Abstract
Purpose
Ulcerative colitis (UC) and colitis-associated colorectal cancer (CAC) is a serious health issue, but etiopathological factors remain unclear. Aldo-keto reductase 1B10 (AKR1B10) is specifically expressed in the colonic epithelium, but down-regulated in colorectal cancer. This study was aimed to investigate the etiopathogenic role of AKR1B10 in UC and CAC.
Experimental design
UC and CAC biopsies (paraffin-embedded sections) and frozen tissues were collected to examine AKR1B10 expression. Aldo-keto reductase 1B8 (the ortholog of human AKR1B10) knockout (AKR1B8 −/−) mice were produced to estimate its role in the susceptibility and severity of chronic colitis and associated dysplastic lesions, induced by dextran sulfate sodium (DSS) at a low dose (2%). Genome-wide Exome sequencing was used to profile DNA damage in DSS-induced colitis and tumors.
Results
AKR1B10 expression was markedly diminished in over 90% of UC and CAC tissues. AKR1B8 deficiency led to reduced lipid synthesis from butyrate and diminished proliferation of colonic epithelial cells. The DSS-treated AKR1B8 −/− mice demonstrated impaired injury repair of colonic epithelium and more severe bleeding, inflammation, and ulceration. These AKR1B8 −/− mice had more severe oxidative stress and DNA damage, and dysplasias were more frequent and at a higher grade in the AKR1B8 −/− mice than in wild type mice. Palpable masses were seen in the AKR1B8 −/− mice only, not in wild type.
Conclusion
AKR1B8 is a critical protein in the proliferation and injury repair of the colonic epithelium and in the pathogenesis of UC and CAC, being a new etiopathogenic factor of these diseases.
Keywords: AKR1B8, AKR1B10, colonic crypt cell, colitis, dysplasia
Introduction
Ulcerative colitis (UC) is a chronic inflammatory disease of the colon characterized by mucosal inflammation (1). The current model for the pathogenesis of UC is that in the colonic mucosa there is an inappropriately robust immune response to commensal bacteria. This is driven at least in part by genetic defects or susceptibility in the host although no single gene has been found to be either necessary or sufficient for the development of UC (2, 3).
Long standing UC is associated with an increased incidence of colorectal cancer (4, 5). The pathogenesis of this colitis-associated cancer (CAC) has some similarities and some differences when compared with the pathogenesis of sporadic colorectal cancer. Colorectal cancers, either sporadic or CAC, develop from a dysplastic precursor lesion. In sporadic colorectal cancer, the dysplastic precursor is usually an adenomatous polyp, a discrete focus of neoplasia; in contrast, CAC develops from dysplastic lesions that can be polyploid, flat, localized or multifocal (6, 7). Like sporadic colorectal cancer, CAC occurs through mutations of somatic cells followed by clonal expansion (5). Many of the cellular defects associated with the development of sporadic colorectal cancer are also seen with CAC. Frequencies of chromosomal instability and microsatellite instability are similar in both (8). However, progression of sporadic colorectal cancer differs from CAC in the timing and frequency of these alterations. For example, loss of adenomatous polyposis coli (APC) function is an early event in the progression of sporadic colorectal cancer, but is less frequent and occurs later in the development of CAC. In contrast, the loss of p53 function is an important early step in progression of CAC, but occurs later in the development of sporadic colorectal cancer (9–11).
CAC progresses from no dysplasia to low grade dysplasia to high grade dysplasia to carcinoma. This progression is primarily driven by somatic mutations that develop in response to chronic inflammation (5, 11). Although it is clear that somatic mutations seen in CAC are related to chronic inflammation, it is not clear how this chronic colonic inflammation leads to the somatic mutations. Inflammatory oxidative stress is a critical factor in the pathogenesis of CAC (10, 12, 13). Overloaded reactive oxygen species (ROS) target a wide range of macromolecules including proteins, DNA and lipids. Reactive oxygen species cause protein dysfunction by interacting with susceptible amino acid residues (14) and induce DNA mutations and breaks by forming 8-oxo-deoxyguanosine (8-oxodG) adducts (13). Free radical-mediated oxidative stress also induces lipid peroxidation and biomembrane damage (15). Lipid peroxides may serve as secondary contributors to the cellular and DNA damage that drive malignant transformation in UC. Lipid peroxides are electrophilic carbonyl compounds and are highly cytotoxic and genotoxic (16–18). By interaction with proteins and DNA, peroxides form covalently modified protein or DNA adducts, leading to protein dysfunction and DNA mutations and breaks. Therefore, cellular and DNA damage induced by oxidative stress provides a mechanistic basis for events, including epithelial damage, genetic instability, and gene mutations which drive the severity of UC and its progression to cancer.
