Abstract
Obesity is becoming the new pediatric epidemic. Non-alcoholic fatty liver disease (NAFLD) is frequently associated with obesity and has become the most common cause of pediatric liver disease. The gut microbiome is the major metabolic organ and determines how calories are processed, serving as a caloric gate and contributing towards the pathogenesis of NAFLD. The goal of this study is to examine gut microbial profiles in children with NAFLD using phylogenetic, metabolomic, metagenomic and proteomic approaches. Fecal samples were obtained from obese children with or without NAFLD and healthy lean children. Stool specimens were subjected to 16S rRNA gene microarray, shotgun sequencing, mass spectroscopy for proteomics and NMR spectroscopy for metabolite analysis. Children with NAFLD had more abundant Gammaproteobacteria and Prevotella and significantly higher levels of ethanol, with differential effects on short chain fatty acids. This group also had increased genomic and protein abundance for energy production with a reduction in carbohydrate and amino acid metabolism and urea cycle and urea transport systems. The metaproteome and metagenome showed similar findings. The gut microbiome in pediatric NAFLD is distinct from lean healthy children with more alcohol production and pathways allocated to energy metabolism over carbohydrate and amino acid metabolism, which would contribute to development of disease.
Keywords: NASH, obesity, gut microbiome, carbohydrate, energy metabolism
This study describes the gut microbial profile of obese children with and without fatty liver and identifies several key metabolites that have potential to contribute to the pathophysiology of this common disease.
This study describes the gut microbial profile of obese children with and without fatty liver and identifies several key metabolites that have potential to contribute to the pathophysiology of this common disease.
INTRODUCTION
The incidence of obesity among children in the United States has increased 3-fold in the last few decades (Kuczmarski et al. 2002; Ogden et al., 2006). The obesity epidemic is paralleled by an increase of non-alcoholic fatty liver disease (NAFLD) in the pediatric population, affecting at least 3% of children and adolescents in the United States and Asia (Strauss et al., 2000; Park et al., 2005). NAFLD is the most common cause of liver disease in children and adolescents in the United States (Schwimmer et al., 2006). The term NAFLD includes a spectrum of histological features from simple steatosis to inflammation with cellular injury to frank cirrhosis. Because the definitive diagnosis requires liver biopsy, which is not feasible in pediatric studies, increased serum aminotransferase levels and increased echogenicity on radiographic studies have been used as diagnostic tools (Patton et al., 2006). Non-alcoholic steatohepatitis (NASH) can potentially progress to cirrhosis, liver failure and hepatocellular carcinoma (White et al., 2012; Stepanova et al., 2013).
The pathophysiology of NAFLD remains unclear, but the gut microbiome is considered an important contributor. The gut microbiome houses bacterial communities that direct host digestion and energy homeostasis (Backhed et al., 2005; Sonnenburg et al., 2005; Flint et al., 2008). The ‘obese microbiota’ is characterized by an increased capacity to harvest energy from the diet (Turnbaugh et al., 2006). Studies on obese mice showed a 50% reduction in Bacteroidetes and an increase in Firmicutes by a comparable amount, demonstrating that ‘obesogenic microbiota’ is distinct from ‘lean microbiota’ (Ley et al., 2005) and that the differences in microbiota composition in mice can determine response to a high-fat diet and whether those mice will develop hepatic steatosis (Le Roy et al., 2013), as well as how the disease will progress (Henao-Mejia et al., 2012). Comparable observations are noted in human studies. Microbiota of obese adults had fewer Bacteroidetes (Mouzaki et al., 2013) and more Firmicutes resulting in difference in Bacteroidetes-to-Firmicutes ratio between obese and non-obese subjects, and the pattern was reversed with weight loss (Ley et al., 2006). Some studies suggest that the ‘manipulation of gut microbial communities could be another approach in the treatment of obesity’ (Jumpertz et al., 2011). Recent studies support this notion and further demonstrate that microbiota can be transmissible and could prevent the development of obesity and obesity-associated metabolic phenotypes in experimental animals (Ridaura et al., 2013; Walker and Parkhill 2013).
The objective of this study was to compare the fecal gut microbiome community structure, composition and function in obese children with and without NAFLD to those of healthy children. Microarray analysis was used to characterize microbial communities based on the 16S rRNA gene. The functional capacity was determined using high-throughput metagenomic sequencing and functional differences were determined using MS proteomic and NMR metabolomic analyses of fecal samples.
METHODS
This study was approved by the Institutional Review Board at the Children's Hospital Los Angeles and Dayton Children's Hospital. Lean healthy children and obese children with and without a clinical diagnosis of NAFLD based on ultrasound findings and elevated transaminases were recruited into the study. Other liver diseases were excluded. We could not definitely exclude early liver disease in the obese group without clinical markers of NAFLD since confirmation of absence of NAFLD in the obese subjects would require a liver biopsy. For ethical issues, we could not obtain liver biopsies in this subset of patients for the purpose of this study.
Body mass index (BMI) for healthy children were less than the 85% for age, and the BMI for obese children with and without clinical NAFLD were greater than the 95% for age. The average aspartate and alanine transaminases were 96 and 157 IU/L, respectively, in the NAFLD group. BMI was calculated as weight in kilograms divided by height in meters squared. AST and ALT were performed through local commercial labs which utilize the modified IFCC method (Lustig et al., 1988). There were no dietary restrictions for any subjects during this study and there was no significant difference in caloric intake, carbohydrate, protein or fat intake between the three groups. None of the children received antibiotics for at least six months prior to the study.
