Abstract
Technological advances in the large scale analysis of human genetics have generated profound insights into possible genetic contributions to chronic diseases including the inflammatory bowel diseases (IBDs), Crohn’s disease and ulcerative colitis. To date, 163 distinct genetic risk loci have been associated with either Crohn’s disease or ulcerative colitis, with a substantial degree of genetic overlap between these 2 conditions. Although many risk variants show a reproducible correlation with disease, individual gene associations only affect a subset of patients, and the functional contribution(s) of these risk variants to the onset of IBD is largely undetermined. Although studies in twins have demonstrated that the development of IBD is not mediated solely by genetic risk, it is nevertheless important to elucidate the functional consequences of risk variants for gene function in relevant cell types known to regulate key physiological processes that are compromised in IBD. This article will discuss IBD candidate genes that are known to be, or are suspected of being, involved in regulating the intestinal epithelial barrier and several of the physiological processes presided over by this dynamic and versatile layer of cells. This will include assembly and regulation of tight junctions, cell adhesion and polarity, mucus and glycoprotein regulation, bacterial sensing, membrane transport, epithelial differentiation, and restitution.
Keywords: epithelial, permeability, inflammation, tight junctions
The use of genome-wide association studies (GWAS) has transformed our understanding of genetic risk factors for inflammatory bowel disease (IBD) and many other chronic inflammatory conditions. These studies have opened new avenues of investigation into pathologic mechanisms involved in IBD pathogenesis such as bacterial sensing, autophagy, and dysregulated immune response signaling pathways as well as the variance of disease phenotypes found in Crohn’s disease (CD) and ulcerative colitis (UC).1 GWAS have also identified susceptibility genes involved in the function of various aspects of the intestinal barrier. A number of excellent review articles have been published describing IBD candidate genes and their potential roles in the pathogenesis of IBD.2–5 This article attempts to update the list of IBD candidate genes that have been identified as having a role in regulating intestinal barrier properties and how they serve to link IBD genetics with this major pathophysiological series of events involved in the onset and progression of IBD. In the interests of clarity, the focus of this article will primarily be on candidate genes affecting barrier function. However, the use of the term “intestinal barrier properties” in this article will incorporate broader aspects of the epithelial barrier including, but not limited to, the strictest definition of “intestinal barrier function,” which reflects changes in intestinal permeability and refers exclusively to altered flux of solutes and fluid across the epithelium.6
INTESTINAL EPITHELIAL BARRIER
The epithelial barrier plays a critical role in maintaining intestinal homeostasis because it lies at the interface between luminal microbes and the host immune system, while also being the first site of exposure to many of the environmental factors that can act as triggers of disease activity.7,8 The epithelium communicates in a bidirectional manner with luminal microbes and the immune system; and therefore, the integrity of the epithelial lining is critical for the appropriate partitioning of interactions between immune cells and microbes.9 The epithelium therefore has the capacity to sense luminal contents i.e., microbial products and through activation of surface receptors such as toll-like receptor signaling pathways, secrete biochemical signals to adjacent lamina propria immune cells.10 Furthermore, luminal antigens are sampled across the epithelial layer thus contributing to the immune surveillance role of the mucosal barrier.11,12 However, excessive uptake of antigens can result in overactivation of the immune system, thus leading to mucosal inflammation.11 In addition to its wider role in innate immune signaling, an essential function of the epithelial lining that allows it to perform a vast array of sophisticated tasks is its ability to form a tightly regulated and selectively permeable barrier. The “barrier” function of the epithelium is absolutely essential to intestinal function. It is required for the ability of the epithelium to absorb nutrients and water under the direction of solute-couple uptake mechanisms. By the same token, a regulated barrier is a prerequisite for the regulated secretion of fluid and digestive enzymes to break down ingested foods and allow digestion and assimilation of nutrients, as well as fluid secretion that serves to lubricate the epithelial surface or wash away noxious agents or pathogens from the epithelial surface.13,14
MUCUS LAYERS AND EPITHELIAL SECRETORY FUNCTIONS
Another essential component of intestinal homeostasis is provided by the secretion of mucus from goblet cells and dispersal of tightly-packed mucus droplets by bicarbonate secretion.15,16 Mucus particle dispersal facilitates formation of appropriately hydrated and expanded mucus layers. The stomach and colon have 2 mucus layers with an inner mucus layer that is 50 to 200 μm thick, firmly attached to the epithelium, and is built around the gel-forming MUC5A mucin protein in the stomach and MUC2 in the intestine that act as a structural skeleton for the inner mucus layer.17 This inner layer can act as a filter to prevent bacteria from accessing the epithelial surface. The outer colonic mucus layer is generated through endogenous protease cleavage of MUC2 resulting in a 4- to 5-fold expansion in volume. This layer is easily removed, and although it has a less defined outer border, it has been reported to be approximately 700 μm in thickness.17–19 In contrast to the colon, the small intestine has only 1 layer of mucus.18 However, the small intestine does exhibit more pronounced release of Paneth cell-derived antimicrobial peptides including cathelicidins, defensins, and bactericidal lectins such as RegIII proteins that help to maintain a relative bacterial sterility on the epithelial surface.2,20,21 Expression of antimicrobial peptides is mostly reduced in IBD. A possible influence of IBD candidate genes on antimicrobial peptide expression was suggested by the decrease in human defensin 5 and human defensin 6 peptides that occurred in both adult and pediatric ileal patients with CD particularly in those with mutations in the CARD15 gene that encodes the bacterial-sensing protein, NOD2.2 In contrast, the cathelicidin LL-37 was observed to be upregulated in active and quiescent UC but not in CD.22 An additional link between antimicrobial peptides and IBD genetics was also identified in one study that reported that a SNP in the gene locus of the C-type lectin, CLEC16A, was associated with patients with CD harboring 3 established CD-associated NOD2/CARD15 mutations.23 Due to space limitations and the scope of the topic, IBD candidate genes involved in Paneth cell function (i.e., KCNN4, XBP1, ATG16L1, etc.) will not be highlighted in this article. Many excellent articles have covered the roles of Paneth cell dysfunction and associated candidate genes in considerable depth, and the reader is referred to a selection of those articles.2,3,24
Secretory IgA
Another important component of the intestinal barrier involves enterocyte transcytosis of secretory immunoglobulin A (sIgA) into the lumen through the polymeric immunoglobulin receptor, which binds to the dimeric (or polymeric) form of IgA on the basal membrane of intestinal epithelial cells (IECs).25 IgA is the most abundant antibody isotype produced in the body and provides mucosal protection through interaction with the pIgR.26 The sIgA promotes the clearance of antigens and pathogenic microorganisms from the intestinal lumen by impeding access to epithelial receptors, thus leading to their entrapment in mucus, and subsequent removal by peristaltic and mucociliary activities. This mechanism has been referred to as “immune exclusion.” Moreover, sIgA has been shown to negate bacterial virulence factors, modify composition of the intestinal microflora, and dampen inflammatory responses to allergens and pathogenic bacteria.9,27–30
Electrolyte and Fluid Transport
Epithelial electrolyte and fluid transport plays a crucial role in the maintenance of intestinal homeostasis.13 As previously mentioned, fluid secretion is required for dispersal of mucus, flushing of intestinal crypts, and dispersal of Paneth cell secretions and antimicrobial peptides. The overall balance of fluid and electrolyte absorption and secretion is fundamental for pH regulation of the luminal microenvironment and can also influence composition of the microbiome.14,31 Moreover, loss of transporters such as sodium-hydrogen exchanger 3 or the Cl-/HCO3-exchanger, downregulated in adenoma, in mouse intestine can alter baseline inflammatory status and predispose to more severe injury after challenge.32–36 Regarding genetic links between electrolyte transporters and IBD, there is 1 report of a SNP in the KCNN4 gene that is associated with ileal CD.37 This gene encodes a calcium-activated K+ channel (KCNN4) that is widely distributed in IECs and has many important transport functions. These include recycling of K+ across the basolateral membrane during colonocyte Cl− secretion, mediating colonocyte apical K+ secretion, and an apparent role in regulating Paneth cell secretion of antimicrobial peptides.37,38 KCNN4 (KCa3.1) is also a major calcium-activated K+ channel in T cells, and inhibition of this channel has been shown to have efficacy in limiting T-lymphocyte–mediated murine colitis.39
EPITHELIAL JUNCTIONS
The intestinal epithelium is composed of a single layer of polarized columnar epithelial cells that are laterally affixed to adjacent epithelial cells by apicolateral tight junctions, adherens junctions, and desmosomes located toward the basal aspect of lateral space.40,41 Although formation of the adherens junctions is essential for appropriate polarization and formation of tight junctions, the tight junctions themselves are the primary regulators of paracellular permeability.42,43 Many excellent review articles have been published describing adherens junctions and tight junctions in great depth, and we refer the reader to a sample of those articles for a more comprehensive appreciation of these structures.41,44–49 Briefly, adherens junctions are formed through interactions between a family of cadherin transmembrane proteins, i.e., E-cadherin that form strong interactions with molecules on adjacent cells such as p120 catenin and β-catenin. These molecules in turn regulate local actin assembly and perijunctional actomyosin ring development.41,50 Adherens junctions are required for assembly of the tight junction, which seals the paracellular space. Tight junctions are composed of at least 2 functionally distinct pathways. First, there is a high-capacity, charge-selective pore pathway that allows passage of small ions and uncharged molecules. In addition, there is a low-capacity leak pathway that permits the flux of larger ions and molecules, regardless of charge. Therefore, the tight junction operates as a selectively permeable barrier, and this seems to operate independently of the number of tight junction strands based on comparative studies in Madin-Darby canine kidney (MDCK) epithelial cell clones with markedly different electrical resistances but identical tight junction strand numbers.49,51 Tight junctions are comprised of integral membrane-spanning proteins such as members of the claudin family, occludin, and immunoglobulin superfamily members such as the junctional adhesion molecules. A variety of other tight junction proteins including integral membrane, peripheral membrane tight junction proteins, and signaling proteins, including a number of kinases involved in tight junction regulation, have also been identified.52,53 A critical feature of these proteins is that many interact with the actomyosin perijunctional ring that forms the key stabilizing structure necessary for tight junction and adherens junction integrity. Arguably, the most important of the transmembrane proteins are members of the claudin family, because they define various aspects of tight junction permeability in a tissue-specific manner. The great variety of claudin family members lends great flexibility to the composition of the tight junctions and their overall influence on tight junction function. For example, although one might intuitively consider expression of a leaky claudin as detrimental to barrier function, loss of expression of claudins 2 and 15 in mice caused deficient Na+ paracellular flow and nutrient uptake, resulting in death from malnutrition.54
Occludin is another integral transmembrane tight junction protein, and it can interact directly with claudins and actin. The role of occludin in tight junction regulation is less clearly understood as occludin knockout (KO) mice show no obvious defect in barrier function.55 However, occludin has been implicated as a protective factor in barrier regulation in a number of studies.56–59 Peripheral membrane proteins, such as zonula occludens 1 and 2 (ZO-1, ZO-2), are crucial to tight junction assembly and maintenance. This is due in part to their multiple domains that facilitate interaction with other integral or regulatory proteins, including claudins, occludin, and actin and thus allow them to act as scaffolding proteins.60–62 Furthermore, these proteins are dynamic and a typical marker of barrier dysfunction altered membrane localization or internalization of tight junction proteins, with or without changes in overall expression.56 Therefore, appropriate localization of tight junction proteins is an essential factor in epithelial homeostasis. Disruption of epithelial barrier function can therefore affect many key physiological functions of the intestine, from protection against infection, to nutrient uptake to appropriate hydration. Thus, it should come as no surprise that a compromised epithelial barrier is a feature of many chronic intestinal conditions including IBD, celiac disease, and type 1 diabetes.63
BARRIER DYSFUNCTION IN IBD
CD and UC, collectively referred to as IBD are believed to arise from disrupted intestinal homeostasis caused by a combination of genetic susceptibility, altered immune interactions with intestinal microbiota, and environmental factors that can initiate or reactivate disease.4,64 A number of studies in patients with IBD have identified that intestinal barrier function is disrupted both in active and in quiescent disease states.65 Indeed, landmark studies identified that increased intestinal permeability to inert tracer molecules such as PEG molecules or 51Cr-EDTA occurs before inflammation and can predict disease relapse in patients with CD.66,67 Moreover, increased permeability has also been documented in siblings of patients with IBD while there is also a case report of an IBD sibling who had increased intestinal permeability without any symptoms of IBD but later went on to develop CD.67–70 Additional methods used to quantify intestinal barrier function in humans have used measuring the intestinal uptake and urinary excretion of the inert sugars probes such as lactulose and mannitol. A number of studies have shown an increase in the lactulose:mannitol ratio in patients with IBD indicative of barrier defects that reflect abnormal tight junction permeability. Increased permeability to lactulose is associated with elevated permeability of the small intestine because lactulose is broken down by colonic bacteria and therefore is not a marker for colonic permeability.71,72 Functional measures of increased intestinal permeability were supported by freeze fracture electron microscopy studies showing a reduction in the overall number of tight junction contacts in addition to disjointed tight junction strand patterns. In addition, another well-documented feature of epithelial tight junctions in tissues from patients with IBD is the altered expression or localization of claudin proteins and occludin in mild to moderately active IBD.73–75
Studies in a number of animal models of IBD have also provided strong supporting evidence for increased intestinal permeability preceding the onset of intestinal inflammation, thus suggesting a prominent role early in disease development. Madsen et al described increased permeability before the onset of intestinal inflammation in the IL-10−/− mouse, whereas a similar phenomenon has been shown in the Samp1/Yit model of Crohn’s ileitis and mdr1a−/− mice.76–78 Intriguingly, increased permeability is necessary but not sufficient for the onset of inflammation as shown in elegant studies using a constitutively active myosin light chain kinase transgenic mouse. Myosin light chain kinase has been shown to represent a final common pathway for activation of barrier “leaks” that are large enough to allow macromolecules to pass through tight junctions between epithelial cells.79–81 These studies support the “two-hit” hypothesis that a barrier defect and a trigger of an inappropriate immune response are required for manifestation of intestinal inflammation.41,81 Indeed, the importance of an intact barrier was further emphasized in studies showing that restoration of barrier function, as assessed by reducing small molecule permeability with the zonulin inhibitor AT-1001, alleviated inflammation in an IL-10−/− mouse model.82 The success of this intervention suggests that this approach of specifically improving barrier function before onset of severe inflammation may have viability as a clinical approach in patients in remission. In addition to established effects in paracellular permeability, there is also evidence that suggests dysfunctional transcellular pathways in IBD. This includes studies on excised intestinal tissues from UC and CD patients mounted in Ussing chambers that demonstrate increased transcellular passage of large molecules such as horseradish perox-idase and ovalbumin.83–85 These studies suggest that increased translocation of luminal contents and bacterial products could be elevated in IBD and thus facilitate increased exposure of lamina propria immune cells to antigenic stimuli that may propagate an inflammatory response. Defects in intestinal barrier function are not restricted to IBD, however, because they are also associated with early stages of disease in celiac disease, type 1 diabetes, and in microscopic colitis, specifically collagenous colitis.63,86
REGULATION OF INTESTINAL BARRIER PROPERTIES BY IBD CANDIDATE GENES
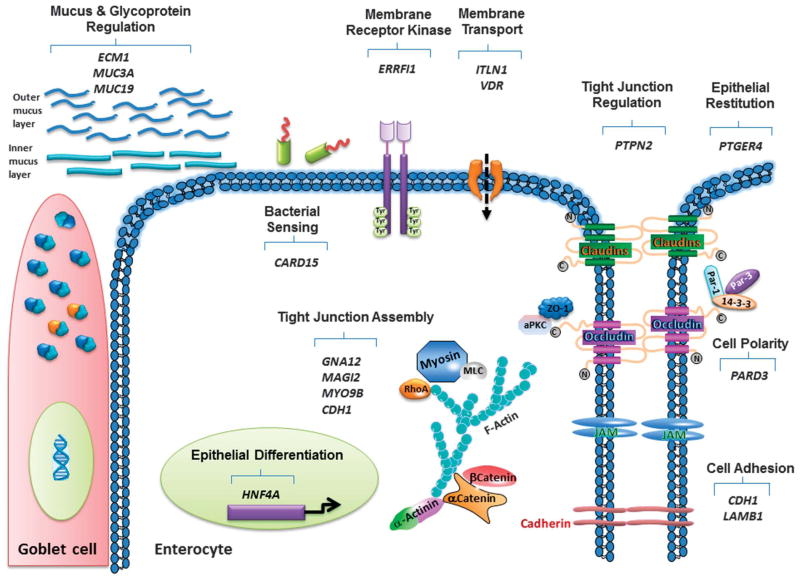
IBD candidate genes are involved in regulating multiple components of the epithelial barrier. Although most of the genes listed have been linked to overall modulation of epithelial and/or intestinal permeability, for the purposes of functional relevance, genes have been broadly grouped according to their primary functions in regulating epithelial properties with the caveat that some genes fall into multiple groups. Genes are presented in Table 1 in the order of their discussion in the text.
TABLE 1.
