Abstract
In anaerobic environments, mutually beneficial metabolic interactions between microorganisms (syntrophy) are essential for oxidation of organic matter to carbon dioxide and methane. Syntrophic interactions typically involve a microorganism degrading an organic compound to primary fermentation by-products and sources of electrons (i.e., formate, hydrogen, or nanowires) and a partner producing methane or respiring the electrons via alternative electron accepting processes. Using a transposon gene mutant library of the sulfate-reducing Desulfovibrio alaskensis G20, we screened for mutants incapable of serving as the electron-accepting partner of the butyrate-oxidizing bacterium, Syntrophomonas wolfei. A total of 17 gene mutants of D. alaskensis were identified as incapable of serving as the electron-accepting partner. The genes identified predominantly fell into three categories: membrane surface assembly, flagellum-pilus synthesis, and energy metabolism. Among these genes required to serve as the electron-accepting partner, the glycosyltransferase, pilus assembly protein (tadC), and flagellar biosynthesis protein showed reduced biofilm formation, suggesting that each of these components is involved in cell-to-cell interactions. Energy metabolism genes encoded proteins primarily involved in H2 uptake and electron cycling, including a rhodanese-containing complex that is phylogenetically conserved among sulfate-reducing Deltaproteobacteria. Utilizing an mRNA sequencing approach, analysis of transcript abundance in wild-type axenic and cocultures confirmed that genes identified as important for serving as the electron-accepting partner were more highly expressed under syntrophic conditions. The results imply that sulfate-reducing microorganisms require flagellar and outer membrane components to effectively couple to their syntrophic partners; furthermore, H2 metabolism is essential for syntrophic growth of D. alaskensis G20.
INTRODUCTION
Anaerobic oxidation of organic compounds, such as alcohols and fatty acids, is thermodynamically unfavorable when protons or carbon dioxide are used as the electron acceptor, unless the H2 partial pressure or formate concentration, respectively, can be maintained at extremely low concentrations (1). Thus, complete degradation of organic matter in methanogenic and some other anaerobic systems requires a microbial consortium composed of two or more microbial species to oxidize the carbon and subsequently remove hydrogen (2–5). This synergistic interaction between different microorganisms is defined as syntrophy. The term was first coined in anaerobic, sulfur-oxidizing phototrophic cocultures (6) but was experimentally verified when the methanogenic, archaean Methanobacterium bryantii strain MOH was separated from an ethanol-oxidizing partner, both of which were present in a culture called Methanobacillus omelianskii, which at the time was believed to be a pure culture (7). This work established the general syntrophic model of two microorganisms mutualistically cooperating by transferring electrons from one species to the other through H2 and/or formate (8). However, direct electron transfer via nanowires has also been described (9). Regardless of the electron transfer mechanism, the keystone of syntrophy requires that the metabolic interactions be mutually beneficial (10). Such is the case in many anaerobic environments where the degradation of alcohols, fatty and alicyclic acids, and aromatic compounds is thermodynamically possible only when the electron-accepting partner maintains very low H2 or formate concentrations (11). Because these compounds are only degraded by syntrophic consortia under methanogenic conditions, the net free energy available from the overall reaction, which is typically small, must be shared between both partners (9, 12–15).
To date, syntrophic organisms have been shown to be either strictly fermentative (e.g., Syntrophus aciditrophicus and Syntrophomonas wolfei) or those capable of syntrophy in the absence of a terminal electron-accepting process (e.g., Geobacter and Desulfovibrio) (9, 16). The electron-accepting organisms are often methanogens (3), but sulfate-reducing (16) and fumarate-reducing organisms (9) have been shown to function efficiently with H2 and/or formate-producing syntrophic metabolizers. Sulfate-reducing bacteria acting as syntrophic electron acceptors are found in bioreactors treating waste streams containing sulfate (17–20) and are likely also present in many other anaerobic environments in which both methanogenesis and sulfate reduction occur together.
Since syntrophic growth is energetically restrictive, the syntrophic partners must adapt to take advantage of this energy-limited, ecological niche. Several studies detected significant alterations in global gene expression of Desulfovibrio species when comparing the lifestyles of a sulfate reducer (with lactate and sulfate) to a syntrophic electron donor (lactate) with a methanogen as the electron-accepting partner (16, 21). Microarray and real-time PCR analyses have revealed a functionally unknown, yet transcriptionally active gene set in Desulfovibrio vulgaris Hildenborough, consisting of three colocated genes, which may be involved in the lifestyle change of D. vulgaris from sulfate respiration to syntrophic metabolism (22). Two of these protein-coding genes contain an iron-sulfur cluster-binding/ATPase domain, while the third encodes an MTH1175-like hypothetical protein. Furthermore, under syntrophic conditions D. vulgaris mutants significantly elevate transcription for genes coding a number of hydrogenases and membrane associated electron transfer functions, including Coo, Hmc, Hyd, and Hyn (21). Mutations in the genes encoding the latter four enzymes impaired or severely limited the ability of D. vulgaris to grow as the syntrophic electron-donating partner but had little effect on growth in axenic cultures. Similarly, 20 mutants in Desulfovibrio alaskensis were identified which were attenuated in the ability to grow as electron-donating partner (16, 23). Genes involved in electron transfer functions, including hyd (Fe-only hydrogenase), qrc (quinone reactive complex), cyc (cytochrome c3), and hyp (NiFe hydrogenase maturation), were shown to be important, as well as a number of other functional and regulatory genes. Many of these mutants were deficient in motility as well as syntrophic growth. Although these studies highlight the genes necessary for Desulfovibrio to function in the role of syntrophic electron producer, little is known of genes necessary for syntrophic growth where Desulfovibrio accepts electrons and respires sulfate.
In order to most efficiently transfer electrons in the form of H2, small interspecies distances are desirable and are facilitated by the formation of cell aggregates (12). In anaerobic wastewater treatment systems, the formation of aggregates is common (13). Syntrophic cultures also can transfer electrons directly between the two species using filamentous structures (9). Aggregates of pure cultures of D. vulgaris occurring in biofilms show a flagellum-like structure, which appears to connect cells together (12, 14). The composition of the latter filaments have not been identified nor has a role for flagella been proven in the syntrophic interaction.
In a previous study, we described a transposon mutant library of 5760 mutants in D. alaskensis G20 created using a mini-Tn10 transposon-bearing plasmid (24). Here, we use the library to identify genes that are important for syntrophic growth of D. alaskensis as the electron acceptor with S. wolfei as the electron donor (17–20). The mutant screen and subsequent gene expression studies allowed us to identify genes necessary for syntrophic interactions. A focus was on the importance of flagella and the role of H2 and formate using enzymes.
MATERIALS AND METHODS
Medium components and culture growth.
A lactate-sulfate (LS) medium was used to grow D. alaskensis G20 and its mutants axenically. Each liter of medium contained 2.0 ml of vitamin solution, 12.5 ml of metal solution, 25 ml of 1 M HEPES, 1.0 g of yeast extract, 16.1 g of NaSO4·10H2O, 5.79 g of sodium lactate, 1.97 g of MgSO4·7H2O, 0.088 g of CaCl2·2H2O, 0.383 g of K2PO4, 1.07 g of NH4Cl, and 0.62 ml of 0.1% resazurin, and the pH was adjusted to 7.2. The vitamin solution contained (per liter) 2 mg of biotin, 2 mg of folic acid, 10 mg of pyridoxine HCl, 5 mg of thiamine HCl, 5 mg of riboflavin, 5 mg of nicotinic acid, 5 mg of calcium d-(+)-pantothenate, 5 mg of cyanocobalamin, 5 mg of p-aminobenzoic acid, 5 mg of thioctic acid, and 10 mg of mercaptoethanesulfonic acid. Metal solution contained (per liter) 12.8 g of nitrilotriacetic acid, 0.42 g of FeSO4·7H2O, 0.1 g of MnCl2·4H2O, 0.17 g of CoCl2·6H2O, 0.02 g of CuCl2·2H2O, 0.21 g of ZnSO4·7H2O, and 0.01 g of Na2MoO4·2H2O. Media were prepared using strict anaerobic procedures (25). Anoxic media (10 ml) were dispensed into serum tubes under N2 and 0.1 ml of 7% sodium bicarbonate and 0.15 ml of 2.5% cysteine-HCl were added after autoclave sterilization. D. alaskensis transposon mutants were grown axenically in LS media with the addition of 1,050 μg of kanamycin sulfate/ml.