Gut microbial 16S rRNA gene microarray
All fecal specimens were homogenized and immediately frozen at −80°C. Triplicate samples of each stool were processed. Total genomic DNA (gDNA) was isolated using ZR Fecal DNA Isolation Kit (Zymo Research Corporation) according to manufacturer's protocol. gDNA was amplified, pooled together, fragmented and then hybridized to Microbiota Array as we previously described (Paliy et al., 2009; Agans et al., 2011; Michail and Kenche 2011; Michail et al., 2012).
High-throughput DNA sequencing
Samples were prepared for sequencing using the standard protocol for the Ion One Touch™ System and loaded onto Ion 318™ chips and run on the Ion Torrent Personal Genome Machine™ (Life Technologies, Carlsbad, CA) under standard conditions. The genomic center at Children's Hospital of Los Angeles is CLIA certified, and all CLIA quality control measures were applied. Shotgun sequencing of whole gDNA were performed on DNA samples without prior 16S rRNA gene amplification.
Proteomic processing
Fecal specimens were thawed at 4°C and microbial cells extracted by differential centrifugation (Apajalahti et al., 2002; Verberkmoes et al., 2009). Microbial cell pellets (∼100 mg wet weight) were washed, and then lysed via probe sonication in wash buffer with 0.25% w/v Rapigest. Lysed samples were centrifuged at 15 K rcf for 5 min, and the protein concentration was determined via bicinchoninic acid colorimetric assay (Chutipongtanate et al., 2012). Thirty mcg were fractionated via SDS-PAGE and in-gel digested overnight with trypsin. Tryptic peptides were purified via Varian C18 OMIX tips and analysed using an Eksigent nanoLC-2D coupled to a Thermo Orbitrap XL tandem mass spectrometer. Human and microbial proteins were identified from raw MS data using Proteome Discoverer and Scaffold software packages in conjunction with the UniProt human proteome and the METADB database, respectively. The METADB database has been previously described by Verberkmoes et al. (2009).
NMR metabolite analysis
Stools were frozen at −80°C until further analysis. Fecal water was extracted using a weighed sample of thawed stool material (200 mg) and adding five volumes of sterile phosphate buffer (1.9 mM Na2HPO4, 8.1 mM NaH2PO4, pH 7.4). The mixture was homogenized by vortex mixing for 5 min. The fecal slurry was centrifuged at 4500 rpm for 10 min and the supernatant was filtered through a GDX filter (Whatman). The filtrate was centrifuged at 16 000 g for 15 min and the supernatant retained for NMR analysis.
NMR studies were conducted using a Varian Inova 600 NMR. Briefly, 550 μl of fecal extract was mixed with 150 μl of a solution of 9 mM trimethylsilylpropionic −2,2,3,3−d4 acid (TSP) in D2O, which was used as a chemical shift and quantification reference. A 1H NMR spectrum was acquired at 600 MHz and 25°C using a NOESY pulse sequence (first increment) with water suppression during the relaxation delay and mixing time (the total interpulse delay ∼ 11 s). Spectral intensities were normalized to the total sum of all metabolite signals and integrated peak areas were measured for specific resonances of interest (known metabolite signals).
Statistical analysis
Quantification of NMR metabolites
Quantification of specific metabolite resonances was accomplished using an interactive spectral deconvolution algorithm in MATLAB adapted from our previously described methods (Anderson et al., 2009, 2011). The deconvolution tool fits a defined spectral region using a combination of tunable baseline shapes (spline, v-shaped, linear or constant) and a Gauss–Lorentz peak-fitting function. Integrated areas for peaks of interest are then output to a text file. All metabolite peak intensities were corrected for equivalent number of protons and normalized relative to the TSP signal intensity; but signals were not corrected for partial T1 saturation. Since all extracts were prepared from an identical stool weight, relative comparisons between samples are fully valid, but absolute quantitative amounts of each metabolite are not reported. Analyses of proton NMR data included multivariate data analyses of the full spectra and of 12 measured fecal metabolites. To obtain average levels of metabolites among all samples within each sample type, weighted mean around median was calculated as we described previously (Agans et al., 2011). P values were adjusted for multiple comparisons using Bonferroni corrections.
Metabolic profiling, sequencing and statistical analysis
We utilized the HUMAnN analysis pipeline together with RTG's mapx translated nucleotide search algorithm to infer community-wide metabolic pathway and gene abundances in the study. Filtered sequence data were searched against the KEGG database with mapx, using the following parameters: w = 8, a = 3 (sensitivity settings equivalent to BLASTX—blastall with default settings) and identity threshold at 70%. Output from mapx was processed by the HUMAnN pipeline (Abubucker et al., 2012).
Proteomic analysis
Proteins were quantitated in Scaffold according to number of assigned spectra, using 95% minimum protein and peptide identification probabilities, and a minimum peptide count of two. Protein quantification was averaged across three samples obtained from three separate stool specimens for control and NAFLD samples, and the resulting values were used for analysis, including functional category mapping via the databases of clusters of orthologous groups (COG).
Microarray statistical analysis
Signals from 852 separate bacteria were obtained from subjects in all groups. The bacteria were classified into 120 different genera, 53 families, 25 orders, 14 classes and 8 phyla. The signal from each bacterium for each patient was then divided by its genus average copy number to obtain an adjusted signal for each bacterium. The 852 adjusted signals were summed and the percent of the total adjusted signal was determined for all bacteria. Adjusted signals were then summed by genus, family, order, class and phylum, and the percentages of the total signal were determined for each hierarchical classification level.
