Abstract
The amidohydrolase superfamily has remarkable functional diversity, with considerable structural and functional annotation of known sequences. In microbes, the recent evolution of several members of this family to catalyze the breakdown of environmental xenobiotics is not well understood. An evolutionary transition from binuclear to mononuclear metal ion coordination at the active sites of these enzymes could produce large functional changes such as those observed in nature, but there are few clear examples available to support this hypothesis. To investigate the role of binuclear-mononuclear active-site transitions in the evolution of new function in this superfamily, we have characterized two recently evolved enzymes that catalyze the hydrolysis of the synthetic herbicides molinate (MolA) and phenylurea (PuhB). In this work, the crystal structures, mutagenesis, metal ion analysis, and enzyme kinetics of both MolA and PuhB establish that these enzymes utilize a mononuclear active site. However, bioinformatics and structural comparisons reveal that the closest putative ancestor of these enzymes had a binuclear active site, indicating that a binuclear-mononuclear transition has occurred. These proteins may represent examples of evolution modifying the characteristics of existing catalysts to satisfy new requirements, specifically, metal ion rearrangement leading to large leaps in activity that would not otherwise be possible.
INTRODUCTION
The amidohydrolase superfamily (AHS) has been extensively studied, with >36,000 attributed sequences since its first classification in 1998, most of which are from bacteria (1, 2). All AHS members have a (β/α)8-barrel structural fold and catalyze metal-dependent hydrolysis reactions (3). The scissile bond cleaved varies between AHS enzymes, with C-O, P-O, P-S, C-N, C-S, and C-Cl bonds all having been reported to be hydrolyzed (4–8). In accordance with the metal-dependent mechanism, a mononuclear or binuclear metal binding site is observed in all AHS enzymes (9, 10). The role of the metal ion(s) in catalysis varies, depending on the substrate being hydrolyzed and the enzyme involved (10, 11). Generally, one or two metals are enlisted to lower the pKa of a catalytic water molecule, favoring the formation of a nucleophilic hydroxide (12). A second metal may polarize the substrate directly, often at a carbonyl or phosphoryl bond, or stabilize a negative charge in the transition state (13, 14).
Enzymes within the AHS can be separated into subtypes on the basis of the specific metal binding ligands at the conserved mononuclear or binuclear metal binding sites. Since the comprehensive review published by Seibert and Raushel in 2005 (9), in which seven subtypes were defined, two additional subtypes have been identified (Table 1) (6, 15). A typical binuclear metal binding site (subtypes I, II, and VI) is characterized by a bridging ligand (generally a glutamate or carboxylated lysine residue) that coordinates both (Mα and Mβ) metal ions (16–18). Mononuclear metal binding ligands are more variable, and the metal ion can be situated at the Mα or Mβ site (subtypes III, IV, V, VI, VII, and IX) (9). Recently identified subtype VIII contains three characterized enzymes, all of which are of interest owing to their activity with recently developed synthetic herbicides. Molinate hydrolase (MolA) was isolated from the bacterium Gulosibacter molinativorax ON4T from areas contaminated with the herbicide molinate (7), while phenylurea hydrolases A (PuhA) and B (PuhB) were isolated from Mycobacterium brisbanense JK1 and are capable of breaking down the herbicide diuron (Fig. 1) (6). All subtype VIII members have a novel Asn-Xaa-His motif in strand one that is distinct from the His-Xaa-His motif seen in most members of the AHS (6, 7, 19). Prior to this work, no structures or definitive mechanisms of subtype VIII members were available, making the catalytic impact of the Asn-Xaa-His motif unknown.
TABLE 1.
The nine current subtypes in the AHS are distinguished by their conserved metal binding ligands
| Subtype | Metal(s) | Metal position(s) | Representative PDB code(s) | Residue(s)a in strand: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||||
| I | Zn, Ni | α, β | 1HZY, 3MTW | HxH | K | H | H | D | |||
| II | Zn | α, β | 1BF6 | HxH | E | H | H | D | |||
| III | Zn, Fe | α | 1A4M, 4DZH | HxH | H | hc | D | ||||
| IV | Fe | β | 1O12 | hxhb | E | H | H | dc | |||
| V | Zn | β | 1M7J | hxhb | C | H | H | dc | |||
| VI | Zn | α, β | 1ITQ | HxD | E | H | H | dc | |||
| VII | 1J5S | HxH | H | dc | |||||||
| VIII | Znd | β | 4WGX, 4WHB | NxH | kc | H | hc | D | |||
| IX | Zn | α | 3LS9 | HxH | H | hc | T | ||||
Uppercase amino acid code letters indicate ligands that fulfill their characteristic roles; lowercase letters indicate amino acids that differ from these roles. Table modified from Khurana et al. (6).
These residues are present in the active site but do not appear to ligate a metal. In some cases, they can accommodate a second metal ion, but it is not required for catalysis.
Hydrogen bonds link these residues to the hydrolytic water molecule.
Despite the previous classification of subtype VIII member MolA as a Co metalloenzyme (7), we have concluded that MolA is more likely to be a Zn metalloenzyme.
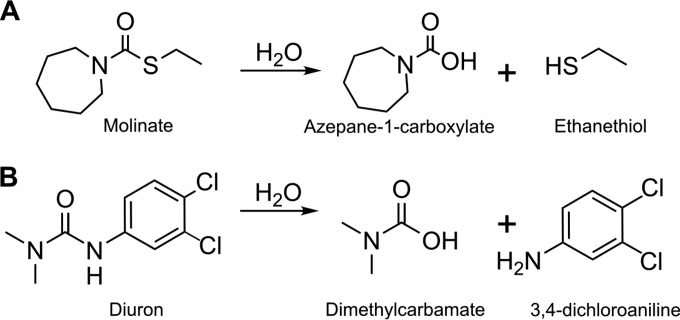
FIG 1.
The hydrolysis reactions catalyzed by MolA on molinate (A) and by PuhB on diuron (B).
Transitions between the binuclear and mononuclear forms of conserved AHS enzymes have previously been observed as a species-dependent trend, without associated differences in substrate specificity (19, 20). N-Acetyl-d-glucosamine-6-phosphate deacetylase (NagA) enzymes derived from Escherichia coli and Thermotoga maritima are mononuclear, whereas Bacillus subtilis-derived NagA is binuclear (19). The transition to mononuclear NagA sites occurs in concert with mutations to the conserved His-Xaa-His motif in strand 1 and is associated with the presence of an additional histidine residue (His143) that is thought to polarize the carbonyl bond of the substrate during nucleophilic attack (19, 21). A residue equivalent to His143 (His171) is observed in binuclear carboxypeptidases of subtype I, where it has a homologous role in the mechanism (22). The alteration of metal-coordinating residues in NagA results in a modified mechanism but has a limited physiological impact beyond a reduced metal ion requirement (21).
Since the most recent classification of the AHS, a trend in the expansion of the defined subtypes can be observed. There are a number of subtypes specialized for ancient functions (II, III, IV, V, VI), from which a number of the newer subtypes have evolved to catalyze the hydrolysis of environmental xenobiotics (I, VIII, IX). For example, subtype I phosphotriesterases break down organophosphate nerve agents and pesticides and have been shown to have evolved from subtype II lactonases (5, 23, 24). Subtype IX triazine hydrolases break down the herbicide atrazine and have been shown to evolve from subtype III deaminases (25, 26).
In the present study, both PuhB and MolA were used as model enzymes to investigate the evolution of xenobiotic-hydrolyzing subtype VIII in the AHS. The structures of MolA and PuhB solved in this work have revealed important features common to both enzymes, in addition to binuclear and mononuclear AHS members. Kinetic analysis has identified the broad substrate promiscuity of MolA, and mutagenesis has identified residues important for catalysis and determination of the substrate specificity of subtype VIII members. A mechanism has been proposed that is based on the crystal structures and predicted metal-coordinating residues, the pH-dependent activity profiles common to both MolA and PuhB, substrate docking, and the known mechanism of structurally similar enzymes. On the basis of sequence analysis, subtype VIII is the latest subtype proposed to diverge from an already described subtype within the AHS in response to synthetic compounds. However, this is the first example of such a drastic change as loss of a second metal to facilitate a change in mechanism. The modification of extant microbial AHS catalysts with mechanisms close to a required reaction may be a pertinent example of nature appropriating existing catalysts in response to environmental pressure, in this case, molinate and diuron contamination.
MATERIALS AND METHODS
Cloning.
A gene encoding molinate hydrolase, molA, was synthesized by DNA 2.0 with NdeI and BamHI restriction sites for cloning and delivered in plasmid pJ204. Following the transformation of E. coli DH5α cells, clones were sequenced and used to inoculate 10 ml of LB. The pJ204 vector was extracted with the QIAprep Spin Miniprep kit (Qiagen, Hilden, Germany) by following the manufacturer's protocol and subcloned via the NdeI and BamHI restriction sites into plasmid pETcc2 (a variant of pET14b) (C. Scott, R. J. Russell, C. W. Coppin, J. G. Oakeshott, and C. J. Jackson, patent application US 13/061,754, 22 September 2011, Australian Patent Office). The ligation product was used to transform electrocompetent E. coli BL21(DE3) (Promega). Constructs containing a tobacco etch virus (TEV) cleavage site in pETMCSIII with NdeI and EcoRI restriction sites were generated for enzymes that were to be used in analyses of kinetics and stability (27).
Expression and purification.