A crotonate minimal medium was used to grow S. wolfei and S. aciditrophicus axenically (26). Each liter of media contained 50 ml of Pfennig I (10.0 g/liter K2HPO4), 50 ml of Pfennig II (8.0 g/liter NaCl, 8.0 g/liter NH4Cl, 6.6 g/liter MgCl2·2H2O, 1.0 g/liter CaCl2·2H2O), 5.0 ml of metal solution, 10.0 ml of vitamin solution, 1 ml of 0.1% resazurin, and 0.4 g of crotonic acid. The metal and vitamin solutions are described above. Serum bottles with 100 ml of crotonate medium were prepared with a N2/CO2 (80/20 [vol/vol]) headspace and reduced with 1.0 ml/liter of 2.5% cysteine-HCl–2.5% sodium sulfide solution. A butyrate-sulfate minimal medium was used to grow cocultures of G20 and S. wolfei, and a similar benzoate-sulfate medium was used to grow G20-S. aciditrophicus cocultures. Media were prepared as for crotonate-minimal medium except 6.6 g/liter NaSO4·10H2O and 2.2 g/liter sodium butyrate or 1.7 g/liter sodium benzoate were substituted for crotonate. When cocultures were grown for RNA-seq analysis, 0.01% yeast extract was added to provide higher growth yields. Cultures of D. alaskensis G20, S. aciditrophicus, and S. wolfei were all grown at their optimal temperature of 37°C. Growth was monitored weekly using a Spec21 spectrophotometer to determine optical density at 600 nm (OD600).
Screening of G20 mutants in coculture with S. wolfei.
A mutant library of 5,760 D. alaskensis G20 mutants providing 1.5-fold coverage of total genes was prepared with a mini-Tn10 transposon-bearing plasmid as described in previous work (24). The transposon was shown to insert randomly. To screen for syntrophic growth deficiency, butyrate medium was inoculated with 0.5 ml of washed cells of each mutant of D. alaskensis (to remove the antibiotic) and 1 ml of a late-log to early-stationary-phase culture of S. wolfei grown on crotonate. Growth was monitored weekly and individual mutants with normal syntrophic growth formed cocultures, which increased in OD600 by >0.1, which was approximately one doubling during a 2- to 3-week growth period. Mutants that exhibited an increase of <0.05 (final OD600) in coculture with S. wolfei were considered to be growth deficient and chosen for further study. Mutants identified during the initial screening were retested in coculture to verify that they were truly deficient in coculture growth. Mutants were then grown in LS medium with or without yeast extract to distinguish mutations related to syntrophy versus those with nutritional or other physiological defects. Mutants that grew poorly in pure culture were not chosen for further study.
Identifying the transposon insertion location.
Total DNA from an overnight culture of each D. alaskensis syntrophy mutant was extracted with a Qiagen DNeasy kit and a two-round arbitrary PCR was performed (24). The PCR products were sequenced and compared to the D. alaskensis genome (accession no. NC_007519) using BLASTN.
Morphological analysis of flagella mutants.
The parent strain G20 and the flagella (flhA) mutant were grown on the semisolid 0.1% LS agar inoculated with a drop of culture to grossly compare relative motility. Plates were incubated at 37°C in an anaerobic growth chamber with an N2:H2 (95:5) atmosphere. Phase-contrast microscopy was also used to determine whether the parent strain and flagella mutants were motile by observing motility in several fields. To determine whether flagella mutants exhibited a structural phenotype, cultures were prepared for scanning electron microscopy by allowing the culture to grow in the presence of glass slides for 30 h. Slides were rinsed with phosphate-buffered saline (PBS; pH 7.2) and immersed in 4% paraformaldehyde in 50 mM sodium cacodylate buffer (pH 7.0) overnight. The slides were then rinsed in water, dehydrated through an ethanol series, and dried using a critical point dryer (Tousimis Autosamdri 814). Samples were then sputter coated with silver and examined on a Zeiss Neon dual-beam-field emission scanning electron microscope. Three fields were photographed.
Quantification of biofilm production.
LS media (15 ml with 1,050 μg of kanamycin/ml added to the mutant culture tubes) were prepared and dispensed into serum tubes, which contained one half of a microscope slide. Individual tubes were then inoculated with the parent strain of G20, flhA, fliF, and tadC mutants and grown to early stationary phase (20 h). Slides were then harvested, rinsed briefly with PBS buffer, and then submerged in a 0.1% crystal violet solution for 10 min. Excess crystal violet was removed from slides with distilled water and then slides were placed into a 20-ml ethanol-acetone (4:1) solution overnight to leach crystal violet adhered to cells and biofilm attached to the slide. Crystal violet was quantified spectrophotometrically at 580 nm. The relative quantity of biofilm formation was expressed as the OD580 of the destain solution/the OD600 of the culture at 20 h. Significant differences in biofilm formation between mutants and the parent strain were determined by using the Student t test.
Operon analysis of flagellar genes.
Operon analysis was done to determine whether transposon insertions interrupted transcription. Total RNA was extracted from the parent strain, and flhA, and fliF mutants using the Qiagen RNeasy kit. Pure cultures grown for 12 h were chilled in a dry ice-ethanol bath, harvested by centrifugation at 4°C, and stabilized with Qiagen RNAprotect reagent. The RNAprotect protocol 1 was followed with enzymatic lysis of cells using lysozyme. RNA was treated with an Ambion Turbo DNA-free kit, and a PCR control reaction was performed to ensure there was no DNA contamination. Purity and adequate yield was confirmed spectrophotometrically. First-strand cDNA synthesis was performed using a Fermentas RevertAid kit with gene-specific primers covering the entirety of the flhA and fliF mutants' operons (Table 1). Primers were designed to span the gaps between each gene in the operon. PCR was performed with the parent strain RNA and each primer set to determine first whether all genes were expressed on each operon and whether the genes were on the same operon. A PCR was then performed on the gaps for the respective operons of flagellar flhA and fliF mutants to determine whether the transposon insertion affected the expression of other genes in the operon.
TABLE 1.
Primers used for gap analysis to determine whether transcription of all of the genes in the operon occurred
| Gap (genes) | Primer sequence (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| Gap BC (flgB and flgC) | TCGTCCACGGCGAAGATCAGGT | TATGGCCCGCCGCCTATGGT |
| Gap CE (fligC and fliE) | GACCTCGCAGCGCGGGTATC | TCCACTCTCGGTTCCGGCGT |
| Gap EF (fliE and fliF) | AGTGATGGAAGCGTACCGCGAAC | CGACCACAGTCAGGCCGACG |
| Gap FG (fliF and fliG) | CACCGGCGAAGAGCGTCTGG | GGCACGGTGTCCAGTTCGACC |
| Gap GH (fliG and fliH) | GCGCGGCCACCATGATCAGA | GGGCTTCTTCACGCAGCCGT |
| Gap HI (fliH and fliI) | ACACCCCCGCGGTGGAGGATA | GGCGCCCAGAGGCGCTTTTA |
| Gap RB (fliR and flhB) | GCAGATGAACCTGCTGATGA | GCCATGGTCACAAAGGTTTT |
| Gap AB (flhB and flhA) | GCCTTGTATAGGCAGGTGGA | GGTAACCAGCAGAAGCGAAG |
| Gap AF (flhA and flhF) | CAGCTGCTCCAGAGGTTTCT | GCCATGATGTGCCCTTTTAT |
Transcriptional analysis of parent and mutant cultures with RNA-seq.
