Abstract
The human genome contains about 1.5 million Alu elements, which are transcribed into Alu RNAs by RNA polymerase III. Their expression is upregulated following stress and viral infection, and they associate with the SRP9/14 protein dimer in the cytoplasm forming Alu RNPs. Using cell-free translation, we have previously shown that Alu RNPs inhibit polysome formation. Here, we describe the mechanism of Alu RNP-mediated inhibition of translation initiation and demonstrate its effect on translation of cellular and viral RNAs. Both cap-dependent and IRES-mediated initiation is inhibited. Inhibition involves direct binding of SRP9/14 to 40S ribosomal subunits and requires Alu RNA as an assembly factor but its continuous association with 40S subunits is not required for inhibition. Binding of SRP9/14 to 40S prevents 48S complex formation by interfering with the recruitment of mRNA to 40S subunits. In cells, overexpression of Alu RNA decreases translation of reporter mRNAs and this effect is alleviated with a mutation that reduces its affinity for SRP9/14. Alu RNPs also inhibit the translation of cellular mRNAs resuming translation after stress and of viral mRNAs suggesting a role of Alu RNPs in adapting the translational output in response to stress and viral infection.
INTRODUCTION
Translation regulation ensures protein homeostasis under normal growth conditions. In addition, modulating the translation efficiency of already existing mRNAs provides a rapid mechanism for adapting protein expression in development, memory formation, synaptic plasticity and in response to stress and apoptotic signals. Although translational control may occur at different steps during protein synthesis, the majority of the mechanisms target translation initiation (1,2).
The canonical pathway for translation initiation proceeds through sequential formation of several initiation complexes (3). Ternary complex and the initiation factors bind to the 40S ribosomal subunit yielding the 43S preinitiation complex, which is then loaded onto the 5′-proximal region of an mRNA and scans for the initiation codon. Once the initiation codon is positioned at the P-site, the 48S initiation complex is assembled. The joining of the 60S ribosomal subunit leads to formation of the 80S initiation complex, which then proceeds to elongation cycles.
Translation initiation is regulated by the orchestrated action of multiple control mechanisms, many of which remain to be elucidated. We have previously shown that the noncoding, retrotransposon-derived Alu RNA in complex with the heterodimeric protein SRP9/14 (forming together the Alu RNP) is able to diminish polysome formation in a wheat germ translation system and thus, most likely interferes with translation initiation (4). These data raised the intriguing possibility that Alu RNPs are involved in a novel translational control pathway.
Alu elements are derived from the 7SL RNA gene (5), which encodes the RNA moiety of the signal recognition particle (SRP), and following retrotransposition Alu elements now make up over 10% of the human genome mass. The modern Alu element is composed of two similar, but not identical monomers named the left arm and the right arm (Figure 1A). Based on mutations at diagnostic positions, Alu elements are divided into three families of different evolutionary ages (8). The most ancient family is named AluJ followed by AluS and AluY. Alu elements can be part of a polymerase II transcription unit and are therefore present in pre-mRNAs and mRNAs. These embedded Alu sequences can influence gene expression in many ways for example by providing alternative splice sites as well as target sites of miRNAs and editing enzymes (6).
Figure 1.
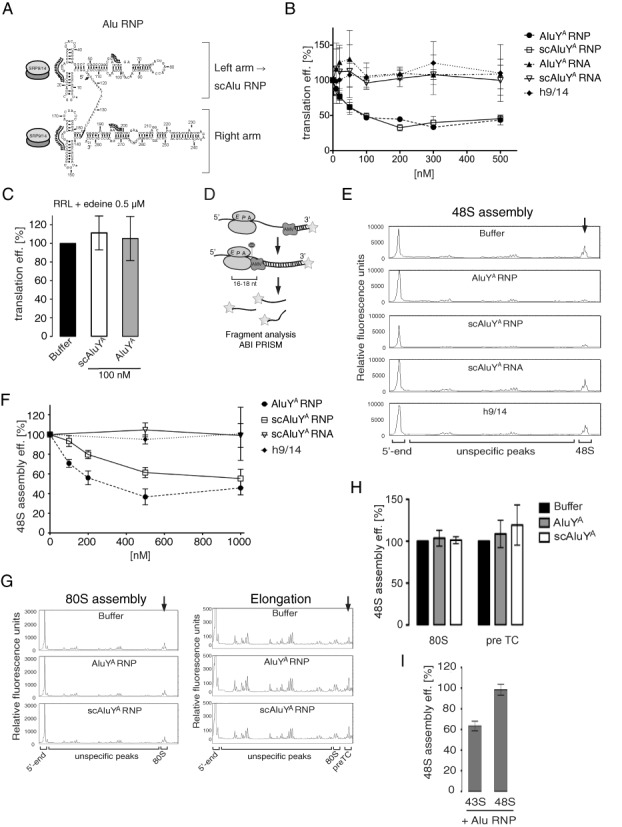
Alu RNPs repress translation initiation by inhibiting 48S complex formation. (A) Secondary structure model of AluYA RNA. Binding sites of h9/14 are indicated by analogy to SRP RNA. Preparation of Alu RNPs is described in Supplementary Figure S1. (B) Translation efficiencies of uncapped pPL mRNA in RRL supplemented with increasing concentrations of Alu RNPs or the h9/14 protein or the AluYA/scAluYA RNAs alone. [35S]-labeled proteins were resolved by SDS-PAGE and quantified by phosphorimager (see Supplementary Figure S2A and B for SDS-PAGE autoradiographs). Translation efficiency was calculated as a percentage of the value obtained in the absence of RNPs. Error bars are shown as SD (n ≥ 3). (C) Translation efficiencies of uncapped pPL mRNA in RRL translation reaction synchronized by 0.5 μM edeine in the presence of 100 nM AluYA/scAluYA RNP. Translation reactions were incubated 3 min at 30°C before the addition of 0.5 μM edeine. Translation mixtures were incubated another 1 min and then supplemented with the RNPs. See also Supplementary Figure S2C. (D) Schematic representation of the toeprint analysis. Reverse transcription by avian myeloblastosis virus reverse transcriptase (AMV) stalls upon encounter with ribosomal complexes. Fluorescently labeled cDNAs were analyzed by capillary electrophoresis. (E) Toeprinting analysis of 48S complexes reconstituted in the presence of 500 nM Alu RNPs, h9/14 or scAluYA RNA. See also Supplementary Figure S2D for toeprint analysis of MVHL-stop mRNA in the absence of 40S and translation factors. (F) Relative efficiency of 48S complex formation in the presence of increasing concentrations of Alu RNPs, h9/14 or scAluYA RNA. To calculate the efficiency of 48S complex assembly, the intensity of the 48S toeprint signal was quantified as described in ‘Materials and Methods’ section. The relative efficiency of 48S complex assembly was expressed as a percentage of the value obtained in the absence of RNPs (buffer). Error bars are shown as SD (n ≥ 3). (G) Toeprint analysis of 80S complexes (left panel) and preTC (right panel) reconstituted in the presence of 500 nM AluYA RNP or 1 μM scAluYA RNP. Quantification is shown in (H). Error bars are shown as SD (n ≥ 3). (I) Relative efficiency of 48S complex assembly in the presence of 500 nM scAluYA RNP added either to the preassembled 43S complexes (43S) or to preassembled 48S complexes (48S). Error bars are shown as SD (n ≥ 3).
Alu elements can also be transcribed by RNA polymerase III into the noncoding dimeric Alu RNA (7), and under normal physiological conditions their expression is thought to be repressed by various cis- and trans-acting factors (8) most likely to keep retrotransposition events at low levels. However, their abundance is increased about 10-fold and higher in response to heat shock and viral infection (6). Relatively little is known about cellular functions of Alu RNA. It has been shown to inhibit pol. II-dependent transcription, and its inhibitory activity was assigned to the right arm (9).
The dimeric Alu RNA transcripts can be further processed into small cytoplasmic Alu (scAlu) RNA by endonucleolytic cleavage (10) (Figure 1A), and Alu and scAlu RNAs have been shown to form complexes comprising SRP9/14 and, for the dimeric Alu RNP, the poly(A)-binding protein (11–13).
SRP9/14 was discovered as a subunit of SRP, which binds the Alu portion of 7SL RNA. The protein is required for the elongation arrest activity of SRP, which ensures efficient protein translocation into the endoplasmic reticulum (ER) in mammalian cells (14). In addition to forming complexes with 7SL RNA and Alu RNA, the majority of SRP9/14 exists as a free protein in primate cells (11).
