue. Despite the decreased ability to identify significant methylation changes in unmethylated genomic contexts due to technical limitations, small changes in promoter gene methylation have been extensively described in several fields [35], including the one of environmental epigenetics [36–39]. Small changes in regional methylation as well as single CpG sites have been described to interfere with gene/protein expression [40] via mechanisms that include alteration of affinity of transcription factors to their binding sites or chromosome looping events [41]. Even when small changes in methylation would not impact gene expression, they might still serve as markers of exposure or outcome [35].
4.1.1 Correlation of the subtelomeric region and mitochondrial genome between tissues
As described above, methylation levels in umbilical cord blood may not always reflect those in the placenta and vice versa. In our study, we observed significant correlations between tissues at the subtelomeric region (D4Z4), and mitochondrial genome (MT-RNR1 and D-loop). Our results expand upon those from a previous study investigating global and gene-specific DNA methylation across multiple tissues in early pregnancy [42] which reported that only the repetitive element AluY8b, used as proxy for global DNA methylation, was correlated between placental tissue and cord blood. However, global methylation levels analyzed by LUMA showed no correlation between the umbilical cord blood and placental tissue [33]. Our observations regarding subtelomeric methylation levels extends previous observations of concordance in telomere length among multiple organs of the human foetus [23, 43]. Together, these data indicate that methylation patterns in mitochondria and repetitive elements, such as D4Z4 and Alu, are likely to reflect a common epigenetic signature between cord blood and placental tissue. Indeed, methylation levels at the ends of chromosomes and the mitochondrial genome may be more susceptible to changes as these ‘hotspots’ are more sensitive to oxidative stress in comparison to other parts of the genome.
4.1.2 Correlation between the maternal and foetal side of placental tissue
The methylation levels of the subtelomeric region and mitochondrial genome correlated between the maternal and foetal side of placental tissue. This was in addition to our findings of a correlation between foetal placental tissue and cord blood, thereby suggesting that the methylation status of these regions was consistent from maternal to foetal side to cord blood. A previous study of placental methylation variation showed good correlation (r = 0.76, p < 0.0001) for LINE-1 between the foetal and maternal sides, but no such correlation existed for stress-related genes [24].
4.2 Within-placenta variation in DNA methylation
Placental tissue, as with cord blood, is composed of a complex population of cells that makes this organ shows high variability in overall DNA methylation compared with other tissues [44]. Cytotrophoblasts and fibroblasts are the two main cell types in the placenta, and most genes exhibit similar promoter methylation patterns. Although some specific genes show differential promoter methylation between these cell types, the methylation status of placental villi mainly reflects the profile of the cytotropoblast cells [5]. Avila and colleagues concluded that the within-placenta variability for several genes generally showed less sample-to-sample variation for DNA methylation than gene expression, and different placental sites and depths showed consistent methylation patterns [25]. Our findings of low variation between four sampled biopsies of most targets is largely consistent with the results of Avila et al. [25] and Non et al. [24], and we can conclude that methylation variation is probably not due to sample location but rather due to cell composition differences between samples. Nevertheless, these authors found that methylation patterns of some genes differed across placental locations, but these differences were of limited magnitude. Since we did not observe differences between sample locations, our chosen biopsy location provides a representative biological measure from each individual placenta. However, future studies on placental methylation may consider pooling multiple sites across the middle region of the placenta to further reduce sampling variability [27].