PuhB was expressed and purified as described previously (6). For MolA expression and subsequent purification, a single colony of E. coli BL21(DE3) cells transformed with a molA construct was used to inoculate TB medium (5 ml) (28). Five milliliters of this small scale culture was used to inoculate 1 liter of LB medium containing 0.5 M NaCl and 1 mM betaine and supplemented with 100 μM CoCl2, CuSO4, ZnSO4, or FeSO4 (29). After incubation at 25°C for 42 h, cells were pelleted by centrifugation and resuspended in buffer A (50 mM NaH2PO4, 10 mM imidazole, 300 mM NaCl, pH 7.5). The resuspended cells were lysed, the lysate was sedimented, and the supernatant was filtered before being loaded onto a 5-ml Ni-nitrilotriacetic acid (NTA) Superflow Column (Qiagen) equilibrated with buffer A. The protein was eluted with an increasing gradient of buffer B (50 mM NaH2PO4, 300 mM imidazole, 300 mM NaCl, pH 7.5). Enzyme-containing fractions were then buffer exchanged into TEV reaction buffer (50 mM NaH2PO4, 1 mM dithiothreitol, pH 8.0). His-tagged TEV protease was added at 1:10 to the enzyme and left overnight at 25°C. The cleaved enzyme was then separated from any uncleaved product and TEV by collecting the flowthrough from a Ni-NTA Superflow column. Before crystallography, the sample was loaded onto a Superdex 200 Hiload 26/6 column (GE Healthcare) equilibrated with buffer C (20 mM HEPES, 50 mM NaCl, 100 μM CoCl2, pH 7.5).
Enzyme kinetics.
Enzyme concentration was determined by measuring absorbance at 280 nm with an extinction coefficient of 43,555 M−1 cm−1, calculated in ExPASy (30). Determination of kinetic parameters of purified MolA was achieved by monitoring changes in the absorbance of triplicate 200-μl reaction mixtures in 96-well plates with a microplate spectrophotometer (Epoch; 0.5-cm path length). Substrates were dissolved in methanol, but the total organic solvent concentration in wells was identical and <2%. The monitoring wavelength and product extinction coefficient (ε) were 405 nm and 18.1 mM−1 cm−1, respectively, for 4-nitrophenol-producing substrates (p-nitrophenyl butyrate, p-nitrophenyl propionate, o-nitrophenyl acetate, paraoxon ethyl, p-nitrophenyl octanoate), 405 nm and 9.62 mM−1 cm−1 for p-nitroaniline-producing l-alanine p-nitroanilide, and 412 nm and 14.15 mM−1 cm−1 for molinate ethiolate and S-ethyl-dipropylthiocarbamate (EPTC); 5 mM 5,5′-dithiobis(2-nitrobenzoic acid) was used as an indicator (31). The hydrolysis of diuron and other phenylurea herbicides was detected with Fast Blue B Salt at 450 nm and 9.25 mM−1 250 μl−1 (32). Reactions were carried out in 20 mM NaH2PO4–150 mM NaCl, pH 7.5. The substrate concentrations used in assays were between 0.01 and 2.0 mM. Autohydrolysis was accounted for with negative controls (reaction mixtures where the buffer [20 mM NaH2PO4, 150 mM NaCl, 100 μM CoCl2, pH 7.5] replaced the enzyme). Initial rates were corrected for autohydrolysis, and kinetic parameters were acquired with the Michaelis-Menten equation in Kaleidagraph; error ranges relate to the standard deviation of data obtained from three independent measurements. To determine the effect of pH on enzyme activity, AMT buffer (50 mM glacial acetic acid, 50 mM morpholineethanesulfonic acid [MES], 100 mM Tris) (33) was used from pH 4.5 to pH 9.5. p-Nitrophenyl butyrate was assayed in triplicate as described above and measured at the isosbestic point (346 nm). The data were fitted to the previously described equation y = (y)max/[(10−pH/10−pKa1) + (10−pKa2/10−pH)] (6), where the apparent activity (y) is log(kcat) and pKa1 and pKa2 are the apparent pKa values of the acidic and basic groups, respectively.
ICP-OES.
Purified molinate hydrolase was desalted with a PD-10 desalting column (GE Healthcare) into metal-free buffer (20 mM MES, 150 mM NaCl, pH 6.5) and then digested into concentrated nitric acid (final concentration, 4%). The sample was analyzed after filtering by inductively coupled plasma optical emission spectrometry (ICP-OES) with a PerkinElmer Optima 2100 DV. In addition to normal calibrating standards, buffer-spiked standards were also used to rule out buffer interference with metal analysis.
Thermal stability analysis.
Circular-dichroism spectra were recorded with an Applied Photophysics Chirascan spectropolarimeter connected to a temperature control unit (TC125; Quantum Northwest) and a circulating-water bath (F3; Fisons Haake). Protein samples in phosphate buffer (20 mM NaH2PO4, 20 mM NaCl, 100 μM CoCl2 or ZnSO4, pH 7.5) were initially scanned to confirm their folded state (1-nm intervals from 180 to 260 nm, 0.5 s per point). Thermal-denaturation experiments were performed in triplicate at 0.5°C intervals as the temperature was increased from 20 to 90°C at a rate of 1°C · min−1 with a scan duration of 24 s per point and measurement at 222 nm.
Metal chelation.
Co-MolA (4.5 μM) was desalted into metal-chelating solution (5 mM 1,10-phenanthroline, 20 mM NaH2PO4, 150 mM NaCl, pH 7.5) with a PD-10 desalting column (GE Healthcare) and left overnight at room temperature before a final desalting step into phosphate buffer suitable for circular dichroism. The activity of the resultant protein was measured and assessed to be below 2% of the original activity of Co-MolA.
Crystallography.
Purified molinate hydrolase was concentrated to 8.4 mg · ml−1 in 20 mM HEPES–50 mM NaCl–100 μM CoCl2 (pH 7.5) buffer. Screens were optimized by hanging-drop vapor diffusion at 4°C. Drops consisting of 2 μl of reservoir solution and 1 μl of MolA were equilibrated over a 500-μl reservoir with EasyXtal-15-Well Tools (Qiagen, Hilden, Germany). Small microcrystals were obtained in 45% (vol/vol) (±)-2-methyl-2,4-pentanediol–0.2 M ammonium acetate–0.1 M bis-Tris, pH 6. These were microseeded by an adaptation of the method of D'Arcy et al. (34). Crystals were crushed with a melted 10-μl pipette tip (Fisher Biotec, Australia). The crushed crystals were used to make dilutions ranging from 1/102 to 1/107 with the reservoir solution. A 0.3-μl volume of a diluted microseed solution was added to a drop of 1.4 μl of reservoir solution and 0.7 μl of protein. An improvement in the size of crystals was achieved by microseeding into 48% (vol/vol) (±)-2-methyl-2,4-pentanediol–0.2 M ammonium acetate–0.1 M bis-Tris, pH 6.0. Data collection was performed with crystals with the mother liquor as the cryoprotectant and the addition of 1 mM cobalt before they were flash cooled in a nitrogen stream at 100 K.
Purified PuhB was concentrated to 12.5 mg · ml−1 in 25 mM HEPES–50 mM KCl, pH 8. Screens were optimized by hanging-drop vapor diffusion at 4°C with drops consisting of 1 μl of reservoir solution and 1 μl of protein. A crystal was obtained in 0.02% (vol/vol) sodium azide–0.2 M MgCl–8% (vol/vol) polyethylene glycol (PEG) monomethyl ether (MME) 550–8% (vol/vol) PEG 20000–0.05 MES at pH 6.5. The cryoprotectant maintains the mother liquor conditions, but the PEG MME 550 content was increased to 22% (vol/vol). The crystals were flash cooled in a nitrogen stream at 100 K.
Data collection and refinement.
MolA and PuhB diffraction data were collected at the Australian Synchrotron at a wavelength of 0.9537 Å. The X-ray diffraction data for both MolA and PuhB were indexed and integrated with the program XDS and merged with XSCALE within the XDS package (35). The merged MolA and PuhB data were imported into the CCP4 program suite (36).
The structure of MolA was solved by molecular replacement by using the coordinates of the protein with Protein Data Bank (PDB) code 3MTW and CHAINSAW to create the model (22, 37). Initial phases were obtained from molecular replacement with the program PHASER (38). This yielded a single solution with four copies in the asymmetric unit. Refinement was performed with REFMAC, and manual rebuilding was done with COOT (39–41).
The structure of PuhB was solved by molecular replacement by using the coordinates of MolA and MOLREP (42). This yielded a single solution with eight copies in the asymmetric unit. Refinement was performed with REFMAC and PHENIX, and manual rebuilding was done with COOT (39–41, 43).
Docking.
AutoDock 4.2 was used for molecular docking (44). The substrates to be docked were designed and generated with the GlycoBioChem ProDRG2 server (45). All rotatable bonds of the substrates and a grid around the binding pocket were defined. An output of potential binding positions based on the calculated reduction in total energy upon binding was generated; the bound poses with the greatest reduction in energy were used for all diagrams.
Bioinformatic analysis.
Amino acid sequences used for the construction of the AHS phylogenetic tree were acquired with structurally characterized members of each subtype as queries (PDB codes 1P1M, 3LS9, 2VHL, 4WGX, 1BF6, 3GU1, 3GIP, 1M7J, 1J5S, 3IAC, 1ITQ, and 3FDG). BLASTP was used to find homologs of these representative subtype query sequences in the nonredundant protein sequence database (46).
The remainder of the structurally characterized AHS subtype members were added to the collection to increase the number of characterized sequences. Structurally characterized triosephosphate isomerase sequences were used as an outgroup for the superfamily. Sequences were aligned with MUSCLE (47). Phylogenetic trees were constructed by the maximum-likelihood method in MEGA and bootstrapped with 100 replicates (48). For this analysis, the WAG rate matrix was used, with empirical amino acid frequencies estimated from the data, rate heterogeneity between sites modeled with the discrete-gamma model with five rate categories, and a fraction of invariant sites included in the model (WAG+I+G+F).
Ancestral reconstruction of subtype VIII, I, and V sequences from the complete AHS phylogenetic tree was performed. The tree was constructed with PhyML by using the model described above (49). Ancestral sequences were reconstructed by the empirical Bayesian method implemented in CODEML in the PAML4 software package (50).
Structurally characterized AHS members were analyzed with sequence similarity networks (SSNs) in Cytoscape (51). The sequences were combined into a BLAST database and blasted against each other to give E values, which allowed sequence similarity to be ranked. An E value cutoff of 1 × E−20 was chosen with reasoning similar to that of previously published work (52).