IBD Candidate Genes Modify Multiple Barrier Components
| Gene | Protein | Function | SNP ID | Locus | CD | UC | Other Diseases | IBD References |
|---|---|---|---|---|---|---|---|---|
| CDH1 | E-cadherin | Major epithelial adherens junction protein | rs12597188; rs10431923; rs9935563; rs1728785 | 16q22 | + | + | GI Cancers; breast cancer | 87–89 |
| LAMB1 | Laminin β1 | Secreted protein involved in adhesion and differentiation | rs886774 | 7q31 | + | Leiomyoma | 87,90 | |
| PARD3 | Par-3 (partition defective-3) | Cell polarity | rs10763976a; rs4379776b | 10p11 | + | Celiac disease; breast cancera | 91–93 | |
| GNA12 | Guanine nucleotide-binding protein α12 | Inhibition of TJ assembly through Src and HSP90 phosphorylation of ZO-1, ZO-2 | rs798502 | 7p22 | + | Schizophrenia | 94,95 | |
| MAGI2 | Membrane-associated guanylate kinase, WW, and PDZ domain-containing protein 2 | Acts as scaffold for assembly of junction proteins. Enhances PTEN dephosphorylation of AKT1 | rs6962966 | 7q21 | + | + | Celiac disease, early; infantile epileptic; encephalopathy | 92,96 |
| MYO9B | Myosin IXB | Actin-based molecular motor | rs1545620; rs2305767 | 19p13 | + | + | Celiac disease | 93,97–99 |
| PTPN2 | TCPTP | Dephosphorylates Tyr residues on kinases. Inhibits IFN-γ induction of claudin-2 | rs2542151; rs1893217; rs7234029 | 18p11 | + | + | Celiac disease; type 1 diabetes | 100–105 |
| HNF4A | Hepatocyte nuclear factor 4α | Transcription factor. Regulates differentiation along crypt-villus axis | rs6017342; rs1884613 | 20q13 | + | + | Mature onset diabetes of the young | 87,106,107 |
| ECM1 | Extracellular matrix protein 1 | Secreted glycoprotein involved in cell proliferation | rs3737240; rs13294 | 1q21 | + | Hyalinosis cutis et mucosae | 108–110 | |
| MUC3A | Mucin 3A | Transmembrane mucin in the protective inner mucus layer | D7S669 | 7q22 | + | + | Epithelial cancers | 111–113 |
| MUC19 | Mucin 19 | Essential secreted mucin in outer mucus layer | rs11175593 | 12q12 | + | Ankylosing spondylitis | 114–116 | |
| ITLN1 | Intelectin-1 | Lectin involved in brush border protection, lactoferrin transport | rs2274910; rs9286879; rs11584383 | 1q23 | + | Type 2 diabetes; Asthma | 117–119 | |
| VDR | Vitamin D receptor | Ca2+ and phosphate absorption; immune regulation | TaqI | 12q12-q14 | + | + | Osteoporosis | 120 |
| PTGER4 | EP4 receptor | Epithelial restitution | rs4495224; rs7720838; rs4613763 | 5p13.1 | + | + | Rheumatoid arthritis | 121–123 |
| CARD15 | NOD2 | Recognition of pathogen-associated molecular patterns, bacterial sensing | 3020insC | 5q31 | + | Graft versus Host disease, Blau Syndrome | 124–126 | |
| ERRFI1 | ErBB receptor feedback inhibitor 1 | Protein product RALT/MIG6 suppresses EGFR signaling | rs35675666 | 1p36 | + | Various cancers | 94,127 |
rs10763976—Strong association with Celiac disease.
brs4379776—Suggestive association with UC.
CELL ADHESION
CDH1
The CDH1 locus at 16q22 comprises 3 genes of which CDH1 is the most plausible candidate gene.87 This locus was identified by the UK IBD Genetics Consortium in 2009 as being associated with UC, and the CDH1 gene encodes the adherens junction protein E-cadherin.88 As mentioned above, E-cadherin plays a critical role in cell–cell adhesion and is also required for tight junction formation. The association with UC was confirmed by McGovern et al in 2010 and in a Dutch cohort by van Sommeren et al.87,89 Muise et al90 identified an association between the CDH1 locus risk haplotypes rs12597188, rs10431923, and rs9935563 with CD. This study also identified that these haplotypes were associated with increased E-cadherin cytoplasmic accumulation in the intestinal epithelium that was likely because of the presence of a novel truncated form of E-cadherin. In complementary in vitro studies using the Caco-2 IEC line and the MDCK renal epithelial cell line, transfection of the truncated E-cadherin in polarized epithelial cells resulted in abnormal intra-cellular accumulation and reduced plasma membrane localization of both E-cadherin and β-catenin. The manifestation of a CDH1 mutation as a functional defect in the form of loss of E-cadherin membrane localization is consistent with our understanding of cytoplasmic internalization of junctional proteins being a common mode of junctional disruption in the setting of inflammation or in response to infection. Evidence for a genetic defect in cadherins and in E-cadherin function leading to altered intestinal homeostasis was generated by studies in mice expressing a dominant-negative N-cadherin, where the function of E-cadherin was perturbed and this resulted in Crohn’s-like inflammation in the small intestine.91
E-cadherin is also a target for invasion by intestinal pathogens such as Candida albicans and Bacteroides fragilis that can cleave E-cadherin. In the case of B. fragilis, this occurs through release of a toxin called fragilysin that cleaves the extracellular domain of E-cadherin thus disrupting adherens junctions and compromising TJ integrity.92 Moreover, Listeria monocytogenes is capable of binding to E-cadherin through the surface protein internalin-A that interacts with E-cadherin and triggers the same signaling cascade induced by homotypic E-cadherin interactions, including the recruitment of α- and β-catenin, myosin VIIa, and the AJ-associated protein vezatin at bacterial sites of entry. Regarding patients harboring CDH1 mutations, defective E-cadherin localization could conceivably facilitate bacterial penetrance across the epithelium through generalized AJ and TJ defects rather than specific targeting of the extracellular domain of E-cadherin as an entry portal because it is likely internalized in these patients and therefore not as accessible as wild-type E-cadherin.
A study by Demetter et al investigating E-cadherin expression at various sites in active IBD indicated a complex pattern of expression where E-cadherin was upregulated in active IBD at sites of focal inflammation and in sites adjacent to ulcerated areas but was actually reduced in areas of ulceration.93 Although it has been speculated that downregulation of E-cadherin expression in cells located at the margins of mucosal ulceration could be an important part of epithelial restitution and repair as cell migration is facilitated by reduced cell adhesion, however, this remains to be verified.5,94 Furthermore, CDH1 mutations are associated with increased proliferation, invasion, and metastasis of multiple epithelial tumors including gastric and colorectal cancer.95–98 The separate associations of CDH1 with IBD and colorectal cancer are intriguing given the significantly higher risk of patients with IBD for the development of colorectal cancer.99,100 Moreover, promoter methylation of the CDH1 locus has been associated with dysplasia in patients with UC. Therefore, epigenetic regulation of the CDH1 promoter by methylation may potentially serve as a biomarker to identify patients at high risk of progression to colorectal cancer.5,101
LAMB1
A UC-specific susceptibility locus was identified in a region on chromosome 7q31, indicating the LAMB1 gene as the strongest corresponding candidate.88,102 The UK study observed peak association at rs886774, whereas the North American study reported a cluster of LAMB1 SNPs at 7q31 with the rs4598195 SNP having the highest association.88,102 However, a study in a Dutch cohort of patients with UC failed to establish an association of the rs6949033 SNP (after correction).87 LAMB1 encodes the laminin β1 subunit, a light chain present in the heterotrimers laminin-1, laminin-2, and laminin-10. Laminins represent the most abundant glycoprotein component of the basement membrane. These αβγ heterotrimeric molecules interact with integrins to create a key cell adhesion network in the intestinal epithelium.103,104 Consequently, laminins are involved in multiple cell functions including adhesion, migration, differentiation, and survival.105
Laminin expression occurs in a heterogeneous manner because segregated expression of laminins has been reported along the crypt-villus axis in human small intestinal basement membrane, with laminin-10 occurring in the upper-crypt/villus and laminin-2 expressed deeper in the crypts.5,106,107 In vitro studies identified that laminin-10 promotes crypt-villus differentiation as assessed by expression of the epithelial brush border enzyme, sucrose-isomaltase.108 In IBD, laminin expression seems to be increased in serum in both UC and CD, and this correlates positively with disease activity.109 Intriguingly, laminin levels in the epithelial basement membrane of colonic crypts in inflamed UC was reduced.110 Although the rs886774 variant of LAMB1 was not associated with CD, a previous study identified altered expression profiles of laminins in ileal crypts of patients with CD with reduced laminin-2 but increased laminin-5 and laminin-10.111,112 These data further suggest heterogeneous modes of regulation of laminin expression in response to inflammatory cytokines. Indeed, regulation of laminin expression in an inflamed environment seems to be quite complex. In vitro studies using the normal embryonic human IEC line, HIEC-6, demonstrated that the proinflammatory cytokines TNF-α and IFN-γ can synergistically potentiate laminin-5 and -10 secretion from these cells and this is in contrast to the overall reduced laminin expression in active UC mentioned above.110,113 The human UC study may suggest that loss of laminin from basement membrane could contribute to overall barrier impairment in UC but in addition that the appearance of laminin in the serum may serve as a biomarker for intestinal barrier dysfunction, although this requires confirmation. LAMB1 also shows a genetic association with uterine leiomyoma (fibroids) with altered expression of LAMB1 identified in the leiomyomatous region, thus suggesting a role for LAMB1 in the genesis and development of leiomyoma.114 Of greater relevance to intestinal cancers is that serum laminin has been identified as a predictive marker in colorectal cancer.115
CELL POLARITY
PARD3
Wapenaar et al uncovered a suggestive association between a cell polarity gene, PARD3, and UC having made a clear association of this gene with celiac disease.116 The SNPs in the PARD3 gene differed between the conditions with rs10763976 significantly associated with celiac disease and rs4379776 falling just short of significance.116 In contrast, Wolters et al117 found no association between PARD3 and either UC or CD in a Canadian cohort. Par-3 regulation of tight junctions was apparent from a study showing that altered Par-3 polarization in vitro or in biopsies from patients with celiac disease was associated with increased expression of pore-forming claudins like claudin-2 and a reduction in the sealing claudins, 3, 5 and 7, and reduced membrane localization of the tight junction-associated regulatory protein ZO-1.118 Interestingly, there was no increase in apoptosis in epithelial cells with impaired Par-3 localization. Although the association of SNPs in the PARD3 gene is stronger with celiac disease rather than IBD, the functional studies on the role of the PARD3 protein product, Par-3, in regulating tight junction protein organization provide valuable insights into the consequences of dysregulation of these genes involved in epithelial cell polarity for intestinal barrier function in disease. Par3 is thought to be a scaffolding protein required for the recruitment of the binding complex of an additional PDZ containing protein, Par6, with an atypical protein kinase C (aPKC) isoform that activates Par6, to sites where its activity is required. For example, Par3 binds directly to junctional adhesion molecules, and this recruits it to the tight junctions in simple epithelial cells.119 In simple epithelial cells, the Par3/Par6/aPKC complex is localized to tight junctions, and it can be recruited to these structures either by the interaction of Par3 with junctional adhesion molecule-1 or by binding of Par6 to the Crumbs (Crb) transmembrane protein complex.119–122 In contrast to Par6, studies in keratinocytes did not implicate a role for Par3 in epithelial wound healing.120 However, a Par3-aPKC complex can also be recruited to the leading edge of migrating MDCK II epithelial cells through association with the multi-PDZ protein PATJ (PALS1-associated TJ protein) and in regulating the polarization of microtubule organizing centers during wound healing.123 Moreover, the Par3-Par6-aPKC polarity complex interacts with the TJ protein occludin to facilitate directional movement at the leading edge of epithelial cells engaged in wound repair.124 This suggests that mutations in PARD3 may also affect the signaling outcomes of occludin-regulated events and compromise wound repair thus weakening the recovery response of the epithelial barrier to injury or inflammation.
TIGHT JUNCTION ASSEMBLY
GNAI2
Heterotrimeric G proteins regulate fundamental cellular properties through their Gα and Gβγ subunits.125 In addition to their well-recognized roles in signaling from plasma membrane receptors, G proteins have been identified as having roles in TJ assembly and barrier function regulation.126 Gα12 is the protein product of GNA12 and is a membrane-bound GTPase that has been shown to directly interact with the TJ accessory protein ZO-1.127 Follow-on studies showed that Gα12 activates Src kinase through a mechanism that uses the heat shock protein 90 (Hsp90) chaperone.128 Src activation causes phosphorylation of ZO-1 and ZO-2, resulting in loss of claudin 1 and occludin interactions with ZO-1. They also showed that endogenous Gα12 was activated during TJ assembly in the absence of receptor agonists, and that thrombin stimulation of endogenous Gα12 can play a role in thrombin-induced permeability increases in intact MDCK monolayers and in delaying TJ assembly after calcium-switch assays.128 GNA12 (rs798502) was identified as the most likely candidate at the 7p22 locus that exhibited a significant association with UC by Anderson et al.129 Although the functional studies cited above suggest that mutations in GNA12 may contribute directly to an epithelial barrier defect, the contribution of GNA12 mutations to UC pathogenesis may also extend to dysregulation of adaptive immune function. Herroeder et al showed that T-cell-specific inactivation of Gα12 and Gα13 in vivo resulted in an increased activity of integrin leukocyte function antigen-1 in murine CD4+ T cells and lymphadenopathy because of increased lymph node entry and enhanced T-cell proliferation. In addition, they observed increased susceptibility of T-cell Gα12 and Gα13 double-deficient mice toward 2 models of T-cell–mediated pathology: contact hypersensitivity to dinitrofluorobenzene as a model for responses to foreign antigen and low-dose streptozotocin-induced diabetes mellitus as a model of autoimmune disease.130
MAGI2
The first study identifying a significant association of the MAGI2 gene with UC was made by Wapenaar et al116 in 2008 in a Dutch cohort. This study had the added significance of identifying a risk association of this gene in celiac disease, and thus an additional shared association between these 2 intestinal inflammatory conditions. The shared genetic overlap between IBD and celiac disease is well recognized, and patients with celiac disease, or gluten sensitive enteropathy, have an approximately 5-fold increased risk of developing IBD.116,131–133 The Wapenaar study focused on 41 genes from the tight junction pathway, including those encoding transmembrane, adaptor, signal transduction, and transcriptional regulatory proteins.116 The association of MAGI2 with both UC and celiac disease occurred at rs6962966 suggesting a similar functional defect in this gene in both conditions. In conflict with these findings, and similar to their data with PARD3 in the same cohort, Wolters et al117 found no association between MAGI2 and either UC or CD. McGovern et al made a very comprehensive study of the MAGI2 gene and its possible association with both CD and UC.134 They genotyped 113 MAGI2 SNPs in 681 cases of CD, 259 cases of UC and 195 controls. The most significant IBD association was in intron 6 (rs2160322) and both UC and CD contributed to this association. This article also made the first associations of MAGI2 with CD in contrast to Wapenaar et al116 who found no significant association with CD. The most significant CD association was with an intron 2 haplotype (rs7785088/rs323149/rs13246026). They also observed highly significant associations with UC in intron 6 (rs7803276/rs7803705) and also significant associations in introns 2, 6 and 20. In assessing links with other IBD biomarkers, they identified significant associations with IgG ASCA, anti-CBir1, and anti-OMPc positive CD. The UC-associated SNP identified by Wapenaar was not tested in the McGovern paper.116,134 The MAGI2 gene is roughly 1.4 megabases consisting of 21 exons that encode a protein of 2410 amino acids, MAGI-2, also known as synaptic scaffolding molecule.134 The authors concluded that they likely identified SNPs associated with disease rather than actual “causative” polymorphisms.134
The MAGI-2 scaffolding protein contains 9 potential protein–protein interaction domains. These include a guanylate kinase-like (Guk) domain, 6 PDZ domains (modular protein interaction domains that bind in a sequence-specific manner to short C-terminal peptides), and 2 WW domains that mediate protein–protein interaction by recognition of proline rich peptide motifs and/or phosphoserine/phosphothreonine containing motifs.135 MAGI-2 is involved in epithelial tight junction assembly.136 Consequently, a defect in MAGI-2 expression or function may contribute to disassembly of protein networks involved in tight junction or adherens junction function with resultant barrier defects. The association of MAGI2 SNPS with immunophenotypic subgroups of CD is intriguing, and the loss of function of this structural scaffolding protein and consequent compromise in barrier properties against intestinal microbes, may explain the association between MAGI2 SNPs and antibodies to microbial antigens.134 In addition, MAGI-2 can inhibit cell migration and proliferation by increasing the stability of the enzyme PTEN (phosphatase and tensin homolog) after binding.137,138 PTEN plays a critical role in the regulation of cell growth and apoptosis, and defects in apoptosis have been implicated in IBD pathogenesis.139 Therefore, in addition to effects on junctional assembly, alterations in the balance of epithelial cell growth versus apoptosis may represent an additional consequence of MAGI2 gene mutations associated with IBD.