4.2.1 Analysis of within-placenta variability in DNA methylation
In our study we used two parameters, Friedman’s statistic and ICC, which provided different information regarding within-placenta variation. An advantage of this combination is that we were able to evaluate the methylation levels across biopsies together with the agreement between biopsies. The higher the ICC, the more reliably a biopsy measure reflects true between-individual differences. Although there is considerable variation in methylation levels between individuals, as seen with the large range of methylation values, all targets showed similar methylation levels across the four biopsies (Friedman’s test). However, both regions of the mitochondrial genome, MT-RNR1 and D-loop, showed borderline significant differences compared between biopsies, although not significant after Bonferroni correction. This observation was not reflected in the ICC, which was fairly high (58% and 61% for MT-RNR1 and D-loop respectively), therefore showing a good agreement between biopsies. One remark on the usage of the ICC as a reliability statistic is that it is usually applied to assess the reproducibility of measurements in replicates, which normally yield high ICC values. In our study we applied the ICC to different biopsies from the same organ, assuming that some biological variation is already present and will therefore yield lower ICCs. Variation across sites in an organ as complex as the placenta is within its nature and even a small deviation in methylation level in one biopsy influences the separate biopsy correlations and subsequently the ICC. The low methylated genes TFAM and SIRT1 lacked correlation between biopsies, possibly as a result of the relatively higher variation on account of the low absolute methylation levels. We did not observe any obvious differences in the histology or cell type composition between the foetal samples sites across the middle region of the placenta nor between placentas.
4.3 Conclusion
The methylation of the subtelomeric region and mitochondrial genome showed concordance between the different tissues (cord blood and the foetal and maternal side of the placenta). However, the other selected candidate targets showed tissue-specific methylation patterns at promoter regions, as exemplified by lack of correlation between cord blood and placental tissue. These data indicate that methylation patterns in mitochondria and repeated elements are likely to reflect a common epigenetic signature between cord blood and placental tissue. Within the placenta, all our selected methylation targets on the mitochondrial-telomere axis showed similar methylation levels, and for these targets it is amenable that sample location does not affect DNA methylation. This information may guide future molecular epidemiological research investigating the epigenetics of targets operating within the core axis of ageing.
Supplementary Material
Acknowledgements
The authors thank the participating women, as well as the staff of the maternity ward (Anja Moors, Christine De Bruyn, Caroline Dielen, Noelie Dony, Julie Faes, Carmen Kerkhofs, Elisabeth Vandendriessche), midwives, and the staff of the clinical laboratory of East-Limburg Hospital in Genk. We thank dr. Timothy Barrow for proofreading the manuscript. The ENVIRONAGE birth cohort is supported by the EU Program “Ideas” (ERC-2012-StG 310898), by the Flemish Scientific Fund (FWO, N1516112/G.0.873.11.N.10) and Bijzonder Onderzoeks Fonds of Hasselt University (BOF). This work was also supported by funding from the National Institute of Environmental Health Sciences (R01ES021733 and R01ES021357). Benedetta Izzi has a postdoctoral fellowship (PDMK/13/163).
Abbreviations
- D4Z4
Non-satellite tandem repeats in the subtelomeric region
- D-loop
Displacement loop
- ICC
Intra-class correlation coefficient
- MT-RNR1
12S ribosomal RNA
- PPARG
Peroxisome proliferator-activated receptor γ
- PPARGC1A
Peroxisome proliferator-activated receptor γ-coactivator 1α
- SIRT1
Sirtuin 1
- TFAM
Mitochondrial transcription factor A
- TP53
Tumour protein 53
Footnotes
The authors declare they have no competing financial interests.