Pure cultures of D. alaskensis, S. wolfei, and S. aciditrophicus and cocultures of S. wolfei with D. alaskensis and S. aciditrophicus with D. alaskensis were grown as described above to mid-log phase in 10-ml tubes. The different syntrophic electron producers were used because they have been shown capable of shuttling electrons differentially (26). After 2 to 3 weeks of initial growth, 2 ml of each coculture was transferred to a tube with 10 ml of medium. The 2- to 3-week incubation and transfer were repeated twice, several tubes were pooled, and 20 ml of the pooled cocultures was used to inoculate a serum bottle containing 100 ml of either butyrate-sulfate or benzoate-sulfate media for S. wolfei-D. alaskensis and S. aciditrophicus-D. alaskensis, respectively. The transfers continued every 2 to 3 weeks with growth determined visually. At each of four subsequent transfers, 80 ml was recovered for RNA extraction, and 20 ml was used for the next transfer. Frozen cells from each coculture were pooled, and RNA was extracted with a Qiagen RNeasy kit. DNA was removed with an Ambion Turbo DNA-free kit, and a MicrobeExpress kit was used to deplete rRNA. RNA was ethanol precipitated into Tris-EDTA buffer prior to rRNA removal. Purified RNA concentration was quantified by using NanoDrop (Thermo Scientific). RNA was shipped on dry ice to the University of Georgia Genomics facility for 100-bp single-end sequencing with Illumina HiSeq.
Processing Illumina reads and genome mapping.
The transcriptome of D. alaskensis G20 and mutant cultures were passed through a previously described pipeline (27). Briefly, each growth condition was represented by a single RNA sample consisting of single samples of pure cultures and pooled cocultures, which were processed independently whereby cDNA reads generated were screened for quality and trimmed using Sickle (https://github.com/ucdavis-bioinformatics/sickle.git). Single-end reads were then mapped with BWA v.06.0 (28, 29) using default settings to described genes from the parent strain D. alaskensis G20 genome (IMG object ID 637000095). Gene abundance was extracted from BAM files with SAMtools v0.1.18 (29). Individual gene abundance was normalized to the number reads recruited to protein coding genes.
RESULTS
Identification and characterization of syntrophy mutants.
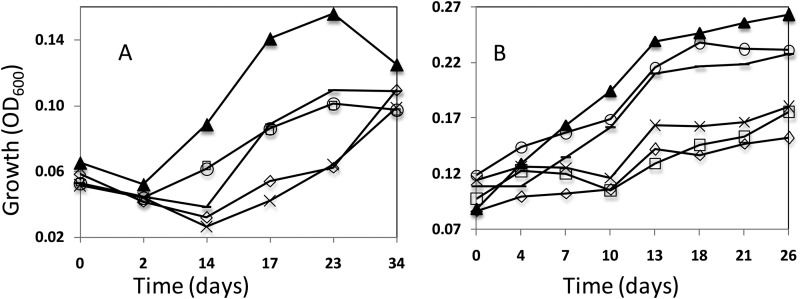
Seventeen genes in D. alaskensis G20 were identified as necessary for syntrophic growth as the electron-accepting partner with S. wolfei as the electron-producing partner (Table 2). These genes were not necessary for growth of G20 in pure culture on lactate and sulfate. Coculture growth curves of mutants and parent strain G20 show the relative growth deficiencies observed for some of the mutants identified (Fig. 1). The lack of coculture growth for the mutants is reproducible. Some of the mutants shown appeared to be growing much slower than the parent strain, while others had little detectable growth. We found that some mutants, which were not able to grow with S. wolfei, grew well with S. aciditrophicus (Fig. 1B; e.g., flhA and tadC), indicating that some of the genes will not have a important phenotypic effect with all electron-donating syntrophs. However, not all mutants were tested with S. aciditrophicus.
TABLE 2.
D. alaskensis G20 genes identified as important for syntrophic growth as the electron-accepting partnera
| Gene type and locus tag | Gene annotation | H2-SO42− growth | Expression fold change in G20b |
||
|---|---|---|---|---|---|
| Coculture with S. aciditrophicus | Coculture with S. wolfei | Pure culture on H2-SO42− | |||
| Surface assembly genes | |||||
| Dde_0215 | Glycosyl transferase, group 2 family protein | ++ | 0.659 | 0.193† | –0.28† |
| Dde_0439 | Glycosyl or glycerophosphate transferase | +++ | 3.06** | 1.85* | 0.04 |
| Dde_0590 | Cardiolipin synthase | +++ | 0.393† | 0.11† | 0.068† |
| Flagellum and pilus assembly genes | |||||
| Dde_0353 | FliF flagellar ring protein | +++ | 3.88** | 2.02** | 0.323 |
| Dde_0380 | FlhA flagellar biosynthesis protein | ++ | 2.79** | 1.80* | –0.19 |
| Dde_2365 | TadC, Flp pilus assembly protein | +++ | ND | ND | 1.22*† |
| Energy metabolism genes | |||||
| Dde_0582 | Electron transport complex RnfD | + | 0.494 | 2.02** | 1.21* |
| Dde_0585 | Electron transport complex RnfA | – | 0.413 | 1.91* | 1.37* |
| Dde_0604 | Pyruvate phosphate dikinase | +++ | –0.51 | 0.36† | 0.0004† |
| Dde_0653 | High-molecular-weight cytochrome c | – | –1.34* | –0.67 | 0.82 |
| Dde_0681 | Fe-S cluster hydrogenase component | +++ | 4.52** | 3.51*† | 2.29**† |
| Dde_2281 | Iron-only hydrogenase | + | –0.079 | –1.99† | 2.87** |
| Dde_2933 | Qrc-quinone reactive complex | – | –0.893 | –0.27 | 0.966* |
| Miscellaneous genes | |||||
| Dde_0304 | Glyoxylase family protein | +++ | 0.032† | 1.20* | 0.321† |
| Dde_2611 | Putative lipoprotein | +++ | –0.983 | 0.51 | –0.038 |
| Dde_3047 | Serine phosphatase | +++ | 3.75** | 2.88* | 0.041† |
| Dde_3305 | Choline glycine betaine transporter | +++ | –0.035 | –0.08 | –0.249 |
Genes are grouped into their functional categories. Whether each mutant was able to grow on H2-sulfate is indicated by the relative amount of growth compared to the parent strain with a “−” indicating no detectable growth and “+++” indicating growth equivalent to that of the parent strain.
The relative RNA expression of individual genes in coculture with S. wolfei, with S. aciditrophicus, or in pure culture on H2-sulfate is given relative to growth in pure culture on lactate-sulfate. Fold regulation values were calculated as the log10(normalized reads for S. aciditrophicus, S. wolfei, or hydrogen-grown G20/normalized reads for G20 lactate-sulfate). †, value calculated with both normalized read values below 1; *, gene abundance that was one sigma unit from the mean; **, gene abundance that was two or more sigma units from the mean, i.e., putatively significant. ND, not determined.
FIG 1.
Growth curves of G20 parent strain and mutants in coculture with S. wolfei (A) and S. aciditrophicus (B). Results for the parent strain D. alaskensis (▲), flhA mutant (–), fliF mutant (×), tadC mutant (○), rnfA mutant (□), and rnfD mutant (◇) are shown.