MYO9B
The MYO9B gene encodes a member of the class IX myosin family of actin-based molecular motor heavy chain proteins. The MYO9B protein contains 4 Isoleucine–glutamine motifs located in the neck domain that bind calmodulin, which serves as a light chain.140,141 The MYO9B complex has a single-headed structure, and it exhibits processive movement on actin filaments. The protein also has rho-GTPase activity that converts active Rho-GTP into inactive Rho-GDP, thereby downregulating Rho-dependent signaling pathways.142 Rho family GTPases have been shown to play important roles in regulating actin filament remodeling and tight junction integrity.52,143,144 In addition to maintaining regulation of paracellular permeability in established TJs, they also have roles in the assembly of intercellular associations and recovery of barrier function.145 Therefore, mutations in MYO9B could affect multiple facets of myosin and actin filament regulation of barrier function. The initial connection between MYO9B and impaired barrier function in chronic intestinal inflammatory disease was made by Monsuur et al146 when they associated MYO9B mutations, in particular, rs2305764 (intron 28) with increased risk of celiac disease. However, a number of smaller follow-up studies failed to show MYO9B association with celiac disease in different European cohorts.147,148 The proposed site of genetic variation on chromosome 19p13 also contains an IBD linkage locus (IBD6). Although a number of studies have investigated the association between MYO9B SNPs and IBD, results have been conflicting. Van Bodegraven et al149 identified association of a nonsynonymous coding SNP (rs1545620) in UC and less strongly in CD across 3 cohorts of European descent (United Kingdom, Dutch, Italian/Canadian). A weaker association with the MYO9B SNP (rs2305764) was also observed. In contrast, Cooney et al150 found that the coding rs1545620 SNP had no significant association with CD or UC, however, a noncoding intronic SNP (rs2305767) had significant association with CD only. Interestingly, evidence of an interaction between multiple MYO9B SNPs and another IBD candidate gene, IL-23R, was observed, whereas epistasis was not observed with ATG16L1, CARD15 genes, or the IBD5 haplotype.150 Further corroboration for an association of 3 MYO9B SNPS (rs2305764, rs2305767, and rs1457092) with IBD and UC but not CD was determined in a Spanish cohort.151
A study in a Canadian cohort found no association of MYO9B SNPs with UC but identified an association of 3 MYO9B SNPs, with rs1457092 being the most prominent, with ileal CD only. Intriguingly, the odds ratio association was in the opposite direction to those identified in the previous studies. By way of possible explanation, Wolters et al117 proposed that these non-GWAS studies may have been underpowered. An additional concern raised by the authors was that none of the MYO9B variants were associated in GWAS.129,152 This suggests that if the disease association with MYO9B is not a false positive, it must be associated with a subphenotype that was not part of the large-scale GWAS. However, the authors advocated that regardless of the direction of the association of MYO9B between the different IBD cohorts discussed, the data from these smaller genetic studies do suggest a possible role for MYO9B in IBD pathogenesis. Functional studies indicated a correlation between MYO9B protein expression and intestinal permeability in patients with type 1 diabetes. This seemed to be related to elevated levels of the protein zonulin in patients with diabetes. Zonulin can promote TJ disassemby and increased levels correlated with increased intestinal permeability in vivo and with an increased MYO9B gene expression in intestinal tissue.153,154 This suggested a possible link between MYO9B expression and intestinal permeability changes that were also supported by data identifying a trend towards abnormal intestinal permeability in patients with CD carrying the rs1545620 risk allele.155,156 More recent studies also indicate a key role for MYO9B in TJ integrity and wound healing. In wounded monolayers of the Caco2BBe IEC line, MYO9B protein localized to the leading edge of lamellipodia of migrating cells. Loss of MYO9B expression by RNAi, or by transfection with a dominant-negative MYO9B C-terminal, results in a decrease in lamellipodia and failure of cells to migrate into the wound.157 These cells also exhibit mislocalization of ZO-1, occludin, and claudin-1 TJ proteins, and increased TRITC-dextran permeability as determined in a microscopy-based assay, thus indicating a barrier defect. Although the possible role of MYO9B in barrier dysfunction in IBD remains to be conclusively demonstrated, however, the provoking findings from genetic studies and indicators from functional studies in other chronic inflammatory diseases make it an attractive candidate for further investigation.
TIGHT JUNCTION REGULATION
PTPN2
The protein tyrosine phosphatase nonreceptor type 2 (PTPN2) gene locus represents the strongest candidate at the IBD-associated locus 18p11. The first identification of PTPN2 as an IBD candidate gene was made in the landmark WTCCC article in 2007.152 The rs2542151 SNP was significantly associated with CD, and this SNP has been the most widely identified PTPN2 SNP associated with IBD. The PTPN2 association with CD was confirmed by a number of studies while Waterman et al observed an association between the rs2542151 SNP and colonic CD thus providing some insight into the association of PTPN2 with inflammation in different intestinal regions.111,129,158–160 Franke et al161 made the initial association of the rs2542151 PTPN2 SNP with UC, although in their study they only observed a weak association of this SNP with CD. Additional insight into associations between PTPN2 SNPs and disease outcomes was provided by Glas et al162 In this study, genotype–phenotype analysis could not identify a clear phenotypic outcome associated with PTPN2 SNPs; however, there was a potential association between the rs7234029 SNP with a stricturing disease phenotype in patients with CD and weak associations of the same SNP with an early onset of CD and UC.162 In silico analysis, it is predicted that the increased CD risk mediated by rs7234029 may be related to a stronger activation of proinflammatory transcription factors such as NF-κB, C/EBP, and E4BP4. A weak association was described between PTPN2 and the autophagy gene, ATG16L1, being a strong IBD candidate gene. Although a functional interaction between these genes and their protein products has not been confirmed, PTPN2 has been shown to play a role in autophagosome formation in IEC lines.163 Furthermore, the rs1893217 SNP, an intronic noncoding SNP that is in complete linkage disequilibrium with the GWAS lead SNP rs2542151, has been shown to exert a functional defect in muramyl dipeptide signaling and autophagosome formation in fibroblasts isolated from patients with CD harboring this SNP.164 This suggests that genes integral to particular pathways may actually represent points of convergence between multiple IBD-associated physiological events and thus increase patient susceptibility through a number of routes. The convergence of PTPN2 with other IBD-associated genes was also highlighted in a study by Yu et al165 who identified association of a putative protective PTPN2 SNP (rs2542152) with CD but also showed that PTPN2 in B-lymphocytes and intestinal tissues was regulated by the IBD-associated transcription factor gene, NK2 transcription factor related, locus 3 (NKX2-3) in vitro. Moreover, the authors also described the presence of putative NKX2-3 binding sites upstream of the PTPN2 transcription start site. The nature of the proposed interaction between NKX2-3 and its regulation of PTPN2 remains to be determined; however, NKX2-3 KO mice exhibit disturbed small intestinal development and reduced epithelial proliferation.165,166
The protein product of PTPN2 is the enzyme T-cell protein tyrosine phosphatase (TCPTP), so called because it was originally cloned from a T-cell cDNA library, although it is expressed ubiquitously.167,168 Colonic TCPTP expression is increased in active CD and particularly so in the epithelium.169–171 A study of potential causal variants in the PTPN2 gene locus identified that 5 associated intronic SNPs affected TCPTP protein expression and transcription factor binding.169 This included rs2542170, rs2847288, and rs1893217, which showed the strongest association in genotyped patients tested in this study. Given that TCPTP expression is upregulated in CD, this may suggest that levels of TCPTP are either insufficient to adequately regulate signaling substrates in an inflamed environment, or that TCPTP enzymatic activity is functionally compromised. The rs1893217 SNP is associated with both CD and UC, but was originally strongly associated with type 1 diabetes.129,172 In addition, this SNP was associated with early-onset pediatric CD.173 PTPN2 SNPs have also been associated with celiac disease thus emphasizing the potential for disruption of major signaling pathways in multiple chronic inflammatory diseases through shared genetic mutations.174 A striking feature of the pathology of these 4 chronic inflammatory diseases, CD, UC, type 1 diabetes, and celiac disease, is that they all feature increased intestinal permeability before the onset of inflammation.71
Two functional variants of TCPTP are expressed that arise from alternative splicing of PTPN2 message. The 48 kD form is targeted to the endoplasmic reticulum by a hydrophobic C-terminus, whereas the 45 kD variant lacks the hydrophobic C-terminus and is targeted to the nucleus by a bipartite nuclear localization sequence that is also expressed in the 48kD form, but this sequence is masked by the hydrophobic C-terminus.175,176 The 45 kD form can exit the nucleus in response to a variety of stimuli to access substrates in the cytoplasm and at the plasma membrane.177 TCPTP substrates include: (1) receptor protein tyrosine kinases such as the epidermal growth factor receptor (EGFR) and the insulin receptor; (2) non-receptor protein tyrosine kinases such as members of the Src family and JAK-1 and -3; and (3) Protein tyrosine kinase substrates such as p52Shc and signal transducers and activators of transcription (STAT)-1, -3, -5 and -6.177–183 Mice globally deficient for Ptpn2 develop a systemic inflammation and die at approximately 5 weeks of age from severe anemia, hematopoietic defects, and the development of progressive systemic inflammatory disease, characterized by increases in circulating proinflammatory cytokines, splenomegaly, diarrhea, and lymphocytic infiltrates in nonlymphoid tissues.184,185 However, features of the inflammation in Ptpn2-deficient mice are strain-dependent.186 Bone marrow chimeric studies indicate that the loss of Ptpn2 in the nonhematopoietic compartment is the critical factor in the severe inflammation these mice develop.185 Furthermore, heterozygous mice lacking 1 Ptpn2 allele show increased susceptibility to challenge with dextran sulfate sodium (DSS).187
Regarding the role(s) of PTPN2/TCPTP in the regulation of barrier function, we have previously shown that knockdown of TCPTP in IECs facilitates a greater barrier defect induced by the proinflammatory cytokine, IFN-γ.171 This was associated with expression of the cation-selective claudin-2 pore that mediates increased paracellular passage of sodium ions. Furthermore, TCPTP-deficient cells also exhibited an enhanced permeability to 10 kD FITC-dextran suggesting additional mechanisms involved in regulating a leak barrier defect, as distinct from a claudin-2 pore effect, are also likely affected. Although TCPTP regulates a number of substrates, it does seem to exert a discreet level of control depending on the signaling context. Specifically, TCPTP regulation of EGFR phosphorylation was studied in response to direct versus indirect activation of the EGFR by epidermal growth factor (EGF) and IFN-γ, that can transactivate the EGFR, respectively.188 Knockdown of TCPTP in T84 colonic epithelial cells accentuated EGF but not IFN-γ-induced EGFR tyrosine residue phosphorylation and enhanced EGF inhibition of Ca2+-stimulated chloride secretion.189 Therefore, this capacity of TCPTP to discriminate between different modes of activation of the same substrate may be of particular consequence about identifying the functional consequences of IBD-associated PTPN2 SNPs. Indeed, this will likely be of great importance in studying the effects of PTPN2 SNPs on signaling events initiated by IBD-associated inflammatory cytokines such as IFN-γ and TNF-α that disrupt barrier function and whose activity is negatively regulated by TCPTP.56,170,171,190
EPITHELIAL DIFFERENTIATION
HNF4A
The gene locus for Hepatocyte nuclear factor 4-alpha (HNF4A) has been positively associated with UC in a number of GWAS. The rs6017342 SNP had the strongest new association in the large Wellcome Trust Case Control Consortium 2 (WTCCC2) study.88 Furthermore, rs6017342 mapped within a recombination hotspot on chromosome 20q13, containing the 3′ untranslated region of the HNF4A gene alone.5 This SNP was also strongly associated with UC in a Dutch cohort.87 Although common variants of HNF4A have previously been shown to affect the risk of type 2 diabetes (rs2144908) and polygenic dyslipidemia (rs1800961), rs6017342 was not in linkage disequilibrium with either of these.152,191 Although HNF4A was not found to be associated with CD in the original WTCCC study, Marcil et al192 identified 6 HNF4A SNPs that were associated with a Canadian pediatric CD cohort, whereas the rs1884613 SNP was significantly associated with both the discovery cohort and a second distinct cohort after correction for multiple testing. This study concluded that the HNF4A gene locus may be a common genetic determinant of childhood-onset CD.
The HNF4A gene has previously been identified as being associated with the disease, maturity onset diabetes of the young. Maturity onset diabetes of the young is a genetically heterogeneous monogenic form of noninsulin-dependent (type 2) diabetes mellitus, characterized by early onset, usually before 25 years of age and often in adolescence or childhood and is distinct from type 1 or juvenile diabetes.193 The HNF4A gene belongs to the nuclear hormone receptor family of ligand-dependent transcription factors, and it encodes the transcription factor hepatocyte nuclear factor-4 alpha, which is expressed in the liver, pancreas, kidney, stomach, and intestine.194 It has a crucial role in mammalian development of the gastrointestinal tract, and expression is found in both absorptive and secretory cell lineages of the intestinal epithelium.195 Hnf4a−/− mouse embryos die before embryonic day (E) 10.5 and show arrested and disrupted gastrulation with the primary defect thought to be impaired visceral endoderm function.196 KO mice lacking functional HNF4α in the developing colon were generated to circumvent embryonic lethality, and although these mice were subject to neonatal lethality, recovery of colonic tissue at E18.5 showed no expression of HNF4α.195 Histologically, these tissues showed absent crypt formation and reduced formation of smooth muscle. However, there was no alteration in transition from pseudostratified to simple columnar epithelium in the Hnf4a KO mice. In support of the development of a normal epithelium, the cell-adhesion protein, E-cadherin, was readily detected between adjoining epithelial cells regardless of the presence of HNF4α with only a modest reduction occurring in Hnf4a KO mouse intestine. This is in contrast to the fetal liver, where loss of HNF4α results in a dramatic reduction of E-cadherin expression.197 Interestingly, although laminin extended throughout the mucosa occupying the spaces between the crypts of control mouse colons, in Hnf4a-null colons, laminin was still clearly detectable but was organized as a lining at the base of the epithelium.195 The authors concluded that the altered localization of laminin was a consequence of defective crypt formation rather than complete absence of the lamina propria.195 This may have relevance to patients with IBD with mutations in HNF4A and an additional IBD candidate gene, LAMB1 (discussed below), that encodes for the laminin β1 subunit. In addition, colonic tissue from Hnf4a-null mice exhibited a reduction in enterocyte cell numbers due to impairment of proliferation and also in goblet cell numbers. The observed loss of goblet cell morphology likely reflected disrupted maturation of mucus granules as opposed to a significant loss of expression of secretory mucin core protein genes. Conditional KO of Hnf4a in IECs using villin-Cre transgenic mice avoided embryonic and neonatal lethality but increased susceptibility to chemical colitis and increased intestinal permeability after treatment with DSS.198 A separate study using a conditional tamoxifen-inducible Hnf4a-villin-Cre KO mouse showed that loss of HNF4α in the small intestines altered Wnt/b-catenin signaling and the balance of crypt proliferation and differentiation. This resulted in increased IEC crypt proliferation, increased permeability, distended tight junctions, and altered tight junction and adherens junction formation. This was associated with decreased ZO-1, claudin-4, and claudin-7 mRNA expression, increased claudin-2 and E-cadherin mislocalization to the cytoplasm.199 Another important function of HNF4A is its role as a transcriptional regulator of ion transporters. Darsigny et al200 demonstrated that loss of HNF4α in mice initiated loss of mucosal homeostasis through a decline in mucosal ion transport rather than the expected increase in permeability. The loss of mucosal homeostasis triggered a chronic inflammatory response in the colon of the mice and a more severe degree of tissue damage in experimental colitis. This served to emphasize the importance of ion transport function in regulating overall intestinal inflammatory status. These mice also displayed increased goblet and enteroendocrine cell numbers in contrast to observations in the developing colon described by Garrison et al.195,201 Regarding patient studies, Ahn et al198 described a decrease in HNF4A in colonic tissues from UC and CD patients using a commercially available IBD tissue qPCR array panel. This correlated with their findings of decreased HNF4α expression in murine DSS colitis.
HNF4A is also linked to colon cancer as 3 SNPs in the human HNF4α protein, 2 of which are in the HNF4α F domain that interacts with the Src kinase SH3 domain, increase phosphor-ylation by Src tyrosine kinase. Src activation is associated with the early stages as well as progression and metastasis.202 The 3 HNF4A SNPs can decrease HNF4α protein stability and function, suggesting that individuals with those variants may be more susceptible to Src-mediated effects.203 Therefore, although HNF4A SNPs may influence the onset of both UC and CD through disruption of mucosal homeostasis through effects on IEC crypt formation and differentiation, as well as mucosal permeability and ion transport, they may in addition further increase the susceptibility of patients with IBD to the development of colorectal cancer.
MUCUS AND GLYCOPROTEIN REGULATION
ECM1
A number of IBD candidate genes have been identified that are involved in glycoprotein regulation. The 1q21.2 gene locus spans 290 kb and contains the gene encoding extracellular matrix protein 1 (ECM1). This gene locus was identified as being selectively associated with UC rather than CD with the rs3737240 and rs13294 SNPs reaching significance.204,205 Further confirmation that the rs13294 SNP in the ECM1 locus was selective for UC was provided in a study of a Dutch patient cohort.206 Regarding the biology of ECM1 and its role in the mucosal barrier, ECM1 has been best characterized in the skin. ECM1 is a secreted glycoprotein that plays an important role in the structural and functional biology of the skin because loss-of-function mutations in ECM1 can lead to the development of genodermatosis lipoid proteinosis. This condition is characterized by reduplication of the skin basement membrane and hyalinization of the underlying dermis.207 ECM1 has been proposed to act as a “biological glue” in the dermis, helping to regulate basement membrane and interstitial collagen fibril macroassembly and growth factor binding.208 Although the function of ECM1 in the intestine is not as well understood, it is possible that it may have a role to play in gut epithelial basement membrane integrity and function. The basement membrane is a specialized structure composed of extracellular matrix components that separates the epithelium from the underlying connective tissue or lamina propria. Moreover, basement membrane can also influence the behavior of epithelial cells by controlling their shape, gene expression, adhesion, migration, proliferation, and apoptosis.209 Therefore, ECM1 may be required for basement membrane structural or functional support of the intestinal crypt niche and consequently the ability of the intestinal epithelium to form an effective barrier. Indeed, the Wnt/beta-catenin signal transduction pathway, which governs epithelial differentiation and has also been implicated in tumorigenesis, has been shown to influence ECM1 expression.210 ECM1 has also been shown to inhibit the activity of matrix metalloproteinase-9, a tissue degrading enzyme that is elevated in active IBD.211 Furthermore, the destructive capacity of MMP-9 outcompetes the beneficial properties of the barrier protective gelatinase, MMP-2.212 Thus in patients with ECM1 SNPs, loss-of-function mutations may facilitate increased tissue destruction by MMP-9 and thus exacerbate intestinal ulceration or scarring in IBD. ECM1 is also associated with cell proliferation, angiogenesis, and differentiation and has been shown to be overexpressed in metastatic epithelial tumors including gastric and colorectal cancer.213 It will be of interest to determine if patients with UC with ECM1 SNPs are at increased risk for development of colorectal cancer versus other UC-associated candidate genes.