In the present study, we have investigated the role of Alu RNPs in regulating translation using both a rabbit reticulocyte lysate and a mammalian translation system reconstituted from purified components. Our studies demonstrate that Alu RNPs interfere with the formation of the 48S complex by a novel mechanism in which Alu RNPs act directly on the 40S ribosomal subunit to prevent its association with the mRNA. Moreover, the SRP9/14 protein dimer constitutes the inhibitory component, which has to be transferred to 40S by the Alu RNA. This mechanism efficiently inhibits cap-dependent as well as IRES-mediated translation initiation. Overexpression of Alu RNA in HEK 293T cells inhibits in an SRP9/14-dependent fashion translation of mRNAs that initiate protein synthesis de novo. Our results are consistent with a physiological role of Alu RNPs in the translational response to viral infection and stress.
MATERIALS AND METHODS
Plasmid construction, in vitro transcription, purification of the proteins, RNPs and ribosomal subunits, aminoacylation of tRNA, ribosome binding assay, cell culture and northern blot are described in the ‘Extended Methods’ section.
In vitro translation in RRL
RRL (Green Hectares, Oregon, USA) was treated with micrococcal nuclease as described (49) with some modifications: micrococcal nuclease concentration was decreased to 75 U/ml final and incubation time was reduced to 10 min at 20°C. Translation reactions containing L-[35S]-methionine (Anawa, Zurich, Switzerland) were carried out in the presence of 100 mM potassium acetate and 1.5 mM magnesium acetate in a final volume of 10 μl at 30°C for 20 min. Translation mix was preincubated with Alu RNPs, Alu RNAs or h9/14 for 3 min at 30°C prior to addition of 125 ng mRNA. For edeine-synchronized translation the reaction programmed with 125 ng of uncapped pPL mRNA was first incubated for 3 min at 30°C and then the initiation was arrested by the addition of edeine to 0.5 μM. The reaction was incubated one more minute before the addition of 100 nM Alu RNPs and translation continued for 15 min. Synthesized polypeptides were resolved by SDS-PAGE and quantified by phosphorimager analysis.
Assembly and analysis of translation complexes
For 43S complex assembly 75 nM 40S ribosomal subunits were incubated with 500 nM eIF2, 100 nM eIF3, 200 nM eIF1, 200 nM eIF1A and 75 nM Met-tRNAiMet for 10 min at 37°C. To assemble the 48S complex on the MVHL-stop mRNA the 43S complex reaction was supplemented with 200 nM eIF4GI738–1116, 200 nM eIF4A, 200 nM eIF4B, 37.5 nM mRNA and incubated at 37°C for another 10 min. For toeprint assays, the mRNA was incubated with fluorescently labeled oligonucleotide for 10 min at 37°C before addition to the assembly reaction. The 80S complex was assembled by incubating the 48S complex with 200 nM eIF5, 200 nM eIF5B and 100 nM 60S subunits at 37°C for 10 min. Peptide elongation reaction and assembly of preTC was performed by incubation of the 80S complex with 300 nM eEF1, 50 nM eEF2 and 100 nM aminoacylated total calf liver tRNA for 10 min at 37°C. All assembly reactions were performed in buffer containing 20 mM Tris–HCl pH 7.5, 120 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 0.25 mM spermidine, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 2 mM adenosine triphosphate (ATP )and 0.2 mM guanosine triphosphate (GTP).
Reaction mixtures for 48S complex assembly on HCV-NS’ mRNA contained 75 nM 40S subunits, 500 nM eIF2, 100 nM eIF3 and 37.5 nM HCV-NS’ mRNA. The CrPV-40S complex was obtained by incubating 75 nM 40S subunits with 75 nM CrPV-VHLM mRNA.
Toeprint assay was performed as described previously (19) except that reverse transcription reaction was incubated for 15 min at 37°C. For quantification, the toeprint signal from the translation complex was expressed as a percentage of the combined intensities of itself, the signal from the 5′-end of the mRNA and all peaks between them. Toeprint signals on CrPV-VHLM RNA were quantified using invariable peaks in the area between AUG and 3′-end of the mRNA. Briefly, the intensity of 48S toeprint was normalized to one of the three different peaks which intensity was constant throughout different experiments. All three normalizations gave identical results.
43S and 48S complexes assembled with [35S]-Met-tRNAiMet and [32P]-MVHL-stop mRNA were subjected to centrifugation through 5–25% sucrose gradient containing 20 mM Tris–HCl pH 7.5, 100 mM potassium acetate, 5 mM MgCl2, 2 mM DTT in a Beckman SW60 rotor at 60000 rpm for 2 h. Fractions were analyzed by scintillation counting.
Transfection, luciferase assay and metabolic labeling
HEK 293T cells were transfected with TransIT-T2020 reagent (Mirus) according to manufacturer's protocol. Luciferase assays were performed 24 h post-transfection using Luciferase Assay Kit (Promega). For estradiol-induced luciferase expression, HEK 293T cells were transiently transfected with a GAL-responsive luciferase reporter plasmid and with a plasmid expressing the protein GAL4 fused to the hormone binding domain of human estrogen receptor. Cells were stimulated with 100 nM estradiol at 24 h post-transfection and bioluminescence was recorded continuously for 24 h. Metabolic labeling with [35S]-methionine/cysteine was performed 48 h post-transfection. After 15 min, pulse labeling cells were washed twice with ice-cold phosphate buffered saline and lysed in buffer containing 20 mM Tris–HCl pH 7.5, 300 mM KCl, 1% Triton X-100, 0.1% SDS, 1 mM EGTA and protease inhibitors. Aliquots of lysates were precipitated with 10% TCA, pellets were boiled for 15 min washed twice with 10% TCA followed by acetone, dissolved in 1% SDS and analyzed by scintillation counting. Where indicated, cells were treated with 500 μM sodium arsenite for 30 min then the media was changed and cells were allowed to recover for 0, 30, 60, 90 and 120 min prior to pulse labeling.
Viral infection
HEK293T cells were transfected with pDL7enh, pDLscAlu or pDL4.5S and 24 h later infected with VSV-PeGFP (50) at a multiplicity of infection of 10 PFU/cell in Dulbecco's modified Eagle's medium (DMEM) 2% fetal bovine serum (FBS). Six hours post-infection, the media was changed and cells were pulse labeled with [35S]-methionine/cysteine for 15 min, proteins were precipitated in 10% TCA, resolved by SDS-PAGE and quantified by phosphorimager analysis.
Statistics
Error bars in the figures represent the standard deviation (SD) or where indicated as the standard error of the mean (SEM). Statistical analysis was performed using an unpaired two-tailed Student's t test. Analysis of the luciferase expression data was done using the paired two-tailed t test.
RESULTS
Alu RNPs inhibit assembly of 48S initiation complex
Expression of the three different families of Alu elements has so far been poorly investigated. Based on cDNA analysis (15,16), the AluY elements are most abundantly expressed. We therefore chose to use in vitro transcribed AluYA and scAluYA RNAs derived from the Alu element in the human α-fetoprotein gene (Table 1) together with human SRP9/14 (h9/14) to assemble AluYA and scAluYA RNPs, respectively. The RNPs were purified by gel filtration chromatography, and the integrity of the RNA moiety was confirmed by denaturing polyacrylamide gel electrophoresis followed by ethidium bromide staining (Supplementary Figure S1B). Western blot analysis demonstrated that the AluYA and the scAluYA RNPs contained, respectively, two and one molecules of h9/14 per molecule of RNA (Supplementary Figure S1C).
Table 1. Alu RNAs and Alu RNPs used in this study.
| RNP | Family | Origin | Protein component |
|---|---|---|---|
| AluYA | Y | Dimeric Alu, human α-fetoprotein gene, NG_023028.1 | h9/14 |
| scAluYA | Y | Left arm Alu, human α-fetoprotein gene, NG_023028.1 | h9/14 |
| scAluYL | Y | Left arm Alu, human low-density lipoprotein receptor gene NM_001195803.1 | h9/14 |
| scAluYa5 | Y | Left arm Alu, Alu Ya5a consensus sequence | h9/14 |
| scAluYf2 | Y | Left arm, Alu Yf2a consensus sequence | h9/14 |
| scAluYj4 | Y | Left arm, Alu Yj4a consensus sequence | h9/14 |
| SA151 | 7SL | SRP-Alu domain, nucleotides 1–99 and 251–299 of the human 7SL RNA gene | h9/14 |
| SA110 | 7SL | SRP-Alu domain, nucleotides 1–74 and 271–299 of the human 7SL RNA gene | h9/14 |
| SA86 | 7SL | SRP-Alu domain, nucleotides 1–64 and 283–299 of the human 7SL RNA gene | h9/14 |
| scAluYA 14A5 | Y | Left arm Alu, human α-fetoprotein gene | h9/14A5 |
| scAluYA 9–3A | Y | Left arm Alu, human α-fetoprotein gene | h9-3A/14 |
| scAluYNF1 | Y | Left arm Alu, human NF1 gene (51) | - |
aConsensus sequences were taken from Repeat Masker library, www.repeatmasker.org.