References
- 1.Constancia M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417(6892):945–948. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- 2.Bourque DK, Avila L, Peñaherrera M, von Dadelszen P, Robinson WP. Decreased Placental Methylation at the H19/IGF2 Imprinting Control Region is Associated with Normotensive Intrauterine Growth Restriction but not Preeclampsia. Placenta. 2010;31(3):197–202. doi: 10.1016/j.placenta.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Blair JD, Yuen RKC, Lim BK, McFadden DE, von Dadelszen P, Robinson WP. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Molecular Human Reproduction. 2013;19(10):697–708. doi: 10.1093/molehr/gat044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulkarni A, Dangat K, Kale A, Sable P, Chavan-Gautam P, Joshi S. Effects of altered maternal folic acid, vitamin B12 and docosahexaenoic acid on placental global DNA methylation patterns in Wistar rats. PLoS ONE. 2011;6(3):e17706. doi: 10.1371/journal.pone.0017706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grigoriu A, Ferreira JC, Choufani S, Baczyk D, Kingdom J, Weksberg R. Cell specific patterns of methylation in the human placenta. Epigenetics. 2011;6(3):368–379. doi: 10.4161/epi.6.3.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herzog E, Galvez J, Roks A, Stolk L, Verbiest M, Eilers P, et al. Tissue-specific DNA methylation profiles in newborns. Clinical Epigenetics. 2013;5(1):8. doi: 10.1186/1868-7083-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ollikainen M, Smith KR, Joo EJ-H, Ng HK, Andronikos R, Novakovic B, et al. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Human Molecular Genetics. 2010;19(21):4176–4188. doi: 10.1093/hmg/ddq336. [DOI] [PubMed] [Google Scholar]
- 8.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464(7288):520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. The Journal of Cell Biology. 2010;191(7):1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 11.Aquilano K, Vigilanza P, Baldelli S, Pagliei B, Rotilio G, Ciriolo MR. Peroxisome Proliferator-activated Receptor γ Co-activator α (PGC-1α) and Sirtuin 1 (SIRT1) Reside in Mitochondria: Possible direct function in mitochondrial biogenesis. Journal of Biological Chemistry. 2010;285(28):21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahin E and DePinho RA. Axis of ageing: telomeres, p53 and mitochondria. Nat Rev Mol Cell Biol. 2012;13(6):397–404. doi: 10.1038/nrm3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee HC and Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37(4):822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Wieser F, Waite L, Depoix C, Taylor RN. PPAR Action in Human Placental Development and Pregnancy and Its Complications. PPAR Res. 2008;2008:527048. doi: 10.1155/2008/527048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci USA. 2011;108(9):3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aloni Y and Attardi G. Expression of the mitochondrial genome in HeLa cells. II. Evidence for complete transcription of mitochondrial DNA. J Mol Biol. 1971;55(2):251–267. doi: 10.1016/0022-2836(71)90195-1. [DOI] [PubMed] [Google Scholar]
- 17.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8(4):299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8(4):416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 19.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet. 2007;39(2):243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 20.Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27(54):6817–6833. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- 21.Gadalla SM, Katki HA, Shebl FM, Giri N, Alter BP, Savage SA. The relationship between DNA methylation and telomere length in dyskeratosis congenita. Aging Cell. 2012;11(1):24–28. doi: 10.1111/j.1474-9726.2011.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boekelheide K, Blumberg B, Chapin RE, Cote I, Graziano JH, Janesick A, et al. Predicting later-life outcomes of early-life exposures. Environ Health Perspect. 2012;120(10):1353–1361. doi: 10.1289/ehp.1204934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuda K, Bardeguez A, Gardner JP, Rodriguez P, Ganesh V, Kimura M, et al. Telomere length in the newborn. Pediatr Res. 2002;52(3):377–381. doi: 10.1203/00006450-200209000-00012. [DOI] [PubMed] [Google Scholar]