MUC3A
A genetic association for mucin genes with IBD has been studied for many years. Satsangi et al214 performed linkage analysis for susceptibility genes in IBDs involving 186 affected sibling pairs and generated evidence for linkage to markers D12S83, D7S669, and D3S1573. The marker D7S669 was located near the human intestinal mucin gene “MUC3,” which is located on chromosome 7q212, and previous reports had indicated a possible mucin abnormality in patients with IBD.215,216 Kyo et al then examined a correlation between “MUC3” and IBD by genotyping of variable number of tandem repeat in the “MUC3” gene.217 Analysis of the 3′ terminal structure showed that MUC3 consists of 2 genes, MUC3A and MUC3B each containing 2 EGF-like domains.218 In addition, their data implied that rare alleles of MUC3A, which have an unusual number of 51-bp repeat units, are associated with risk of UC and CD.217,218 The authors made the interesting postulation that these rare alleles in MUC3A could encode mucin proteins with conformational alterations. Given that O-glycosylation and N-glycosylation are dependent on the structure of the mucin protein, a conformational alteration of the protein could result in decreased glycosylation.219 This in turn could lead to increased sensitivity to intestinal bacterial proteases and mucin degradation thus contributing to barrier disruption.219,220 This gains particular importance given that MUC3A is now recognized as a membrane-bound mucin as opposed to the original belief that MUC3 was secreted. In addition, colonic mucin is heavily sulfated and this helps prevent degradation by bacterial proteases. A significant loss of mucin sulfation has been reported in patients with UC, and this may also be related to the possible under-glycosylation of MUC3A protein associated with rare alleles.217,221 MUC3A and MUC3B are expressed in the small intestine, colon, and the gall bladder.222 In addition to other established barrier properties of mucin proteins such as epithelial restitution, an interesting degree of cross-regulation has been observed for MUC3 and the cystic fibrosis transmembrane conductance regulator (CFTR) transporter that is responsible for secretion of chloride and bicarbonate ions.223 Therefore, it is possible that defects in the MUC3 gene may have consequences for other proteins critically involved in additional aspects of intestinal epithelial homeostasis such as ion transporters.
MUC19
Danoy et al224 first postulated a potential link between a SNP in the MUC19 gene locus and IBD. The rs11175593 SNP was identified in the locus of MUC19 and LRRK2 genes. Follow-up studies investigated nonsynonymous coding variants in the MUC19 and LRRK2 genes in a case-control sample comprising CD cases aged younger than 18 years at diagnosis.225 Three MUC19 SNPS (rs11564245, Asp/His; rs4768261, Ser/Phe; and rs2933353, Glu/Ala) were confirmed as being risk associated with CD with rs4768261 retaining significance after corrections. None of the LRRK2 SNPs were associated with CD, thus leading the authors to conclude that the GWAS signal at chr 12 position 22 could be accounted for by the MUC19 gene locus. Rivas et al226 also identified a rare variant highlighted in MUC19 (p.V56M on chromosome 12, position 39226476) that was independent of the common associated GWAS variants. Muc19 protein was originally identified as a secretory product of murine sublingual mucous cells.227 The association of MUC19 with CD and UC is intriguing as a detailed study by Das et al228 in the mouse demonstrated that although MUC19 transcript and protein are readily detectable in mucus glands of the oral cavity, MUC19 is not detectable in the stomach, small intestine, or colon. This raises the question to the relevance and pathophysiological role of MUC19 mutations. Although patients with CD can exhibit extraintestinal manifestations including oral ulcers, the contribution of mutations in this gene to UC are more puzzling given the lack of transcript and protein expression in normal colon, albeit these studies were conducted using mouse rather than human colon.229 Whether a defect in MUC19 at mucosal sites proximal to the GI tract may have an influence on the pathology of UC remains to be determined.
MEMBRANE TRANSPORT
ITLN1
Human intelectin-1 (hITLN-1) is a Ca2+-dependent, D-galactosyl-specific lectin (120 kD) expressed in Paneth and goblet cells of the small intestine and is believed to serve a protective role in the innate immune response to parasite infection.230 It recognizes galactofuranosyl residues found in the cell wall of various microorganisms but not in mammalian tissues.231 Human ITLN1 (120-kDa) seems to have quite different properties from its murine counterpart. Although both hITLN-1 and mITLN-1 recognize galactofuranosyl residues, these intelectins have different quaternary structures and saccharide-binding specificities with human intelectin-1, a disulfide-linked trimer, whereas the mouse homolog is a monomer.232 The intelectin gene was first cloned from intestinal paneth cells and was suggested to be involved in host defense against microorganisms including Trichinella spiralis.233,234 Intelectin has since been identified as a receptor for the iron-binding glycoprotein, lactoferrin (LF) and is involved in regulating uptake, subcellular localization, and release of immunochemically detectable LF by intestinal epithelial Caco-2 cells.235,236 Intelectin-1 has been identified as sharing 100% structural homology with an adipokine, Omentin, a secretory protein that is highly expressed in visceral adipose tissue relative to subcutaneous adipose tissue.237 In vitro studies have shown that intelectin/omentin increases insulin signal transduction and enhances insulin-stimulated glucose transport in isolated human adipocytes. Furthermore, low levels of circulating intelectin/omentin are associated with obesity-induced metabolic dysfunction such as insulin resistance and glucose intolerance while it has also been indicated to play an anti-inflammatory role by reducing TNF-α induction of COX-2 in vascular endothelial cells.238,239 Therefore, it may play a paracrine or endocrine role in modulating insulin sensitivity and is consequently of great interest in the study of diabetes.238 A small patient-based study indicated intelectin-1 as an autoantigen in intestinal mucosa isolated from patients with CD perhaps providing additional avenues of investigation for the functional contribution of this gene to IBD.240 A follow-up study identified that both serum and colonic omentin levels (protein and mRNA, respectively) were reduced in active CD, and it was suggested that reduced omentin may represent a more robust biomarker of active disease than C-reactive protein.241 A genetic association of the ITLN1 gene with IBD was identified in a GWAS study by Barrett et al111 who uncovered a risk association of the rs2274910 SNP at position 1q23 with CD. Interestingly, this SNP was associated with a positive outcome of biologic therapy in the GOIA2 study from a Portuguese CD cohort.242 However, Shaffer et al243 did not identify an association of a novel single nucleotide missense polymorphism (Val109Asp) in white patients with IBD in a German cohort. Moreover, an ITLN1 SNP was also associated with increased risk of asthma.244 Although the exact contribution of Intelectin-1 to barrier properties of the gut remains to be resolved, its role in the uptake of LF may be critical given the anti-inflammatory effects of LF and its capacity to reduce both bacterial infection, and the effects of LPS, in murine models.245,246
Vitamin D Receptor
The vitamin D receptor (VDR) is a member of the steroid receptor family and mediates the effects of the active metabolite 1,25(OH)2 vitamin D3 by regulating transcription of a number of different cellular genes. An 1,25(OH)2 vitamin D3 acts as a pleiotropic endocrine hormone and influences many physiological processes.247,248 An example of the essential role of vitamin D in human health is that severe vitamin D deficiency leads to rickets because 1,25(OH)2 vitamin D3 is essential for adequate calcium and phosphate absorption from the intestine and hence for bone formation.249 Vitamin D also has numerous effects on the immune system, and the VDR is expressed by monocytes and activated B and T lymphocytes, whereas 1,25 (OH)2 vitamin D3 is synthesized by activated macrophages and can act in a local paracrine manner similar to other cytokines.250 Of note, 1,25(OH)2 vitamin D3 activates monocytes and macrophages but suppresses lymphocyte proliferation, immunoglobulin production, NF-κB activation, and the production of IL-2, IL-12, and IFN-γ cytokines.251–253 The linked, commonly occurring, single nucleotide polymorphisms at the 3′ end of VDR (BsmI, ApaI, and TaqI) 11 and the exon 2 splice site FokI polymorphism12 were initially associated with bone mineral density and osteoporosis in a number of different populations. The VDR gene is localized on chromosome 12 at a region linked to IBD in a number of populations.214,254 Simmons et al248 were the first to identify a genetic association between CD susceptibility and the VDR gene. Although the TaqI “tt” genotype was found to be associated with CD, no association was found with UC. The TaqI polymorphism is an A to C base substitution at codon 352 of exon 8 of VDR but this does not produce an amino acid coding change. The functional consequence of this polymorphism for IBD remains to be determined. Although controversial, it has been shown using a VDR-luciferase reporter gene construct that the “t” allele may be associated with increased levels of mRNA production in vitro; however, conflicting data also suggest that this polymorphism leads to reduced VDR mRNA or has no effect on VDR expression.255,256
Subsequent studies have confirmed the TaqI association with IBD and identified associations of other polymorphisms with IBD. The BsmI VDR gene polymorphism has been associated with UC in Jewish Ashkenazi patients.257 Xue et al conducted a meta-analysis in Asian and European subjects and identified that a significant increase in CD risk for Europeans carrying TaqI “tt” genotype and a significant decrease in CD risk for all carriers of the Apal “a” allele.258 In Asian subjects, the VDR FokI polymorphism seemed to confer susceptibility to UC, whereas in males, the TaqI “tt” genotype was associated with susceptibilities to both UC and CD. Studies in an Iranian IBD cohort showed significant association of the FokI polymorphism with CD and UC.259 In contrast to the observations described above, a study in a large cohort of 1359 Irish patients showed no significant association of any of the 4 common VDR polymorphisms with IBD, although the Fokl allele did approach significance.260 Although VDR plays a crucial role in calcium uptake and consequent bone formation, VDR polymorphisms were not indicators for osteoporosis in patients with IBD with low body mass index being the only independent risk factor.261
VDR was confirmed as a protective factor in intestinal homeostasis in studies where Vdr KO mice were crossed with other transgenic mouse models of colitis. In the CD45RB transfer model of IBD, CD4+/CD45RBhigh T cells from Vdr KO mice induced more severe colitis than wild-type CD4+/CD45RBhigh T cells. The second model of IBD investigated was the spontaneous colitis that occurs in IL-10 KO mice. Vdr/IL-10 double KO mice developed accelerated IBD and 100% mortality by 8 weeks of age. At 8 weeks of age, all of the Vdr and IL-10 single KO mice were healthy.262 A clear influence of vitamin D on intestinal epithelial barrier function was demonstrated by Kong et al263 who showed that Vdr ko mice were more susceptible to DSS and developed more severe mucosal barrier defects. Intriguingly, the colonic ulceration observed in Vdr−/− mice was preceded by a greater loss of intestinal transepithelial electric resistance (TER) compared with Vdr+/+ mice after 3 days of DSS. Confocal and electron microscopy analysis also revealed disruption of epithelial junctions in Vdr−/− mice after 3-day DSS treatment. Therefore, Vdr−/− mice were much more susceptible to DSS-induced mucosal injury than Vdr+/+ mice. In cell cultures, 1,25(OH)2 vitamin D3 increased expression of ZO-1, claudin-1 and E-cadherin in Caco-2 monolayers, increased TER, and restricted TER loss and E-cadherin relocalization by DSS. The effect of 1,25(OH)2 vitamin D3 was shown to be VDR-dependent because these effects were lost in VDR siRNA-transfected cells. These observations suggest that VDR plays a critical role in mucosal barrier homeostasis by preserving the integrity of junction complexes and the healing capacity of the colonic epithelium.263 Confirmation that epithelial VDR is important for mucosal integrity was demonstrated in a study showing that intestinal epithelial-specific Vdr KO mice were more susceptible to DSS colitis characterized by greater loss of body weight, increased tissue damage, and inflammation. These mice also displayed decreased levels of bile acids in feces.264 Although the contribution of VDR dysfunction to intestinal inflammation is yet to be determined, there is suggestive evidence that a loss of calcium uptake could, in part, explain some of the defects associated with loss of VDR activity. The influence of calcium on intestinal homeostasis was demonstrated in a study where calcium supplementation reduced inflammation and strengthened the mucosal barrier in the transgenic HLA-B27 rat model of colitis.265 Regarding targeting of VDR for therapeutic purposes, a synthetic form of 1,25(OH)2 vitamin D3, 1alpha,25 (OH)2-16-ene-20-cyclopropyl-vitamin D(3) (BXL-62) is a VDR agonist that has shown potent anti-inflammatory activity with the benefit of low-calcemic activity. BXL-62 reduced the severity of DSS colitis in mice and caused a greater induction of VDR target genes, and inhibition of proinflammatory cytokine production, than 1,25(OH)2 vitamin D3 in peripheral blood mononuclear cells isolated from patients with IBD.266
EPITHELIAL RESTITUTION
PTGER4
A number of GWAS have identified an association of PTGER4 with IBD. In a GWAS analyzing more than 318,000 SNPs, Libioulle et al267 identified a 250 kb region on chromosome 5p13.1 contributing to CD susceptibility. The gene located closest to the associated region is PTGER4, which encodes for the EP4 prostaglandin receptor that binds prostaglandin E2 (PGE2). The disease-associated alleles (rs4495224 and rs7720838) strongly correlated with increased EP4 expression levels. The 5p13.1 association was validated in a follow-up GWAS identifying the rs1553575 SNP as the most prominent, while also indicating a higher frequency of the 5p13.1 signal in females versus males.267,268 An association of PTGER4, specifically the rs4613763 variant in the 5p13.1 region, with UC was first reported by McGovern et al.89 In a large study of a German cohort, Glas et al confirmed association of the risk alleles rs4495224 and rs7720838 with CD, whereas PTGER4 association with CD was also identified in the large meta-analysis by Jostins et al.1,269 Furthermore, in silico analysis identified predicted rs4495224 and rs7720838 as essential parts of binding sites for the transcription factors NF-κB and XBP1 with higher binding scores for carriers of the CD risk alleles. The authors suggested that this may provide an explanation of how these SNPs might contribute to increased PTGER4 expression.269 EP4 receptors are expressed in lamina propria immune cells and while constitutively expressed in human colonic epithelium, they are upregulated in IBD.270,271 In vivo studies with Ptger4−/− mice showed that they develop more severe DSS colitis than wild-type mice. Moreover, treatment with EP4-selective agonists enhanced epithelial viability and regeneration thus exerting protection against colitis in these mice.270,272,273 However, the roles of EP4 receptors in inflammation are likely cell-type specific as a proinflammatory role for EP4 has been described in models of rheumatoid arthritis or experimental autoimmune encephalitis, although EP4 has also been shown to drive the differentiation of Th1 cells and proliferation of Th17.274–276 In addition, PGE2 upregulates IL-8 production in T84 colonic epithelial cells through EP4 interaction, further suggesting a proinflammatory role of PGE2 activation of EP4.277
Regarding the possible influence of EP4 on barrier function, an in vitro study using Caco-2 IECs showed that PGE2 can increase paracellular permeability through EP4 or EP1 receptor activation.278 The EP4-dependent effect occurred in a cAMP-Protein kinase A-dependent manner and resulted in redistribution of the tight junction proteins occludin and ZO-1, as well as the perijunctional actin ring. These effects were also associated with an increase in myosin light chain kinase activity. Lejeune et al showed that in healthy colonic mucosa and T84 IECs, EP4 receptors were localized on apical plasma membrane of epithelial cells at the tip of mucosal folds, whereas, in patients with IBD and in rats with DSS-induced colitis, they were diffusely overexpressed throughout the mucosa. Apical administration of PGE2 to T84 monolayers caused a decrease in barrier function, which was alleviated by an EP4 receptor antagonist.279 However, PGE2 may also have a protective/restorative role on barrier function after injury. Gookin et al showed in an ex vivo chemical injury model of Ussing chamber-mounted porcine ileum that endogenous prostaglandin production or exogenous PGE2 administration rescued TER by osmotic-driven collapse of the paracellular space, and this required Cl− secretion. These studies suggested that mucosal restitution is ineffective in the absence of PG-mediated paracellular space closure.280 Therefore, the exact role(s) of the EP4 receptor in the regulation of barrier function are likely context-dependent and will require careful elucidation as to the suitability of this target for therapeutic intervention in IBD.
BACTERIAL SENSING
CARD15
Mutations in the CARD15 gene were the first to achieve significant association with IBD and have been associated most strongly with ileal CD.281,282 The protein product of CARD15, nucleotide-binding oligomerization domain-containing 2 (NOD2), is known to play a critical role as a microbial sensor in innate immune cells including epithelial cells and has been extensively reviewed elsewhere.283 Regarding a role for CARD15/NOD2 in barrier function, Buhner et al284 identified that CARD15 mutations were associated with increased intestinal permeability in vivo. In a study of 128 patients with quiescent CD, 129 first degree relatives (CD-R), 66 nonrelated household members (CD-NR), and 96 healthy controls, they analyzed the 3 most common CARD15 polymorphisms (R702W, G908R, and 3020insC) and correlated their subjects genetic profile with intestinal permeability as determined by the lactulose/mannitol ratio. Intestinal permeability was significantly increased in CD and CD-R groups compared with CD-NR and controls. Perhaps most strikingly, in healthy first degree relatives, high mucosal permeability was associated with the CARD15 3020insC mutation. These data provided the first indication that an established IBD-associated genetic mutation could contribute to, or was at least associated with, increased intestinal permeability in patients with IBD. This conclusion was in part supported by a study identifying that patients with CD with NOD2 variants had increased levels of tissue endotoxin as determined by immunohistochemistry compared with NOD2 wild-type patients with CD. Although a decrease in adherens junction proteins and altered molecular ratios of tight junction proteins were observed in inflamed versus noninflamed CD tissues, an association between CARD15 SNPs and altered junctional protein levels was not investigated.285 Because a number of mechanisms could be involved in the increased tissue endotoxin levels, including translocation across intestinal epithelium, the impact of CARD15 mutations on junctional proteins is yet to be fully defined.