To examine the effects of AluYA and scAluYA RNPs on translation we used a rabbit reticulocyte lysate (RRL) programmed with bovine preprolactin (pPL) mRNA. Addition of either AluYA or scAluYA RNPs diminished the translation efficiencies of pPL mRNA in a similar, dose-dependent manner whereas the addition of RNA or protein alone had no effect (Figure 1B). Translation efficiency was reduced to 50% in the presence of 100 nM AluYA or scAluYA RNPs and, surprisingly, remained unchanged up to 5-fold higher RNP concentrations indicating that the inhibitory effect reached saturation.
In contrast, Alu RNPs were unable to inhibit the synthesis of pPL when added to translation reactions that were after a short incubation treated with 0.5 μM edeine (‘Materials and Methods’ section). Edeine inhibits initiation but allows translation of already formed initiation complexes (17) (Figure 1C). This shows experimentally that Alu RNPs interfere with initiation in RRL translation reactions.
We next studied the effect of Alu RNPs on the assembly of different translation complexes from purified mammalian translation factors and high-salt purified ribosomal subunits (18). The assembly efficiency was determined by primer extension inhibition (toeprinting) using fluorescently labeled DNA primers and capillary electrophoresis (Figure 1D) (19,20).
To study the effect of Alu RNPs on 48S complex assembly, we combined initiation factors, 40S ribosomal subunits, methionyl-tRNAiMet, ATP, GTP and Alu RNPs prior to addition of the mRNA (Material and methods). In the presence of either AluYA or scAluYA RNPs a dose-dependent decrease in the efficiency of 48S complex formation was observed reminiscent of the dose-dependent decrease of translation observed in RRL. Like in RRL, the RNA or protein components alone had no effect on 48S complex formation (Figure 1E and F). Interestingly, the inhibition seen for AluYA RNP was stronger than for scAluYA RNP suggesting that both arms contribute individually to the activity (Figure 1F). This difference was not observed in the RRL, probably because the less stable right arm of the AluYA RNP was partially degraded during the translation reaction (4).
We further analyzed the effect of Alu RNPs on 80S complex assembly and elongation. To this end, Alu RNPs were added to the reconstituted 48S and 80S complexes before forming 80S and pre-termination complexes (preTC), respectively ((‘Materials and Methods’ section). We observed no significant changes in the efficiencies of 80S initiation complex formation and of preTC formation in the presence of AluYA and scAluYA RNPs (Figure 1G and H).
These results demonstrated that Alu RNPs inhibit translation initiation in mammalian translation systems consistent with the differences observed in the polysome profiles using the wheat germ translation system (4). The inhibitory effect occurs prior to formation of the 48S complex. In the reconstituted mammalian system, the inhibition was maximal at 5-fold higher particle concentration (500 nM) than in RRL (100 nM), which we attributed to absolute and relative differences in the abundances of active ribosomes and initiation factors as compared to RRL. Moreover, a fraction of the Alu particles might be inactivated by nonspecific binding to components present at elevated and imbalanced concentrations in the reconstituted system.
AluY RNA but not the Alu portion of 7SL RNA confers inhibitory activity to the Alu RNP
As mentioned before Alu RNPs are ancestrally related to the Alu domain of SRP. However, the latter inhibits nascent chain elongation and has no function in translation initiation. We wanted to examine whether the Alu domain alone, separated from the S domain of SRP, was able to inhibit translation initiation. We synthesized RNA representing truncated versions of the 7SL Alu domain (SA86, SA110 and SA151, Supplementary Figure S3A, Table 1). Furthermore, we produced AluY RNAs representing the different subfamilies of the genomic AluY elements to generalize our findings on AluY RNPs (Table 1, Supplementary Figure S3C). Particles were purified and analyzed as before and their RNA and protein contents were consistent with a 1:1 stoichiometry of the complexes (Supplementary Figure S3E).
The effect of the AluYL and SA110 RNPs on translation efficiency was monitored at different concentrations in the pPL-primed RRL. The scAluYL RNP decreased the translation efficiency in a concentration-dependent way reminiscent of the results obtained for the scAluYA RNP (Figures 1B and 2A). In contrast, the SA110 RNP had no effect at all concentrations tested. Analysis of the activities of three additional AluY RNPs and of SA86 and SA151 RNPs showed that all AluY RNPs inhibited translation whereas SA151 and SA86 RNPs had no effect (Figure 2B).
Figure 2.
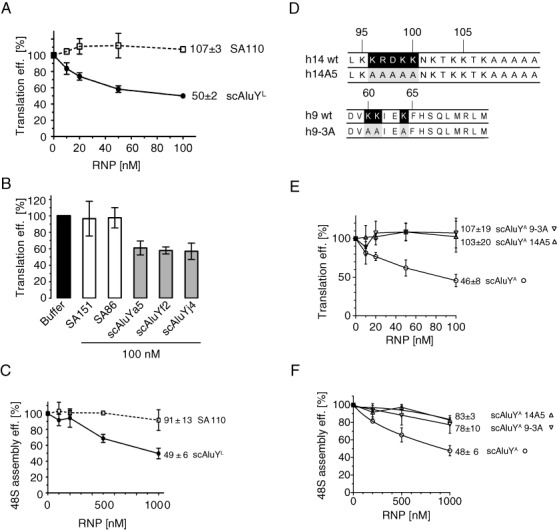
The inhibitory activity of the Alu RNP depends on the identity of its RNA component and the presence of the two positively charged patches in SRP9/14. (A) Translation efficiency of uncapped pPL mRNA in RRL supplemented with increasing concentrations of the SA110 or the scAluYL RNP. Error bars are shown as SD (n ≥ 3). See also Supplementary Figure S3B. (B) Translation efficiency of uncapped pPL mRNA in RRL supplemented with 100 nM SA151, SA86, scALuYa5, scAluYf2 or scAluYj4 RNPs. Error bars are shown as SD (n ≥ 3). See also Supplementary Figure S3D. (C) Relative efficiency of 48S complex formation in the presence of increasing concentrations of SA110 or scAluYL RNPs. Error bars are shown as SD (n ≥ 3). (D) Mutations introduced in the h14 and h9 proteins. Numbers correspond to the human sequence. Amino acid residues of interest are shown in black. Mutated residues are shown in grey. (E) Translation efficiency of uncapped pPL mRNA in RRL supplemented with increasing concentrations of scAluYA, scAluYA 14A5 or scAluYA 9-3A RNPs. Error bars are shown as SD (n ≥ 3). See also Supplementary Figure S4B. (F) Relative efficiency of 48S complex formation in the presence of increasing concentrations of scAluYA, scAluYA 14A5 or scAluYA 9-3A RNPs. Error bars are shown as SD (n ≥ 3).
We chose two representative RNPs, scAluYL and the SA110 RNPs, to determine the effects on 48S complex formation. We observed a concentration-dependent decrease in the formation of the 48S complex for the scAluYL RNP whereas the SA110 RNP had no detectable effect at all concentrations (Figure 2C).
Hence, despite a remarkable sequence similarity between the SRP Alu RNA and the AluY RNAs, only the AluY RNPs, and not the SRP Alu domain, possess the inhibitory activity.
The functional activity of the Alu RNP depends on specific amino acid residues in SRP9/14, which are also required for elongation arrest activity of SRP
Mutational analysis of SRP9/14 revealed that it contains two positively charged patches of amino acid residues, a basic pentapeptide in the C-terminal region of SRP14 and three lysines in SRP9 (Figure 2D, residues labeled in black) that are essential for the elongation arrest activity of the SRP (14,21). The two patches are located in proximity in SRP9/14 and were therefore proposed to form together a functional minidomain (21). We decided to investigate the possibility that the same amino acid residues may also be required for Alu RNP-mediated inhibition of translation. We assembled scAluYA RNPs with the mutated proteins h9/14A5 and h9-3A/14 (Figure 2D). The scAluYA RNA formed stable complexes with both mutated protein dimers (Supplementary Figure S4A).
When added to pPL-primed RRL the scAlu RNPs containing h9-3A/14 and h9/14A5 completely failed to inhibit translation (Figure 2E), and each particle had a strongly reduced capacity to inhibit formation of the 48S complex (Figure 2F).
These results demonstrated that SRP9/14 is essential for the inhibitory activity of the Alu RNP, and that the functional activity of the Alu RNP involves the same minidomain in SRP9/14 as the elongation arrest activity of SRP.