Protein structure accession numbers.
The protein structures determined in this study have been submitted to the PDB and assigned accession numbers 4WGX and 4WHB.
RESULTS
Three-dimensional structures of MolA and PuhB.
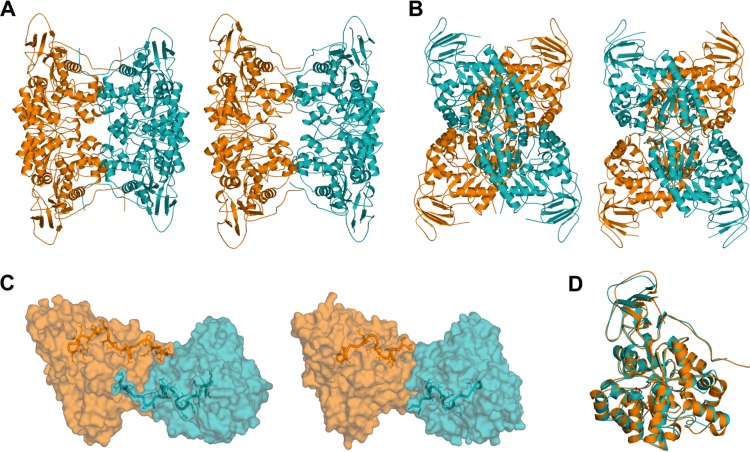
The crystal structures of MolA and PuhB were determined to resolutions of 2.29 and 2.96 Å, respectively (4WGX, 4WHB) (Table 2). The tertiary and quaternary structures of these proteins are shown in Fig. 2. MolA and PuhB consist of two domains, the characteristic (β/α)8 barrel domain present in all members of the AHS and an accessory domain consisting of a β sandwich. The two enzymes are organized into a cruciform assembly, a homotetramer in a dimer-of-dimers arrangement. The dimers of the assembly interact via an extensive set of salt bridges and electrostatic interactions, including residues in the domain-swapped extended C terminus (Fig. 2). Charged glutamate (Glu88), aspartate (Asp347, Asp349, Asp353), and arginine (Arg86, Arg167, Arg172, Arg459) residues interact to form salt bridges across the MolA dimer-dimer interface. Similar interactions involving fewer residues are seen in PuhB (Glu88, Asp351, Asp355, Arg86, Arg164, Arg169) (see Fig. S1 in the supplemental material). This could explain the somewhat looser PuhB assembly inferred from the comparatively larger central cavity of the complex than from the smaller cavity of MolA (Fig. 2A).
TABLE 2.
Refinement statistics
| Parameter | Valuea found for crystal: |
|
|---|---|---|
| MolA | PuhB | |
| Space group | I212121 | P2 |
| Unit-cell parameters | ||
| a (Å) | 126.43 | 77.37 |
| b (Å) | 226.32 | 100.52 |
| c (Å) | 265.38 | 238.55 |
| β (°) | 90, 90, 90 | 90, 90.37, 90 |
| Wavelength (Å) | 0.9537 | 0.9537 |
| Resolution range (Å) | 48.94–2.29 (2.33–2.29) | 39.60–2.96 (3.02–2.96) |
| No. of unique reflections | 170,085 | 75,147 |
| Completeness (%) | 99.9 (99.0) | 99.2 (99.8) |
| Multiplicity | 20.3 (13.5) | 5.6 (5.4) |
| Rmerge(I)b | 0.182 (2.201) | 0.201 (0.683) |
| Rpimc | 0.059 (0.893) | 0.135 (0.474) |
| Mean I/σ(I) ratio | 16.3 (1.6) | 8.3 (2.6) |
| CC1/2d | 0.998 (0.564) | 0.981 (0.752) |
| Refinement | ||
| No. of reflections used | 170,085 | 75,147 |
| Resolution range (Å) | 48.94–2.29 (2.33–2.29) | 39.60–2.96 (3.02–2.96) |
| RWork/RFreee | 0.1761/0.2032 (0.2917/0.3272) | 0.2438/0.2806 (0.3510/0.4233) |
| RMS deviations | ||
| Bond length (Å) | 0.008 | 0.015 |
| Bond angle (°) | 1.18 | 1.76 |
| PDB ID | 4WGX | 4WHB |
Values in parentheses are for the highest-resolution shell.
Rmerge(I) = Rmerge(I) = (Σhkl Σj |Ihkl,j − 〈Ihkl〉|)/(Σhkl Σj Ihkl,j), where 〈Ihkl〉 is the average intensity of j symmetry-related observations of reflections with Miller indices hkl.
Rpim = (Σhkl Σj |Ihkl,j − 〈Ihkl〉|)/(Σhkl Σj Ihkl,j).
CC1/2 = percentage of correlation between intensities from random half-data sets.
RWork = Σhkl|F(obs) − F(calc)|/Σhkl|F(obs)|; 5% of the data that were excluded from the refinement were used to calculate RFree.
FIG 2.
A cruciform quaternary structure is common to MolA and PuhB. (A) Front views of MolA (left) and PuhB (right). (B) Side views of MolA (left) and PuhB (right). (C) C-terminal domain swapping allows extensive salt bridge and hydrogen bond formation across the dimer-dimer interface, which stabilizes the cruciform assembly. MolA has a large C-terminal domain, which enables significant interactions (left). PuhB has a shorter C terminus, which could explain its looser assembly than that of MolA (right). (D) The overall topologies of both enzymes align well. MolA, orange; PuhB, teal.
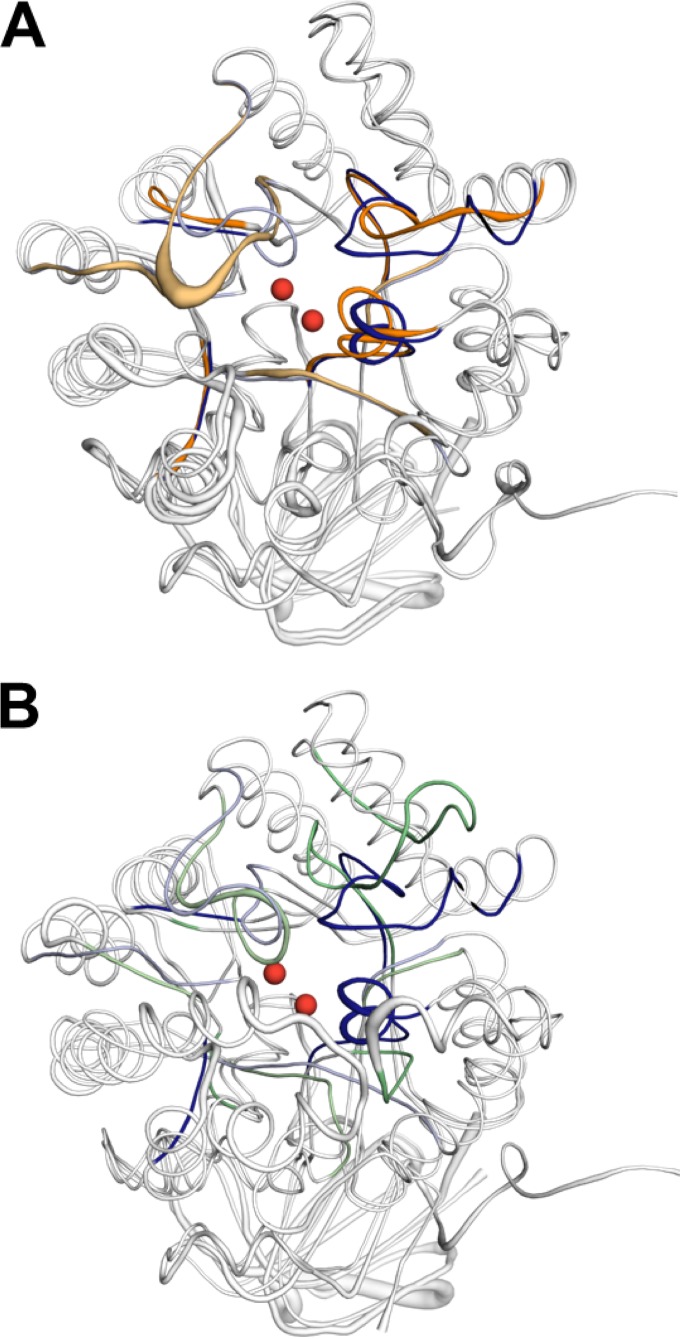
The structures of the enzymes overlap extensively, with a root mean square diameter of 1.3 Å for the Cα-aligned structures, as expected, given the overall amino acid identity of 51% (53). Despite the overall structural similarity, there is significant divergence in the positions and lengths of active-site loops 3 and 4 (Fig. 3A). Differences in loop geometry have been suggested to have a marked effect on substrate specificity in members of the AHS (9, 54). The different loop geometries observed between MolA and PuhB are caused by a five-amino-acid insertion in loop 4 of PuhB, compared to the equivalent loop 4 in MolA, in addition to a three-amino-acid deletion in loop 3 of PuhB, compared to the equivalent loop in MolA. The longer loop 4 of PuhB allows an additional lobe adjacent to the active-site entrance to be formed. This lobe may act as a lid domain that can be used to control the binding of substrates, as is found in other enzymes such as phosphoenolpyruvate carboxykinase (55). The active-site entrances of the respective enzymes also differ, as the active-site entry of MolA is positioned toward the dimer-dimer interface, whereas the active-site entry of PuhB is positioned outward.
FIG 3.
(A) Loop geometry is the greatest structural difference between aligned MolA (blue) and PuhB (orange). Loops 3 and 4 exhibit the greatest divergence in geometry. The basis of the divergence in loop 3 is a three-amino-acid deletion in PuhB compared to the equivalent loop in MolA. Loop 4 has a five-amino-acid insertion in PuhB compared to MolA, which creates a significant topology difference. (B) Loop geometry is significantly different between MolA (blue) and subtype I carboxypeptidase Cc2672 (PDB code 3MTW) (green). Loops 1, 3, 4, and 8 all have very different arrangements. The two catalytic zinc ions of Cc2672 are shown as red spheres in both structures.