MEMBRANE RECEPTOR KINASE REGULATION
ERRFI1
A significant association of the ErbB receptor feedback inhibitor 1 (ERRFI1) gene with UC was indicated in a GWAS by Anderson et al.129 The identified SNP, rs35675666, lies on chromosome 1p36. The UC-associated locus is shared by a number of genes in addition to ERRFI1. These include TNFRSF9 (tumor necrosis factor receptor superfamily member 9 or CD137), UTS2 (Urotensin-2), and PARK7 (Parkinson disease autosomal recessive, early onset 7).129 Of these genes, ERRFI1 is the most viable candidate to have an impact on epithelial barrier function. The ERR-FI1 gene encodes mitogen-inducible gene 6 (Mig-6) protein. Mig-6 selectively controls EGFR-family signaling and downstream MAPK activation by a negative feedback mechanism because EGF can actually induce Mig-6 expression. Impairment of Mig-6 attenuation of EGFR family signaling may lead to increased cell proliferation and tumor formation.286–288 Furthermore, on ErbB receptor family ligand deprivation, Mig6 dissociates from the ErbB receptor and binds to and activates the tyrosine kinase c-Abl to trigger p73-dependent apoptosis in mammary epithelial cells. This was also shown to occur in ErrfI1 siRNA knockdown epithelial cells.289 These data suggest that ErrfI1 loss-of-function may predispose to epithelial apoptosis, which could in turn compromise barrier function. In addition, EGFR family members play critical roles in intestinal epithelial homeostasis including growth, restitution, wound healing, and transport and barrier functions.290–294 Although EGFR activation has been shown to have “positive” roles about the outcomes of these physiological processes for intestinal homeostasis, EGFR activation has been shown in a number of studies to participate in deleterious events that compromise barrier function. This involves transactivation of the EGFR in response to agents such as zonulin, oxidants, and bile acids.295–297 Therefore, context-dependent altered regulation of EGFR signaling may have important consequences for epithelial barrier function.
CONCLUSIONS
GWAS have generated profound insights into the genetic contribution to IBD and the genomic regions that contain risk factors for IBD. Translation of these genetic markers into identification of their influence on biological processes that contribute to the pathogenesis of IBD is the next challenge to an increased understanding of how IBD occurs. IBD risk involves multigenic contributions, each with a relatively modest effect size. Therefore, identification of functional consequences of multiple risk loci that collectively contribute to affect individual or intersecting signaling pathways will lead to significant advances in determining how broad physiological events such as regulation of barrier function and autophagy are affected in IBD. Functional studies of IBD genes associated with barrier function will also help to identify specific barrier regulatory mechanisms affected in CD versus UC (Fig. 1). Although GWAS, fine-mapping, and epigenetic characterization together with follow-up functional studies will greatly enhance our understanding of IBD pathology, an additional goal is to use these risk markers for better patient classification, diagnosis, and potentially treatment. Initial efforts offer promise because a recent study successfully generated a diagnostic tool incorporating serological markers, genetic variants, and markers of inflammation into a computational algorithm to examine patterns of combinations of markers to first identify patients with IBD and second to differentiate patients with CD from those with UC. Two of the 4 genetic markers used, ECM1 and NKX2-3, are associated with aspects of barrier function regulation as described above. The incorporation of genetic markers, along with markers of inflammation, gave superior efficacy in diagnosing patients with IBD and discriminating between CD and UC than serological markers alone.298 In addition, expression of tight junction proteins such as claudin-2 and claudin-8 serves as a valuable indicator of the treatment response to the anti–TNF-α agent, Infliximab, in responder versus nonresponder UC patients.299 Therefore, studies of barrier function-associated IBD genetic risk factors will likely enhance not just our understanding of IBD pathophysiology, but also contribute to better diagnosis and treatment.
FIGURE 1.
IBD candidate genes regulate multiple aspects of epithelial barrier integrity. IBD candidate genes are involved in regulating multiple components of the epithelial barrier. Although most of the genes listed have been linked to overall modulation of epithelial/intestinal permeability; here, genes are broadly categorized with specific aspects of epithelial barrier properties that they have been identified as influencing.
Acknowledgments
Supported by NIH NIDDK R01 DK091281.
The author is grateful to the following colleagues for helpful discussions and suggestions in the preparation of this manuscript: Dermot P. McGovern (Cedars-Sinai Medical Center); Christian Y. Lytle (UC Riverside); Meera G. Nair (UC Riverside); and Frances M. Sladek (UC Riverside).
Footnotes
The author has no conflicts of interest to disclose.
References
- 1.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jager S, Stange EF, Wehkamp J. Inflammatory bowel disease: an impaired barrier disease. Langenbecks Arch Surg. 2013;398:1–12. doi: 10.1007/s00423-012-1030-9. [DOI] [PubMed] [Google Scholar]
- 3.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGuckin MA, Eri R, Simms LA, et al. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm Bowel Dis. 2009;15:100–113. doi: 10.1002/ibd.20539. [DOI] [PubMed] [Google Scholar]
- 5.Thompson AI, Lees CW. Genetics of ulcerative colitis. Inflamm Bowel Dis. 2011;17:831–848. doi: 10.1002/ibd.21375. [DOI] [PubMed] [Google Scholar]
- 6.Odenwald MA, Turner JR. Intestinal permeability defects: is it time to treat? Clinical gastroenterology and hepatology. Clin Gastroenterol Hepatol. 2013;11:1075–1083. doi: 10.1016/j.cgh.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCole DF, Barrett KE. Varied role of the gut epithelium in mucosal homeostasis. Curr Opin Gastroenterol. 2007;23:647–654. doi: 10.1097/MOG.0b013e3282f0153b. [DOI] [PubMed] [Google Scholar]
- 9.Goto Y, Kiyono H. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol Rev. 2012;245:147–163. doi: 10.1111/j.1600-065X.2011.01078.x. [DOI] [PubMed] [Google Scholar]
- 10.Cario E. Toll-like receptors in inflammatory bowel diseases: a decade later. Inflamm Bowel Dis. 2010;16:1583–1597. doi: 10.1002/ibd.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahan S, Roth-Walter F, Arnaboldi P, et al. Epithelia: lymphocyte interactions in the gut. Immunol Rev. 2007;215:243–253. doi: 10.1111/j.1600-065X.2006.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDole JR, Wheeler LW, McDonald KG, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCole DF, Barrett KE. Epithelial transport and gut barrier function in colitis. Curr Opin Gastroenterol. 2003;19:578–582. doi: 10.1097/00001574-200311000-00011. [DOI] [PubMed] [Google Scholar]
- 14.Montrose MH, Keely SJ, Barrett KE. Electrolyte secretion and absorption: small intestine and colon. In: Yamada T, editor. Textbook of Gastro-enterology. 4. Philadelphia, PA: Lippincott Williams & Wilkins; 2003. pp. 308–339. [Google Scholar]
- 15.Garcia MA, Yang N, Quinton PM. Normal mouse intestinal mucus release requires cystic fibrosis transmembrane regulator-dependent bicarbonate secretion. J Clin Invest. 2009;119:2613–2622. doi: 10.1172/JCI38662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinton PM. Cystic fibrosis: impaired bicarbonate secretion and mucoviscidosis. Lancet. 2008;372:415–417. doi: 10.1016/S0140-6736(08)61162-9. [DOI] [PubMed] [Google Scholar]
- 17.Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012;15:57–62. doi: 10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson ME, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawashima H. Roles of the gel-forming MUC2 mucin and its O-glycosylation in the protection against colitis and colorectal cancer. Biol Pharm Bull. 2012;35:1637–1641. doi: 10.1248/bpb.b12-00412. [DOI] [PubMed] [Google Scholar]
- 20.Canny G, Levy O, Furuta GT, et al. Lipid mediator-induced expression of bactericidal/permeability-increasing protein (BPI) in human mucosal epithelia. Proc Natl Acad Sci U S A. 2002;99:3902–3907. doi: 10.1073/pnas.052533799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cash HL, Whitham CV, Behrendt CL, et al. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schauber J, Rieger D, Weiler F, et al. Heterogeneous expression of human cathelicidin hCAP18/LL-37 in inflammatory bowel diseases. Eur J Gastroenterol Hepatol. 2006;18:615–621. doi: 10.1097/00042737-200606000-00007. [DOI] [PubMed] [Google Scholar]
- 23.Marquez A, Varade J, Robledo G, et al. Specific association of a CLEC16A/KIAA0350 polymorphism with NOD2/CARD15(-) Crohn’s disease patients. Eur J Hum Genet. 2009;17:1304–1308. doi: 10.1038/ejhg.2009.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VanDussen KL, Liu TC, Li D, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology. 2014;146:200–209. doi: 10.1053/j.gastro.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol Rev. 2005;206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 26.Cerutti A, Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apter FM, Lencer WI, Finkelstein RA, et al. Monoclonal immunoglobulin A antibodies directed against cholera toxin prevent the toxin-induced chloride secretory response and block toxin binding to intestinal epithelial cells in vitro. Infect Immunity. 1993;61:5271–5278. doi: 10.1128/iai.61.12.5271-5278.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantis NJ, Rol N, Corthesy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekirov I, Russell SL, Antunes LC, et al. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 30.Stokes CR, Soothill JF, Turner MW. Immune exclusion is a function of IgA. Nature. 1975;255:745–746. doi: 10.1038/255745a0. [DOI] [PubMed] [Google Scholar]
- 31.Talbot C, Lytle C. Segregation of Na/H exchanger-3 and Cl/HCO3 exchanger SLC26A3 (DRA) in rodent cecum and colon. Am J Physiol Gastrointest Liver Physiol. 2010;299:G358–G367. doi: 10.1152/ajpgi.00151.2010. [DOI] [PubMed] [Google Scholar]
- 32.Kiela PR, Laubitz D, Larmonier CB, et al. Changes in mucosal homeostasis predispose NHE3 knockout mice to increased susceptibility to DSS-induced epithelial injury. Gastroenterology. 2009;137:965–975. e961–e910. doi: 10.1053/j.gastro.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larmonier CB, Laubitz D, Thurston RD, et al. NHE3 modulates the severity of colitis in IL-10-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2011;300:G998–G1009. doi: 10.1152/ajpgi.00073.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker NM, Simpson JE, Brazill JM, et al. Role of down-regulated in adenoma anion exchanger in HCO3- secretion across murine duodenum. Gastroenterology. 2009;136:893–901. doi: 10.1053/j.gastro.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker NM, Simpson JE, Yen PF, et al. Down-regulated in adenoma Cl/HCO3 exchanger couples with Na/H exchanger 3 for NaCl absorption in murine small intestine. Gastroenterology. 2008;135:1645–1653. e1643. doi: 10.1053/j.gastro.2008.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao F, Juric M, Li J, et al. Loss of downregulated in adenoma (DRA) impairs mucosal HCO3(-) secretion in murine ileocolonic inflammation. Inflamm Bowel Dis. 2012;18:101–111. doi: 10.1002/ibd.21744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simms LA, Doecke JD, Roberts RL, et al. KCNN4 gene variant is associated with ileal Crohn’s Disease in the Australian and New Zealand population. Am J Gastroenterol. 2010;105:2209–2217. doi: 10.1038/ajg.2010.161. [DOI] [PubMed] [Google Scholar]
- 38.Flores CA, Melvin JE, Figueroa CD, et al. Abolition of Ca2+-mediated intestinal anion secretion and increased stool dehydration in mice lacking the intermediate conductance Ca2+-dependent K+ channel Kcnn4. J Physiol. 2007;583:705–717. doi: 10.1113/jphysiol.2007.134387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di L, Srivastava S, Zhdanova O, et al. Inhibition of the K+ channel KCa3.1 ameliorates T cell-mediated colitis. Proc Natl Acad Sci U S A. 2010;107:1541–1546. doi: 10.1073/pnas.0910133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powell DW. Barrier function of epithelia. Am J Physiol. 1981;241:G275–G288. doi: 10.1152/ajpgi.1981.241.4.G275. [DOI] [PubMed] [Google Scholar]
- 41.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 42.Ivanov AI, Parkos CA, Nusrat A. Cytoskeletal regulation of epithelial barrier function during inflammation. Am J Pathol. 2010;177:512–524. doi: 10.2353/ajpath.2010.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch S, Nusrat A. Dynamic regulation of epithelial cell fate and barrier function by intercellular junctions. Ann N Y Acad Sci. 2009;1165:220–227. doi: 10.1111/j.1749-6632.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 44.Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol. 2009;1:a002584. doi: 10.1101/cshperspect.a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol Renal Physiol. 2008;295:F867–F876. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez-Mariscal L, Tapia R, Chamorro D. Crosstalk of tight junction components with signaling pathways. Biochim Biophys Acta. 2008;1778:729–756. doi: 10.1016/j.bbamem.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 47.Gunzel D, Fromm M. Claudins and other tight junction proteins. Compr Physiol. 2012;2:1819–1852. doi: 10.1002/cphy.c110045. [DOI] [PubMed] [Google Scholar]
- 48.Nusrat A, Turner JR, Madara JL. Molecular physiology and pathophysiology of tight junctions. IV. Regulation of tight junctions by extracellular stimuli: nutrients, cytokines, and immune cells. Am J Physiol Gastrointest Liver Physiol. 2000;279:G851–G857. doi: 10.1152/ajpgi.2000.279.5.G851. [DOI] [PubMed] [Google Scholar]
- 49.Shen L, Weber CR, Raleigh DR, et al. Tight junction pore and leak pathways: a dynamic duo. Annu Rev Physiol. 2011;73:283–309. doi: 10.1146/annurev-physiol-012110-142150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. 2008;1778:660–669. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claude P. Morphological factors influencing transepithelial permeability: a model for the resistance of the zonula occludens. J Membr Biol. 1978;39:219–232. doi: 10.1007/BF01870332. [DOI] [PubMed] [Google Scholar]
- 52.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 53.Steed E, Balda MS, Matter K. Dynamics and functions of tight junctions. Trends Cell Biol. 2010;20:142–149. doi: 10.1016/j.tcb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 54.Wada M, Tamura A, Takahashi N, et al. Loss of claudins 2 and 15 from mice causes defects in paracellular Na+ flow and nutrient transport in gut and leads to death from malnutrition. Gastroenterology. 2013;144:369–380. doi: 10.1053/j.gastro.2012.10.035. [DOI] [PubMed] [Google Scholar]
- 55.Saitou M, Furuse M, Sasaki H, et al. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Sadi R, Boivin M, Ma T. Mechanism of cytokine modulation of epithelial tight junction barrier. Front Biosci (Landmark Ed) 2009;14:2765–2778. doi: 10.2741/3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchiando AM, Shen L, Graham WV, et al. Caveolin-1-dependent occludin endocytosis is required for TNF-induced tight junction regulation in vivo. J Cell Biol. 2010;189:111–126. doi: 10.1083/jcb.200902153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Itallie CM, Fanning AS, Holmes J, et al. Occludin is required for cytokine-induced regulation of tight junction barriers. J Cell Sci. 2010;123:2844–2852. doi: 10.1242/jcs.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu AS, McCarthy KM, Francis SA, et al. Knockdown of occludin expression leads to diverse phenotypic alterations in epithelial cells. Am J Physiol Cell Physiol. 2005;288:C1231–C1241. doi: 10.1152/ajpcell.00581.2004. [DOI] [PubMed] [Google Scholar]
- 60.Furuse M, Itoh M, Hirase T, et al. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nusrat A, Chen JA, Foley CS, et al. The coiled-coil domain of occludin can act to organize structural and functional elements of the epithelial tight junction. J Biol Chem. 2000;275:29816–29822. doi: 10.1074/jbc.M002450200. [DOI] [PubMed] [Google Scholar]
- 62.Van Itallie CM, Anderson JM. Occludin confers adhesiveness when expressed in fibroblasts. J Cell Sci. 1997;110:1113–1121. doi: 10.1242/jcs.110.9.1113. [DOI] [PubMed] [Google Scholar]
- 63.Meddings J. The significance of the gut barrier in disease. Gut. 2008;57:438–440. doi: 10.1136/gut.2007.143172. [DOI] [PubMed] [Google Scholar]
- 64.Meddings J. What role does intestinal permeability have in IBD pathogenesis? Inflamm Bowel Dis. 2008;14(suppl 2):S138–S139. doi: 10.1002/ibd.20719. [DOI] [PubMed] [Google Scholar]