For statistical analyses, all adjusted signals were transformed using the natural logarithmic transformation. Comparisons between groups were made with repeated measures analysis of variance. P values were adjusted for multiple comparisons using Bonferroni corrections.
RESULTS
Demographic characteristics
All three groups of children had similar characteristics except for a higher BMI (Table 1) in the two NAFLD and the obese children without evidence of NAFLD. All children with NAFLD had evidence of fatty liver change on ultrasonography and elevated AST and ALT levels. Three children underwent liver biopsy in our cohort; two had evidence of NASH and the third had steatosis without NASH. Some of the obese children with or without NAFLD had coexisting functional abdominal pain. Family history of obesity was observed in 78, 69 and 31% of children with NAFLD, obese without NAFLD and healthy children, respectively, while 34, 15 and 4% had a family history of fatty liver.
Table 1.
Characteristics of healthy children and obese children with NAFLD.
| Healthy | NAFLD | Obese no evidence | |
|---|---|---|---|
| Healthy | NAFLD | of NAFLD | |
| Variable | n = 26 | n = 13 | n = 11 |
| Gender | |||
| Male | 14 (53.8) | 6 (46.0) | 8 (72.7) |
| Female | 12 (46.2) | 7 (54.0) | 3 (27.3) |
| Race | |||
| White | 26 (100) | 12 (92.4) | 11 (100) |
| Other | 0 (0) 1 | (7.6) | 0 (0) |
| Age (y) | 13.3 ± 2.7 | 13.6 ± 3.0 | 13.2 + 3.8 |
| Height (cm) | 155.5 ± 16.9 | 164.4 ± 18.0 | 159.2 + 19.1 |
| BMI (kg/m2) | 19.4 ± 3.7a | 40.8 ± 4.0b | 31.2 + 4.5c |
Values listed are total for the variable (percent of total value n) or Means ± SD.
Across rows different superscript letters indicate means that are significantly different (P < 0.01) using analysis of variance (ANOVA) with Bonferroni correction.
Lack of superscript letters indicate means that are not significantly different, P > 0.05
16S rRNA gene analysis of bacterial populations
Differences exist at all levels when comparing the microbiota profiles of the NAFLD and healthy groups. Table 2 demonstrates changes noted at phylum and class levels. At the phylum level, children with NAFLD harbor more Gamma- and Epsilonproteobacteria than the healthy group. At the genus level, children with NAFLD harbor a more abundant member of the phylum Bacteroidetes—Prevotella, which is detected at lower levels in healthy children as well as obese children without evidence of NAFLD (Table 2). There was no difference in Firmicutes and Bacteroidetes or their ratio in any of the groups. However, obese children without NAFLD had lower Clostridia and higher Alphaproteobacteria.
Table 2.
Microbiota profile in healthy and obese children with and without fatty liver. Percent (mean ± SD) by phylum and class/genus.
| Phylum | Class (subclass)/[genus] | No. of species | Healthy (n = 26) | NAFLD (n = 13) | Obese without evidence of NAFLD (n = 11) |
|---|---|---|---|---|---|
| Actinobacteria | Actinobacteria (Actinobacteridae) | 20 | 1.02 ± 0.12 | 1.10 ± 0.34 | 1.14+0.22 |
| Actinobacteria (Coriobacteridae) | 9 | 1.05 ± 0.11a | 1.13 ± 0.22b | 1.04 + 0.28a | |
| Bacteroidetes | Bacteroidia | 142 | 14.97 ± 2.43 | 16.59 ± 5.9 | 16.28 + 2.77 |
| [Prevotella] | 25 | 0.19 + 0.042a | 7 + 0.13b | 0.21 + 0.03a | |
| Firmicutes | Bacilli | 20 | 1.55 ± 0.33 | 1.61 ± 0.65 | 1.61 + 0.49 |
| Clostridia | 564 | 76.66 ± 2.51a | 74.81 ± 5.83ab | 74.65 + 2.72b | |
| Erysipelotrichi | 39 | 2.24 ± 0.39a | 0.30 ± 0.03b | 2.49 + 0.27a | |
| Fusobacteria | Fusobacteria | 3 | 0.13 ± 0.01a | 0.15 ± 0.09b | 0.14 + 0.07a |
| Lentisphaerae | Lentisphaeria | 1 | 0.06 ± 0.01 | 0.06 ± 0.03 | 0.08 + 0.01 |
| Proteobacteria | Alphaproteobacteria | 9 | 0.17 ± 0.03a | 0.10 ± 0.34b | 0.20 + 0.03a |
| Betaproteobacteria | 18 | 0.61 ± 0.15 | 0.52 ± 0.26 | 0.70 + 0.28 | |
| Deltaproteobacteria | 5 | 0.30 ± 0.04 | 0.28 ± 0.06 | 0.30 + 0.05 | |
| Epsilonproteobacteria | 6 | 0.27 ± 0.04a | 0.30 ± 0.03b | 0.27 + 0.03a | |
| Gammaproteobacteria | 11 | 0.66 ± 0.14a | 0.73 ± 0.10b | 0.77 + 0.09c | |
| Spirochaetes | Spirochaetia | 4 | 0.13 ± 0.01 | 0.13 ± 0.04 | 0.13 + 0.02 |
| Verrucomicrobia | Verrucomicrobiae | 1 | 0.07 ± 0.05 | 0.04 ± 0.03 | 0.05 + 0.04 |
Across rows, different superscript letters indicate means that are significantly different (P < 0.05) using analysis of variance (ANOVA) with Bonferroni correction.