Alu RNPs prevent association of the 43S complex with the mRNA
Next, we wanted to address the question whether Alu RNPs interfere with the formation of the 48S complex, which is to say with the recruitment of the mRNA to the 43S complex, or whether they already inhibit 43S complex formation. To this end, we reconstituted 43S and 48S complexes from purified components in the presence of the AluYA or scAluYA RNPs and as a negative control we used the SA110 RNP. Formation of 43S and 48S complexes was performed in the presence of [35S]-Met-tRNAiMet or [32P]-labeled MVHL-stop mRNA, respectively, and the assembly reactions were fractionated by sucrose gradient centrifugation to monitor the distribution of the radioactive components.
None of the Alu RNPs affected the association of [35S]-Met-tRNAiMet with the 40S subunit (Figure 3A). In contrast, a significant reduction in [32P]-labeled mRNA was found to comigrate with 40S subunits in the presence of AluYA and scAluYA RNPs and a corresponding increase of free RNA was observed in the upper fractions (Figure 3B). As also observed in the toeprint assay (Figure 1E), the effect was more pronounced for the AluYA RNP than for the scAluYA RNP. Addition of the inactive SA110 RNP to the assembly reaction did not affect the association of the mRNA with the 40S subunit.
Figure 3.
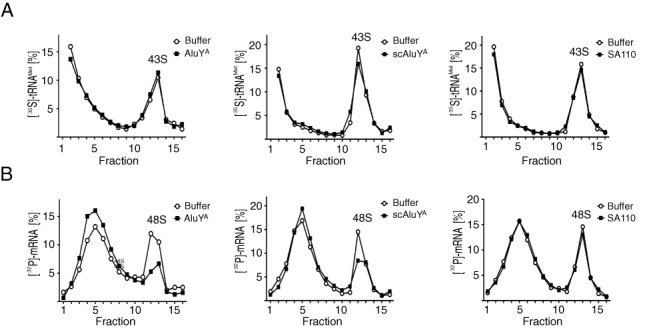
Alu RNPs do not affect 43S complex formation but prevent its recruitment to the mRNA. (A, B) 43S and 48S complexes were reconstituted in the presence of 0.5 μM AluYA, scAluYA or SA110 RNPs and either [35S]-Met-tRNAiMet (A) or [32P]-labeled MVHL-stop mRNA (B). Reconstitution reactions were fractionated on 5–20% sucrose gradients. Results are expressed as percentage of total cpm across all fractions. All experiments were repeated twice.
To confirm that Alu RNPs inhibited initiation after 43S complex formation, we added the scAluYA RNP to the preassembled 43S complexes prior to addition of mRNA. The intensity of the 48S toeprint was strongly decreased (Figure 1I). In contrast, the addition of the scAluYA RNP to the assembled 48S complexes did not cause its dissociation.
Taken together these results indicate that Alu RNPs prevent the association of the 43S complex with the mRNA. Furthermore, Alu RNPs cannot displace the mRNA, once the 48S complex is assembled.
Alu RNPs inhibit initiation of capped mRNAs with different 5′ UTRs
So far, we used uncapped mRNAs in the experiments. However, translation of mRNAs in vivo usually depends on the cap structure. In addition, most translation regulators affect mRNA recruitment to the 43S complex by interacting with the 5′ or 3′ UTR of the mRNA. To investigate whether the cap structure and the 5′ UTR affect the inhibitory activity of Alu RNPs, we generated reporter constructs in which the Firefly luciferase coding sequence (Fluc) was fused to the 5′ UTRs of the β-globin (β-glo), β-actin (β-act) and LINE-1 (L1) mRNAs. The β-glo and β-act mRNAs have relatively short 5′ UTRs (53 and 84 nucleotides, respectively) while the L1 5′ UTR is GC-rich and 900 nucleotides in length. Uncapped and capped transcripts of the three reporter constructs were translated in RRL in the presence of AluYA and scAluYA RNPs. Translation efficiencies were not significantly different between the capped and uncapped transcripts and between the reporter mRNAs comprising different 5′ UTRs (Figure 4A). They were reduced by 40–60% in the presence of both Alu RNPs (Figure 4A). Hence, Alu RNPs can inhibit cap-dependent translation of mRNAs with different 5′ UTRs.
Figure 4.
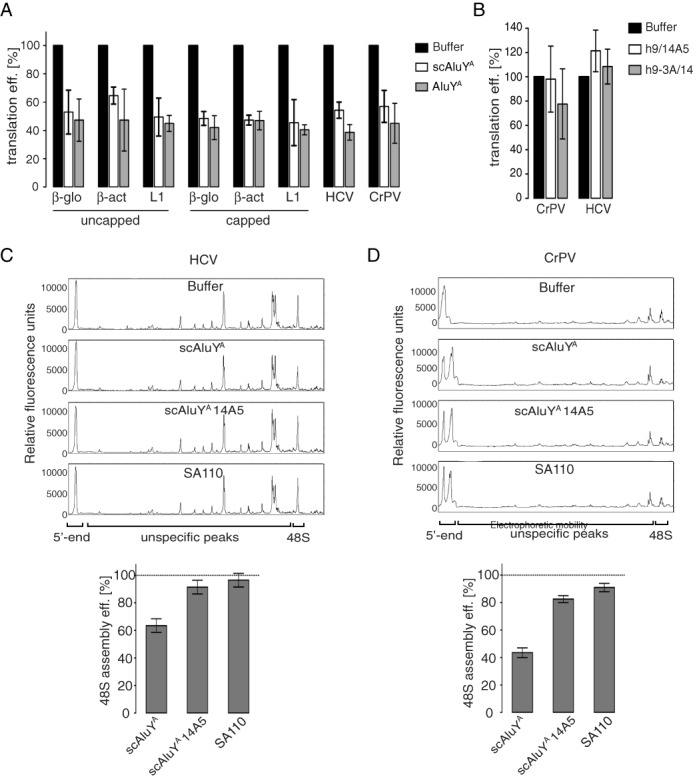
Alu RNPs inhibit cap-dependent and IRES-dependent translation. Translation efficiencies in RRL of the indicated reporter mRNAs in the presence of (A) 100 nM AluYA or scAluYA RNP; (B) 100 nM scAluYA 14A5 RNP or scAluYA 9–3A RNP. The translation efficiency of each mRNA in the absence of the RNP (buffer) was set to 100%. Error bars are shown as SD (n ≥ 3). See also Supplementary Figure S5A. (C) Toeprint analysis of 48S complex assembly on HCV-NS’ mRNA (23) in the presence of 1 μM Alu RNPs as indicated. Quantification is shown in lower panel. Dashed line indicates 48S complex assembly in the absence of RNPs, which was set to 100%. Error bars are shown as SD (n ≥ 3). (D) Toeprint analysis of 48S complex assembly on CrPV-VHLM mRNA in the presence of 0.5 μM Alu RNPs. Quantification is shown in lower panel. Dashed line indicates 48S complex assembly in the absence of RNPs, which was set to 100%. Error bars are shown as SD (n ≥ 3). See also Supplementary Figure S5B.
Alu RNPs inhibit IRES-dependent translation initiation
Many viruses have evolved alternative mechanisms of translation initiation that use internal ribosome entry sites (IRES) in the 5′-UTRs of viral mRNAs. IRES-mediated initiation is cap-independent and bypasses the requirements for certain initiation factors.
To determine whether Alu RNPs could also block IRES-mediated translation initiation, we made reporter constructs containing IRES sequences from the hepatitis C virus (HCV) and the cricket paralysis virus (CrPV) upstream of Fluc. Formation of the 48S complex on HCV IRES is dependent on eIF3 and either eIF2/eIF5 or eIF5B, whereas the CrPV IRES directly recruits the 40S subunit to the start codon without the need for initiation factors (22).
Both AluYA and scAluYA RNPs reduced the translation efficiencies of both reporter mRNAs by approximately 50% (Figure 4A). Furthermore, the Alu RNPs containing the mutated h9/14A5 or h9–3A/14 did not significantly inhibit CrPV- and HCV-mediated translation (Figure 4B).
We also performed 48S complex assembly on CrPV-VHLM (see Supplementary Methods) and the HCV-NS’ (23) mRNAs containing the HCV and the CrPV IRES, respectively. In both cases, 48S complex formation was specifically inhibited by the scAluYA RNP whereas the SA110 RNP and the scAluYA 14A5 RNP, the negative controls, did not significantly reduce the formation of 48S initiation complexes (Figure 4C and D).
We conclude that the mechanism of translation inhibition by Alu RNPs is neither mRNA template- nor initiation factor-dependent, which suggested to us that the Alu RNP might act directly on the 40S subunit.