- 65.Teshima CW, Dieleman LA, Meddings JB. Abnormal intestinal permeability in Crohn’s disease pathogenesis. Ann N Y Acad Sci. 2012;1258:159–165. doi: 10.1111/j.1749-6632.2012.06612.x. [DOI] [PubMed] [Google Scholar]
- 66.Arnott ID, Kingstone K, Ghosh S. Abnormal intestinal permeability predicts relapse in inactive Crohn disease. Scand J Gastroenterol. 2000;35:1163–1169. doi: 10.1080/003655200750056637. [DOI] [PubMed] [Google Scholar]
- 67.Wyatt J, Vogelsang H, Hubl W, et al. Intestinal permeability and the prediction of relapse in Crohn’s disease. Lancet. 1993;341:1437–1439. doi: 10.1016/0140-6736(93)90882-h. [DOI] [PubMed] [Google Scholar]
- 68.Hollander D, Vadheim CM, Brettholz E, et al. Increased intestinal permeability in patients with Crohn’s disease and their relatives. A possible etiologic factor. Ann Intern Med. 1986;105:883–885. doi: 10.7326/0003-4819-105-6-883. [DOI] [PubMed] [Google Scholar]
- 69.Irvine EJ, Marshall JK. Increased intestinal permeability precedes the onset of Crohn’s disease in a subject with familial risk. Gastroenterology. 2000;119:1740–1744. doi: 10.1053/gast.2000.20231. [DOI] [PubMed] [Google Scholar]
- 70.May GR, Sutherland LR, Meddings JB. Is small intestinal permeability really increased in relatives of patients with Crohn’s disease? Gastroenterology. 1993;104:1627–1632. doi: 10.1016/0016-5085(93)90638-s. [DOI] [PubMed] [Google Scholar]
- 71.Arrieta MC, Bistritz L, Meddings JB. Alterations in intestinal permeability. Gut. 2006;55:1512–1520. doi: 10.1136/gut.2005.085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Menzies IS, Laker MF, Pounder R, et al. Abnormal intestinal permeability to sugars in villous atrophy. Lancet. 1979;2:1107–1109. doi: 10.1016/s0140-6736(79)92507-8. [DOI] [PubMed] [Google Scholar]
- 73.Marin ML, Greenstein AJ, Geller SA, et al. A freeze fracture study of Crohn’s disease of the terminal ileum: changes in epithelial tight junction organization. Am J Gastroenterol. 1983;78:537–547. [PubMed] [Google Scholar]
- 74.Weber CR, Raleigh DR, Su L, et al. Epithelial myosin light chain kinase activation induces mucosal interleukin-13 expression to alter tight junction ion selectivity. J Biol Chem. 2010;285:12037–12046. doi: 10.1074/jbc.M109.064808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeissig S, Burgel N, Gunzel D, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Madsen K, Cornish A, Soper P, et al. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–591. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 77.Olson TS, Reuter BK, Scott KG, et al. The primary defect in experimental ileitis originates from a nonhematopoietic source. J Exp Med. 2006;203:541–552. doi: 10.1084/jem.20050407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Resta-Lenert S, Smitham J, Barrett KE. Epithelial dysfunction associated with the development of colitis in conventionally housed mdr1a-/- mice. Am J Physiol Gastrointest Liver Physiol. 2005;289:G153–G162. doi: 10.1152/ajpgi.00395.2004. [DOI] [PubMed] [Google Scholar]
- 79.Clayburgh DR, Barrett TA, Tang Y, et al. Epithelial myosin light chain kinase-dependent barrier dysfunction mediates T cell activation-induced diarrhea in vivo. The J Clinical Investigation. 2005;115:2702–2715. doi: 10.1172/JCI24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shen L, Black ED, Witkowski ED, et al. Myosin light chain phosphorylation regulates barrier function by remodeling tight junction structure. J Cell Sci. 2006;119:2095–2106. doi: 10.1242/jcs.02915. [DOI] [PubMed] [Google Scholar]
- 81.Su L, Shen L, Clayburgh DR, et al. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551–563. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arrieta MC, Madsen K, Doyle J, et al. Reducing small intestinal permeability attenuates colitis in the IL10 gene-deficient mouse. Gut. 2009;58:41–48. doi: 10.1136/gut.2008.150888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salim SY, Soderholm JD. Importance of disrupted intestinal barrier in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:362–381. doi: 10.1002/ibd.21403. [DOI] [PubMed] [Google Scholar]
- 84.Schurmann G, Bruwer M, Klotz A, et al. Transepithelial transport processes at the intestinal mucosa in inflammatory bowel disease. Int J Colorectal Dis. 1999;14:41–46. doi: 10.1007/s003840050181. [DOI] [PubMed] [Google Scholar]
- 85.Soderholm JD, Streutker C, Yang PC, et al. Increased epithelial uptake of protein antigens in the ileum of Crohn’s disease mediated by tumour necrosis factor alpha. Gut. 2004;53:1817–1824. doi: 10.1136/gut.2004.041426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Camilleri M, Madsen K, Spiller R, et al. Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterology and motility. Neurogastroenterol Motil. 2012;24:503–512. doi: 10.1111/j.1365-2982.2012.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Sommeren S, Visschedijk MC, Festen EA, et al. HNF4alpha and CDH1 are associated with ulcerative colitis in a Dutch cohort. Inflamm Bowel Dis. 2011;17:1714–1718. doi: 10.1002/ibd.21541. [DOI] [PubMed] [Google Scholar]
- 88.Consortium UIG, Barrett JC, Lee JC, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McGovern DP, Gardet A, Torkvist L, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muise AM, Walters TD, Glowacka WK, et al. Polymorphisms in E-cadherin (CDH1) result in a mis-localised cytoplasmic protein that is associated with Crohn’s disease. Gut. 2009;58:1121–1127. doi: 10.1136/gut.2008.175117. [DOI] [PubMed] [Google Scholar]
- 91.Hermiston ML, Gordon JI. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science. 1995;270:1203–1207. doi: 10.1126/science.270.5239.1203. [DOI] [PubMed] [Google Scholar]
- 92.Bonazzi M, Cossart P. Impenetrable barriers or entry portals? the role of cell-cell adhesion during infection. J Cell Biol. 2011;195:349–358. doi: 10.1083/jcb.201106011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Demetter P, De Vos M, Van Damme N, et al. Focal up-regulation of E-cadherin-catenin complex in inflamed bowel mucosa but reduced expression in ulcer-associated cell lineage. Am J Clin Pathol. 2000;114:364–370. doi: 10.1093/ajcp/114.3.364. [DOI] [PubMed] [Google Scholar]
- 94.Karayiannakis AJ, Syrigos KN, Efstathiou J, et al. Expression of catenins and E-cadherin during epithelial restitution in inflammatory bowel disease. J Pathol. 1998;185:413–418. doi: 10.1002/(SICI)1096-9896(199808)185:4<413::AID-PATH125>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 95.Hazan RB, Qiao R, Keren R, et al. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 96.Houlston RS, Webb E, Broderick P, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klucky B, Mueller R, Vogt I, et al. Kallikrein 6 induces E-cadherin shedding and promotes cell proliferation, migration, and invasion. Cancer Res. 2007;67:8198–8206. doi: 10.1158/0008-5472.CAN-07-0607. [DOI] [PubMed] [Google Scholar]
- 98.Machado JC, Oliveira C, Carvalho R, et al. E-cadherin gene (CDH1) promoter methylation as the second hit in sporadic diffuse gastric carcinoma. Oncogene. 2001;20:1525–1528. doi: 10.1038/sj.onc.1204234. [DOI] [PubMed] [Google Scholar]
- 99.Beaugerie L, Svrcek M, Seksik P, et al. Risk of colorectal high-grade dysplasia and cancer in a prospective observational cohort of patients with inflammatory bowel disease. Gastroenterology. 2013;145:166–175. e168. doi: 10.1053/j.gastro.2013.03.044. [DOI] [PubMed] [Google Scholar]
- 100.Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 2001;48:526–535. doi: 10.1136/gut.48.4.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Azarschab P, Porschen R, Gregor M, et al. Epigenetic control of the E-cadherin gene (CDH1) by CpG methylation in colectomy samples of patients with ulcerative colitis. Genes Chromosomes Cancer. 2002;35:121–126. doi: 10.1002/gcc.10101. [DOI] [PubMed] [Google Scholar]
- 102.Silverberg MS, Cho JH, Rioux JD, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Teller IC, Beaulieu JF. Interactions between laminin and epithelial cells in intestinal health and disease. Expert Rev Mol Med. 2001;3:1–18. doi: 10.1017/S1462399401003623. [DOI] [PubMed] [Google Scholar]
- 104.Timpl R, Brown JC. Supramolecular assembly of basement membranes. BioEssays. 1996;18:123–132. doi: 10.1002/bies.950180208. [DOI] [PubMed] [Google Scholar]
- 105.Miner JH, Yurchenco PD. Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol. 2004;20:255–284. doi: 10.1146/annurev.cellbio.20.010403.094555. [DOI] [PubMed] [Google Scholar]
- 106.Beaulieu JF, Vachon PH. Reciprocal expression of laminin A-chain isoforms along the crypt-villus axis in the human small intestine. Gastroenterology. 1994;106:829–839. doi: 10.1016/0016-5085(94)90740-4. [DOI] [PubMed] [Google Scholar]
- 107.Simon-Assmann P, Duclos B, Orian-Rousseau V, et al. Differential expression of laminin isoforms and alpha 6-beta 4 integrin subunits in the developing human and mouse intestine. Dev Dyn. 1994;201:71–85. doi: 10.1002/aja.1002010108. [DOI] [PubMed] [Google Scholar]
- 108.Vachon PH, Beaulieu JF. Extracellular heterotrimeric laminin promotes differentiation in human enterocytes. Am J Physiol. 1995;268:G857–G867. doi: 10.1152/ajpgi.1995.268.5.G857. [DOI] [PubMed] [Google Scholar]
- 109.Koutroubakis IE, Petinaki E, Dimoulios P, et al. Serum laminin and collagen IV in inflammatory bowel disease. J Clin Pathol. 2003;56:817–820. doi: 10.1136/jcp.56.11.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schmehl K, Florian S, Jacobasch G, et al. Deficiency of epithelial basement membrane laminin in ulcerative colitis affected human colonic mucosa. Int J Colorectal Dis. 2000;15:39–48. doi: 10.1007/s003840050006. [DOI] [PubMed] [Google Scholar]
- 111.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bouatrouss Y, Herring-Gillam FE, Gosselin J, et al. Altered expression of laminins in Crohn’s disease small intestinal mucosa. Am J Pathol. 2000;156:45–50. doi: 10.1016/S0002-9440(10)64704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Francoeur C, Escaffit F, Vachon PH, et al. Proinflammatory cytokines TNF-alpha and IFN-gamma alter laminin expression under an apoptosis-independent mechanism in human intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2004;287:G592–G598. doi: 10.1152/ajpgi.00535.2003. [DOI] [PubMed] [Google Scholar]
- 114.Saito E, Okamoto A, Saito M, et al. Genes associated with the genesis of leiomyoma of the uterus in a commonly deleted chromosomal region at 7q22. Oncol Rep. 2005;13:469–472. [PubMed] [Google Scholar]
- 115.Saito N, Kameoka S. Serum laminin is an independent prognostic factor in colorectal cancer. Int J Colorectal Dis. 2005;20:238–244. doi: 10.1007/s00384-004-0676-3. [DOI] [PubMed] [Google Scholar]
- 116.Wapenaar MC, Monsuur AJ, van Bodegraven AA, et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. 2008;57:463–467. doi: 10.1136/gut.2007.133132. [DOI] [PubMed] [Google Scholar]
- 117.Wolters VM, Xu W, Zhao X, et al. Replication of genetic variation in the MYO9B gene in Crohn’s disease. Hum Immunol. 2011;72:592–597. doi: 10.1016/j.humimm.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 118.Schumann M, Gunzel D, Buergel N, et al. Cell polarity-determining proteins Par-3 and PP-1 are involved in epithelial tight junction defects in coeliac disease. Gut. 2012;61:220–228. doi: 10.1136/gutjnl-2011-300123. [DOI] [PubMed] [Google Scholar]
- 119.Ebnet K, Suzuki A, Horikoshi Y, et al. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM) EMBO J. 2001;20:3738–3748. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Helfrich I, Schmitz A, Zigrino P, et al. Role of aPKC isoforms and their binding partners Par3 and Par6 in epidermal barrier formation. J Invest Dermatol. 2007;127:782–791. doi: 10.1038/sj.jid.5700621. [DOI] [PubMed] [Google Scholar]
- 121.Hurd TW, Fan S, Liu CJ, et al. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol. 2003;13:2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 122.Lemmers C, Michel D, Lane-Guermonprez L, et al. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Molecular Biology of the Cell. 2004;15:1324–1333. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shin K, Wang Q, Margolis B. PATJ regulates directional migration of mammalian epithelial cells. EMBO Reports. 2007;8:158–164. doi: 10.1038/sj.embor.7400890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Du D, Xu F, Yu L, et al. The tight junction protein, occludin, regulates the directional migration of epithelial cells. Developmental Cell. 2010;18:52–63. doi: 10.1016/j.devcel.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 125.Neer EJ. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 126.Balda MS, Gonzalez-Mariscal L, Contreras RG, et al. Assembly and sealing of tight junctions: possible participation of G-proteins, phospholipase C, protein kinase C and calmodulin. J Membr Biol. 1991;122:193–202. doi: 10.1007/BF01871420. [DOI] [PubMed] [Google Scholar]
- 127.Meyer TN, Schwesinger C, Denker BM. Zonula occludens-1 is a scaffolding protein for signaling molecules. Galpha(12) directly binds to the Src homology 3 domain and regulates paracellular permeability in epithelial cells. J Biol Chem. 2002;277:24855–24858. doi: 10.1074/jbc.C200240200. [DOI] [PubMed] [Google Scholar]
- 128.Sabath E, Negoro H, Beaudry S, et al. Galpha12 regulates protein interactions within the MDCK cell tight junction and inhibits tight-junction assembly. J Cell Sci. 2008;121:814–824. doi: 10.1242/jcs.014878. [DOI] [PubMed] [Google Scholar]
- 129.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Herroeder S, Reichardt P, Sassmann A, et al. Guanine nucleotide-binding proteins of the G12 family shape immune functions by controlling CD4+ T cell adhesiveness and motility. Immunity. 2009;30:708–720. doi: 10.1016/j.immuni.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 131.Cottone M, Marrone C, Casa A, et al. Familial occurrence of inflammatory bowel disease in celiac disease. Inflamm Bowel Dis. 2003;9:321–323. doi: 10.1097/00054725-200309000-00006. [DOI] [PubMed] [Google Scholar]
- 132.Schedel J, Rockmann F, Bongartz T, et al. Association of Crohn’s disease and latent celiac disease: a case report and review of the literature. Int J Colorectal Dis. 2005;20:376–380. doi: 10.1007/s00384-004-0661-x. [DOI] [PubMed] [Google Scholar]
- 133.Tursi A, Giorgetti GM, Brandimarte G, et al. High prevalence of celiac disease among patients affected by Crohn’s disease. Inflamm Bowel Dis. 2005;11:662–666. doi: 10.1097/01.mib.0000164195.75207.1e. [DOI] [PubMed] [Google Scholar]
- 134.McGovern DP, Taylor KD, Landers C, et al. MAGI2 genetic variation and inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:75–83. doi: 10.1002/ibd.20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ingham RJ, Colwill K, Howard C, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol. 2005;25:7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu X, Hepner K, Castelino-Prabhu S, et al. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc Natl Acad Sci U S A. 2000;97:4233–4238. doi: 10.1073/pnas.97.8.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hu Y, Li Z, Guo L, et al. MAGI-2 Inhibits cell migration and proliferation via PTEN in human hepatocarcinoma cells. Arch Biochem Biophys. 2007;467:1–9. doi: 10.1016/j.abb.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 138.Valiente M, Andres-Pons A, Gomar B, et al. Binding of PTEN to specific PDZ domains contributes to PTEN protein stability and phosphorylation by microtubule-associated serine/threonine kinases. J Biol Chem. 2005;280:28936–28943. doi: 10.1074/jbc.M504761200. [DOI] [PubMed] [Google Scholar]
- 139.Zeissig S, Bojarski C, Buergel N, et al. Downregulation of epithelial apoptosis and barrier repair in active Crohn’s disease by tumour necrosis factor alpha antibody treatment. Gut. 2004;53:1295–1302. doi: 10.1136/gut.2003.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Inoue A, Saito J, Ikebe R, et al. Myosin IXb is a single-headed minus-end-directed processive motor. Nat Cell Biol. 2002;4:302–306. doi: 10.1038/ncb774. [DOI] [PubMed] [Google Scholar]
- 141.Post PL, Tyska MJ, O’Connell CB, et al. Myosin-IXb is a single-headed and processive motor. J Biol Chem. 2002;277:11679–11683. doi: 10.1074/jbc.M111173200. [DOI] [PubMed] [Google Scholar]
- 142.Muller RT, Honnert U, Reinhard J, et al. The rat myosin myr 5 is a GTPase-activating protein for Rho in vivo: essential role of arginine 1695. Mol Biol Cell. 1997;8:2039–2053. doi: 10.1091/mbc.8.10.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bruewer M, Hopkins AM, Hobert ME, et al. RhoA, Rac1, and Cdc42 exert distinct effects on epithelial barrier via selective structural and biochemical modulation of junctional proteins and F-actin. Am J Physiol Cell Physiol. 2004;287:C327–C335. doi: 10.1152/ajpcell.00087.2004. [DOI] [PubMed] [Google Scholar]
- 144.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 145.Hopkins AM, Walsh SV, Verkade P, et al. Constitutive activation of Rho proteins by CNF-1 influences tight junction structure and epithelial barrier function. J Cell Sci. 2003;116:725–742. doi: 10.1242/jcs.00300. [DOI] [PubMed] [Google Scholar]