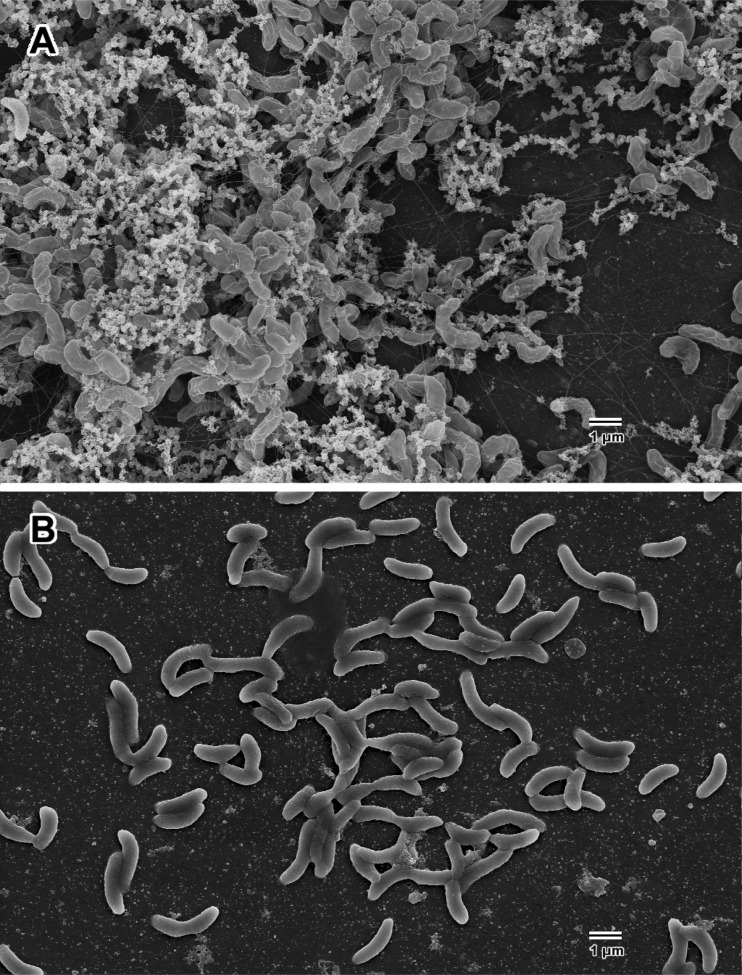
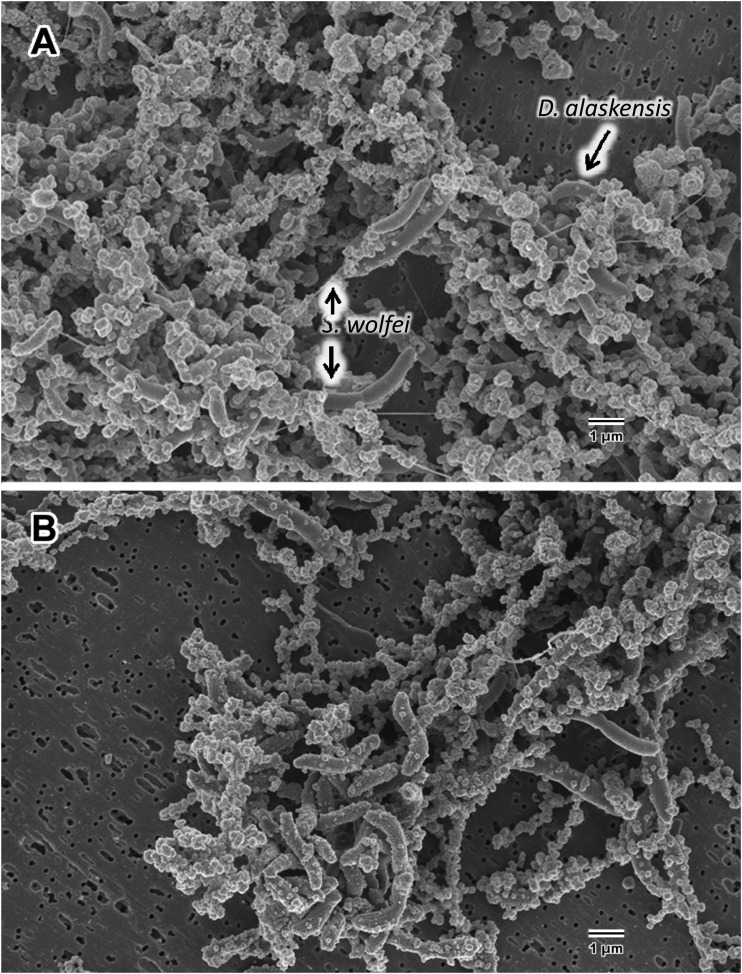
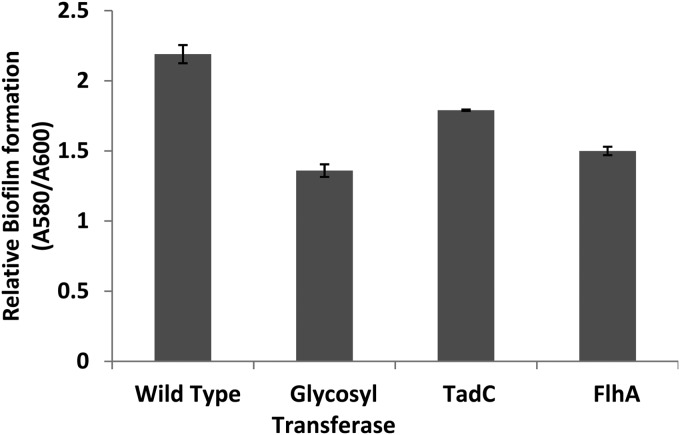
Strain G20 genes necessary for syntrophic growth with S. wolfei had annotated functions that fell into five functional categories: glycosyltransferases, flagellum and pilus synthesis, energy metabolism, cell wall/membrane synthesis and transport, and gene regulation (serine phosphatase). Mutants involved with flagellum and pilus synthesis and biofilm formation (glycosyltransferase genes) were tested further for potential phenotype changes. Microscopic visualization showed that the flhA and fliF mutants had little to no motility. When grown without shaking on H2 and sulfate, the parent strain formed a turbid layer on the surface, which was not observed with the flhA mutant, indicating that the mutant is unable to move toward the source of H2. Scanning electron microscopy (SEM) confirmed the absence of flagella in flhA and fliF mutants (see Fig. 2 for the flhA mutant), whereas flagella were clearly visible on cells of the parent strain in pure culture (Fig. 2) and in coculture with S. wolfei (Fig. 3). To determine whether the transposon eliminated downstream expression of genes in the flh and fli operons, we performed gap analysis, amplifying non-protein-coding regions of RNA transcripts. The flhA gene mutant is in the third gene of an operon with the first four genes involved in flagellar biosynthesis. The fliF mutation is in the fourth gene in a seven-gene flagellar protein operon. The first three gaps in the flh operon and the six gaps in the fli operon were PCR amplifiable from cDNA, indicating that mutants were capable of generating full transcripts despite the transposon insertion. Thus, all genes in these two operons are expressed, although an insertion in one of the genes (flhA or fliF, respectively) disrupted the cell's ability to assemble a functional flagellum. Significantly decreased biofilm production in flhA, tadC, and glycosyltransferase mutants relative to the parent strain was also observed (Fig. 4). Similarly, we observed by SEM a dearth of extracellular polymeric substance in the flhA mutant compared to parent strain biofilms (Fig. 2).
FIG 2.
SEM images of glass slide biofilms formed by G20 (A) and the flhA mutant (B).