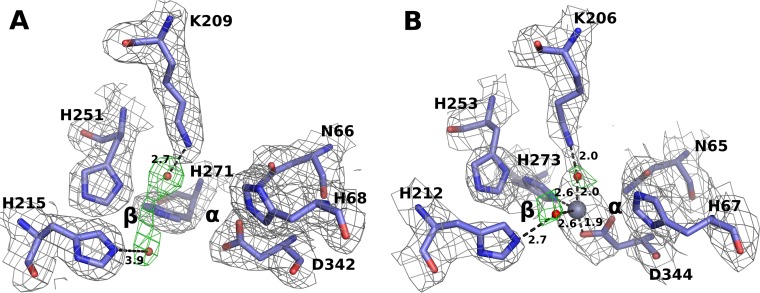
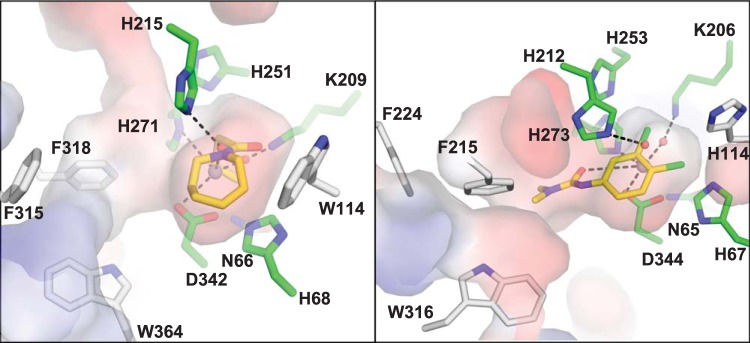
Significant electron density is observed in the crystal structure of PuhB, consistent with metal ion occupancy in the active site (Fig. 4). Although the metal ion coordination distances are not exact at this resolution, Asp334, His273, a water molecule coordinated to His212, and a water molecule coordinated to Lys206 coordinate the zinc ion in a distorted tetrahedral arrangement commonly accepted by zinc metalloenzymes (56). Water molecules in the coordination sphere are consistent with the presence of electron density and the homologous water coordination observed in other AHS structures (Fig. 4) (19, 22). The replacement of a carboxylated lysine in a binuclear metalloenzyme with a water molecule coordinated to the lysine upon conversion to a mononuclear enzyme has a precedent, having previously been observed in the laboratory evolution of a binuclear phosphotriesterase to a mononuclear phosphotriesterase (57). Future work could employ extended X-ray absorption fine structure or electron paramagnetic resonance analysis to define the fine details of the coordination site (58, 59).
FIG 4.
The metal binding sites of apo-MolA (A) and zinc-bound PuhB (B) are extremely similar; minor reorientations are observed for proposed metal-coordinating residues in both structures. Electron density (2Fo-dFc) is shown in gray, contoured at 1σ. OMIT electron density maps are shown in green and were generated in the absence of water molecules (mFo-dFc), contoured at 4.0σ (MolA) and 2.5 σ (PuhB). The zinc ion is shown as a gray sphere, and water molecules are shown as red spheres.
We were unable to solve the crystal structure of MolA in complex with a metal ion, although the apoenzyme crystallized readily. However, the apoenzyme structure, solved at higher resolution, has all of the metal ion-coordinating ligands in common with PuhB and is consistent with the metal ion-bound state observed in PuhB. Specifically, Lys209 coordinates to a water molecule, as does His215; one can see that metalation of the active site would involve minor conformational changes in Lys209, His271, His215, and Asp342.
Metal ion composition.
Previous atomic absorption analyses of both PuhB and MolA have identified these enzymes as zinc and cobalt mononuclear enzymes, respectively, with 1:1 metal enzyme stoichiometry for active enzymes (6, 7). Typically, enzymes classified within subtype I, to which MolA bears the most similarity, contain two divalent cations that are coordinated by four histidine residues, an aspartate, a bridging carboxylated lysine residue, and a hydroxide ion (9, 22). The optimal and physiologically relevant metal ion composition of enzymes is very difficult to establish, with many proteins having tight and loose binding sites, which might become occupied only in the presence of exogenous metal ions and substrate (60–62). Accordingly, we tested whether the catalytic rate of these enzymes could be increased by using different concentrations of exogenous metal ions and exogenous carbonate (see Fig. S2 in the supplemental material) (63). These results confirm that maximum activity is achieved with a single metal ion. We observe that apo-MolA is 4°C more stable than the holoenzyme (see Table S1 in the supplemental material). Moreover, metal reconstitution was not possible, despite many attempts. Thus, it appears that the metal ion acts more as a prosthetic group that is incorporated during folding than as an exchangeable cofactor.
The previous classification of MolA as a Co metalloenzyme is surprising, given the relatively low abundance of cobalt in the environment and the lack of any other “native” cobalt-dependent metalloenzymes in the AHS (7, 64). Therefore, the metal-dependent activity of MolA was investigated. Whole-cell assays were used to determine what metals should be used for future analysis. The metals tested were Fe, Cu, Co, Zn, Mn, and Ni on the basis of the most environmentally prevalent metals. The Fe-, Cu-, Zn-, and Co-treated forms exhibited detectable activity and were tested further.
The cobalt form of MolA (Co-MolA) had the highest activity with the thiocarbamate molinate (Table 3). As Raushel and colleagues have demonstrated with phosphotriesterase, it is not uncommon for cobalt metalloenzymes to have higher activity than their zinc counterparts, particularly with “softer,” sulfur-containing substrates (65, 66). The main reason for the previous classification of MolA as a Co metalloenzyme was its greater activity with the sulfur-containing substrates with which it was tested (7). Indeed, phosphotriesterase exhibits higher activity with both cobalt and cadmium on some substrates, despite being a native Zn metalloenzyme (66). Additionally, while zinc-, iron-, or copper-supplemented growth medium resulted in full metal ion incorporation, cobalt supplementation did not lead to complete occupancy of the metal ion at the active site of MolA (Table 3). Fe2+- and Cu2+-MolA were an order of magnitude less active than Zn2+- and Co2+-MolA. The assay performed with Fe-MolA was performed in the presence of tris(2-carboxyethyl)phosphine to prevent oxidation to the Fe3+ form. If there were a preference for a pentacoordinate transition state, then iron and copper would not be as well suited as zinc and cobalt (67, 68). Given the bioavailability of zinc, the difficulty of obtaining complete cobalt incorporation, and that the zinc form of MolA has activity comparable to that of the cobalt form with p-nitrophenyl propionate, paraoxon ethyl, and l-alanine p-nitroanilide (Table 4), it seems likely that MolA will exist most commonly as a Zn2+ metalloenzyme in nature.
TABLE 3.
Metal equivalentsa and corresponding activity of MolA with molinate
| Enzyme treatmentb | No. of metal equivalents |
Activity with molinate |
|||||
|---|---|---|---|---|---|---|---|
| Cobalt | Copper | Iron | Zinc | Km (μM) ± SE | kcat (s−1) ± SE | kcat/Km (s−1 M−1) ratio | |
| Iron | 0.00 | 0.03 | 1.15 | 0.02 | 46 ± 18 | 1.1 × 10−3 ± 0.2 × 10−4 | 23.3 |
| Copper | 0.00 | 1.23 | 0.01 | 0.01 | 56 ± 4.6 | 1.7 × 10−3 ± 2.0 × 10−4 | 32.2 |
| Cobalt | 0.37 | 0.11 | 0.01 | 0.02 | 14 ± 0.7 | 33 × 10−3 ± 4.8 × 10−4 | 2,380 |
| Zinc | 0.00 | 0.04 | 0.01 | 0.97 | 55 ± 2.5 | 29 × 10−3 ± 2.5 × 10−4 | 520 |
Metal concentration relative to protein concentration as determined by ICP-OES. All proteins were desalted into metal-free buffer before analysis.
MolA was expressed in medium supplemented with the respective metals at 100 μM.
TABLE 4.
Kinetic characterization of the cobalt and zinc forms of MolA
| Substrate and enzyme form | Km (μM) ± SE | kcat (s−1) ± SE | kcat/Km (s−1 M−1) ratio |
|---|---|---|---|
| p-Nitrophenyl propionate | |||
| Co | 655 ± 68 | 3.4 × 10−2 ± 5.0 × 10−3 | 53 |
| Zn | 307 ± 64 | 1.0 × 10−2 ± 8.8 × 10−4 | 34 |
| p-Nitrophenyl butyrate | |||
| Co | 218 ± 63 | 6.1 × 10−1 ± 9.3 × 10−2 | 29 × 102 |
| Zn | 351 ± 95 | 1.4 × 10−1 ± 1.2 × 10−2 | 43 × 101 |
| Molinate | |||
| Co | 14.1 ± 0.7 | 3.3 × 10−2 ± 4.8 × 10−4 | 24 × 102 |
| Zn | 54.9 ± 2.5 | 2.9 × 10−2 ± 2.5 × 10−3 | 52 × 101 |
| Ethiolate | |||
| Co | 1,960 ± 470 | 8.2 × 10−2 ± 1.9 × 10−3 | 42 |
| Zn | NAa | ||
| Paraoxon ethyl | |||
| Co | 622 ± 33 | 7.9 × 10−3 ± 1.8 × 10−4 | 13 |
| Zn | 209 ± 33 | 2.0 × 10−3 ± 1.3 × 10−4 | 10 |
| l-Alanine p-nitroanilide | |||
| Co | 495 ± 30 | 3.5 × 10−4 ± 4.0 × 10−5 | 0.7 |
| Zn | 403 ± 218 | 5.0 × 10−4 ± 1.2 × 10−4 | 1.5 |
NA, not applicable.
Kinetic properties of MolA.