Nuclear magnetic resonance measurements of small metabolites
Data were analysed by spectral integration of a few specific signals assigned to various chemicals of interest, shown in Table 3. Some short chain fatty acids (SCFA), specifically formate, acetate, and valerate were less abundant in the NAFLD group relative to healthy controls. However, other SCFA (butyrate and propionate) were unaffected. The NAFLD group also had significantly higher levels of ethanol than healthy children. Obese children without evidence of NAFLD had higher lysine metabolite compared to healthy children. There was no statistical difference observed in lactate among the three groups.
Table 3.
Relative metabolite levels measured by 1H NMR of fecal water extracts from obese children with NAFLD, obese children without evidence of NAFLD and normal healthy children. Values are Mean ± SE of integrated signal intensities (arbitrary units) normalized to TSP standard.
| METABOLITE | Healthy | Obese with NAFLD | Obese without evidence of NAFLD |
|---|---|---|---|
| (n = 22) | (n = 11) | (n = 6) | |
| Ethanol | 15.3 ± 4.1a | 32.1 ± 9.8b | 12.4 ± 4.2ab |
| Uracil | 7.2 ± 0.8 | 6.1 ± 1.6 | 8.9 ± 1.8 |
| Acetate | 550.0 ± 47.1a | 417.9 ± 44.7b | 497.5 ± 95.8ab |
| Formate | 7.0 ± 0.5a | 4.2 ± 1.1b | 7.0 ± 1.0ab |
| Glucose | 17.8 ± 5.1 | 23.4 ± 3.0 | 27.4 ± 7.6 |
| Fumarate | 1.4 ± 0.2 | 1.4 ± 0.5 | 1.6 ± 0.5 |
| Lactate | 17.9 ± 2.7 | 11.7 ± 2.3 | 15.1 ± 1.6 |
| Alanine | 34.9 ± 5.2ab | 27.0 ± 3.9a | 42.9 ± 8.1ab |
| Butyrate | 221.0 ± 30.5 | 226.0 ± 36.8 | 260.1 ± 75.9 |
| Proprionate | 250.2 ± 30.9 | 227.3 ± 44.0 | 249.3 ± 45.4 |
| Valerate | 19.9 ± 3.0a | 9.9 ± 2.3b | 12.2 ± 2.2ab |
| Lysine | 44.3 ± 8.1a | 62.8 ± 16.1ab | 81.7 ± 29.6b |
Across rows, different superscript letters indicate means that are significantly different (P < 0.05) using analysis of variance (ANOVA) with Bonferroni correction.
Shotgun gut microbial metabolite sequencing
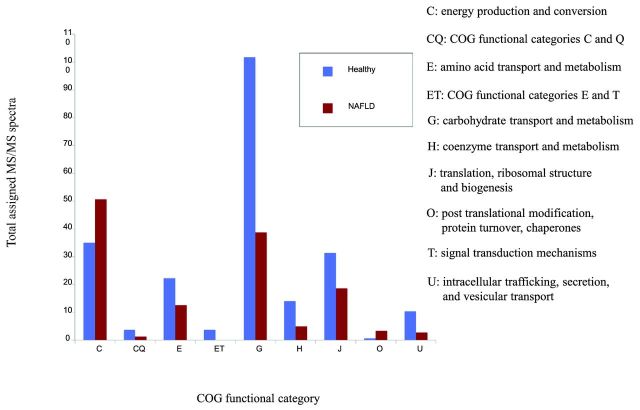
Subsequently, we identified metabolic pathways that are exclusively present in the healthy (n = 11) or NAFLD groups (n = 7, Tables 4A and B and Tables S4 and 5 in the Supporting Information). The average number of sequences obtained for each sample was 2 850 000 sequences with a GC content of 51 ± 4. We also noted pathways that were more abundant in either population. There are more pathways for energy metabolism and lipid synthesis and fewer pathways for amino acid metabolism, especially in relation to the urea cycle and elimination of ammonia in the NAFLD group. Methane oxidation was found to be many folds higher in children with NAFLD compared to healthy children. This finding suggests that energy metabolism in the gut microbiome of children with NAFLD is much more efficient than in healthy children. This is also supported by the proteomic findings of more pathways dedicated to energy production (Fig. 2).
Table 4A.
Shotgun genomics: pathways identified exclusively in children with NAFLD.
| Metabolic pathway | KEGG description |
|---|---|
| M00030: lysine biosynthesis, 2-oxoglutarate => 2-aminoadipate => lysine | Amino acid metabolism: amino acid synthesis |
| M00032: lysine degradation, lysine =>saccharopine =>acetoacetyl-CoA | Amino acid metabolism: amino acid degradation |
| M00142: complex I (NADH dehydrogenase), NADH dehydrogenase I | Energy metabolism; ATP synthesis |
| M00319: manganese/zinc/iron transport system | ABC transporters-iron and B12 TRANSPORT |
Table 4B.
Shotgun genomics: pathways more abundant in NAFLD.
| Metabolic pathway | Fold change | KEGG description |
|---|---|---|
| M00174: methane oxidation, methylotroph, methane => CO2 | 4.87E+05 | Energy metabolism |
| M00287: PTS system, galactosamine-specific II component | 111 | Galactose metabolism |
| M00082: fatty acid biosynthesis, initiation | 6.67 | Lipid synthesis |
| M00012: glyoxylate cycle | 5.56 | Carbohydrate and Lipid metabolism |
| M00096: C5 isoprenoid biosynthesis, non-mevalonate pathway | 3.23 | Carbohydrate and Lipid metabolism |
Figure 2.