FIG 3.
SEM image of a syntrophic culture of S. wolfei with G20 (A) and the flhA mutant (B).
FIG 4.
Relative amounts of biofilm formation by flhA (Dde_0380), tadC (Dde_2365), and glycosyltransferase (Dde_0215) mutants involved in cell surface component production.
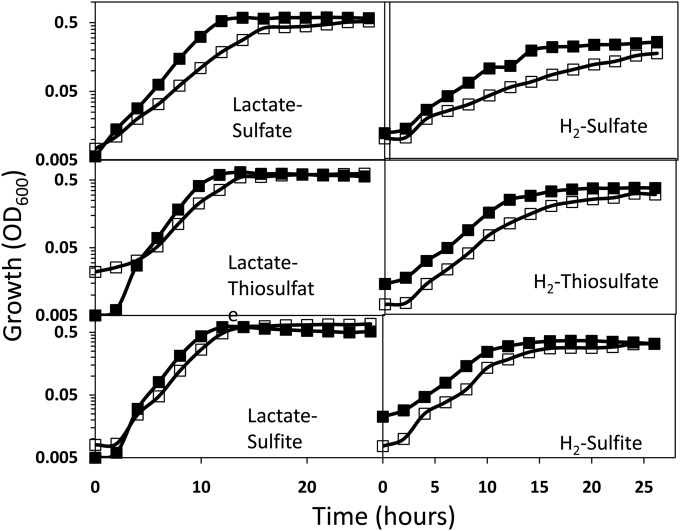
Energy metabolism mutants identified under syntrophic growth conditions fell into two primary categories based on whether or not the mutant grew on H2-sulfate in pure culture (Table 2). Although several of these mutants have been described previously (iron-only hydrogenase and Qrc-quinone reactive complex (16, 23), we also identified a putative multiprotein complex needed for growth of D. alaskensis G20 as the electron-accepting partner. Mutation to the gene Dde_0681, annotated as an 4Fe-4S cluster hydrogenase component was incapable of syntrophic coupling. Pure culture growth experiments showed that the mutant grew at a lower growth rate on H2 or lactate with sulfate than the wild-type strain, but its growth rate was similar to that of the wild-type when thiosulfate or sulfite were the electron acceptors (Fig. 5). Upon further inspection, this gene is within a putative operon (Dde_0678-83) containing genes annotated as a molybdopterin and selenocysteine containing 4Fe-4S cytochrome protein, a formate dehydrogenase gamma subunit (membrane spanning), and two nonduplicated rhodanese-like proteins. Homologous operons are found in many other sequenced Desulfovibrio species listed on the IMG website (see Fig. S1 in the supplemental material). The molybdopterin, cytochrome, and rhodaneses are likely translocated to the periplasm and linked to the membrane, since signal peptides were identified in all three genes and membrane-spanning helices were detected in the cytochrome and rhodanese protein sequences (Dde_0679). The first rhodanese gene (Dde_0679) contains three rhodanese pfams in its deduced amino acid sequence, suggesting a true rhodanese protein. In contrast, the second rhodanese gene (Dde_0678) does not contain a transmembrane helix and only contains a single rhodanese domain and a single thioredoxin-like superfamily domain in its deduced amino acid sequence. The formate dehydrogenase gamma subunit does not contain a signal peptide but has four predicted transmembrane helices. The putative Fe-S hydrogenase subunit did not contain either membrane components or signal peptides.
FIG 5.
Growth of the parent strain of D. alaskensis (■) and the Dde_0681 mutant (□; FeS-binding protein) with different combinations of electron donors and acceptors.
In addition to this putative complex, the syntrophic mutant screen also identified several previously studied energy usage genes, including the Fe-only hydrogenase mutant, the quinone reactive complex (qrc) mutant (16, 23) and the high-molecular-weight cytochrome c complex (hmc) mutant (30). Fe-hydrogenase hmc and qrc mutants, in addition to being syntrophically deficient, had little to no growth with H2/SO42− in the present study, although the parent strain is capable of growing on H2 or formate with sulfate (Table 2). Previously, Fe-hydrogenase (hyd), and qrc mutants were also shown to be deficient in growth on formate plus sulfate (16). We also identified two mutants in the rnf operon. Rnf is an electron transfer protein that has been shown in several bacteria to couple the oxidation of reduced ferredoxin to the reduction of NAD+ (31, 32). This process has been shown to generate a Na+ ion gradient in Acetobacterium woodii and a proton gradient in Clostridium ljungdahlii (33). The need for Rnf in Desulfovibrio coupled to the inability of the rnf mutant to grow on H2 suggests that Rnf may be involved in a pathway for H2 metabolism.
Expression of genes identified in parent strain G20 under syntrophic conditions.
Expression profiling using RNA-seq analysis was done to confirm whether the genes identified as important for syntrophy as the electron-accepting partner were transcriptionally active and abundant under syntrophic growth in the parent strain G20. Among the 17 genes identified during the syntrophy screen, six were induced >2-fold during syntrophic growth, in most cases under both syntrophic coculture conditions (Table 2). These included one of the glycosyltransferase genes (Dde_0439), flagellar protein genes (Dde_0353 and Dde_0380), rnfD (Dde_0582), the hydrogenase component gene (Dde_0681), and the serine phosphatase gene (Dde_3047) (Table 2). Due to limitations in sequencing of multiple replicates, a traditional statistical analysis could not be performed. To identify potentially significantly, upregulated genes, we calculated z-scores for all genes under each condition. All six of these genes were greater than two sigma units from the mean z-scores, indicating that these genes are most likely significantly more expressed during syntrophy and are among the most highly expressed genes under syntrophic conditions.
Based on the results from the screening studies, we investigated transcription of several putative operons, specifically those involved in flagellar proteins, chemotaxis, biofilm and cell surfaces, energy metabolism, and hydrogen and formate consumption. This allowed us to explore the role of genes related to those identified during the mutant screen. The results are presented for the expression of the putative operons to simplify the presentation of the results (Table 3). The most dramatically upregulated group of genes were the flagellum structural and biosynthesis genes, suggesting an important role for flagella during syntrophy. Moreover, a variety of chemotaxis-related genes are ∼2-fold upregulated, indicating an involvement of chemotaxis during syntrophic growth. Several biofilm and cell surface genes were also upregulated. This includes several glycosyltransferase operons, as well as a highly expressed phage shock protein gene, perhaps involved in dealing with stress (34).
TABLE 3.