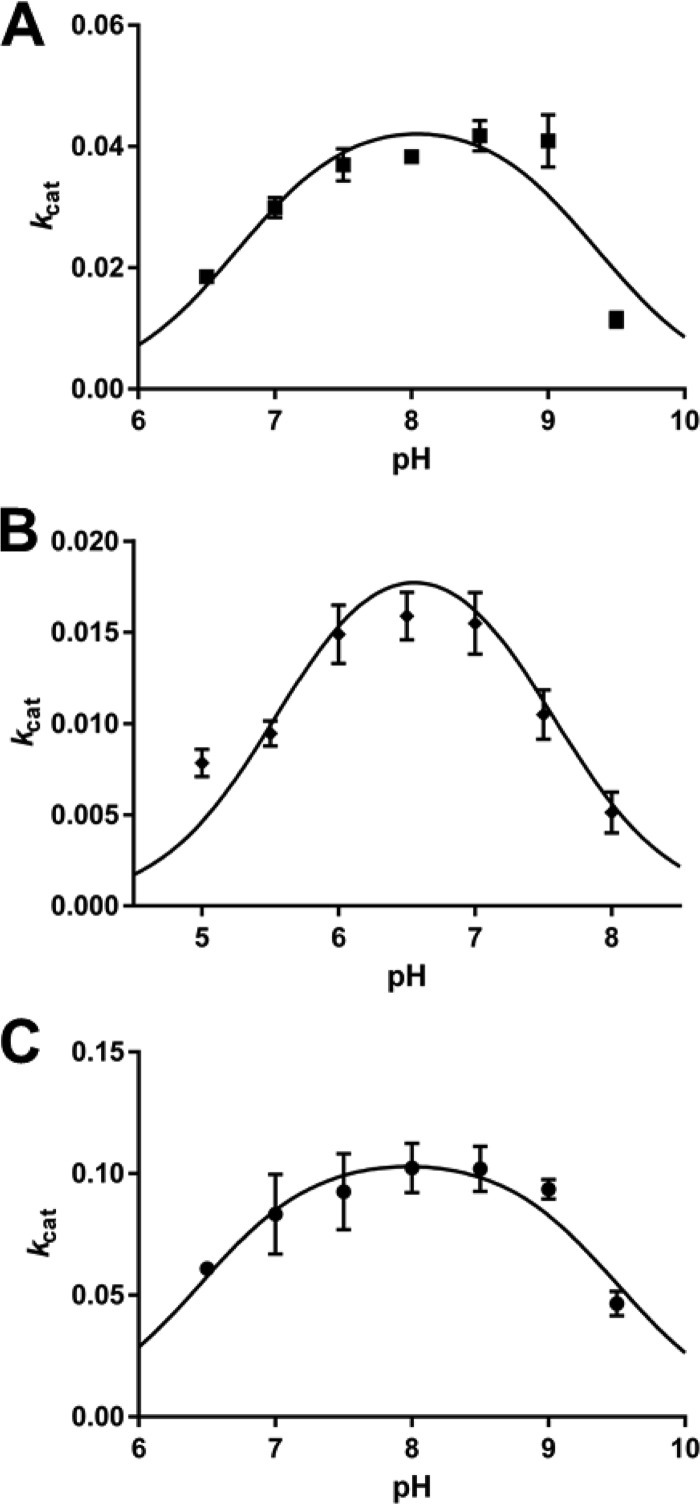
The pH-dependent activities of Co-MolA, Zn-MolA, and Cu-MolA were investigated in order to determine the impact of metal substitution on MolA activity and the acidic and basic pKa values for the reaction (pKa1 and pKa2). The impact of environmental pH on the preferential formation of a cobalt or zinc metalloenzyme is negligible, as the pKa values of cobalt and zinc are extremely similar and result in little difference in the corresponding pKa1 and pKa2 values of both metal forms (Table 5; Fig. 5) (69). The Zn-MolA pH activity curves and pKa values bear a strong resemblance to those of PuhB and indicate a shared mechanism (6). In contrast, the maximum activity of the copper form of MolA is shifted down 1.5 pH units to pH 6.5. This reflects the pKa of copper (7.3) versus those of cobalt (9.8) and zinc (9.0) (69). Likewise, the pKa1 and pKa2 values are both shifted down by the same amount. This result shows that both acidic and basic regions of the curve are dependent on the metal ion. The sharp increase in activity from pH 6 to 8 in Zn- and Co-MolA (pH 4.5 to 6 in Cu-MolA) likely corresponds to the generation of a nucleophilic hydroxide by the metal ion with assistance from an amino acid side chain acting as a general base (His215) since this value is significantly lower than the pKa of water-Co2+ (Fig. 5). Similarly, the second pKa value (pH 6.5 to 8.5 in Zn/Co-MolA and pH 8 to 10 in Cu-MolA) also shows metal ion dependence, in this case being consistent with the mechanism requiring a water molecule to be maintained in a protonated state to act as a general acid. Again, this is likely to be stabilized through further interaction with a positively charged amino acid side chain (Lys209). A detailed catalytic mechanism is proposed in the Discussion.
TABLE 5.
Dissociation constants for the hydrolysis of p-nitrophenyl butyrate by the cobalt, zinc, and copper forms of MolA
| Enzyme | pKa1 | pKa2 |
|---|---|---|
| Cobalt MolA | 6.45 | 9.50 |
| Zinc MolA | 6.73 | 9.36 |
| Copper MolA | 5.56 | 7.56 |
FIG 5.
The pH-dependent activities (kcat) of Zn-, Cu-, and Co-MolA on p-nitrophenyl butyrate, from which dissociation constants were generated. The activities of Zn-MolA (A; pKa1, 6.73; pKa2, 9.36), Cu-MolA (B; pKa1, 5.56; pKa2, 7.56), and Co-MolA (C; pKa1, 6.45; pKa2, 9.50) on p-nitrophenyl butyrate are shown. Points are averages of three replicates, and error bars represent the standard deviations of three independent replicates.
To further probe the mechanism of hydroxide generation in subtype VIII members, a histidine residue suspected to be involved in the mechanism was mutated to asparagine (His215Asn). Although the protein was stable and expressed, no activity was observed, confirming the essential role of this residue in the catalytic mechanism. Additionally, a MolA variant where the subtype VIII Asn-Xaa-His motif was reverted to the otherwise conserved AHS His-Xaa-His motif was tested. This variant exhibited very poor expression and unmeasurable activity, suggesting that the Asn-Xaa-His dyad and associated mononuclear active site are essential for molinate hydrolase activity.
Substrate range of MolA and PuhB.
The substrate specificity of MolA was probed with 27 different compounds. Of these, 16 were substrates for MolA and 6 had significant activity from which kinetic parameters could be calculated (Table 4; see Fig. S3 in the supplemental material). The hydrolysis of one thiocarbamate, molinate, has previously been determined with the cobalt enzyme, but with much lower activity than in this study (7). We believe that this might have been due to leaching of metal ions from the protein. Two other thiocarbamates, ethiolate and EPTC, could be hydrolyzed, although with lower and unmeasurable kcat values, respectively.
MolA displays notable substrate promiscuity and is able to catalyze the hydrolysis of esters, amides, and phosphotriester compounds. In fact, MolA has a higher kcat value with the highly activated carboxyl ester p-nitrophenyl butyrate than with molinate (Table 4). The detectable levels of activity with diuron are consistent with the high structural similarity between MolA and PuhB. The low levels of activity with l-alanine p-nitroanilide are of note, as the substrate binding sites of MolA and its closest structural homolog, the arginine/lysine carboxypeptidase Cc2672, differ considerably.
PuhB also exhibits substrate promiscuity, as it is able to cleave the amide bond of linuron at a rate comparable to that of the ester bond of its carbamate analogue (6). The phosphotriester ethyl paraoxon can also be hydrolyzed by PuhB, but the phosphothionate methyl parathion cannot; this is also seen in MolA, where catalytic parameters can be determined only for ethyl paraoxon. The observed catalytic promiscuity of both MolA and PuhB supports a theory that these enzymes are general hydrolases and their specificity for thiocarbamate or amide bonds is a result of an idealized substrate binding pocket for these synthetic compounds (6).
Probing the structural determinants of MolA and PuhB substrate specificity.
MolA exhibits a binding site that is remarkably complementary to that of molinate (Fig. 6A). Key residues are placed to interact and create an ideal binding pocket for the ring of molinate, in particular, Trp114. The characteristic AHS catalytic residues are well positioned to interact with the carbonyl and thiol groups of molinate. The active site of PuhB is similar to that of MolA (Fig. 6B), the majority of the differences being at the entry tunnel to the active site; some of these residues were therefore targeted to probe their effect on MolA substrate specificity.
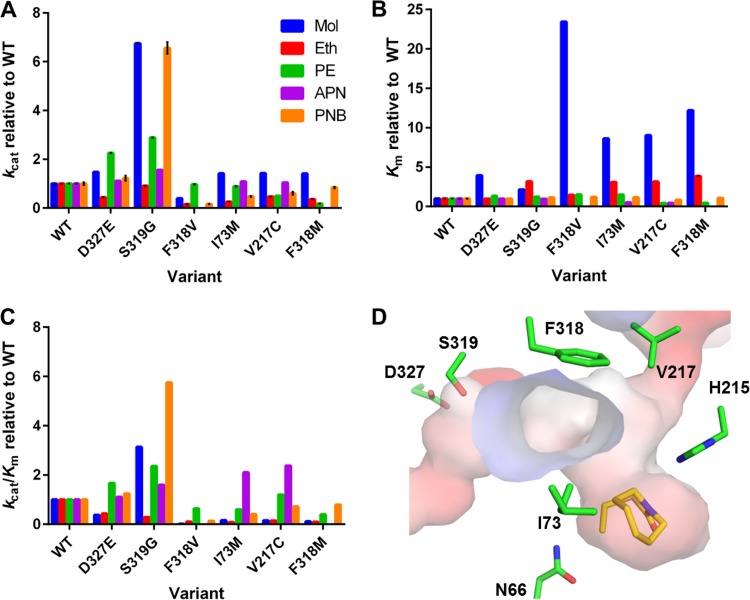
FIG 6.