Alu RNA promotes functional binding of SRP9/14 to 40S subunits
In parallel experiments in our laboratory, we have recently discovered that SRP9/14 can bind directly to 40S subunits and that binding to 40S is required for stress granule localization. Furthermore, we found that binding of SRP9/14 to 40S subunits and Alu RNA is mutually exclusive (24). However, translation initiation is only inhibited by the Alu RNP and not by the protein alone, and both the RNA and protein moieties are essential for the inhibitory activity of the Alu RNP. To study the roles of the RNA and the protein components of the Alu RNP in inhibiting initiation, we performed binding experiments. High salt purified 40S and 60S subunits were incubated with the scAluYA RNP and with h9/14 alone, and the complexes fractionated on sucrose gradients.
Upon addition of the scAluYA RNP to either of the two subunits, we found one third of h9/14 comigrating with 40S subunits whereas less than 10% comigrated with the 60S subunits (Figure 5A). Remarkably, the scAluYA RNA migrated exclusively in the top fractions consistent with the notion that the 40S-bound h9/14 was dissociated from the Alu RNA. In contrast, free h9/14 bound quantitatively and nonselectively to both the 40S and the 60S subunits (Figure 5B).
Figure 5.
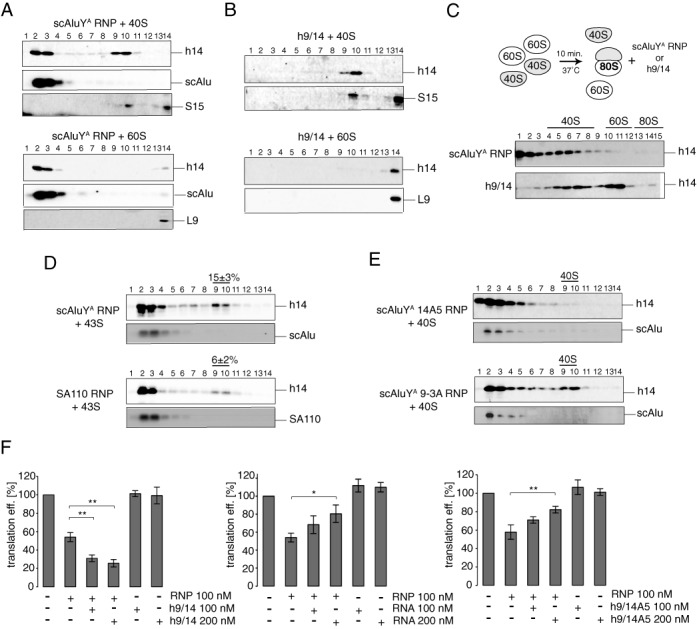
Alu RNA is required for the specific delivery of SRP9/14 to an inhibitory site in the 40S subunit. Binding of scAluYA RNP (A) or h9/14 (B) to the 40S (top panel) or 60S (bottom panel) subunits. Equimolar amounts of RNP were incubated with ribosomal subunits and the binding reactions were fractionated on 5–20% sucrose gradients. Fractions 13 and 14 contain 40S dimers. (C) Binding of scAluYA RNP (upper panel) or h9/14 (lower panel) to the reassembled 80S ribosomes. To form 80S ribosomes, equimolar amounts of 40S and 60S were incubated at 37°C for 10 min. Alu RNP was added and the binding reaction was incubated for another 10 min and fractionated on 10–30% sucrose gradients. 40S- and 60S-containing fractions were identified by probing for S15 and L9, respectively. (D) Binding of scAluYA RNP (left panel) or SA110 RNP (right panel) to the 43S complex. SRP9/14 in ribosomal fractions was quantified from Western blots against h14. The signal intensities in 40S-containing fractions (9 and 10) were expressed as a percentage of the total signal intensity across all fractions. Data are shown as mean ± SD (n ≥ 3). (E) Binding of scAluYA 14A5 RNP (top panel) or scAluYA 9-3A RNP (bottom panel) to 40S subunits. (F) Translation efficiency of pPL mRNA in RRL supplemented with 100 or 200 nM h9/14 (left panel), scAluYA RNA (middle panel) or h9/14A5 (right panel). scAluYA RNP were added to 100 nM. Error bars are shown as SD (n ≥ 3). *P < 0.05, **P < 0.01 by Student's t-test. See also Supplementary Figure S7.
We next examined the binding of the scAluYA RNP and h9/14 to 80S ribosomes. To this end, 80S ribosomes were allowed to associate from high salt purified 40S and 60S subunits for 10 min before the addition of the Alu RNP or h9/14. Only a fraction of the subunits assembled into 80S ribosomes in the presence of 1.5 mM magnesium acetate and individual subunits were therefore still present. Upon the addition scAluYA RNP we found h9/14 to be exclusively associated with 40S subunits whereas in the absence of the Alu RNA the protein bound nonselectively and was equally distributed between 40S and 60S containing fractions (Figure 5C). Hence, the presence of the Alu RNA appears to confer binding specificity to h9/14. This is further supported by the fact that all h9/14 bound quantitatively to 40S subunits even when present in a 2-fold molar excess over 40S indicating that it has more than one binding site on the 40S subunit (Supplementary Figure S6). In contrast the presence of 2-fold molar excess of scAluYA RNP over 40S did not increase the amount of the 40S-bound h9/14 suggesting that in this case the binding is saturable and occurs most likely to one specific site on the ribosome.
We also incubated 43S complexes reconstituted from purified components with scAluYA RNP. We found 15% of SRP9/14 comigrating with the 43S complex (Figure 5D, top panel). The amount of SRP9/14 bound to 40S was reduced ∼3-fold in the sample containing the inactive SA110 RNP (Figure 5D, bottom panel). The correlation between the ability of Alu RNPs to promote binding of h9/14 to the 40S subunit and the corresponding capacity to inhibit translation initiation was fully consistent with the notion that the RNA component of the Alu RNP acted as an assembly factor to promote functional binding of SRP9/14 to 40S subunits.
Alu RNA is not part of the inactive 40S complex
The previous results suggested that the functional association of SRP9/14 with the 40S subunit mediated by the Alu RNA is sufficient to render 40S incompetent for initiation. If indeed the continuous association of the Alu RNA were not required to inactivate the 40S subunit, the released RNA might serve in another round of targeted delivery of free SRP9/14 to 40S. To assess this hypothesis, we supplemented the RRL translation reaction containing scAluYA RNP with an equal amount or a 2-fold excess of h9/14. The presence of excess h9/14 significantly enhanced the inhibitory activity of the scAluYA RNP, while the protein alone at the same concentrations had no effect on translation (Figure 5F, left panel). Hence, the Alu RNA is not part of the inactive 40S complex.
Next, we examined whether an excess of Alu RNA over Alu RNP affected the activity of the Alu RNP. An equal amount or a 2-fold excess of the scAluYA RNA over the scAluYA RNP attenuated the inhibition (Figure 5F, middle panel). These results are consistent with the mutually exclusive and reversible binding of SRP9/14 to either the Alu RNA or to a functionally important site in the 40S subunit. The competition between 40S and Alu RNA may also explain why the inhibitory effect of Alu RNPs saturated at around 50% (Figure 1B and F). The targeted transfer of SRP9/14 from the Alu RNP to 40S leads to the release of free RNA and to the concomitant decrease in the amount of 40S subunits available for SRP9/14 binding. Saturation is reached when 40S subunits are no longer able to compete with the Alu RNA for SRP9/14 binding.
Functional role of SRP14 and SRP9 in 40S inactivation
Next, we examined whether the inactive scAluYA 14A5 and scAluYA 9-3A RNPs were able to mediate binding of the mutated proteins to the 40S subunit. Both Alu RNPs were incubated individually with 40S subunits and the complexes analyzed by sucrose gradients (Figure 5E). As observed in similar experiments with the protein alone (24), h9/14A5 was absent from the 40S fractions whereas h9-3A/14 bound as efficiently as the wild type protein to 40S subunits. These results indicated that SRP9/14 binding to the 40S subunit was required but not sufficient for Alu RNP-mediated inhibition of 48S complex formation. Supplementing the translation reactions containing scAluYA RNP with equimolar amounts of h9/14A5 did not significantly change the inhibitory activity of the scAluYA RNP (Figure 5F, right panel). However, a 2-fold excess of h9/14A5 over the scAluYA RNP diminished the inhibitory effect. Binding of h9/14 to Alu RNAs is characterized by high on and off-rates (25). In the presence of an excess of h9/14A5 over the wild-type protein, the latter was presumably exchanged with the mutated protein forming the inactive scAluYA 14A5 RNP, which lead to a loss in the inhibitory activity of the RNP.