Relative expression of D. alaskensis G20 operons involved in specific cellular functions that were highly expressed during syntrophya
| Operon range | Operon annotation | Expression fold change vs axenic G20 |
Avg normalized expression |
|||
|---|---|---|---|---|---|---|
| Coculture with S. aciditrophicus | Coculture with S. wolfei | On H2-SO42− | S. aciditrophicus coculture | Lactate SO42− | ||
| Flagellum-related genes | ||||||
| Dde_0350-56 | Fli flagellar components | 18.74 | 5.76 | 1.19 | 31.7 | 1.69 |
| Dde_0378-86 | Flh flagellar biosynthesis proteins | 2.32 | 1.35 | 0.82 | 8.65 | 3.72 |
| Dde_1119-20 | Fli flagellar components | 5.33 | 2.05 | 1.09 | 9.98 | 1.87 |
| Dde_1501 | Flagellin | 4.53 | 1.63 | 0.69 | 7.03 | 1.55 |
| Dde_1502 | Flagellin | 3.08 | 1.9 | 1.27 | 57.5 | 18.6 |
| Dde_1570 | Flagellin | 21.7 | 8.6 | 4.24 | 28.8 | 1.32 |
| Dde_1709 | Flagellin | 10.14 | 3.91 | 1.06 | 62.8 | 6.2 |
| Dde_2705-08 | Fli flagellar protein | 3.7 | 1.06 | 0.9 | 5.85 | 1.58 |
| Dde_3149-59 | Flg flagellar components operon | 11.0 | 3.54 | 1.09 | 22.2 | 2.02 |
| Dde_3582-90 | Fli flagellar biosynthesis proteins | 6.35 | 3.06 | 1.04 | 8.21 | 1.29 |
| Chemotaxis-related genes | ||||||
| Dde_0281 | Chemotaxis protein | 2.29 | 0.68 | 0.70 | 7.63 | 3.33 |
| Dde_0369 | Methyl-accepting chemotaxis protein | 2.37 | 0.23 | 0.89* | 1.40 | 0.59 |
| Dde_1322 | Methyl-accepting chemotaxis protein | 1.69 | 1.27* | 1* | 1.05 | 0.62 |
| Dde_1665 | Methyl-accepting chemotaxis protein | 2.07 | 1.57 | 1.68 | 3.25 | 1.57 |
| Dde_2411 | Chemoreceptor protein A | 1.82 | 1.87 | 1.23 | 1.89 | 1.04 |
| Dde_2814 | Methyl-accepting chemotaxis protein | 1.56 | 1.63 | 1.22 | 1.32 | 0.85 |
| Dde_2857 | Chemoreceptor protein A | 2.19 | 2.06 | 1.32 | 27.1 | 12.3 |
| Dde_2968 | Methyl-accepting chemotaxis protein | 2.03 | 2.22 | 0.93* | 1.34 | 0.66 |
| Biofilm-, pilus-, and cell surface-related genes | ||||||
| Dde_0215-17 | Glycosyltransferase | 2.22 | 1.09 | 0.84 | 3.11 | 1.40 |
| Dde_0337 | Glycosyltransferase, group 1 family | 2.01 | 0.53 | 0.68* | 2.04 | 1.01 |
| Dde_0425–28 | Glycosyltransferase operon with a GDP-fucose synthetase | 3.97 | 1.45 | 0.72 | 4.65 | 1.17 |
| Dde_0438-39 | Glycosyl or glycerophosphate transferase | 13.97 | 6.03 | 1.14 | 24.3 | 1.74 |
| Dde_0590 | Cardiolipin synthase | 1.32* | 1.08* | 1.05* | 0.98 | 0.75 |
| Dde_3224-26 | Phage shock protein | 25.7 | 33.4 | 1.03 | 23.1 | 0.90 |
| Dde_2358-70 | Tad pilus operon | 2.6* | 1.7* | 0.95* | 0.30 | 0.12 |
| Alcohol dehydrogenase- heterodisulfide reductase | ||||||
| Dde_3523 | Alcohol dehydrogenase, iron-containing | 260.3 | 60.81 | 11.60 | 118.7 | 0.46 |
| Dde_3524-30 | Heterodisulfide reductase | 79.8 | 12.2 | 4.5 | 7.61 | 0.095 |
| Hydrogenases | ||||||
| Dde_0656-57 | Fe-S cluster binding proteins | 10.78 | 4.24 | 1.87 | ||
| Dde_0081-82 | Hydrogenase (Fe-only) | 0.87 | 0.93 | 10.5 | 0.25 | 0.29 |
| Dde_2134-35 | Hydrogenase (Ni-Fe-Se) | 0.78 | 1.23 | 3.79 | 7.59 | 9.69 |
| Dde_2137-38 | Hydrogenase (Ni-Fe) | 43.56 | 47.13 | 5.17 | 12.5 | 0.29 |
| Dde_3754-56 | Hydrogenase (Ni-Fe) | 5.92 | 3.30 | 1.10 | 0.76 | 0.13 |
| Formate dehydrogenases | ||||||
| Dde_0678-83 | Rhodanese/formate dehydrogenase/oxidoreductase operon | 22.39 | 11.58 | 3.43 | 2.68 | 0.12 |
| Dde_0706-09 | Formate dehydrogenase formation protein | 8.90 | 6.81 | 2.66 | 4.74 | 0.53 |
| Dde_0715-18 | Formate dehydrogenase operon | 46.07 | 31.33 | 6.05 | 17.8 | 0.39 |
The relative expression for multiple gene operons was determined as the average change for all of the genes in the operon when grown with S. wolfei, with S. aciditrophicus, or in pure culture on H2-sulfate relative to growth in pure culture on lactate-sulfate. *, a ratio calculated where both samples being compared have low expression (<1 normalized read) values.
The most highly differentially expressed region of the genome during syntrophic growth was the region from Dde_3523 to Dde_3530. This region encodes an alcohol dehydrogenase, the heterodisulfide reductase complex, and a putative FAD, the selenocysteine-containing NAD+-binding protein complex. The alcohol dehydrogenase has previously been shown necessary for ethanol-dependent growth in D. vulgaris (35). This gene is situated next to a large heterodisulfide reductase operon that is also highly upregulated (45- to 105-fold). Without additional information, it is difficult to say why these genes are highly upregulated. It is possible that the physiological change associated with slow growth or with the decreased level of yeast extract could have induced these changes, or that ethanol is an interspecies intermediate during syntrophic growth.
Hydrogen and formate are important electron transport metabolites for S. wolfei and S. aciditrophicus (26). However, under syntrophic conditions, the D. alaskensis Fe-only hydrogenase was downregulated (Table 3) but this operon was highly expressed in axenic H2-SO4 growth. The Ni-Fe-Se hydrogenase was the most transcriptionally abundant periplasmic hydrogenase during axenic growth on H2. However, of the three periplasmic hydrogenases only the two Ni-Fe hydrogenases (Dde_2137-38, Dde_3754-56) were significantly upregulated when cells were grown syntrophically. No significant upregulation of the hmc operon was detected, but it was transcriptionally active under all growth conditions, suggesting this cytochrome is important in metabolism of strain G20. When grown syntrophically the formate dehydrogenases are likely involved in formate oxidation, as this is a primary electron shuttle for both S. wolfei and S. aciditrophicus. One of four predicted fdh operons in strain G20 was upregulated during syntrophic growth (Dde_0715-18), as well as a nearby formate dehydrogenase formation protein (Dde_0706-09), indicating the importance of formate oxidation during syntrophy.
DISCUSSION
Syntrophic growth typically involves electron transfer, typically mediated by H2 and formate (36–39). Because most organisms growing syntrophically can use or produce both H2 and formate, it has been difficult to determine which compound is preferred for electron transfer. H2 has traditionally been thought to be the electron transfer currency. However, the use of formate facilitates electron transfer at much higher rates, due to its much higher diffusivity in aqueous solutions (39). Several studies have provided either chemical evidence (38), proteomic (40), or transcriptional (26, 41) support for a contribution of formate uptake, along with H2 uptake during methanogenic syntrophic interactions. Several of the aforementioned studies used Methanospirillum hungatei in coculture with the S. wolfei or Syntrophobacter fumaroxidans. The results suggest that M. hungatei can adjust for different relative levels of formate and H2 produced. Here, we present evidence that the same is true for D. alaskensis during syntrophic growth with both S. wolfei and S. aciditrophicus.