- 146.Monsuur AJ, de Bakker PI, Alizadeh BZ, et al. Myosin IXB variant increases the risk of celiac disease and points toward a primary intestinal barrier defect. Nat Genet. 2005;37:1341–1344. doi: 10.1038/ng1680. [DOI] [PubMed] [Google Scholar]
- 147.Amundsen SS, Monsuur AJ, Wapenaar MC, et al. Association analysis of MYO9B gene polymorphisms with celiac disease in a Swedish/Nor-wegian cohort. Hum Immun. 2006;67:341–345. doi: 10.1016/j.humimm.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 148.Hunt KA, Monsuur AJ, McArdle WL, et al. Lack of association of MYO9B genetic variants with coeliac disease in a British cohort. Gut. 2006;55:969–972. doi: 10.1136/gut.2005.086769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.van Bodegraven AA, Curley CR, Hunt KA, et al. Genetic variation in myosin IXB is associated with ulcerative colitis. Gastroenterology. 2006;131:1768–1774. doi: 10.1053/j.gastro.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 150.Cooney R, Cummings JR, Pathan S, et al. Association between genetic variants in myosin IXB and Crohn’s disease. Inflamm Bowel Dis. 2009;15:1014–1021. doi: 10.1002/ibd.20885. [DOI] [PubMed] [Google Scholar]
- 151.Nunez C, Oliver J, Mendoza JL, et al. MYO9B polymorphisms in patients with inflammatory bowel disease. Gut. 2007;56:1321–1322. doi: 10.1136/gut.2007.121905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fasano A, Not T, Wang W, et al. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet. 2000;355:1518–1519. doi: 10.1016/S0140-6736(00)02169-3. [DOI] [PubMed] [Google Scholar]
- 154.Wang W, Uzzau S, Goldblum SE, et al. Human zonulin, a potential modulator of intestinal tight junctions. J Cell Sci. 2000;113:4435–4440. doi: 10.1242/jcs.113.24.4435. [DOI] [PubMed] [Google Scholar]
- 155.Latiano A, Palmieri O, Valvano MR, et al. The association of MYO9B gene in Italian patients with inflammatory bowel diseases. Aliment Pharmacol Ther. 2008;27:241–248. doi: 10.1111/j.1365-2036.2007.03551.x. [DOI] [PubMed] [Google Scholar]
- 156.Sapone A, de Magistris L, Pietzak M, et al. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes. 2006;55:1443–1449. doi: 10.2337/db05-1593. [DOI] [PubMed] [Google Scholar]
- 157.Chandhoke SK, Mooseker MS. A role for myosin IXb, a motor-RhoGAP chimera, in epithelial wound healing and tight junction regulation. Mol Biol Cell. 2012;23:2468–2480. doi: 10.1091/mbc.E11-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Waterman M, Xu W, Stempak JM, et al. Distinct and overlapping genetic loci in Crohn’s disease and ulcerative colitis: correlations with pathogenesis. Inflamm Bowel Dis. 2011;17:1936–1942. doi: 10.1002/ibd.21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weersma RK, Stokkers PC, Cleynen I, et al. Confirmation of multiple Crohn’s disease susceptibility loci in a large Dutch-Belgian cohort. Am J Gastroenterol. 2009;104:630–638. doi: 10.1038/ajg.2008.112. [DOI] [PubMed] [Google Scholar]
- 161.Franke A, Balschun T, Karlsen TH, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 162.Glas J, Wagner J, Seiderer J, et al. PTPN2 gene variants are associated with susceptibility to both Crohn’s disease and ulcerative colitis supporting a common genetic disease background. PLoS One. 2012;7:e33682. doi: 10.1371/journal.pone.0033682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Scharl M, Wojtal KA, Becker HM, et al. Protein tyrosine phosphatase nonreceptor type 2 regulates autophagosome formation in human intestinal cells. Inflamm Bowel Dis. 2012;18:1287–1302. doi: 10.1002/ibd.21891. [DOI] [PubMed] [Google Scholar]
- 164.Scharl M, Mwinyi J, Fischbeck A, et al. Crohn’s disease-associated polymorphism within the PTPN2 gene affects muramyl-dipeptide-induced cytokine secretion and autophagy. Inflamm Bowel Dis. 2012;18:900–912. doi: 10.1002/ibd.21913. [DOI] [PubMed] [Google Scholar]
- 165.Yu W, Hegarty JP, Berg A, et al. PTPN2 is associated with Crohn’s disease and its expression is regulated by NKX2-3. Dis Markers. 2012;32:83–91. doi: 10.3233/DMA-2011-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 167.Cool DE, Tonks NK, Charbonneau H, et al. cDNA isolated from a human T-cell library encodes a member of the protein-tyrosine-phosphatase family. Proc Natl Acad Sci U S A. 1989;86:5257–5261. doi: 10.1073/pnas.86.14.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tiganis T, Bennett AM. Protein tyrosine phosphatase function: the substrate perspective. Biochem J. 2007;402:1–15. doi: 10.1042/BJ20061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Marcil V, Mack DR, Kumar V, et al. Association between the PTPN2 gene and Crohn’s disease: dissection of potential causal variants. Inflamm Bowel Dis. 2013;19:1149–1155. doi: 10.1097/MIB.0b013e318280b181. [DOI] [PubMed] [Google Scholar]
- 170.Scharl M, McCole DF, Weber A, et al. Protein tyrosine phosphatase N2 regulates TNFalpha-induced signalling and cytokine secretion in human intestinal epithelial cells. Gut. 2011;60:189–197. doi: 10.1136/gut.2010.216606. [DOI] [PubMed] [Google Scholar]
- 171.Scharl M, Paul G, Weber A, et al. Protection of epithelial barrier function by the Crohn’s disease associated gene protein tyrosine phosphatase n2. Gastroenterology. 2009;137:2030–2040. e2035. doi: 10.1053/j.gastro.2009.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Amre DK, Mack DR, Morgan K, et al. Susceptibility loci reported in genome-wide association studies are associated with Crohn’s disease in Canadian children. Aliment Pharmacol Ther. 2010;31:1186–1191. doi: 10.1111/j.1365-2036.2010.04294.x. [DOI] [PubMed] [Google Scholar]
- 174.Smyth DJ, Plagnol V, Walker NM, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ibarra-Sanchez MJ, Simoncic PD, Nestel FR, et al. The T-cell protein tyrosine phosphatase. Semin Immunol. 2000;12:379–386. doi: 10.1006/smim.2000.0220. [DOI] [PubMed] [Google Scholar]
- 176.Lorenzen JA, Dadabay CY, Fischer EH. COOH-terminal sequence motifs target the T cell protein tyrosine phosphatase to the ER and nucleus. J Cell Biol. 1995;131:631–643. doi: 10.1083/jcb.131.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tiganis T. PTP1B and TCPTP–nonredundant phosphatases in insulin signaling and glucose homeostasis. FEBS J. 2013;280:445–458. doi: 10.1111/j.1742-4658.2012.08563.x. [DOI] [PubMed] [Google Scholar]
- 178.Galic S, Hauser C, Kahn BB, et al. Coordinated regulation of insulin signaling by the protein tyrosine phosphatases PTP1B and TCPTP. Mol Cell Biol. 2005;25:819–829. doi: 10.1128/MCB.25.2.819-829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mattila E, Pellinen T, Nevo J, et al. Negative regulation of EGFR signalling through integrin-alpha1beta1-mediated activation of protein tyrosine phosphatase TCPTP. Nat Cell Biol. 2005;7:78–85. doi: 10.1038/ncb1209. [DOI] [PubMed] [Google Scholar]
- 180.Simoncic PD, Lee-Loy A, Barber DL, et al. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr Biol. 2002;12:446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 181.ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tiganis T, Bennett AM, Ravichandran KS, et al. Epidermal growth factor receptor and the adaptor protein p52Shc are specific substrates of T-cell protein tyrosine phosphatase. Mol Cell Biol. 1998;18:1622–1634. doi: 10.1128/mcb.18.3.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.van Vliet C, Bukczynska PE, Puryer MA, et al. Selective regulation of tumor necrosis factor-induced Erk signaling by Src family kinases and the T cell protein tyrosine phosphatase. Nat Immunol. 2005;6:253–260. doi: 10.1038/ni1169. [DOI] [PubMed] [Google Scholar]
- 184.Heinonen KM, Nestel FP, Newell EW, et al. T-cell protein tyrosine phosphatase deletion results in progressive systemic inflammatory disease. Blood. 2004;103:3457–3464. doi: 10.1182/blood-2003-09-3153. [DOI] [PubMed] [Google Scholar]
- 185.You-Ten KE, Muise ES, Itie A, et al. Impaired bone marrow microenvironment and immune function in T cell protein tyrosine phosphatase-deficient mice. J Exp Med. 1997;186:683–693. doi: 10.1084/jem.186.5.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wiede F, Chew SH, van Vliet C, et al. Strain-dependent differences in bone development, myeloid hyperplasia, morbidity and mortality in ptpn2-deficient mice. PLoS One. 2012;7:e36703. doi: 10.1371/journal.pone.0036703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hassan SW, Doody KM, Hardy S, et al. Increased susceptibility to dextran sulfate sodium induced colitis in the T cell protein tyrosine phosphatase heterozygous mouse. PLoS One. 2010;5:e8868. doi: 10.1371/journal.pone.0008868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Uribe JM, McCole DF, Barrett KE. Interferon-gamma activates EGF receptor and increases TGF-alpha in T84 cells: implications for chloride secretion. American J Physiol Gastrointest Liver Physiol. 2002;283:G923–931. doi: 10.1152/ajpgi.00237.2002. [DOI] [PubMed] [Google Scholar]
- 189.Scharl M, Rudenko I, McCole DF. Loss of protein tyrosine phosphatase N2 potentiates epidermal growth factor suppression of intestinal epithelial chloride secretion. Am J Physiol Gastrointest Liver Physiol. 2010;299:G935–G945. doi: 10.1152/ajpgi.00106.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.McCole DF. Regulation of epithelial barrier function by the inflammatory bowel disease candidate gene, PTPN2. Ann N Y Acad Sci. 2012;1257:108–114. doi: 10.1111/j.1749-6632.2012.06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Marcil V, Sinnett D, Seidman E, et al. Association between genetic variants in the HNF4A gene and childhood-onset Crohn’s disease. Genes Immun. 2012;13:556–565. doi: 10.1038/gene.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yamagata K, Furuta H, Oda N, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 194.Sladek FM, Zhong WM, Lai E, et al. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 195.Garrison WD, Battle MA, Yang C, et al. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207–1220. doi: 10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chen WS, Manova K, Weinstein DC, et al. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 197.Parviz F, Matullo C, Garrison WD, et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 198.Ahn SH, Shah YM, Inoue J, et al. Hepatocyte nuclear factor 4 alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:908–920. doi: 10.1002/ibd.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cattin AL, Le Beyec J, Barreau F, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol. 2009;29:6294–6308. doi: 10.1128/MCB.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Darsigny M, Babeu JP, Dupuis AA, et al. Loss of hepatocyte-nuclear-factor-4alpha affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS One. 2009;4:e7609. doi: 10.1371/journal.pone.0007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Babeu JP, Darsigny M, Lussier CR, et al. Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am J Physiol Gastrointest Liver Physiol. 2009;297:G124–G134. doi: 10.1152/ajpgi.90690.2008. [DOI] [PubMed] [Google Scholar]
- 202.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22:337–358. doi: 10.1023/a:1023772912750. [DOI] [PubMed] [Google Scholar]
- 203.Chellappa K, Jankova L, Schnabl JM, et al. Src tyrosine kinase phos-phorylation of nuclear receptor HNF4alpha correlates with isoform-specific loss of HNF4alpha in human colon cancer. Proc Natl Acad Sci U S A. 2012;109:2302–2307. doi: 10.1073/pnas.1106799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Anderson CA, Massey DC, Barrett JC, et al. Investigation of Crohn’s disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology. 2009;136:523–529. e523. doi: 10.1053/j.gastro.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fisher SA, Tremelling M, Anderson CA, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Festen EA, Stokkers PC, van Diemen CC, et al. Genetic analysis in a Dutch study sample identifies more ulcerative colitis susceptibility loci and shows their additive role in disease risk. Am J Gastroenterol. 2010;105:395–402. doi: 10.1038/ajg.2009.576. [DOI] [PubMed] [Google Scholar]
- 207.Sercu S, Zhang M, Oyama N, et al. Interaction of extracellular matrix protein 1 with extracellular matrix components: ECM1 is a basement membrane protein of the skin. J Invest Dermatol. 2008;128:1397–1408. doi: 10.1038/sj.jid.5701231. [DOI] [PubMed] [Google Scholar]
- 208.Chan I, South AP, McGrath JA, et al. Rapid diagnosis of lipoid proteinosis using an anti-extracellular matrix protein 1 (ECM1) antibody. J Dermatol Sci. 2004;35:151–153. doi: 10.1016/j.jdermsci.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 209.Li AC, Thompson RP. Basement membrane components. J Clin Pathol. 2003;56:885–887. doi: 10.1136/jcp.56.12.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kenny PA, Enver T, Ashworth A. Receptor and secreted targets of Wnt-1/beta-catenin signalling in mouse mammary epithelial cells. BMC Cancer. 2005;5:3. doi: 10.1186/1471-2407-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fujimoto N, Terlizzi J, Aho S, et al. Extracellular matrix protein 1 inhibits the activity of matrix metalloproteinase 9 through high-affinity protein/protein interactions. Exp Dermatol. 2006;15:300–307. doi: 10.1111/j.0906-6705.2006.00409.x. [DOI] [PubMed] [Google Scholar]
- 212.Garg P, Vijay-Kumar M, Wang L, et al. Matrix metalloproteinase-9-mediated tissue injury overrides the protective effect of matrix metalloproteinase-2 during colitis. Am J Physiol Gastrointest Liver Physiol. 2009;296:G175–G184. doi: 10.1152/ajpgi.90454.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang L, Yu J, Ni J, et al. Extracellular matrix protein 1 (ECM1) is over-expressed in malignant epithelial tumors. Cancer Lett. 2003;200:57–67. doi: 10.1016/s0304-3835(03)00350-1. [DOI] [PubMed] [Google Scholar]
- 214.Satsangi J, Parkes M, Louis E, et al. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet. 1996;14:199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 215.Buisine MP, Desreumaux P, Debailleul V, et al. Abnormalities in mucin gene expression in Crohn’s disease. Inflamm Bowel Dis. 1999;5:24–32. doi: 10.1097/00054725-199902000-00004. [DOI] [PubMed] [Google Scholar]
- 216.Tysk C, Riedesel H, Lindberg E, et al. Colonic glycoproteins in mono-zygotic twins with inflammatory bowel disease. Gastroenterology. 1991;100:419–423. doi: 10.1016/0016-5085(91)90211-3. [DOI] [PubMed] [Google Scholar]
- 217.Kyo K, Parkes M, Takei Y, et al. Association of ulcerative colitis with rare VNTR alleles of the human intestinal mucin gene, MUC3. Hum Mol Genet. 1999;8:307–311. doi: 10.1093/hmg/8.2.307. [DOI] [PubMed] [Google Scholar]
- 218.Kyo K, Muto T, Nagawa H, et al. Associations of distinct variants of the intestinal mucin gene MUC3A with ulcerative colitis and Crohn’s disease. J Hum Genet. 2001;46:5–20. doi: 10.1007/s100380170118. [DOI] [PubMed] [Google Scholar]
- 219.Van den Steen P, Rudd PM, Dwek RA, et al. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 220.Variyam EP, Hoskins LC. Mucin degradation in human colon ecosystems. Degradation of hog gastric mucin by fecal extracts and fecal cultures. Gastroenterology. 1981;81:751–758. [PubMed] [Google Scholar]
- 221.Raouf AH, Tsai HH, Parker N, et al. Sulphation of colonic and rectal mucin in inflammatory bowel disease: reduced sulphation of rectal mucus in ulcerative colitis. Clin Sci (Lond) 1992;83:623–626. doi: 10.1042/cs0830623. [DOI] [PubMed] [Google Scholar]
- 222.Williams SJ, Munster DJ, Quin RJ, et al. The MUC3 gene encodes a transmembrane mucin and is alternatively spliced. Biochem Biophys Res Commun. 1999;261:83–89. doi: 10.1006/bbrc.1999.1001. [DOI] [PubMed] [Google Scholar]
- 223.Pelaseyed T, Hansson GC. CFTR anion channel modulates expression of human transmembrane mucin MUC3 through the PDZ protein GOPC. J Cell Sci. 2011;124:3074–3083. doi: 10.1242/jcs.076943. [DOI] [PubMed] [Google Scholar]
- 224.Danoy P, Pryce K, Hadler J, et al. Association of variants at 1q32 and STAT3 with ankylosing spondylitis suggests genetic overlap with Crohn’s disease. PLoS Genet. 2010;6:e1001195. doi: 10.1371/journal.pgen.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kumar V, Mack DR, Marcil V, et al. Genome-wide association study signal at the 12q12 locus for Crohn’s disease may represent associations with the MUC19 gene. Inflamm Bowel Dis. 2013;19:1254–1259. doi: 10.1097/MIB.0b013e318281f454. [DOI] [PubMed] [Google Scholar]
- 226.Rivas MA, Beaudoin M, Gardet A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Fallon MA, Latchney LR, Hand AR, et al. The sld mutation is specific for sublingual salivary mucous cells and disrupts apomucin gene expression. Physiol Genomics. 2003;14:95–106. doi: 10.1152/physiolgenomics.00151.2002. [DOI] [PubMed] [Google Scholar]
- 228.Das B, Cash MN, Hand AR, et al. Tissue distibution of murine Muc19/smgc gene products. J Histochem Cytochem. 2010;58:141–156. doi: 10.1369/jhc.2009.954891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Plauth M, Jenss H, Meyle J. Oral manifestations of Crohn’s disease. An analysis of 79 cases. J Clin Gastroenterol. 1991;13:29–37. doi: 10.1097/00004836-199102000-00008. [DOI] [PubMed] [Google Scholar]