The substrate binding pockets and entry tunnels of MolA (A) and PuhB (B). Residues that may impact the differences in substrate specificity between the two enzymes (white sticks) and the conserved catalytic residues (green sticks) are shown. MolA has a tryptophan (Trp364) and two phenylalanine residues (Phe315, Phe318) that act as a hydrophobic wall to direct substrates into the active site. PuhB has a tryptophan (Trp316) and two phenylalanine residues (Phe215, Phe224) positioned to form a larger tunnel entry. In the binding pocket of MolA, the ring of molinate can interact with Trp114; in PuhB, this residue is His114. Molinate and diuron (yellow) were docked into the active sites of MolA (A) and PuhB (B), respectively, with AutoDock (44), and the water molecules and metals observed in PuhB are displayed.
Ser319Gly and Asp327Glu variants were designed in order to make the MolA binding site more accommodating to larger substrate molecules, with the Cc2672 binding pocket as a guide. Phe318Met/Val, Ile73Met, and Val217Cys variants were also designed in order to increase the size of the substrate binding cavity, in addition to increasing flexibility. The previously described His215Asn mutation was designed to investigate the mechanism of molinate hydrolase catalysis. Asn66His was designed in order to determine whether the obvious reversion of the unique Asn-Xaa-His motif to His-Xaa-His would allow a binuclear complex to form or change any kinetic parameters.
Ile73Met, Val217Cys, Phe318Met/Val, and Asp327Glu all resulted in reduced binding affinity for molinate (8- to 23-fold) (Fig. 7; see Table S2 in the supplemental material). This, in combination with the high kcat/Km ratio of wild-type MolA with molinate, indicates that the substrate binding site is already well optimized for molinate binding and catalysis. The largest decrease in binding affinity for molinate occurred when Phe318 was mutated, despite the fact that Phe318 is not in the active site. It seems that Phe318 acts in concert with another phenylalanine residue (Phe315), forming a hydrophobic wall that directs molinate into the active site and constricts its shape. By vastly reducing the size (Phe318Val) or increasing the flexibility (Phe318Met) of this residue, we decreased its ability to bind and catalyze the hydrolysis of most substrates.
FIG 7.
Kinetic parameters of variants used to probe the structural determinants of MolA substrate specificity. (A) kcat values of variants relative to those of wild-type (WT) MolA with the substrates molinate (Mol), ethiolate (Eth), paraoxon ethyl (PE), l-alanine p-nitroanilide (APN), and p-nitrophenyl butyrate (PNB). (B) Km values of variants relative to those of the wild type with the same substrates. (C) kcat/Km ratios of variants relative to those of the wild type. (D) Positions of the respective mutations (green) in the structure of MolA docked with molinate (yellow) in AutoDock (44). Full kinetic parameters are shown in Table S2 in the supplemental material.
Only one variant exhibited an increase in the kcat/Km ratio with many substrates, the Ser319Gly variant. Ser319 is the only residue to form a hydrogen bond with loop 7, which aids in forming the hydrophobic active-site tunnel. Loss of this hydrogen bonding interaction likely allows more conformational freedom of loop 7, which would allow for greater expansion and contraction of the hydrophobic tunnel leading to the active site. This is reflected in the increased Km, which reflects a more open active site.
Sequence analysis.
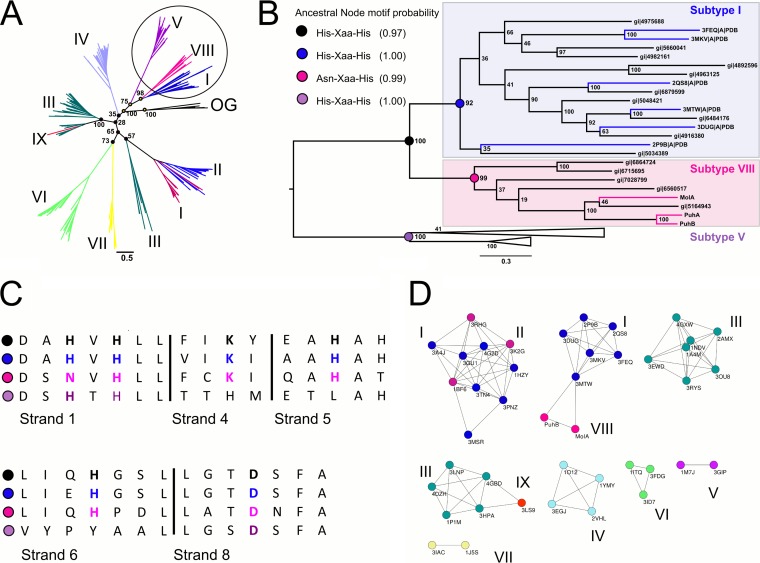
A phylogeny of the AHS was inferred by maximum-likelihood methods (Fig. 8A). The aligned AHS sequences all separated into their respective subtypes in the phylogenetic tree, with the exception of subtype IX, which groups with the subtype III deaminases from which it has evolved (25, 26) and subtype I phosphotriesterases, from the subtype II lactonases from which they have evolved (23, 24). Bootstrap values surrounding the nodes of interest (I, VIII) were excellent and provided a useful outgroup (V) for the subtype I and VIII ancestral reconstruction performed. Ancestral reconstruction was performed to determine whether the ancestral hydrolase from which subtype VIII has evolved was binuclear (His-Xaa-His motif) or mononuclear (Asn-Xaa-His motif) (Fig. 8B). Comparing the conserved metal binding motifs in subtypes VIII, I, and V (Asn-Xaa-His, His-Xaa-His, and His-Xas-His, respectively) in the context of this tree suggests that divergence of subtype VIII was accompanied by a transition from binuclear to mononuclear metal binding. This was supported by computational reconstruction of the ancestral metal binding motif, which showed that the ancestor of subtypes I and VIII had the His-Xaa-His dyad (posterior probability of 0.97) associated with binuclear metal binding while maintaining the rest of the subtype I metal-coordinating residues (Fig. 8B and C).
FIG 8.
(A) Phylogenetic tree of the nine current subtypes in the AHS. The three subtypes used for ancestral reconstruction and relevant bootstrap values are highlighted. (OG, outgroup) (B) The subtree used to determine the ancestral sequences of subtype VIII and I members. Reconstruction of the metal-binding motif in the ancestor of subtype VIII and I members (black node) provides evidence that this ancestor was binuclear Colored branches indicate characterized enzymes. (C) Sequence alignment of the four ancestral nodes showing the conservation of the metal-coordinating residues. (D) SSNs of structurally characterized AHS enzymes. Each node represents a single sequence, and each edge represents a connection between two sequences with a BLAST E value of >1 × E−20. The subtypes are color coded, and the PDB codes of the enzymes are shown.
SSNs provide a potentially better way to represent the entire AHS, as the bootstrap values separating two of the subtypes in the phylogenetic tree are poor. There is excellent agreement between the phylogenetic and SSN representations of the AHS (Fig. 8C). According to the phylogenetic tree, subtypes I and III are polyphyletic, which is also consistent with the SSN. The three subtype pairs that do not separate in the SSN (1 and VIII, I and II, and III and IX) also show close association in the phylogenetic tree (Fig. 8A and D), as explained in the Discussion.
DISCUSSION
Metal ion rearrangement for a new mechanism.
While the evolution of some enzymes in the AHS, such as phosphotriesterases from lactonases, has occurred via point mutations and loop insertions, the evolution of MolA and PuhB has occurred in concert with a much more dramatic event, the transition from a binuclear to a mononuclear active site. For the mutation that caused this alteration (His-Xaa-His to Asn-Xaa-His) to be accommodated in evolution, it must have significantly improved subtype VIII activity and benefitted the bacteria from which these enzymes are derived.
The leaving group of molinate is ethanethiol, which has a high pKa value (10.6); this represents the instability of the anion and suggests that the reaction will require protonation of the leaving group and stabilization of the significant negative charge that will develop in the transition state to proceed. In contrast, the amino acid leaving group of an ancestral carboxypeptidase would be significantly better, owing to dual ionizable groups (carboxy pKa and amino pKb), with the difference between the two groups generally on the order of 7 (70). Accordingly, the active site of a subtype VIII ancestral enzyme would not require an amino acid to protonate the leaving group. The most significant effect of the loss of the metal ion is the loss of carboxylation of the lysine residue, as two metal ions are required to stabilize the carboxylate. Loss of carboxylation enables a water molecule to bridge the metal ion and the lysine, stabilizing the water molecule so that it can act as a proton donor; this water coordination is observed clearly in MolA and implied by the PuhB structure (Fig. 4). Thus, we propose a mechanism in which a nucleophile is generated in a metal ion-dependent manner, represented by the increase in activity in the acidic region of the pH-activity curve, in concert with catalytically essential His215 (His212) acting as a general base (Fig. 9). The leaving group is then protonated from a metal ion-coordinated water molecule stabilized by Lys209 (Lys205), which results in the loss of activity in the basic region of the curve, as that water molecule becomes deprotonated. In summary, the loss of the metal ion enabled the enzyme to dramatically change its catalytic mechanism and degrade these new synthetic compounds.
FIG 9.