Our results showed that all hydrogenases were transcriptionally active under both axenic and syntrophy growth experiments (Table 3). The two Ni-Fe (Dde_2137-38, Dde_3754-56) and the Ni-Fe-Se (Dde_2134-35) hydrogenases were more abundant than the iron-only hydrogenase during syntrophic growth. However, the two Ni-Fe hydrogenases were markedly more abundant under syntrophic conditions than in axenic cultures, where the Ni-Fe-Se hydrogenase was most abundant. Pure culture studies with D. vulgaris showed that expression of the Ni-Fe hydrogenase did not vary with different H2 levels in the headspace and when one of the two Ni-Fe hydrogenases was functionally disrupted, there was little influence on growth (42), providing evidence that the Ni-Fe and Ni-Fe-Se hydrogenase may be functionally equivalent in pure cultures. Studies in D. vulgaris determining protein levels as well as activity have shown that the Ni-Fe-Se hydrogenase can be the most abundant periplasmic hydrogenase in the cell under a variety of growth conditions (43). We also observed high levels of the Ni-Fe-Se hydrogenase in pure and cocultures (Table 3). However, we observed a >40-fold increase in expression levels of one of the Ni-Fe hydrogenases during syntrophic growth, increasing its mRNA concentration above that of the Ni-Fe-Se hydrogenase. Since the major periplasmic hydrogenases all are capable of reducing the same cadre of electron carriers and the Ni-Fe and Ni-Fe-Se behave similarly in terms of forward and backward activity (44), the increased abundance of Ni-Fe hydrogenase in syntrophy cultures suggests that it may have a unique function during syntrophic growth. Although we cannot fully explain the expression behavior of the Ni-Fe hydrogenase genes, we speculate that the binding affinities for hydrogen by their proteins may be higher than for the Ni-Fe-Se hydrogenases. Some Ni-Fe hydrogenases have been shown to have very high H2 binding affinities (Km = 30 to 130 nM) (45). Thus, under syntrophic conditions where the fermentative partner is inhibited quickly by H2 accumulation (46), a high-affinity hydrogenase would maintain sufficiently low hydrogen partial pressures and thereby keep its partner's metabolism uninhibited by thermodynamic constraints.
Although no specific formate dehydrogenases were identified as essential to syntrophic coupling, we did identify a novel operon (Dde_0683-Dde_0678) containing an fdh gamma subunit that was needed for syntrophic growth and was highly upregulated under syntrophic conditions. Interestingly, the operon structure is highly conserved among sulfate-reducing Deltaproteobacteria but is not universally present among genome sequenced members of this group (see Fig. S1 in the supplemental material). Dde_0682/0683 is predicted to be a selenocysteine-containing molybdopterin oxidoreductase. Dde_0683 has the signal peptide and also a domain for cytochrome cbb3. Dde_0682 has the molybdopterin-binding domains and FeS clusters as do many molybdopterin oxidoreductases. Interestingly, this operon contains two, putative rhodanese genes (Dde_0678/0679), which could function as thiosulfate reductases. The presence of the two genes, one of which contains a single sulfane-binding domain (Dde_0678), suggests a potential role as sulfane transferases. Dde_0681 is the FeS protein that likely links the catalytic unit (Dde_0682/0683 gene products) to the membrane protein (Dde_0680 gene product, cytochrome b). Almost all of the homologous membrane subunits are annotated as the formate dehydrogenase gamma subunit, since the transmembrane-helix sequences are conserved and were first described in the Fdh. The physiological data suggest that this complex may be involved in the reduction of sulfate under both heterotrophic and autotrophic conditions. However, more work is necessary to fully elucidate this novel, operon's function.
Previous work has shown cellular aggregation is commonplace in methanogenic systems (21) and, in our parent strain syntrophy cocultures, we observed similar small, visible aggregates. However, flagellum mutants did not produce a similar phenotype, suggesting that flagella are integral components of syntrophic cell aggregates (Fig. 2) (12). Expression experiments show that expression of flagellin gene encoding the main protein in flagella increased by 3- to 20-fold (Tables 1 and 2) during syntrophic growth relative to pure culture growth. Although gene expression does not directly equate to proportionate amounts of protein, this marked increase in expression of flagellin and related proteins by the “hydrogen user” suggests a specific role for flagella under syntrophic conditions. Given the increased abundance of chemotaxis genes under syntrophy conditions (Table 3), increased production of flagella would facilitate chemotaxis to the syntrophic partner. Once a partner cell is located, the increased flagellum production could be used to maintain specific distances for efficient interspecies electron transfer, via H2 or formate, and sustained production (30) would help stabilize the subsequent biofilm. Although our data do not specifically address cell-to-cell communication, strain G20 could initiate syntrophic coupling via the flagella, as previous work has shown that syntrophic systems respond transcriptionally to the presence of flagella (47). However, more work is needed to elucidate whether these communication mechanisms are present in G20.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the Physical Biosciences program, DOE Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division grant number DE-FG02-06ER15769.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03358-14.
REFERENCES
- 1.McInerney MJ, Bryant MP, Hespell RB, Costerton JW. 1981. Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic, syntrophic, fatty acid-oxidizing bacterium. Appl Environ Microbiol 41:1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L, Sieber J, Struchtemeyer CG, Bhattacharyya A, Campbell JW, Gunsalus RP. 2007. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci U S A 104:7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJM, Schink B, Rohlin L, Gunsalus RP. 2008. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann N Y Acad Sci 1125:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- 4.Schink B, Friedrich M. 1994. Energetics of syntrophic fatty-acid oxidation. FEMS Microbiol Rev 15:85–94. doi: 10.1111/j.1574-6976.1994.tb00127.x. [DOI] [Google Scholar]
- 5.Stams AJM. 1994. Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie van Leeuwenhoek 66:271–294. doi: 10.1007/BF00871644. [DOI] [PubMed] [Google Scholar]
- 6.Biebl H, Pfennig N. 1978. Growth yields of green sulfur bacteria in mixed cultures with sulfur and sulfate-reducing bacteria. Arch Microbiol 117:9–16. doi: 10.1007/BF00689344. [DOI] [Google Scholar]
- 7.Bryant MP, Wolin EA, Wolin MJ, Wolfe RS. 1967. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch Microbiol 59:20–31. [DOI] [PubMed] [Google Scholar]
- 8.Schink B. 1992. Syntrophism among prokaryotes, p 276–299. In Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH (ed), The prokaryotes. Springer-Verlag, New York, NY. [Google Scholar]
- 9.Summers ZM, Fogarty HE, Leang C, Franks AE, Malvankar NS, Lovley DR. 2010. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330:1413–1415. doi: 10.1126/science.1196526. [DOI] [PubMed] [Google Scholar]
- 10.Sieber JR, McInerney MJ, Gunsalus RP. 2012. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 11.Schink B, Stams AJM. 2003. Syntrophism among procaryotes, p 471–493. In Dworkin M. (ed), The procaryotes: an evolving electronic resource for the microbial community, release 3.14. Springer-Verlag, New York, NY. [Google Scholar]
- 12.Ishii S, Kosaka T, Hori K, Hotta Y, Watanabe K. 2005. Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus. Appl Environ Microbiol 71:7838–7845. doi: 10.1128/AEM.71.12.7838-7845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Gil G, Lens PN, Van Aelst A, Van As H, Versprille AI, Lettinga G. 2001. Cluster structure of anaerobic aggregates of an expanded granular sludge bed reactor. Appl Environ Microbiol 67:3683–3692. doi: 10.1128/AEM.67.8.3683-3692.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark ME, Edelmann RE, Duley ML, Wall JD, Fields MW. 2007. Biofilm formation in Desulfovibrio vulgaris Hildenborough is dependent upon protein filaments. Environ Microbiol 9:2844–2854. doi: 10.1111/j.1462-2920.2007.01398.x. [DOI] [PubMed] [Google Scholar]