Increased cellular levels of Alu RNA decrease synthesis of reporter proteins in mammalian cells
We next wanted to determine whether in the cell an increase in Alu RNP levels could influence translation efficiency. We chose to express the short scAlu RNA in HEK 293T cells, because it is more stable than Alu RNA (26) and because it lacks the inhibitory activities in Pol II-dependent transcription and in PKR activation (9,27,28). The overexpressed scAluYNF1 RNA (Table 1) is expected to bind SRP9/14, since human cells have a large pool of free SRP9/14 (11). As controls, we expressed 7SL RNA and 4.5S RNA, a 90 nucleotide-long, rodent-specific RNA, which fails to bind SRP9/14 (29). Expression of the RNAs was confirmed by northern blot analysis (Figure 6B) which showed that levels of scAluYNF1 RNA and 7SL RNA were increased 5–10 and 1.5–2-fold respectively, compared to the corresponding endogenous RNA levels.
Figure 6.
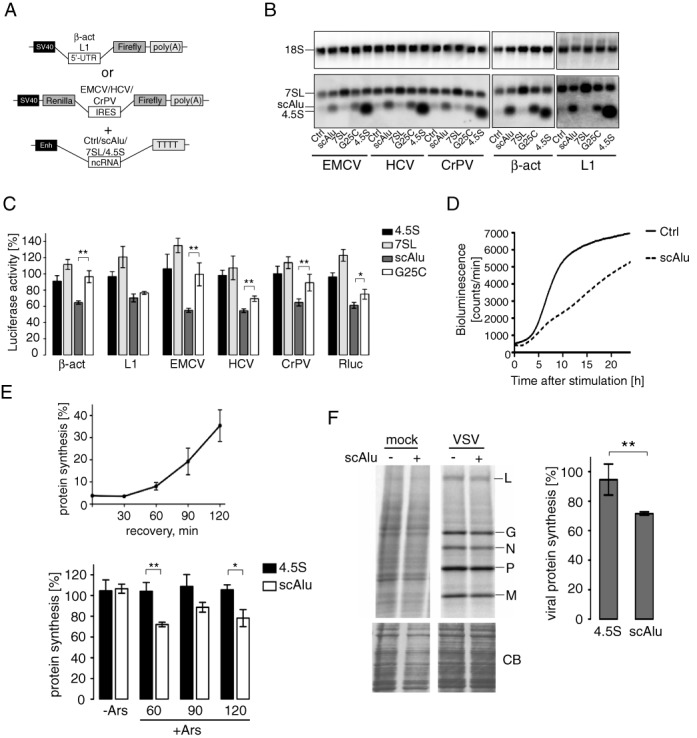
Expression of scAlu RNA reduces the translational capacity of HEK 293T cells. (A) Constructs used for luciferase reporter assay. Sequences of ncRNAs were cloned downstream of the 7SL gene enhancer element (Enh). (B) Northern blot analysis of transiently transfected HEK 293T cells. Membranes containing 0.5 μg of total RNA per lane were hybridized with [32P]-labeled oligonucleotides complementary to 4.5S RNA, 7SL RNA, scAluYNF1 RNA and 18S RNA (loading control). (C) Translation efficiency of reporter mRNAs in HEK 293T cells expressing different ncRNAs. Luciferase activity was measured 24 h post-transfection, normalized to μg of protein in the cell lysate and expressed as a percentage of the value obtained in control cells (ctrl) transfected with the empty vector pDL7Enh. Error bars are shown as SEM (n ≥ 5). *P < 0.05, **P < 0.01 by Student's t-test. See Supplementary Figure S8A for northern blot analysis of reporter mRNAs. (D) Bioluminescence recordings of HEK 293T cells expressing the estradiol-induced luciferase reporter gene along with pDLscAlu (scAlu) or pDL7enh (Ctrl) following stimulation with 100 nM estradiol. (E) Relative total protein synthesis in HEK 293T cells after arsenite treatment (top panel). Control cells were treated with 500 μM sodium arsenite for 30 min at 48 h post-transfection, allowed to recover for the indicated times and pulse labeled with [35S]-methionine/cysteine for 15 min. 100 μg of total protein was precipitated in 10% TCA, [35S]-incorporation was determined by scintillation counting and expressed as a percentage of that in untreated cells. Error bars are shown as SEM (n ≥ 5). Bottom panel shows total protein synthesis in untreated cells (-Ars) and cells expressing scAluYNF1 or 4.5S RNA treated for 30 minutes with 500 μM sodium arsenite and allowed to recover for 60, 90 and 120 min (+Ars). Total protein synthesis is expressed as a percentage of that in control cells measured in parallel. Error bars are shown as SEM (n ≥ 5). *P < 0.05, **P < 0.01 by Student's t-test. Expression of scAluYNF1 RNA is shown in Supplementary Figure S8D. (F) Representative autoradiograph following SDS-PAGE of VSV-infected cell lysates. HEK 293T cells expressing scAluYNF1, 4.5S RNA or control cells were infected with VSV for 6 h, pulse labeled with [35S]-methionine/cysteine for 15 min and 100 μg of total protein was displayed by 5–20% gradient SDS-PAGE. Bands corresponding to viral proteins are indicated. Coomassie-blue staining (bottom panel) was used to check uniform loading. Quantification of the viral protein synthesis is shown on the right. Viral protein synthesis in cells expressing scAluYNF1 RNA or 4.5S RNA was calculated as the sum of the intensities of the viral protein bands and expressed as a percentage of the control, which was set to 100%. Error bars are shown as SD (n ≥ 3). **P < 0.01 by Student's t-test. Expression of scAluYNF1 RNA is shown in Supplementary Figure S8E.
The effect on translation was assessed by co-transfection of plasmids expressing the non-coding RNAs (ncRNAs) together with reporter plasmids expressing the Fluc downstream of different 5′ UTRs (Figure 6A). We also studied IRES-dependent protein synthesis using bicistronic genes in which the Renilla (Rluc) and Firefly luciferase coding sequences were separated by an IRES element.
Expression of the Fluc was reduced for all reporter plasmids co-expressed with the scAluYNF1 RNA (Figure 6C). Similarly, the cap-dependent translation of Rluc from the upstream open reading frame of the bicistronic mRNAs was also diminished (Figure 6C). In contrast, the negative controls, 4.5S and 7SL RNAs, did not significantly affect luciferase expression, although higher levels of 7SL RNA had a slight tendency to increase it. Northern blot analysis revealed that each reporter plasmid produced an mRNA of the expected size, which was present in comparable amounts in the samples expressing ncRNAs (Supplementary Figure S8A).
To confirm that the inhibitory effect was dependent on h9/14, we expressed a mutated scAluYNF1 RNA where C25 replaced G25. This mutation causes a 60-fold decrease in affinity for h9/14 (30). Notably, the mutation does not completely abolish complex formation, since the dissociation constant of the complex with the mutated RNA remains rather high (nanomolar range). The mutated RNA was expressed at comparable levels as the wild type scAluYNF1 RNA (Figure 6B). For five out of six reporter mRNAs tested, luciferase expression levels were significantly higher in cells expressing the mutated scAluYNF1 RNA than in cells expressing the wild type RNA (Figure 6C). Thus, the G25C mutation diminished the inhibitory activity as expected if, as observed in the in vitro experiments, Alu RNA binding to h9/14 was required for translation inhibition in vivo. Only the L1-dependent Fluc expression was still strongly repressed. Presumably, low levels of scAlu RNPs might be sufficient to inhibit translation of this mRNA.
We next studied the effect of Alu expression on global cellular translation by measuring total [35S]-methionine/cysteine incorporation following a 15 min pulse, and somewhat surprisingly, no significant changes were seen in [35S]-incorporation in response to scAlu RNA expression (Figure 6E, lower panel, −Ars). Additionally, protein analysis by SDS-PAGE did not reveal any noticeable changes in the expression of specific proteins at high levels of scAluYNF1 RNA (Supplementary Figure S8B). We therefore reasoned that the Alu RNP-mediated decrease in active 40S subunits might severely hamper protein synthesis from mRNAs starting translation de novo such as the newly synthesized reporter mRNAs whereas protein synthesis from polysome-associated mRNAs remained largely unaffected. Ribosomal subunits are presumably recycled via a closed-loop structure in already existing polysomes (44). To strengthen this hypothesis we performed two experiments. First, we expressed an estradiol-inducible luciferase reporter plasmid in HEK 293T cells along with the scAluYNF1 RNA-expressing or the control plasmid. Fluc mRNA synthesis was induced by the addition of estradiol to the culture medium at 24 h post-transfection and luciferase activity was monitored during the next 24 h. While luciferase expression increased rapidly in control cells, it was strongly delayed in scAluYNF1 RNA-expressing cells (Figure 6D) consistent with our working hypothesis that reduced concentrations of active 40S subunits hampered de novo initiation (Figure 6D).