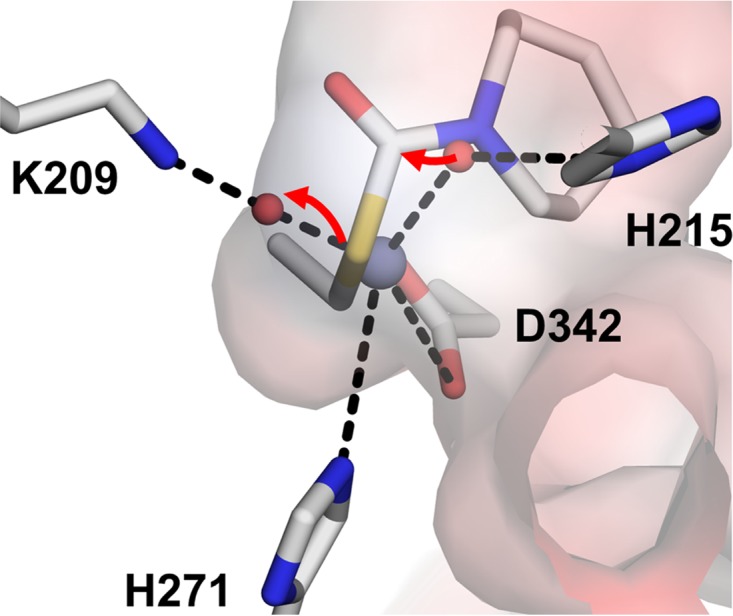
(A) The proposed reaction mechanism of molinate hydrolase acting on docked molinate, with the catalytic metal and water molecules from the aligned structure of PuhB. The nucleophilic hydroxide is generated in a metal ion-dependent manner in concert with His215 acting as a general base. The leaving group is then protonated from a metal-coordinated water molecule stabilized by Lys209. The metal ion can act to stabilize the negative charge that develops at the carbonyl oxygen in the transition state. Docking was performed in AutoDock (44).
Evolution in response to synthetic compounds.
Since the comprehensive review published by Seibert and Raushel in 2005, two new subtypes have been identified as belonging to the AHS (9), subtype IX, consisting of triazine hydrolase, and subtype VIII, two members of which were structurally characterized for the first time in this work. An updated depiction of the AHS is shown in Fig. 8 and described in Table 1. Three subtypes (I, II, III) have new divergent subtypes associated with them (VIII, I, IX) that have been shown to hydrolyze recently created synthetic chemicals. Newly divergent subtypes of subtypes IV, V, VI, and VII are not observed. Previous work has already described the likely evolution of phosphotriesterases (I) from lactonases (II), explaining their lack of separation in the phylogenetic tree and SSN (Fig. 8) (5, 23, 24). Triazine hydrolases (subtype IX) catalyze hydrolytic dechlorinations from s-triazine rings, a reaction similar to the hydrolytic deaminations from diazine ring systems that deaminases (subtype III enzymes) catalyze. Evidence of the evolution of triazine hydrolase from deaminases has now been established, which again explains their lack of separation in both the phylogenetic and SSN representations of the AHS (25, 26). It is therefore of note that subtypes VIII and I group together in the SSN and are closely associated in the phylogenetic tree.
The observed polyphyly of subtypes I and III is likely to represent the evolution of these subgroups from more than one ancestor toward substrate binding pockets idealized for different substrate functionalities. Phosphotriesterases (I) and triazine hydrolases (IX) represent a direct evolutionary trajectory from existing subtypes II and III, respectively, through the mutation of conserved metal binding ligands. Computational reconstruction of the ancestral metal binding motif of the ancestor common to subtypes VIII and I indicates that the ancestor of these subtypes would have been binuclear. Therefore, divergence of subtype VIII was associated with a binuclear-to-mononuclear metal binding transition, through loss of the conserved His-Xaa-His dyad. It could be hypothesized that the elimination of a second metal binding site in subtype VIII has prevented the carboxylation of Lys209 (Lys205), observed in subtype I enzymes, which has resulted in a change of mechanism, enabling the hydrolysis of recently introduced xenobiotics.
Although it cannot be proven that enzymes such as phosphotriesterase, the molinate and diuron hydrolases, and triazine hydrolase have evolved in response to the introduction of synthetic compounds, we can argue that it is a plausible hypothesis. The observations that these enzymes were all isolated from contaminated areas and that all provide a selective advantage to the organism, often allowing them to use the xenobiotic as a source of nutrients, suggest that the activity of these enzymes will be under positive selective pressure to evolve. The work presented here demonstrates that loss of a metal ion and rearrangement of the active site are one plausible route by which enzymes could acquire new activities such as the molinate and diuron hydrolases prior to further specialization. Recent work centered on the evolution of metalloenzymes has identified the many different ways metal-dependent activity can be tuned along evolutionary trajectories. Alterations from the “native” metal of a metalloenzyme is consistently seen to push promiscuous activity to be more prominent (71, 72). Beyond direct metal preference changes, work by Ben-David and coworkers on paraoxonase-1 clearly showed how an alternative binding mode of a catalytic calcium ion could have been the initiator of divergence in enzymatic activity (73). The likelihood of transitions between mononuclear and binuclear metal binding sites in the evolutionary trajectory of the AHS has already been hypothesized (73). MolA and PuhB may represent a transition from a binuclear carboxypeptidase ancestor, where loss of the second metal binding site enables the efficient hydrolysis of molinate and diuron. Further work is required to fully establish the structural and mechanistic determinants of evolutionary transitions between the mononuclear and binuclear proteins of this superfamily.
Supplementary Material
ACKNOWLEDGMENTS
We thank Viki Withers and Thomas Carruthers for assistance with ICP-OES and Benjamin Clifton for assistance with phylogenetic analysis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04016-14.
REFERENCES
- 1.Akiva E, Brown S, Almonacid DE, Barber AE, Custer AF, Hicks MA, Huang CC, Lauck F, Mashiyama ST, Meng EC. 2014. The structure-function linkage database. Nucleic Acids Res 42:D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holm L, Sander C. 1997. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins Struct Funct Genet 28:72–82. [PubMed] [Google Scholar]
- 3.Williams L, Nguyen T, Li Y, Porter TN, Raushel FM. 2006. Uronate isomerase: a nonhydrolytic member of the amidohydrolase superfamily with an ambivalent requirement for a divalent metal ion. Biochemistry 45:7453–7462. doi: 10.1021/bi060531l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapir N, Pedersen C, Gil O, Strong L, Seffernick J, Sadowsky MJ, Wackett LP. 2006. TrzN from Arthrobacter aurescens TC1 is a zinc amidohydrolase. J Bacteriol 188:5859–5864. doi: 10.1128/JB.00517-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elias M, Dupuy J, Merone L, Mandrich L, Porzio E, Moniot S, Rochu D, Lecomte C, Rossi M, Masson P. 2008. Structural basis for natural lactonase and promiscuous phosphotriesterase activities. J Mol Biol 379:1017–1028. doi: 10.1016/j.jmb.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Khurana J, Jackson C, Scott C, Pandey G, Horne I, Russell R, Herlt A, Easton C, Oakeshott J. 2009. Characterization of the phenylurea hydrolases A and B: founding members of a novel amidohydrolase subgroup. Biochem J 418:431–441. doi: 10.1042/BJ20081488. [DOI] [PubMed] [Google Scholar]
- 7.Duarte M, Ferreira-da-Silva F, Lünsdorf H, Junca H, Gales L, Pieper DH, Nunes OC. 2011. Gulosibacter molinativorax ON4T molinate hydrolase, a novel cobalt-dependent amidohydrolase. J Bacteriol 193:5810–5816. doi: 10.1128/JB.05054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson CJ, Liu J-W, Coote ML, Ollis DL. 2005. The effects of substrate orientation on the mechanism of a phosphotriesterase. Org Biomol Chem 3:4343–4350. doi: 10.1039/b512399b. [DOI] [PubMed] [Google Scholar]
- 9.Seibert CM, Raushel FM. 2005. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 10.Gerlt JA, Babbitt PC. 2001. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem 70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 11.Babtie A, Tokuriki N, Hollfelder F. 2010. What makes an enzyme promiscuous? Curr Opin Chem Biol 14:200–207. doi: 10.1016/j.cbpa.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 12.Liaw S-H, Chen S-J, Ko T-P, Hsu C-S, Chen C-J, Wang AH-J, Tsai Y-C. 2003. Crystal structure of d-aminoacylase from Alcaligenes faecalis DA1: a novel subset of amidohydrolases and insights into the enzyme mechanism. J Biol Chem 278:4957–4962. doi: 10.1074/jbc.M210795200. [DOI] [PubMed] [Google Scholar]
- 13.Jackson CJ, Foo J-L, Kim H-K, Carr PD, Liu J-W, Salem G, Ollis DL. 2008. In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase. J Mol Biol 375:1189–1196. doi: 10.1016/j.jmb.2007.10.061. [DOI] [PubMed] [Google Scholar]
- 14.Liao RZ, Yu JG, Raushel FM, Himo F. 2008. Theoretical investigation of the reaction mechanism of the dinuclear zinc enzyme dihydroorotase. Chemistry 14:4287–4292. doi: 10.1002/chem.200701948. [DOI] [PubMed] [Google Scholar]
- 15.Jackson CJ, Coppin CW, Carr PD, Aleksandrov A, Wilding M, Sugrue E, Ubels J, Paks M, Newman J, Peat TS, Russell RJ, Field M, Weik M, Oakeshott JG, Scott C. 2014. 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization. Appl Environ Microbiol 80:4003–4011. doi: 10.1128/AEM.00916-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benning MM, Shim H, Raushel FM, Holden HM. 2001. High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta. Biochemistry 40:2712–2722. doi: 10.1021/bi002661e. [DOI] [PubMed] [Google Scholar]
- 17.Buchbinder JL, Stephenson RC, Dresser MJ, Pitera JW, Scanlan TS, Fletterick RJ. 1998. Biochemical characterization and crystallographic structure of an Escherichia coli protein from the phosphotriesterase gene family. Biochemistry 37:5096–5106. doi: 10.1021/bi971707+. [DOI] [PubMed] [Google Scholar]
- 18.Nitanai Y, Satow Y, Adachi H, Tsujimoto M. 2002. Crystal structure of human renal dipeptidase involved in β-lactam hydrolysis. J Mol Biol 321:177–184. doi: 10.1016/S0022-2836(02)00632-0. [DOI] [PubMed] [Google Scholar]
- 19.Hall RS, Brown S, Fedorov AA, Fedorov EV, Xu C, Babbitt PC, Almo SC, Raushel FM. 2007. Structural diversity within the mononuclear and binuclear active sites of N-acetyl-d-glucosamine-6-phosphate deacetylase. Biochemistry 46:7953–7962. doi: 10.1021/bi700544c. [DOI] [PubMed] [Google Scholar]