Total assigned MS/MS spectra for COG-catalogued proteins that were found to be differentially expressed by a factor of at least three. C: energy production and conversion; CQ: COG functional categories C and Q (secondary metabolites biosynthesis, transport and catabolism); E: amino acid transport and metabolism; ET: COG functional categories E and T (signal transduction mechanisms); G: carbohydrate transport and metabolism; H: coenzyme transport and metabolism; J: translation, ribosomal structure and biogenesis; O: post translational modifications, protein turnover, chaperones; U: intracellular trafficking, secretion and vesicular support.
Metaproteomics
Noting differences in metabolism at the genomic level through sequencing, we attempted to identify such associations at the protein level using proteomics. The bacterial proteins identified were classified into COG functional categories. The most common COG functional categories identified in all groups were related to carbohydrate and energy metabolism (Fig. 1). The abundance of proteins expressed in different groups was compared, while the total number of proteins for each analysis was the same. There are significantly higher numbers of COG pathways for energy production in the NAFLD than in the healthy group (Fig. 2). This is also consistent with the metagenomic findings described above. The healthy group has more pathways allocated for amino acid and carbohydrate metabolism, but these two categories are still highly represented within the NAFLD group. Because more pathways are devoted to energy production, the NAFLD group appears to have fewer metabolic pathways for carbohydrate and amino acid metabolism, which may account for the effects seen on SCFA. COG categories more highly represented in healthy children include intracellular trafficking, translation and ribosomal biogenesis, inorganic ion transport and metabolism (Fig. 2). Proteins expressed 3-fold higher in NAFLD children that are relevant to energy metabolism include NAD-dependent aldehyde dehydrogenases and glyceraldehyde-3-phosphate dehydrogenase/erythrose-4-phosphate dehydrogenase (Table S2 in the Supporting Information).
Figure 1.
Distribution of COG functional categories in healthy children and obese children with NAFLD.
Through the human database, we identified human proteins more highly expressed in the NAFLD group (Table S3 in the Supporting Information). Alpha amylase 1 and pancreatic triacylglycerol lipase are more highly expressed in the NAFLD children and are relevant to carbohydrate and lipid metabolism.
DISCUSSION
The human gut microbiome is composed of a complex structure of trillions of organisms that contribute to the processing of different components of our diet such as carbohydrates, fats and proteins thus serving as the gate that permits energy and caloric retention and subsequent development of obesity. Both genetics and environment play a role in obesity and its sequela, such as NAFLD; however, genetic susceptibility cannot fully explain the epidemic in the United States. Because adherence to lifestyle changes is difficult, it is important to explore alternative targets for therapy. As we uncover more about the gut microbiome and its role in host metabolism and disease, its relevance in the obesity epidemic increases.
Microbiome and host metabolism
The gut microbiota influences host metabolism through a variety of mechanisms. Gut microbiota ferment indigestible carbohydrates, producing SCFA. SCFA are precursors in lipid production and gluconeogenesis, activate hormones that inhibit gut motility, increase nutrient absorption and stimulate appetite (Machado and Cortez-Pinto 2012). Frost and colleagues demonstrate that acetate activates acetyl-CoA carboxylase and changes in the expression profiles of regulatory neuropeptides that cause appetite suppression (Frost et al., 2014). The microbiome influences expression of Angiopoietin-like protein 4 and AMP-activated protein kinase, two important regulatory factors in fatty acid production and adipose tissue deposition (Backhed et al., 2004, 2007). In addition, there is evidence that the gut microbiota can direct increases in hepatic triglyceride production (Backhed et al., 2004). When the gut microbiota is exposed to a high-fat diet, a higher plasma lipopolysaccharide level is achieved. This state has been termed ‘metabolic endotoxemia’, which promotes liver steatosis, inflammation and fibrogenesis (Cani and Delzenne 2009; Laugerette et al., 2011; Zhu et al., 2013). With such a wide range of implications, altering the gut microbiota can be useful in preventing metabolic consequences (Delzenne and Cani 2008). This concept has already proven its efficacy in experimental animals (Ridaura et al., 2013).
The emerging field of metagenomics and metaproteomics permits studying the functions of microbial communities by sampling directly from their habitat (Maron et al., 2007). Analysing fecal samples from lean children and obese children with NAFLD, we identified differences in bacterial populations, gene and protein expression, and metabolite levels. We observed an increase in bacterial subgroups responsible for SCFA and ethanol production and a shift towards energy metabolism in obese children with NAFLD. Although some SCFA were observed to be less abundant, ethanol was significantly higher in the stool from obese children with NAFLD.
Gut microbial phylogenetic differences
There are significant differences among the gut bacterial populations in healthy and obese children with and without NAFLD. Namely, increases in the genus Prevotella and phylum Gammaproteobacteria have clinical importance and were found to be increased in children with NAFLD but not healthy or obese children without evidence of NAFLD. Prevotella is known to play a role in carbohydrate fermentation, producing exogenous SCFA (Salminen et al., 1998; Pouteau et al., 2003; Zhang et al., 2009). The present pediatric study, however, demonstrates the complexity of the relationship between gut microbes and metabolites, as differential effects were observed in SCFA. While we observed a decrease in some SCFA, none of the SCFA measured were found to be elevated in the NAFLD groups. Zhang et al. have highlighted the potential importance of Prevotellaceae in the obesity-associated microbiota, suggesting that H2-producing members of this family coexist with H2-oxidizing methanogenic Archaea in the gastrointestinal tracts of obese individuals (Zhang et al., 2009). This syntrophic relationship may increase host energy extraction from indigestible carbohydrates, as an increase in H2-oxidizing methanogenesis facilitates fermentation. Therefore, Prevotellaceae populations may be an important factor in the association between obesity, fatty liver, ethanol and SCFA production and the gut microbiota. This is consistent with adult studies that suggest that Prevotella populations are under-represented in non-obese subjects (Hoyles and McCartney 2009). However, in our pediatric population, obese children without NAFLD did not have a statistically significant increase in Prevotella suggesting that its role is more specific in NAFLD rather than obesity in general. We do not detect significant differences in the distribution of Firmicutes and Bacteroidetes between healthy and NAFLD groups as prior adult studies have demonstrated (Ley et al., 2006; Jumpertz et al., 2011). This may be related to the patient population being younger and early in the stages of obesity compared to adults.