Second, we monitored the effect of scAluYNF1 RNA expression on the translation efficiency of endogenous mRNAs after disassembly of the existing pool of polysomes by treatment with sodium arsenite (31). As expected, arsenite treatment caused a dramatic 96–97% decrease in cellular protein synthesis (Figure 6E, top panel). At 60 min after arsenite removal protein synthesis levels remained below 10% compared to normal cells and polysomes were hardly detectable (Supplementary Figure S8C). We then determined translation rates at 60, 90 and 120 min after removal of arsenite for cells expressing either the scAluYNF1 or the 4.5S RNA, and the values were normalized to the corresponding values for cells transfected with a control vector (Figure 6E, bottom panel). A reduction in translation rate was specifically seen in cells expressing scAluNF1 RNA while cells expressing 4.5S RNA were unaffected. Hence, high scAlu RNA levels delayed translation initiation of endogenous mRNAs following disassembly of polysomes.
Viral protein synthesis is inhibited in cells expressing Alu RNA
It was previously reported that Alu RNA levels increase in response to viral infection and that these RNAs accumulate in the cytoplasm as SRP9/14 containing RNPs (32,33). We used the vesicular stomatitis virus (VSV), a prototype negative strand RNA virus, as a model to investigate the effects of scAluYNF1 RNA expression on viral protein synthesis. VSV induces a shutdown of host translation by interfering with eIF4F complex formation (34). Although VSV mRNAs are capped and do not contain IRES sequences, their translation proceeds via an unconventional mechanism, which bypasses the need of eIF4F for initiation (35). HEK 293T cells were transiently transfected with plasmids expressing either scAluYNF1 or the 4.5S control RNA. After 48 h, cells were infected with VSV (MOI = 10) and harvested 6 h later by which time the host translation machinery was reprogrammed by VSV mRNAs. In cells overexpressing scAluYNF1 RNA we observed a reduction in viral protein synthesis by approximately 25% compared to cells expressing 4.5S RNA (Figure 6F). This demonstrates that Alu RNPs are able to suppress translation of the VSV mRNAs in the cell following infection.
DISCUSSION
This study describes a novel mechanism of translation regulation affecting expression of all mRNAs including viral mRNAs with IRES. We present evidence that Alu RNPs inhibit translation by interacting directly with the 40S subunit, although only SRP9/14 remains bound to 40S whereas the Alu RNA is released and can act in another round of SRP9/14 binding to 40S. Binding of SRP9/14 prevents the recruitment of the 43S complex to the mRNA, possibly because the entry of the mRNA into the small subunit is directly or indirectly blocked. In cells, this mechanism interferes with translation initiation on viral and reporter mRNAs, and cellular mRNAs that resume the translation cycle after stress-induced shutdown of protein synthesis. The same mechanism of translation regulation is likely to be used in rodents and tree shrews, which also contain Alu-like elements in their genomes.
The unexpected feature in the Alu RNP-mediated translation inhibition is the role of the Alu RNA in the targeted delivery of SRP9/14 to a functional site in the 40S subunit. The Alu RNA is not part of the inactive 40S-SRP9/14 complexes and is therefore able to function in several rounds of SRP9/14 targeting to the 40S subunit (Figure 5F). This conclusion is supported by a parallel study from our laboratory (24). In this study, we show that upon addition of the 40S subunit to an Alu RNP immobilized on beads via its RNA moiety, a fraction of SRP9/14 dissociates within minutes from the RNA to bind to the 40S subunit. Indeed, SRP9/14 appears to equilibrate between the two binding partners (Figure 5A, C, D and (24)) suggesting that the functional binding site of SRP9/14 in 40S has a similar affinity for SRP9/14 as Alu RNA. The capacity of SRP9/14 to exchange rapidly between binding partners has already previously been observed (25) and is consistent with a high off rate of the SRP14-9 fusion protein from a 7SL-Alu domain RNA (36).
The competition between Alu RNA and 40S for SRP9/14 binding would explain why RNPs comprising Alu RNAs representing the Alu domain of 7SL RNA, which has the highest affinity for SRP9/14 (29,30), did not confer an inhibitory activity to the RNP. The affinity of 40S for SRP9/14 would be insufficient to compete with these RNAs for SRP9/14 binding. A plausible explanation for the mutually exclusive binding of SRP9/14 to Alu RNA or to 40S subunits is to presume that the basic β-sheet formed by the two proteins, which represents the major binding domain for Alu RNA (37), will also be involved in binding the 40S subunit, most likely via the 18S ribosomal RNA. The targeted delivery of SRP9/14 to a functionally important site in 40S also predicts that the Alu RNA itself transiently interact with the 40S subunit guiding SRP9/14 to its proper binding site.
The essential feature in the protein for 40S-binding and for initiation inhibition is the C-terminal pentapeptide of SRP14 (Figure 2E and F). Short basic oligopeptides are frequently present in ribosomal and ribosome-binding proteins and have been shown to bind ribosomal RNA (38,39) (Supplementary Figure S9). The three residues K60, K61 and K64 in SRP9 are essential for the elongation arrest function of SRP (21) and are also required for 40S subunit inactivation (Figure 2E and F). However, they are not required for 40S binding. Interestingly, in SRP-ribosome- nascent chain complex these residues are located in close proximity to 40S (40). Further studies will be required to determine the binding sites of SRP9/14 on the 40S subunit and whether the protein uses the same binding site in either function. Our data showed that SRP9/14 can be delivered to the 43S complex (Figures 1I and 5D) but not to the 80S ribosome (Figure 5C). This suggests that protein-binding site is not occupied by the ternary complex, eIF3, eIF1 and 1A but is blocked by the presence of the 60S subunit. The protein SRP9/14 alone had no inhibitory activity, which is consistent with earlier observations that high levels of SRP9/14 are well tolerated in primate cells (11,41)
Only few regulatory factors acting directly on the 40S subunit have so far been described: the acute respiratory syndrome coronavirus nsp1 protein and the proapoptotic protein Reaper (42,43). As for SRP9/14, the binding sites of these proteins in the 40S subunit remain to be revealed.
In cells, detectable effects on translation were observed for mRNAs initiating translation de novo. This may be because mRNAs in polysomes can adopt a closed-loop conformation due to the interaction of the scaffold factor eIFG4 with the poly(A)-binding protein (44). Due to facilitated ribosome recycling within these structures, translation might be less sensitive to 40S inactivation by SRP9/14. Following stress and certain viral infections, polysomes are disassembled and translation undergoes major reprogramming with certain mRNAs engaging de novo in protein synthesis (45,46) increasing the sensitivity of their translation to 40S inactivation.
Expression levels of Alu RNA are kept low to minimize the detrimental effects of frequent retrotransposition events in primate cells (6). In HeLa cells, Alu RNAs are about 500-fold less abundant than ribosomes and it is therefore unlikely that Alu RNPs inactivate an important fraction of 40S subunits. However, expression of specific Alu RNAs can be increased in certain tissues, like NDM29 RNA and BC200 RNA in the nervous tissue (6), and a transient increase in Alu RNA expression has been observed in response to viral infection and to various stresses (32,33,47). In addition, Alu RNA can act in several rounds of targeted delivery of the protein (Figure 5F) thereby multiplying the effect of its upregulation. The SRP9/14 is already present and Alu RNA levels increase within one hour and decline to normal levels within ∼7 h after heat shock (47). Like other regulatory mechanisms in translation, the inhibitory action of the Alu RNP is therefore rapid and transient. It is therefore conceivable that lowering the pool of active 40S subunits might contribute to shaping the translational response in these adaptive processes and thereby also participate in cellular defense mechanisms used against viral infection. A recent study, which shows that HCV translation was more strongly affected by reduced cellular levels of 40S than cellular translation, supports this hypothesis (48).
This study introduces a novel concept that a noncoding RNA shuttles an RNA-binding protein to its functional binding site on a large molecular complex thereby regulating its activity. It is conceivable that this concept is not restricted to translational control but is applicable to a multitude of cellular processes and more examples of RNA-mediated protein delivery might be discovered in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Drs Sergey Dmitriev for the pGL3R luciferase reporter plasmids, Jean Gruenberg for VSV-PeGFP, Dominique Belin for edeine and the genomic platform of the University of Geneva for capillary electrophoresis services. We thank Kinsey Maundrell, Didier Picard and Ueli Schibler for reading the manuscript and for insightful advice.