- 15.Jackson BE, McInerney MJ. 2002. Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415:454–456. doi: 10.1038/415454a. [DOI] [PubMed] [Google Scholar]
- 16.Li XZ, McInerney MJ, Stahl DA, Krumholz LR. 2011. Metabolism of H2 by Desulfovibrio alaskensis G20 during syntrophic growth on lactate. Microbiology 157:2912–2921. doi: 10.1099/mic.0.051284-0. [DOI] [PubMed] [Google Scholar]
- 17.Colleran E, Finnegan S, Lens P. 1995. Anaerobic treatment of sulfate-containing waste streams. Antonie van Leeuwenhoek Int J 67:29–46. doi: 10.1007/BF00872194. [DOI] [PubMed] [Google Scholar]
- 18.Elferink S, Boschker HTS, Stams AJM. 1998. Identification of sulfate reducers and Syntrophobacter sp. in anaerobic granular sludge by fatty-acid biomarkers and 16S rRNA probing. Geomicrobiol J 15:3–17. doi: 10.1080/01490459809378058. [DOI] [Google Scholar]
- 19.Elferink S, Vorstman WJC, Sopjes A, Stams AJM. 1998. Characterization of the sulfate-reducing and syntrophic population in granular sludge from a full-scale anaerobic reactor treating papermill wastewater. FEMS Microbiol Ecol 27:185–194. doi: 10.1111/j.1574-6941.1998.tb00536.x. [DOI] [Google Scholar]
- 20.Elferink SJWH, Visser A, Pol LWH, Stams AJM. 1994. Sulfate reduction in methanogenic bioreactors. FEMS Microbiol Rev 15:119–136. doi: 10.1111/j.1574-6976.1994.tb00130.x. [DOI] [Google Scholar]
- 21.Walker CB, He ZL, Yang ZK, Ringbauer JA, He Q, Zhou JH, Voordouw G, Wall JD, Arkin AP, Hazen TC, Stolyar S, Stahl DA. 2009. The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J Bacteriol 191:5793–5801. doi: 10.1128/JB.00356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholten JC, Culley DE, Brockman FJ, Wu G, Zhang WW. 2007. Evolution of the syntrophic interaction between Desulfovibrio vulgaris and Methanosarcina barkeri: involvement of an ancient horizontal gene transfer. Biochem Biophys Res Commun 352:48–54. doi: 10.1016/j.bbrc.2006.10.164. [DOI] [PubMed] [Google Scholar]
- 23.Li XZ, Luo QW, Wofford NQ, Keller KL, McInerney MM, Wall J, Krumholz LR. 2009. A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J Bacteriol 191:2675–2682. doi: 10.1128/JB.01814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groh JL, Luo Q, Ballard JD, Krumholz LR. 2005. A method adapting microarray technology for signature-tagged mutagenesis of Desulfovibrio desulfuricans G20 and Shewanella oneidensis MR-1 in anaerobic sediment survival experiments. Appl Environ Microbiol 71:7064–7074. doi: 10.1128/AEM.71.11.7064-7074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balch WE, Wolfe RS. 1976. New approach to cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (Hs-Com)-dependent growth of Methanobacterium-ruminantium in a pressurized atmosphere. Appl Environ Microbiol 32:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sieber JR, Le HM, McInerney MJ. 2014. The importance of hydrogen and formate transfer for syntrophic fatty, aromatic and alicyclic metabolism. Environ Microbiol 16:177–188. doi: 10.1111/1462-2920.12269. [DOI] [PubMed] [Google Scholar]
- 27.Baker BJ, Sheik CS, Taylor CA, Jain S, Bhasi A, Cavalcoli JD, Dick GJ. 2013. Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling. ISME J 7:1962–1973. doi: 10.1038/ismej.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi M, Pollock WBR, Reij MW, Keon RG, Fu R, Voordouw G. 1993. The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J Bacteriol 175:4699–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boiangiu CD, Jayamani E, Brügel D, Herrmann G, Kim J, Forzi L, Hedderich R, Vgenopoulou I, Pierik AJ, Steuber J, Buckel W. 2005. Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J Mol Microbiol Biotechnol 10:105–119. doi: 10.1159/000091558. [DOI] [PubMed] [Google Scholar]
- 32.Biegel E, Muller V. 2010. Bacterial Na+-translocating ferredoxin: NAD(+) oxidoreductase. Proc Natl Acad Sci U S A 107:18138–18142. doi: 10.1073/pnas.1010318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tremblay PL, Zhang T, Dar SA, Leang C, Lovley DR. 2012. The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin:NAD+ oxidoreductase essential for autotrophic growth. mBio 4:e00406-12. doi: 10.1128/mBio.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brissette JL, Russel M, Weiner L, Model P. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc Natl Acad Sci U S A 87:862–866. doi: 10.1073/pnas.87.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haveman SA, Brunelle V, Voordouw JK, Voordouw G, Heidelberg JF, Rabus R. 2003. Gene expression analysis of energy metabolism mutants of Desulfovibrio vulgaris Hildenborough indicates an important role for alcohol dehydrogenase. J Bacteriol 185:4345–4353. doi: 10.1128/JB.185.15.4345-4353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stams AJ, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol 7:568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 37.Stams AJ, de Bok FA, Plugge CM, van Eekert MH, Dolfing J, Schraa G. 2006. Exocellular electron transfer in anaerobic microbial communities. Environ Microbiol 8:371–382. doi: 10.1111/j.1462-2920.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- 38.Thiele JH, Zeikus JG. 1988. Control of interspecies electron flow during anaerobic digestion: significance of formate transfer versus hydrogen transfer during syntrophic methanogenesis in flocs. Appl Environ Microbiol 54:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boone DR, Johnson RL, Liu Y. 1989. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and its implications in the measurement of Km for H2 or formate uptake. Appl Environ Microbiol 55:1735–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt A, Muller N, Schink B, Schleheck D. 2013. A proteomic view at the biochemistry of syntrophic butyrate oxidation in Syntrophomonas wolfei. PLoS One 8:e56905. doi: 10.1371/journal.pone.0056905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bok FAM, Luijten MLGC, Stams AJM. 2002. Biochemical evidence for formate transfer in syntrophic propionate-oxidizing cocultures of Syntrophobacter fumaroxidans and Methanospirillum hungatei. Appl Environ Microbiol 68:4247–4252. doi: 10.1128/AEM.68.9.4247-4252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caffrey SA, Park HS, Voordouw JK, He Z, Zhou J, Voordouw G. 2007. Function of periplasmic hydrogenases in the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J Bacteriol 189:6159–6167. doi: 10.1128/JB.00747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valente FAA, Almeida CC, Pacheco I, Carita J, Saraiva LM, Pereira IAC. 2006. Selenium is involved in regulation of periplasmic hydrogenase gene expression in Desulfovibrio vulgaris Hildenborough. J Bacteriol 188:3228–3235. doi: 10.1128/JB.188.9.3228-3235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valente FM, Oliveira AS, Gnadt N, Pacheco I, Coelho AV, Xavier AV, Teixeira M, Soares CM, Pereira IA. 2005. Hydrogenases in Desulfovibrio vulgaris Hildenborough: structural and physiologic characterisation of the membrane-bound [NiFeSe] hydrogenase. J Biol Inorg Chem 10:667–682. doi: 10.1007/s00775-005-0022-4. [DOI] [PubMed] [Google Scholar]
- 45.Greening C, Berney M, Hards K, Cook GM, Conrad R. 2014. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci U S A 111:4257–4261. doi: 10.1073/pnas.1320586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McInerney MJ, Bryant MP. 1981. Basic principles of bioconversions in anaerobic digestion and methanogenesis, p 277–296. In Sofer SS, Zaborsky OR (ed), Biomass conversion processes for energy and fuels. Plenum Press, Inc, New York, NY. [Google Scholar]
- 47.Shimoyama T, Kato S, Ishii S, Watanabe K. 2009. Flagellum mediates symbiosis. Science 323:1574–1574. doi: 10.1126/science.1170086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.