- 24.Non AL, Binder AM, Barault L, Rancourt RC, Kubzansky LD, Michels KB. DNA methylation of stress-related genes and LINE-1 repetitive elements across the healthy human placenta. Placenta. 2012;33(3):183–187. doi: 10.1016/j.placenta.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avila L, Yuen RK, Diego-Alvarez D, Peñaherrera MS, Jiang R, Robinson WP. Evaluating DNA methylation and gene expression variability in the human term placenta. Placenta. 2010;31(12):1070–1077. doi: 10.1016/j.placenta.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Vilahur N, Baccarelli AA, Bustamante M, Agramunt S, Byun HM, Fernandez MF, et al. Storage conditions and stability of global DNA methylation in placental tissue. Epigenomics. 2013;5(3):341–348. doi: 10.2217/epi.13.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogg K, Price EM, Robinson WP. Improved reporting of DNA methylation data derived from studies of the human placenta. Epigenetics. 2014;9(3):333–337. doi: 10.4161/epi.27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen BG, Godderis L, Pieters N, Poels K, Kiciński M, Cuypers A, et al. Placental DNA hypomethylation in association with particulate air pollution in early life. Particle and Fibre Toxicology. 2013;10:22. doi: 10.1186/1743-8977-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssen BG, Munters E, Pieters N, Smeets K, Cox B, Cuypers A, et al. Placental Mitochondrial DNA Content and Particulate Air Pollution during in Utero Life. Environ Health Perspect. 2012;120(9):1346–1352. doi: 10.1289/ehp.1104458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adibi JJ, Hauser R, Williams PL, Whyatt RM, Thaker HM, Nelson H, et al. Placental biomarkers of phthalate effects on mRNA transcription: application in epidemiologic research. Environ Health. 2009;8:20. doi: 10.1186/1476-069X-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byun H-M, Panni T, Motta V, Hou L, Nordio F, Apostoli P, et al. Effects of airborne pollutants on mitochondrial DNA Methylation. Particle and Fibre Toxicology. 2013;10(1):18. doi: 10.1186/1743-8977-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi SH, Worswick S, Byun HM, Shear T, Soussa JC, Wolff EM, et al. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int J Cancer. 2009;125(3):723–729. doi: 10.1002/ijc.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nomura Y, Lambertini L, Rialdi A, Lee M, Mystal EY, Grabie M, et al. Global Methylation in the Placenta and Umbilical Cord Blood From Pregnancies With Maternal Gestational Diabetes, Preeclampsia, and Obesity. Reproductive Sciences. 2013;00(0):1–7. doi: 10.1177/1933719113492206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, et al. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269(5227):1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 35.Michels KB, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10(10):949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- 36.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA Methylation Patterns in Subjects Exposed to Low-Dose Benzene. Cancer Research. 2007;67(3):876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 37.Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. 450K Epigenome-Wide Scan Identifies Differential DNA Methylation in Newborns Related to Maternal Smoking during Pregnancy. Environ Health Perspect. 2012;120(10):1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, Rahman M, et al. Prenatal Arsenic Exposure and DNA Methylation in Maternal and Umbilical Cord Blood Leukocytes. Environ Health Perspect. 2012;120(7):1061–1066. doi: 10.1289/ehp.1104173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J, Tang D, et al. Maternal Exposure to Polycyclic Aromatic Hydrocarbons and 5'-CpG Methylation of Interferon-gamma in Cord White Blood Cells. Environ Health Perspect. 2012;120(8):1195–1200. doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izzi B, Francois I, Labarque V, Thys C, Wittevrongel C, Devriendt K, et al. Methylation defect in imprinted genes detected in patients with an Albright's hereditary osteodystrophy like phenotype and platelet Gs hypofunction. PLoS One. 2012;7(6):e38579. doi: 10.1371/journal.pone.0038579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, et al. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104(30):12377–12382. doi: 10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armstrong DA, Lesseur C, Conradt E, Lester BM, Marsit CJ. Global and gene-specific DNA methylation across multiple tissues in early infancy: implications for children's health research. The FASEB Journal. 2014;28(5):2088–2097. doi: 10.1096/fj.13-238402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Youngren K, Jeanclos E, Aviv H, Kimura M, Stock J, Hanna M, et al. Synchrony in telomere length of the human fetus. Hum Genet. 1998;102(6):640–643. doi: 10.1007/s004390050755. [DOI] [PubMed] [Google Scholar]
- 44.Yuen RK, Avila L, Peñaherrera MS, von Dadelszen P, Lefebvre L, Kobor MS, et al. Human placental-specific epipolymorphism and its association with adverse pregnancy outcomes. PLoS ONE. 2009;4(10):e7389. doi: 10.1371/journal.pone.0007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.