- 20.Edwards BF, Fernando R, Martin PD, Grimley E, Cordes M, Vaishnav A, Brunzelle JS, Evans HG, Evans DR. 2013. The mononuclear metal center of type-I dihydroorotase from Aquifex aeolicus. BMC Biochem 14:36. doi: 10.1186/1471-2091-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall RS, Xiang DF, Xu C, Raushel FM. 2007. N-Acetyl-d-glucosamine-6-phosphate deacetylase: substrate activation via a single divalent metal ion. Biochemistry 46:7942–7952. doi: 10.1021/bi700543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang DF, Patskovsky Y, Xu C, Fedorov AA, Fedorov EV, Sisco AA, Sauder JM, Burley SK, Almo SC, Raushel FM. 2010. Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily. Biochemistry 49:6791–6803. doi: 10.1021/bi100897u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afriat L, Roodveldt C, Manco G, Tawfik DS. 2006. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry 45:13677–13686. doi: 10.1021/bi061268r. [DOI] [PubMed] [Google Scholar]
- 24.Elias M, Tawfik DS. 2012. Divergence and convergence in enzyme evolution: parallel evolution of paraoxonases from quorum-quenching lactonases. J Biol Chem 287:11–20. doi: 10.1074/jbc.R111.257329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Udiković-Kolić N, Scott C, Martin-Laurent F. 2012. Evolution of atrazine-degrading capabilities in the environment. Appl Microbiol Biotechnol 96:1175–1189. doi: 10.1007/s00253-012-4495-0. [DOI] [PubMed] [Google Scholar]
- 26.Noor S, Taylor MC, Russell RJ, Jermiin LS, Jackson CJ, Oakeshott JG, Scott C. 2012. Intramolecular epistasis and the evolution of a new enzymatic function. PLoS One 7:e39822. doi: 10.1371/journal.pone.0039822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neylon C, Brown SE, Kralicek AV, Miles CS, Love CA, Dixon NE. 2000. Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: a model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry 39:11989–11999. doi: 10.1021/bi001174w. [DOI] [PubMed] [Google Scholar]
- 28.Tartof KD, Hobbs CA. 1987. Improved media for growing plasmid and cosmid clones. Focus 9:12. [Google Scholar]
- 29.Oganesyan N, Ankoudinova I, Kim S-H, Kim R. 2007. Effect of osmotic stress and heat shock in recombinant protein overexpression and crystallization. Protein Expr Purif 52:280–285. doi: 10.1016/j.pep.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 31.Schofield DA, DiNovo AA. 2010. Generation of a mutagenized organophosphorus hydrolase for the biodegradation of the organophosphate pesticides malathion and demeton-S. J Appl Microbiol 109:548–557. doi: 10.1111/j.1365-2672.2010.04672.x. [DOI] [PubMed] [Google Scholar]
- 32.Grant DF, Bender DM, Hammock BD. 1989. Quantitative kinetic assays for glutathione S-transferase and general esterase in individual mosquitoes using an EIA reader. Insect Biochem 19:741–751. doi: 10.1016/0020-1790(89)90055-3. [DOI] [Google Scholar]
- 33.Ellis KJ, Morrison JF. 1982. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol 87:405–426. [DOI] [PubMed] [Google Scholar]
- 34.D'Arcy A, Villard F, Marsh M. 2007. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr Sect D Biol Crystallogr 63:550–554. doi: 10.1107/S0907444907007652. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W. 2010. XDS. Acta Crystallogr Sect D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collaborative CP. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr 50:760. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.Stein N. 2008. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J Appl Crystallogr 41:641–643. doi: 10.1107/S0021889808006985. [DOI] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr Sect D Biol Crystallogr 60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30:1022–1025. doi: 10.1107/S0021889897006766. [DOI] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuttelkopf AW, Van Aalten DM. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr Sect D Biol Crystallogr 60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 46.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 50.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 51.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. 2010. Cytoscape Web: an interactive web-based network browser. Bioinformatics 26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uberto R, Moomaw EW. 2013. Protein similarity networks reveal relationships among sequence, structure, and function within the cupin superfamily. PLoS One 8:e74477. doi: 10.1371/journal.pone.0074477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holm L, Park J. 2000. DaliLite workbench for protein structure comparison. Bioinformatics 16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 54.Afriat-Jurnou L, Jackson CJ, Tawfik DS. 2012. Reconstructing a missing link in the evolution of a recently diverged phosphotriesterase by active-site loop remodeling. Biochemistry 51:6047–6055. doi: 10.1021/bi300694t. [DOI] [PubMed] [Google Scholar]
- 55.Johnson TA, Holyoak T. 2010. Increasing the conformational entropy of the Ω-loop lid domain in phosphoenolpyruvate carboxykinase impairs catalysis and decreases catalytic fidelity. Biochemistry 49:5176–5187. doi: 10.1021/bi100399e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCall KA, Huang C, Fierke CA. 2000. Function and mechanism of zinc metalloenzymes. J Nutr 130(5S Suppl):1437S–1446S. [DOI] [PubMed] [Google Scholar]
- 57.Tokuriki N, Jackson CJ, Afriat-Jurnou L, Wyganowski KT, Tang R, Tawfik DS. 2012. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat Commun 3:1257. doi: 10.1038/ncomms2246. [DOI] [PubMed] [Google Scholar]
- 58.Griese JJ, Roos K, Cox N, Shafaat HS, Branca RM, Lehtiö J, Gräslund A, Lubitz W, Siegbahn PE, Högbom M. 2013. Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein. Proc Natl Acad Sci U S A 110:17189–17194. doi: 10.1073/pnas.1304368110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bobyr E, Lassila JK, Wiersma-Koch HI, Fenn TD, Lee JJ, Nikolic-Hughes I, Hodgson KO, Rees DC, Hedman B, Herschlag D. 2012. High-resolution analysis of Zn2+ coordination in the alkaline phosphatase superfamily by EXAFS and X-ray crystallography. J Mol Biol 415:102–117. doi: 10.1016/j.jmb.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson C, Carr P, Kim H, Liu J, Herrald P, Miti N, Schenk G, Smith C, Ollis D. 2006. Anomalous scattering analysis of Agrobacterium radiobacter phosphotriesterase: the prominent role of iron in the heterobinuclear active site. Biochem J 397:501–508. doi: 10.1042/BJ20060276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hadler KS, Tanifum EA, Yip SH-C, Mitic N, Guddat LW, Jackson CJ, Gahan LR, Nguyen K, Carr PD, Ollis DL. 2008. Substrate-promoted formation of a catalytically competent binuclear center and regulation of reactivity in a glycerophosphodiesterase from Enterobacter aerogenes. J Am Chem Soc 130:14129–14138. doi: 10.1021/ja803346w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lai W-L, Chou L-Y, Ting C-Y, Kirby R, Tsai Y-C, Wang AH-J, Liaw S-H. 2004. The functional role of the binuclear metal center in d-aminoacylase one-metal activation and second-metal attenuation. J Biol Chem 279:13962–13967. doi: 10.1074/jbc.M308849200. [DOI] [PubMed] [Google Scholar]
- 63.Hong S-B, Kuo JM, Mullins LS, Raushel FM. 1995. CO2 is required for the assembly of the binuclear metal center of phosphotriesterase. J Am Chem Soc 117:7580–7581. doi: 10.1021/ja00133a046. [DOI] [Google Scholar]
- 64.Kabata-Pendias A. 2010. Trace elements in soils and plants. CRC Press, Boca Raton, FL. [Google Scholar]
- 65.Omburo GA, Kuo JM, Mullins LS, Raushel F. 1992. Characterization of the zinc binding site of bacterial phosphotriesterase. J Biol Chem 267:13278–13283. [PubMed] [Google Scholar]
- 66.Rochu D, Viguié N, Renault F, Crouzier D, Froment M, Masson P. 2004. Contribution of the active-site metal cation to the catalytic activity and to the conformational stability of phosphotriesterase: temperature-and pH-dependence. Biochem J 380:627–633. doi: 10.1042/BJ20031861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rubino JT, Franz KJ. 2012. Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function. J Inorg Biochem 107:129–143. doi: 10.1016/j.jinorgbio.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 68.Holm RH, Kennepohl P, Solomon EI. 1996. Structural and functional aspects of metal sites in biology. Chem Rev 96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 69.Sillén LG, Martell AE. 1971. Stability constants of metal ion complexes: supplement 1. Special publication no. 25. Royal Society of Chemistry, London, United Kingdom. [Google Scholar]
- 70.Sigel A. 2001. Metal ions in biological systems: volume 38. Probing of proteins by metal ions and their low-molecular-weight complexes. CRC Press, Boca Raton, FL. [Google Scholar]
- 71.Daumann LJ, McCarthy BY, Hadler KS, Murray TP, Gahan LR, Larrabee JA, Ollis DL, Schenk G. 2013. Promiscuity comes at a price: catalytic versatility vs efficiency in different metal ion derivatives of the potential bioremediator GpdQ. Biochim Biophys Acta 1834:425–432. doi: 10.1016/j.bbapap.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 72.Sánchez-Moreno I, Iturrate L, Martín-Hoyos R, Jimeno ML, Mena M, Bastida A, García-Junceda E. 2009. From kinase to cyclase: an unusual example of catalytic promiscuity modulated by metal switching. Chembiochem 10:225–229. doi: 10.1002/cbic.200800573. [DOI] [PubMed] [Google Scholar]
- 73.Ben-David M, Wieczorek G, Elias M, Silman I, Sussman JL, Tawfik DS. 2013. Catalytic metal ion rearrangements underline promiscuity and evolvability of a metalloenzyme. J Mol Biol 425:1028–1038. doi: 10.1016/j.jmb.2013.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.