FUNDING
Swiss National Science Foundation [31003A-143844], Swiss-Russian Science and Technology Program, Canton of Geneva, Program on Molecular and Cellular Biology of the Russian Academy of Sciences (to E.A.). Funding for open access charge: Canton of Geneva.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershey J.W., Sonenberg N., Mathews M.B. Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a011528. doi:10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aitken C.E., Lorsch J.R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 4.Hasler J., Strub K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res. 2006;34:2374–2385. doi: 10.1093/nar/gkl246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quentin Y. Origin of the Alu family: a family of Alu-like monomers gave birth to the left and the right arms of the Alu elements. Nucleic Acids Res. 1992;20:3397–3401. doi: 10.1093/nar/20.13.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger A., Strub K. Multiple roles of Alu-related noncoding RNAs. Prog. Mol. Subcell. Biol. 2011;51:119–146. doi: 10.1007/978-3-642-16502-3_6. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrman S.A., Deininger P.L., LaPorte P., Friedmann T., Geiduschek E.P. Analysis of transcription of the human Alu family ubiquitous repeating element by eukaryotic RNA polymerase III. Nucleic Acids Res. 1981;9:6439–6456. doi: 10.1093/nar/9.23.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy-Engel A.M. LINEs, SINEs and other retroelements: do birds of a feather flock together. Front. Biosci. (Landmark Ed.) 2012;17:1345–1361. doi: 10.2741/3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yakovchuk P., Goodrich J.A., Kugel J.F. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maraia R.J., Driscoll C.T., Bilyeu T., Hsu K., Darlington G.J. Multiple dispersed loci produce small cytoplasmic Alu RNA. Mol. Cell. Biol. 1993;13:4233–4241. doi: 10.1128/mcb.13.7.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bovia F., Fornallaz M., Leffers H., Strub K. The SRP9/14 subunit of the signal recognition particle (SRP) is present in more than 20-fold excess over SRP in primate cells and exists primarily free but also in complex with small cytoplasmic Alu RNAs. Mol. Biol. Cell. 1995;6:471–484. doi: 10.1091/mbc.6.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang D.Y., Nelson B., Bilyeu T., Hsu K., Darlington G.J., Maraia R.J. A human Alu RNA-binding protein whose expression is associated with accumulation of small cytoplasmic Alu RNA. Mol. Cell. Biol. 1994;14:3949–3959. doi: 10.1128/mcb.14.6.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West N., Roy-Engel A.M., Imataka H., Sonenberg N., Deininger P. Shared protein components of SINE RNPs. J. Mol. Biol. 2002;321:423–432. doi: 10.1016/s0022-2836(02)00542-9. [DOI] [PubMed] [Google Scholar]
- 14.Lakkaraju A.K., Mary C., Scherrer A., Johnson A.E., Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaikh T.H., Roy A.M., Kim J., Batzer M.A., Deininger P.L. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J. Mol. Biol. 1997;271:222–234. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- 16.Macia A., Munoz-Lopez M., Cortes J.L., Hastings R.K., Morell S., Lucena-Aguilar G., Marchal J.A., Badge R.M., Garcia-Perez J.L. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol. Cell. Biol. 2011;31:300–316. doi: 10.1128/MCB.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak M., Shatkin A.J. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J. Biol. Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- 18.Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 19.Shirokikh N.E., Alkalaeva E.Z., Vassilenko K.S., Afonina Z.A., Alekhina O.M., Kisselev L.L., Spirin A.S. Quantitative analysis of ribosome-mRNA complexes at different translation stages. Nucleic Acids Res. 2010;38:e15. doi: 10.1093/nar/gkp1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gould P.S., Bird H., Easton A.J. Translation toeprinting assays using fluorescently labeled primers and capillary electrophoresis. Biotechniques. 2005;38:397–400. doi: 10.2144/05383ST02. [DOI] [PubMed] [Google Scholar]
- 21.Mary C., Scherrer A., Huck L., Lakkaraju A.K., Thomas Y., Johnson A.E., Strub K. Residues in SRP9/14 essential for elongation arrest activity of the signal recognition particle define a positively charged functional domain on one side of the protein. RNA. 2010;16:969–979. doi: 10.1261/rna.2040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson S.R. Tricks an IRES uses to enslave ribosomes. Trends Microbiol. 2012;20:558–566. doi: 10.1016/j.tim.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds J.E., Kaminski A., Kettinen H.J., Grace K., Clarke B.E., Carroll A.R., Rowlands D.J., Jackson R.J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger A., Ivanova E., Gareau C., Scherrer A., Mazroui R., Strub K. Direct binding of the Alu binding protein dimer SRP9/14 to 40S ribosomal subunits promotes stress granule formation and is regulated by Alu RNA. Nucleic Acids Res. 2014;42:11203–11217. doi: 10.1093/nar/gku822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas Y., Bui N., Strub K. A truncation in the 14 kDa protein of the signal recognition particle leads to tertiary structure changes in the RNA and abolishes the elongation arrest activity of the particle. Nucleic Acids Res. 1997;25:1920–1929. doi: 10.1093/nar/25.10.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W.M., Maraia R.J., Rubin C.M., Schmid C.W. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994;22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu W.M., Ballard R., Carpick B.W., Williams B.R., Schmid C.W. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 1998;18:58–68. doi: 10.1128/mcb.18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mariner P.D., Walters R.D., Espinoza C.A., Drullinger L.F., Wagner S.D., Kugel J.F., Goodrich J.A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 29.Bovia F., Wolff N., Ryser S., Strub K. The SRP9/14 subunit of the human signal recognition particle binds to a variety of Alu-like RNAs and with higher affinity than its mouse homolog. Nucleic Acids Res. 1997;25:318–326. doi: 10.1093/nar/25.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett E.A., Keller H., Mills R.E., Schmidt S., Moran J.V., Weichenrieder O., Devine S.E. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farny N.G., Kedersha N.L., Silver P.A. Metazoan stress granule assembly is mediated by P-eIF2alpha-dependent and -independent mechanisms. RNA. 2009;15:1814–1821. doi: 10.1261/rna.1684009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang D.Y., Hsu K., Maraia R.J. Monomeric scAlu and nascent dimeric Alu RNAs induced by adenovirus are assembled into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panning B., Smiley J.R. Activation of expression of multiple subfamilies of human Alu elements by adenovirus type 5 and herpes simplex virus type 1. J. Mol. Biol. 1995;248:513–524. doi: 10.1006/jmbi.1995.0239. [DOI] [PubMed] [Google Scholar]
- 34.Connor J.H., Lyles D.S. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 2002;76:10177–10187. doi: 10.1128/JVI.76.20.10177-10187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee A.S., Burdeinick-Kerr R., Whelan S.P. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc. Natl. Acad. Sci. U.S.A. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weichenrieder O., Kapp U., Cusack S., Strub K. Identification of a minimal Alu RNA folding domain that specifically binds SRP9/14. RNA. 1997;3:1262–1274. [PMC free article] [PubMed] [Google Scholar]
- 37.Weichenrieder O., Wild K., Strub K., Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- 38.Dudek J., Greiner M., Muller A., Hendershot L.M., Kopsch K., Nastainczyk W., Zimmermann R. ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat. Struct. Mol. Biol. 2005;12:1008–1014. doi: 10.1038/nsmb1007. [DOI] [PubMed] [Google Scholar]
- 39.Muller L., de Escauriaza M.D., Lajoie P., Theis M., Jung M., Muller A., Burgard C., Greiner M., Snapp E.L., Dudek J., et al. Evolutionary gain of function for the ER membrane protein Sec62 from yeast to humans. Mol. Biol. Cell. 2010;21:691–703. doi: 10.1091/mbc.E09-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halic M., Becker T., Pool M.R., Spahn C.M., Grassucci R.A., Frank J., Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 41.Chang D.Y., Sasaki-Tozawa N., Green L.K., Maraia R.J. A trinucleotide repeat-associated increase in the level of Alu RNA-binding protein occurred during the same period as the major Alu amplification that accompanied anthropoid evolution. Mol. Cell. Biol. 1995;15:2109–2116. doi: 10.1128/mcb.15.4.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colon-Ramos D.A., Shenvi C.L., Weitzel D.H., Gan E.C., Matts R., Cate J., Kornbluth S. Direct ribosomal binding by a cellular inhibitor of translation. Nat. Struct. Mol. Biol. 2006;13:103–111. doi: 10.1038/nsmb1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lokugamage K.G., Narayanan K., Huang C., Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012;86:13598–13608. doi: 10.1128/JVI.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amrani N., Ghosh S., Mangus D.A., Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh D., Mathews M.B., Mohr I. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 2013;5:a012351. doi: 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamasaki S., Anderson P. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 2008;20:222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu W.M., Chu W.M., Choudary P.V., Schmid C.W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J.Y., Su W.C., Jeng K.S., Chang T.H., Lai M.M. Attenuation of 40S ribosomal subunit abundance differentially affects host and HCV translation and suppresses HCV replication. PLoS Pathog. 2012;8:e1002766. doi: 10.1371/journal.ppat.1002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson R.J., Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- 50.Das S.C., Nayak D., Zhou Y., Pattnaik A.K. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 2006;80:6368–6377. doi: 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