Alcohol production
Furthermore, both Gammaproteobacteria (Ren et al., 2007) and Prevotella have been associated with endogenous alcohol production, which could account for the high-ethanol signal detected in the NAFLD group. Our findings represent one of the early pediatric studies describing this phenomenon and is consistent with animal studies that demonstrate production of ethanol in microbiota of obese mice with fatty liver, which was attenuated with Neomycin (Cope et al., 2000). Endogenous alcohol production through bacterial fermentation of dietary carbohydrates may contribute to the pathogenesis of fatty liver by delivering a constant source of oxidative stress. This is demonstrated in studies that describe high-blood ethanol levels in obese mice models and in patients with NAFLD (Cope et al., 2000; Volynets et al., 2012; Zhu et al., 2013). Zhu and Baker showed that children and adolescents with NASH had more alcohol-producing bacteria, higher serum ethanol levels and increased gene expression of pathways involved in alcohol catabolism (Zhu et al., 2013). Furthermore, obesity and fat deposition in the liver potentiate the damaging effects of alcohol on the liver (Xu et al., 2011). The abundance of Gammaproteobacteria classes in the NAFLD groups provides clues for future mechanistic studies that could elucidate the initial pathological changes that contribute to the development of NAFLD.
Energy metabolism
Our data show that the NAFLD group has a greater proportion of pathways dedicated to energy production and conversion and a smaller proportion for amino acid and carbohydrate transport and metabolism in comparison to the lean group. The metaproteome and metagenome data show similar findings. Looking at specific protein expression, NAD-dependent aldehyde dehydrogenase is 3-fold higher in the NAFLD group. This protein can impact glycolysis, gluconeogenesis, fatty acid and amino acid metabolism. But more importantly, it may play a direct role in the production of ethanol from acetaldehyde and is likely responsible for the elevated ethanol levels observed in the NAFLD group. The predominance of these pathways supports the idea that obese microbiota are active in energy extraction. Experimental models showed that less energy was lost in the stool in obese mice than their lean littermates, suggesting more efficient energy extraction by the obese microbiome. Conventionalized mice actually had a higher metabolic rate than their germ-free counterparts. Interestingly, despite a higher metabolic rate, there was no accompanying increase in tissue high-energy phosphate stores in conventionalized mice, implying the presence of inefficient metabolism (Backhed et al., 2004; Turnbaugh et al., 2006). We have evidence that the NAFLD microbiome is highly equipped for energy metabolism but cannot explain how the energy is utilized or allocated. Obese children with NAFLD had similar dietary intake compared to healthy children yet developed obesity. This may be explained on the basis of harboring a microbiome that is more efficient in making and preserving calories as noted in the findings of this study showing that gut microbial energy conservation and production was higher allowing the gut microbiome to conserve calories for the host with subsequent weight gain.
Carbohydrate and lipid metabolism
Although the NAFLD group has a smaller proportion of functional groups dedicated to carbohydrate transport and metabolism, these pathways still represent one of the principal functional groups in NAFLD children. This finding has been suggested by previous animal and human studies. Comparative metagenomic analysis of genetically lean and obese mice showed that the cecal microbiome from obese mice was enriched in enzymes involved in carbohydrate digestion (Ley et al., 2006). In a human study comparing metabolic pathways of the microbiome of lean and obese twins, the obese human gut was enriched in pathways involved in microbial processing of carbohydrates (Turnbaugh et al., 2009).
From our human database analysis, there is a 3-fold increase in pancreatic triacylglycerol lipase and alpha amylase 1 in the NAFLD group. Pancreatic triacylglycerol lipase is central in the digestion and absorption of dietary fat, and alpha amylase 1 is responsible for carbohydrate digestion. Alpha amylase 1 inhibitors are being studied to reduce the risk of heart attacks, treat diabetes type II and glucose intolerance (Chiasson et al., 2003). Amylase inhibition is known to induce carbohydrate tolerance, satiety and weight loss (Laugerette et al., 2011). Pancreatic triacylglycerol lipase is the main enzyme responsible for the hydrolysis and absorption of triacylglycerol from our diet. Lipid hydrolases are also targets for pharmacotherapy; Orlistat is a gastrointestinal lipase inhibitor which results in a reduction in the absorption of exogenous fat (Finer et al., 2000). These differences have clinical consequences and may contribute to obesity and NAFLD. The significance of pancreatic triacylglycerol lipase is further noted in mice deficient in pancreatic triacylglycerol lipase and consuming a Western diet and those who were found to have lower fasting cholesterol levels. Because of their reduced dietary lipid absorption, these mice had a significantly reduced susceptibility to high-fat/high-cholesterol diet-induced obesity (Gilham et al., 2007).