- 230.Wrackmeyer U, Hansen GH, Seya T, et al. Intelectin: a novel lipid raft-associated protein in the enterocyte brush border. Biochemistry. 2006;45:9188–9197. doi: 10.1021/bi060570x. [DOI] [PubMed] [Google Scholar]
- 231.Tsuji S, Uehori J, Matsumoto M, et al. Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall. J Biol Chem. 2001;276:23456–23463. doi: 10.1074/jbc.M103162200. [DOI] [PubMed] [Google Scholar]
- 232.Tsuji S, Yamashita M, Nishiyama A, et al. Differential structure and activity between human and mouse intelectin-1: human intelectin-1 is a disulfide-linked trimer, whereas mouse homologue is a monomer. Glycobiology. 2007;17:1045–1051. doi: 10.1093/glycob/cwm075. [DOI] [PubMed] [Google Scholar]
- 233.Komiya T, Tanigawa Y, Hirohashi S. Cloning of the novel gene intelectin, which is expressed in intestinal paneth cells in mice. Biochem Biophys Res Commun. 1998;251:759–762. doi: 10.1006/bbrc.1998.9513. [DOI] [PubMed] [Google Scholar]
- 234.Pemberton AD, Knight PA, Gamble J, et al. Innate BALB/c enteric epithelial responses to Trichinella spiralis: inducible expression of a novel goblet cell lectin, intelectin-2, and its natural deletion in C57BL/10 mice. J Immunol. 2004;173:1894–1901. doi: 10.4049/jimmunol.173.3.1894. [DOI] [PubMed] [Google Scholar]
- 235.Akiyama Y, Oshima K, Kuhara T, et al. A lactoferrin-receptor, intelectin 1, affects uptake, sub-cellular localization and release of immunochemically detectable lactoferrin by intestinal epithelial Caco-2 cells. J Biochem. 2013;154:437–448. doi: 10.1093/jb/mvt073. [DOI] [PubMed] [Google Scholar]
- 236.Suzuki YA, Shin K, Lonnerdal B. Molecular cloning and functional expression of a human intestinal lactoferrin receptor. Biochemistry. 2001;40:15771–15779. doi: 10.1021/bi0155899. [DOI] [PubMed] [Google Scholar]
- 237.Schaffler A, Neumeier M, Herfarth H, et al. Genomic structure of human omentin, a new adipocytokine expressed in omental adipose tissue. Biochim Biophys Acta. 2005;1732:96–102. doi: 10.1016/j.bbaexp.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 238.de Souza Batista CM, Yang RZ, Lee MJ, et al. Omentin plasma levels and gene expression are decreased in obesity. Diabetes. 2007;56:1655–1661. doi: 10.2337/db06-1506. [DOI] [PubMed] [Google Scholar]
- 239.Yamawaki H, Kuramoto J, Kameshima S, et al. Omentin, a novel adipocytokine inhibits TNF-induced vascular inflammation in human endothelial cells. Biochem Biophys Res Commun. 2011;408:339–343. doi: 10.1016/j.bbrc.2011.04.039. [DOI] [PubMed] [Google Scholar]
- 240.Zhou Z, Liu H, Gu G, et al. Immunoproteomic to identify antigens in the intestinal mucosa of Crohn’s disease patients. PLoS One. 2013;8:e81662. doi: 10.1371/journal.pone.0081662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lu Y, Zhou L, Liu L, et al. Serum omentin-1 as a disease activity marker for Crohn’s disease. Dis Markers. 2014:194–202. doi: 10.1155/2014/162517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Duraes C, Machado JC, Portela F, et al. Phenotype-genotype profiles in Crohn’s disease predicted by genetic markers in autophagy-related genes (GOIA study II) Inflamm Bowel Dis. 2013;19:230–239. doi: 10.1002/ibd.23007. [DOI] [PubMed] [Google Scholar]
- 243.Schaffler A, Zeitoun M, Wobser H, et al. Frequency and significance of the novel single nucleotide missense polymorphism Val109Asp in the human gene encoding omentin in Caucasian patients with type 2 diabetes mellitus or chronic inflammatory bowel diseases. Cardiovasc Diabetol. 2007;6:3. doi: 10.1186/1475-2840-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Pemberton AD, Rose-Zerilli MJ, Holloway JW, et al. A single-nucleotide polymorphism in intelectin 1 is associated with increased asthma risk. J Allergy Clin Immunol. 2008;122:1033–1034. doi: 10.1016/j.jaci.2008.08.037. [DOI] [PubMed] [Google Scholar]
- 245.Doursout MF, Horton H, Hoang L, et al. Lactoferrin moderates LPS-induced hypotensive response and gut injury in rats. Int Immunopharmacol. 2013;15:227–231. doi: 10.1016/j.intimp.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 246.Flores-Villasenor H, Canizalez-Roman A, Velazquez-Roman J, et al. Protective effects of lactoferrin chimera and bovine lactoferrin in a mouse model of enterohaemorrhagic Escherichia coli O157:H7 infection. Biochem Cell Biol. 2012;90:405–411. doi: 10.1139/o11-089. [DOI] [PubMed] [Google Scholar]
- 247.Carlberg C, Campbell MJ. Vitamin D receptor signaling mechanisms: integrated actions of a well-defined transcription factor. Steroids. 2013;78:127–136. doi: 10.1016/j.steroids.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Simmons JD, Mullighan C, Welsh KI, et al. Vitamin D receptor gene polymorphism: association with Crohn’s disease susceptibility. Gut. 2000;47:211–214. doi: 10.1136/gut.47.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Renkema KY, Alexander RT, Bindels RJ, et al. Calcium and phosphate homeostasis: concerted interplay of new regulators. Ann Med. 2008;40:82–91. doi: 10.1080/07853890701689645. [DOI] [PubMed] [Google Scholar]
- 250.Koeffler HP. Vitamin D: myeloid differentiation and proliferation. Haematol Blood Transfus. 1985;29:409–417. doi: 10.1007/978-3-642-70385-0_84. [DOI] [PubMed] [Google Scholar]
- 251.D’Ambrosio D, Cippitelli M, Cocciolo MG, et al. Inhibition of IL-12 production by 1,25-dihydroxyvitamin D3. Involvement of NF-kappaB downregulation in transcriptional repression of the p40 gene. J Clin Invest. 1998;101:252–262. doi: 10.1172/JCI1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lemire JM, Archer DC, Beck L, et al. Immunosuppressive actions of 1,25-dihydroxyvitamin D3: preferential inhibition of Th1 functions. J Nutr. 1995;125:1704S–1708S. doi: 10.1093/jn/125.suppl_6.1704S. [DOI] [PubMed] [Google Scholar]
- 253.Rook GA, Steele J, Fraher L, et al. Vitamin D3, gamma interferon, and control of proliferation of Mycobacterium tuberculosis by human monocytes. Immunology. 1986;57:159–163. [PMC free article] [PubMed] [Google Scholar]
- 254.Duerr RH, Barmada MM, Zhang L, et al. Linkage and association between inflammatory bowel disease and a locus on chromosome 12. Am J Hum Genet. 1998;63:95–100. doi: 10.1086/301929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Morrison NA, Qi JC, Tokita A, et al. Prediction of bone density from vitamin D receptor alleles. Nature. 1994;367:284–287. doi: 10.1038/367284a0. [DOI] [PubMed] [Google Scholar]
- 256.Verbeek W, Gombart AF, Shiohara M, et al. Vitamin D receptor: no evidence for allele-specific mRNA stability in cells which are heterozygous for the Taq I restriction enzyme polymorphism. Biochem Biophys Res Commun. 1997;238:77–80. doi: 10.1006/bbrc.1997.7239. [DOI] [PubMed] [Google Scholar]
- 257.Dresner-Pollak R, Ackerman Z, Eliakim R, et al. The BsmI vitamin D receptor gene polymorphism is associated with ulcerative colitis in Jewish Ashkenazi patients. Genet Test. 2004;8:417–420. doi: 10.1089/gte.2004.8.417. [DOI] [PubMed] [Google Scholar]
- 258.Xue LN, Xu KQ, Zhang W, et al. Associations between vitamin D receptor polymorphisms and susceptibility to ulcerative colitis and Crohn’s disease: a meta-analysis. Inflamm Bowel Dis. 2013;19:54–60. doi: 10.1002/ibd.22966. [DOI] [PubMed] [Google Scholar]
- 259.Naderi N, Farnood A, Habibi M, et al. NOD2 exonic variations in Iranian Crohn’s disease patients. Int J Colorect Dis. 2011;26:775–781. doi: 10.1007/s00384-011-1145-4. [DOI] [PubMed] [Google Scholar]
- 260.Hughes DJ, McManus R, Neary P, et al. Common variation in the vitamin D receptor gene and risk of inflammatory bowel disease in an Irish case-control study. Eur J Gastroenterol Hepatol. 2011;23:807–812. doi: 10.1097/MEG.0b013e328349283e. [DOI] [PubMed] [Google Scholar]
- 261.Noble CL, McCullough J, Ho W, et al. Low body mass not vitamin D receptor polymorphisms predict osteoporosis in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2008;27:588–596. doi: 10.1111/j.1365-2036.2008.03599.x. [DOI] [PubMed] [Google Scholar]
- 262.Froicu M, Weaver V, Wynn TA, et al. A crucial role for the vitamin D receptor in experimental inflammatory bowel diseases. Mol Endocrinol. 2003;17:2386–2392. doi: 10.1210/me.2003-0281. [DOI] [PubMed] [Google Scholar]
- 263.Kong J, Zhang Z, Musch MW, et al. Novel role of the vitamin D receptor in maintaining the integrity of the intestinal mucosal barrier. Am J Physiol Gastrointest Liver Physiol. 2008;294:G208–G216. doi: 10.1152/ajpgi.00398.2007. [DOI] [PubMed] [Google Scholar]
- 264.Kim JH, Yamaori S, Tanabe T, et al. Implication of intestinal VDR deficiency in inflammatory bowel disease. Biochim Biophys Acta. 2013;1830:2118–2128. doi: 10.1016/j.bbagen.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Schepens MA, Schonewille AJ, Vink C, et al. Supplemental calcium attenuates the colitis-related increase in diarrhea, intestinal permeability, and extracellular matrix breakdown in HLA-B27 transgenic rats. J Nutr. 2009;139:1525–1533. doi: 10.3945/jn.109.105205. [DOI] [PubMed] [Google Scholar]
- 266.Laverny G, Penna G, Vetrano S, et al. Efficacy of a potent and safe vitamin D receptor agonist for the treatment of inflammatory bowel disease. Immunol Lett. 2010;131:49–58. doi: 10.1016/j.imlet.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 267.Libioulle C, Louis E, Hansoul S, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Franke A, Hampe J, Rosenstiel P, et al. Systematic association mapping identifies NELL1 as a novel IBD disease gene. PLoS One. 2007;2:e691. doi: 10.1371/journal.pone.0000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Glas J, Seiderer J, Czamara D, et al. PTGER4 expression-modulating polymorphisms in the 5p13.1 region predispose to Crohn’s disease and affect NF-kappaB and XBP1 binding sites. PLoS One. 2012;7:e52873. doi: 10.1371/journal.pone.0052873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Nitta M, Hirata I, Toshina K, et al. Expression of the EP4 prostaglandin E2 receptor subtype with rat dextran sodium sulphate colitis: colitis suppression by a selective agonist, ONO-AE1-329. Scand J Immunol. 2002;56:66–75. doi: 10.1046/j.1365-3083.2002.01096.x. [DOI] [PubMed] [Google Scholar]
- 271.Wallace JL. Prostaglandin biology in inflammatory bowel disease. Gastroenterol Clin North Am. 2001;30:971–980. doi: 10.1016/s0889-8553(05)70223-5. [DOI] [PubMed] [Google Scholar]
- 272.Jiang GL, Nieves A, Im WB, et al. The prevention of colitis by E Prostanoid receptor 4 agonist through enhancement of epithelium survival and regeneration. J Pharmacol Exp Ther. 2007;320:22–28. doi: 10.1124/jpet.106.111146. [DOI] [PubMed] [Google Scholar]
- 273.Kabashima K, Saji T, Murata T, et al. The prostaglandin receptor EP4 suppresses colitis, mucosal damage and CD4 cell activation in the gut. J Clin Invest. 2002;109:883–893. doi: 10.1172/JCI14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Chen Q, Muramoto K, Masaaki N, et al. A novel antagonist of the prostaglandin E(2) EP(4) receptor inhibits Th1 differentiation and Th17 expansion and is orally active in arthritis models. Br J Pharmacol. 2010;160:292–310. doi: 10.1111/j.1476-5381.2010.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Clark P, Rowland SE, Denis D, et al. MF498 [N-{[4-(5,9-Diethoxy-6-oxo-6,8-dihydro-7H-pyrrolo[3,4-g]quinolin-7-yl)-3-methylbe nzyl]sulfonyl}-2-(2-methoxyphenyl)acetamide], a selective E prostanoid receptor 4 antagonist, relieves joint inflammation and pain in rodent models of rheumatoid and osteoarthritis. J Pharmacol Exp Ther. 2008;325:425–434. doi: 10.1124/jpet.107.134510. [DOI] [PubMed] [Google Scholar]
- 276.Yao C, Sakata D, Esaki Y, et al. Prostaglandin E2-EP4 signaling promotes immune inflammation through Th1 cell differentiation and Th17 cell expansion. Nat Med. 2009;15:633–640. doi: 10.1038/nm.1968. [DOI] [PubMed] [Google Scholar]
- 277.Yu Y, Chadee K. Prostaglandin E2 stimulates IL-8 gene expression in human colonic epithelial cells by a posttranscriptional mechanism. J Immunol. 1998;161:3746–3752. [PubMed] [Google Scholar]
- 278.Rodriguez-Lagunas MJ, Martin-Venegas R, Moreno JJ, et al. PGE2 promotes Ca2+-mediated epithelial barrier disruption through EP1 and EP4 receptors in Caco-2 cell monolayers. Am J Physiol Cell Physiol. 2010;299:C324–C334. doi: 10.1152/ajpcell.00397.2009. [DOI] [PubMed] [Google Scholar]
- 279.Lejeune M, Leung P, Beck PL, et al. Role of EP4 receptor and prostaglandin transporter in prostaglandin E2-induced alteration in colonic epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1097–G1105. doi: 10.1152/ajpgi.00280.2010. [DOI] [PubMed] [Google Scholar]
- 280.Gookin JL, Galanko JA, Blikslager AT, et al. PG-mediated closure of paracellular pathway and not restitution is the primary determinant of barrier recovery in acutely injured porcine ileum. Am J Physiol Gastrointest Liver Physiol. 2003;285:G967–G979. doi: 10.1152/ajpgi.00532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 282.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 283.Henckaerts L, Vermeire S. NOD2/CARD15 disease associations other than Crohn’s disease. Inflamm Bowel Dis. 2007;13:235–241. doi: 10.1002/ibd.20066. [DOI] [PubMed] [Google Scholar]
- 284.Buhner S, Buning C, Genschel J, et al. Genetic basis for increased intestinal permeability in families with Crohn’s disease: role of CARD15 3020insC mutation? Gut. 2006;55:342–347. doi: 10.1136/gut.2005.065557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Kosovac K, Brenmoehl J, Holler E, et al. Association of the NOD2 genotype with bacterial translocation via altered cell-cell contacts in Crohn’s disease patients. Inflamm Bowel Dis. 2010;16:1311–1321. doi: 10.1002/ibd.21223. [DOI] [PubMed] [Google Scholar]
- 286.Ferby I, Reschke M, Kudlacek O, et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- 287.Fiorentino L, Pertica C, Fiorini M, et al. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol Cell Biol. 2000;20:7735–7750. doi: 10.1128/mcb.20.20.7735-7750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hackel PO, Gishizky M, Ullrich A. Mig-6 is a negative regulator of the epidermal growth factor receptor signal. Biol Chem. 2001;382:1649–1662. doi: 10.1515/BC.2001.200. [DOI] [PubMed] [Google Scholar]
- 289.Hopkins S, Linderoth E, Hantschel O, et al. Mig6 is a sensor of EGF receptor inactivation that directly activates c-Abl to induce apoptosis during epithelial homeostasis. Dev Cell. 2012;23:547–559. doi: 10.1016/j.devcel.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Banan A, Fields JZ, Zhang Y, et al. Key role of PKC and Ca2+ in EGF protection of microtubules and intestinal barrier against oxidants. Am J Physiol Gastrointest Liver Physiol. 2001;280:G828–G843. doi: 10.1152/ajpgi.2001.280.5.G828. [DOI] [PubMed] [Google Scholar]
- 291.Frey MR, Golovin A, Polk DB. Epidermal growth factor-stimulated intestinal epithelial cell migration requires Src family kinase-dependent p38 MAPK signaling. J Biol Chem. 2004;279:44513–44521. doi: 10.1074/jbc.M406253200. [DOI] [PubMed] [Google Scholar]
- 292.McCole DF, Rogler G, Varki N, et al. Epidermal growth factor partially restores colonic ion transport responses in mouse models of chronic colitis. Gastroenterology. 2005;129:591–608. doi: 10.1016/j.gastro.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 293.Wright NA, Pike C, Elia G. Induction of a novel epidermal growth factor-secreting cell lineage by mucosal ulceration in human gastrointestinal stem cells. Nature. 1990;343:82–85. doi: 10.1038/343082a0. [DOI] [PubMed] [Google Scholar]
- 294.Yamaoka T, Frey MR, Dise RS, et al. Specific epidermal growth factor receptor autophosphorylation sites promote mouse colon epithelial cell chemotaxis and restitution. Am J Physiol Gastrointest Liver Physiol. 2011;301:G368–G376. doi: 10.1152/ajpgi.00327.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Forsyth CB, Banan A, Farhadi A, et al. Regulation of oxidant-induced intestinal permeability by metalloprotease-dependent epidermal growth factor receptor signaling. J Pharmacol Exp Ther. 2007;321:84–97. doi: 10.1124/jpet.106.113019. [DOI] [PubMed] [Google Scholar]
- 296.Raimondi F, Santoro P, Barone MV, et al. Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Am J Physiol Gastrointest Liver Physiol. 2008;294:G906–G913. doi: 10.1152/ajpgi.00043.2007. [DOI] [PubMed] [Google Scholar]
- 297.Tripathi A, Lammers KM, Goldblum S, et al. Identification of human zonulin, a physiological modulator of tight junctions, as prehaptoglobin-2. Proc Natl Acad Sci U S A. 2009;106:16799–16804. doi: 10.1073/pnas.0906773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Plevy S, Silverberg MS, Lockton S, et al. Combined serological, genetic, and inflammatory markers differentiate non-IBD, Crohn’s disease, and ulcerative colitis patients. Inflamm Bowel Dis. 2013;19:1139–1148. doi: 10.1097/MIB.0b013e318280b19e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Toedter G, Li K, Sague S, et al. Genes associated with intestinal permeability in ulcerative colitis: changes in expression following infliximab therapy. Inflamm Bowel Dis. 2012;18:1399–1410. doi: 10.1002/ibd.22853. [DOI] [PubMed] [Google Scholar]