CONCLUSION
Our study highlights the complexity of the gut microbial ecology as it contributes to the development of obesity and fatty liver in the pediatric population. It demonstrates that no one technology alone can demonstrate the complexity of its structure or the mechanisms by which gut bacteria influence metabolism and energy balance. Looking at the picture from multiple perspectives—phylogenetic, metabolomic, metagenomic and proteomic–helps elucidate the role the gut microbiome plays in the development of NAFLD. We have yet to determine the mechanisms resulting in NAFLD, and our study adds to existing data that the process is multifactorial. This is one of few studies that focus specifically on the pediatric population. Our population is unique because we are evaluating children with early stages of NAFLD, which may be different from long-standing liver disease seen in adults. We also studied obese children without clinical features of NAFLD, revealing similarities between the two obese groups. These observations support the idea of a continuum in the diagnosis of fatty liver disease and progression to clinical NAFLD. For future studies, it is important to study obese patients without NAFLD as a separate group to identify potential factors that lead to NAFLD in some obese patients but not others and to pinpoint the early changes in the microbiome that result in NAFLD. Our study identifies differences in bacterial subtypes in NAFLD patients that have previously been associated with increased ethanol production and SCFA fermentation, which may have implications for liver damage and increased energy extraction. Our metagenomic and proteomic data show more bacterial pathways devoted to energy production and conversion that also supports the idea of providing more energy to the host. Results of this study may potentially provide a tool to distinguish children with NAFLD who have a gut microbial profile that is more efficient in processing energy and harbors bacteria that are capable of producing ethanol and SCFA. These findings can also help shape future interventional efforts to modify the microbiome in a way that would reverse the offending metabolite profile in NAFLD.
SUPPLEMENTARY DATA
Acknowledgments
The authors wish to thank Adrienne Stolfi for her assistance with some of the statistical analyses, Sean McCann, Harsha Kenche and Daniel Homer for technical assistance and Dr Tim Triche for his assistance with genomics. Part of the initial work was performed while first author was affiliated with Wright State University.
FUNDING
This work was supported by the National Institutes of Health grants NCCAM AT003423 and NICHD HD065575 and funding through the Clinical and Translational Science Institute (CTSI), Children's Hospital Los Angeles and MemorialCare Foundation.
Conflict of interest statement. None declared.
REFERENCES
- Abubucker S, Segata N, Goll J, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agans R, Rigsbee L, Kenche H, et al. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77:404–12. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PE, Raymer ML, Kelly BJ, et al. Characterization of 1H NMR spectroscopic data and the generation of synthetic validation sets. Bioinformatics. 2009;25:2992–3000. doi: 10.1093/bioinformatics/btp540. [DOI] [PubMed] [Google Scholar]
- Anderson PEM, Mahle DA, Doom TE, et al. Dynamic adaptive binning: an improved quantification technique for NMR spectroscopic data. Metabolomics. 2011;7:179–90. [Google Scholar]
- Apajalahti JH, Kettunen H, Kettunen A, et al. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl Environ Microb. 2002;68:4986–95. doi: 10.1128/AEM.68.10.4986-4995.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. P Natl Acad Sci USA. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Backhed F, Manchester JK, Semenkovich CF, et al. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. P Natl Acad Sci USA. 2007;104:979–84. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Design. 2009;15:1546–58. doi: 10.2174/138161209788168164. [DOI] [PubMed] [Google Scholar]
- Chiasson JL, Josse RG, Gomis R, et al. Acarbose treatment and the risk of cardiovascular disease and hypertension in patients with impaired glucose tolerance: the STOP-NIDDM trial. JAMA. 2003;290:486–94. doi: 10.1001/jama.290.4.486. [DOI] [PubMed] [Google Scholar]
- Chutipongtanate S, Watcharatanyatip K, Homvises T, et al. Systematic comparisons of various spectrophotometric and colorimetric methods to measure concentrations of protein, peptide and amino acid: detectable limits, linear dynamic ranges, interferences, practicality and unit costs. Talanta. 2012;98:123–9. doi: 10.1016/j.talanta.2012.06.058. [DOI] [PubMed] [Google Scholar]
- Cope K, Risby T, Diehl AM. Increased gastrointestinal ethanol production in obese mice: implications for fatty liver disease pathogenesis. Gastroenterology. 2000;119:1340–7. doi: 10.1053/gast.2000.19267. [DOI] [PubMed] [Google Scholar]
- Delzenne NM, Cani PD. [Gut microflora is a key player in host energy homeostasis] Med Sci (Paris) 2008;24:505–10. doi: 10.1051/medsci/2008245505. [DOI] [PubMed] [Google Scholar]
- Finer N, James WP, Kopelman PG, et al. One-year treatment of obesity: a randomized, double-blind, placebo-controlled, multicentre study of orlistat, a gastrointestinal lipase inhibitor. Int J Obes Relat Metab Disorder. 2000;24:306–13. doi: 10.1038/sj.ijo.0801128. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, et al. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–31. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Frost G, Sleeth ML, Sahuri-Arisoylu M, et al. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun. 2014;5:3611. doi: 10.1038/ncomms4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilham D, Labonte ED, Rojas JC, et al. Carboxyl ester lipase deficiency exacerbates dietary lipid absorption abnormalities and resistance to diet-induced obesity in pancreatic triglyceride lipase knockout mice. J Biol Chem. 2007;282:24642–9. doi: 10.1074/jbc.M702530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyles L, McCartney AL. What do we mean when we refer to Bacteroidetes populations in the human gastrointestinal microbiota? FEMS Microbiol Lett. 2009;299:175–83. doi: 10.1111/j.1574-6968.2009.01741.x. [DOI] [PubMed] [Google Scholar]
- Jumpertz R, Le DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczmarski RJ, Ogden CL, Guo SS, et al. CDC Growth Charts for the United States: methods and development. Vital Health Stat. 2002;11:1–190. [PubMed] [Google Scholar]
- Laugerette F, Vors C, Peretti N, et al. Complex links between dietary lipids, endogenous endotoxins and metabolic inflammation. Biochimie. 2011;93:39–45. doi: 10.1016/j.biochi.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Le Roy T, Llopis M, Lepage P, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–94. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. P Natl Acad Sci USA. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Lustig V, Papanastasiou-Diamandis A, Goldberg DM. Evaluation of commercially formulated aspartate aminotransferase and alanine aminotransferase activity determinations by the Scandinavian Committee on Enzymes and IFCC methods as modified for use with automated enzyme analysers. Clin Biochem. 1988;21:283–90. doi: 10.1016/s0009-9120(88)80082-1. [DOI] [PubMed] [Google Scholar]
- Machado MV, Cortez-Pinto H. Gut microbiota and nonalcoholic fatty liver disease. Ann Hepatol. 2012;11:440–9. [PubMed] [Google Scholar]
- Maron PA, Ranjard L, Mougel C, et al. Metaproteomics: a new approach for studying functional microbial ecology. Microb Ecol. 2007;53:486–93. doi: 10.1007/s00248-006-9196-8. [DOI] [PubMed] [Google Scholar]
- Michail S, Durbin M, Turner D, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. 2012;18:1799–8. doi: 10.1002/ibd.22860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michail S, Kenche H. Gut microbiota is not modified by randomized, double-blind, placebo-controlled trial of VSL#3 in diarrhea-predominant irritable bowel syndrome. Probiotics Antimicrob Proteins. 2011;3:1–7. doi: 10.1007/s12602-010-9059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouzaki M, Comelli EM, Arendt BM, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–7. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Curtin LR, et al. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006;295:1549–55. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- Paliy O, Kenche H, Abernathy F, et al. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl Environ Microb. 2009;75:3572–9. doi: 10.1128/AEM.02764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Han JH, Choi KM, et al. Relation between elevated serum alanine aminotransferase and metabolic syndrome in Korean adolescents. Am J Clin Nutr. 2005;82:1046–51. doi: 10.1093/ajcn/82.5.1046. [DOI] [PubMed] [Google Scholar]
- Patton HM, Sirlin C, Behling C, et al. Pediatric nonalcoholic fatty liver disease: a critical appraisal of current data and implications for future research. J Pediatr Gastr Nutr. 2006;43:413–27. doi: 10.1097/01.mpg.0000239995.58388.56. [DOI] [PubMed] [Google Scholar]
- Pouteau E, Nguyen P, Ballevre O, et al. Production rates and metabolism of short-chain fatty acids in the colon and whole body using stable isotopes. Proc Nutr Soc. 2003;62:87–93. doi: 10.1079/PNS2003208. [DOI] [PubMed] [Google Scholar]
- Ren N, Xing D, Rittmann BE, et al. Microbial community structure of ethanol type fermentation in bio-hydrogen production. Environ Microbiol. 2007;9:1112–25. doi: 10.1111/j.1462-2920.2006.01234.x. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen S, Bouley C, Boutron-Ruault MC, et al. Functional food science and gastrointestinal physiology and function. Brit J Nutr. 1998;80(Supplement 1):S147–71. doi: 10.1079/bjn19980108. [DOI] [PubMed] [Google Scholar]
- Schwimmer JB, Deutsch R, Kahen T, et al. Prevalence of fatty liver in children and adolescents. Pediatrics. 2006;118:1388–93. doi: 10.1542/peds.2006-1212. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–9. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Stepanova M, Rafiq N, Makhlouf H, et al. Predictors of all-cause mortality and liver-related mortality in patients with non-alcoholic fatty liver disease (NAFLD) Digest Dis Sci. 2013;58:3017–23. doi: 10.1007/s10620-013-2743-5. [DOI] [PubMed] [Google Scholar]
- Strauss RS, Barlow SE, Dietz WH. Prevalence of abnormal serum aminotransferase values in overweight and obese adolescents. J Pediatr. 2000;136:727–33. [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Verberkmoes NC, Russell AL, Shah M, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–89. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- Volynets V, Kuper MA, Strahl S, et al. Nutrition, intestinal permeability, and blood ethanol levels are altered in patients with nonalcoholic fatty liver disease (NAFLD) Digest Dis Sci. 2012;57:1932–41. doi: 10.1007/s10620-012-2112-9. [DOI] [PubMed] [Google Scholar]
- Walker AW, Parkhill J. Microbiology. Fighting obesity with bacteria. Science. 2013;341:1069–70. doi: 10.1126/science.1243787. [DOI] [PubMed] [Google Scholar]
- White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin Gastroenterol H. 2012;10:1342–59 e2. doi: 10.1016/j.cgh.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lai KK, Verlinsky A, et al. Synergistic steatohepatitis by moderate obesity and alcohol in mice despite increased adiponectin and p-AMPK. J Hepatol. 2011;55:673–82. doi: 10.1016/j.jhep.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, DiBaise JK, Zuccolo A, et al. Human gut microbiota in obesity and after gastric bypass. P Natl Acad Sci USA. 2009;106:2365–70. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–9. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]