Aldo-keto reductase 1B10 (AKR1B10), also known as aldose reductase-like-1 (ARL-1), is a multifunctional protein (19). It functions as a monomeric NADPH-dependent enzyme that efficiently reduces cytotoxic and carcinogenic α, β-unsaturated carbonyl compounds to less toxic alcoholic forms, protecting host cells from carbonyl damage (19–21). AKR1B10 also functions as an important regulator of de novo fatty acid/lipid synthesis. Through physical association, ARK1B10 prevents the ubiquitination and proteasomal degradation of acetyl-CoA carboxylase-α (ACCA), a rate limiting enzyme in the de novo fatty acid synthesis, thus promoting fatty acid synthesis (22, 23). Aldo-keto reductase 1B8 (AKR1B8) is the ortholog of human AKR1B10 in mouse (24). AKR1B10 and AKR1B8 are both primarily expressed in the colon and small intestine and have similar functions in carbonyl detoxification and lipid biosynthesis (19, 24). In the intestine, AKR1B10 eliminates carbonyl compounds and thus plays a protective role to the colonic mucosa. AKR1B10 may also promote fatty acid/lipid synthesis in the colonic mucosa and thus facilitate the constant renewal of cryptic cells. The loss of AKR1B10 might be expected to increase the severity of UC and DNA instability, thus accelerating the development of dysplasia in UC patients. In this study, we addressed the question of whether the AKR1B10 plays a role in blocking the development of dysplasia in UC. To address this question, we produced an AKR1B8 deficient (AKR1B8 −/−) mouse strain and studied DSS-induced colitis and CAC. Our results showed an important role for AKR1B8 in the pathogenesis of UC and CAC.
Materials and Methods
Ethics statement
IRB protocols were approved by Springfield Committee for Research Involving Human subjects. Animal protocols were approved by Southern Illinois University School of Medicine Laboratory Animal Care and Use Committee (LACUC).
Human sample procurement
Two groups of human specimens were procured with approved IRB protocols. One is paraffin-embedded sections of biopsies (n=79) from Department of Pathology at Memorial Medical Center, Springfield. These sections were used for immunohistochemistry of AKR1B10 expression. The other is the frozen surgical specimens procured from the Tissue Bank in Simmons Cancer Institute at the Southern Illinois University. These frozen specimens were used for Western blot and RT-PCR analyses of AKR1B10 expression.
AKR1B8 −/− mouse generation and DSS treatments
Heterozygous AKR1B8 mutant C57BL/6 mice were generated in Deltagen Research Laboratory (San Diego, CA). In the animal facility at Southern Illinois University School of Medicine, the heterozygous pairs were used to generate homozygous AKR1B8 −/− and littermate wild type mice for experimental studies. Animals were housed at 24 ± 0.5°C, 50 ± 10% humidity and 12 h of light from 08:00 to 20:00 with free access to regular commercial diet and tap water.
Chronic colitis in mice was induced using 2% dextran sulfate sodium (DSS, 35,000–50,000 kDa; MP Biomedicals, CA) for 4 cycles (Fig. S3) (25, 26).. Each cycle consisted of 7 days of DSS in drink water, followed by 14 days of DSS-free water. Animals were monitored daily for body weight, occult and gross rectal bleeding, and diarrhea for disease activity (Table S4). At indicated time points, mice were euthanized and the entire large intestine (cecum, colon and rectum) was excised and measured. The colons were fixed in 10 % formaldehyde buffer for transverse sections or “Swiss rolls”, or were frozen in liquid nitrogen and stored at −80°C. Palpable tumors were counted, measured, and processed for paraffin blocks or snap frozen.
Histological evaluations
“Swiss rolls” were sectioned at 4µm and stained with hematoxylin and eosin (H&E). Crypt length was defined as cell number per crypt by counting 50 integrated crypts per mouse. Inflammation and neoplastic lesions were evaluated blindly by an experienced investigator and a pathologist. Inflammation was graded from 0 to 4 upon histopathology of colonic mucosa (Table S5). Neoplastic lesions were estimated and counted as described in Table S6.
Immunohistochemistry
Paraffin-embedded sections were deparaffinized using a standard technique. Antigens were retrieved in 10 mM citrate buffer (pH 6.0) and endogenous peroxidase was blocked in 3% H2O2. Immunohistochemistry was carried out using a staining kit (Life technologies, CA).
BrdU labeling assays
Mice received a single intraperitoneal injection of a mixture of 5’-bromo-2’-deoxyuridine (BrdU, 120 mg/kg) and 5’-fluoro-2’-deoxyuridine (12 mg/kg). At 1, 24 and 48 hours after injection, respectively, animals were euthanized, and the whole colons were collected and fixed in formalin for paraffin embedding and immunohistochemistry. Number and location of BrdU positive cells in integrated crypts were assessed.
Western blot
Colonic mucosa was scraped, washed by PBS, and homogenized in 50 µl Roche buffer on ice. Homogenates were cleared up by centrifugation at 4°C, 14,000g for 10 min. Proteins (50–100 µg) in supernatants were separated on 10–12% SDS-PAGE and blotted onto nitrocellulose membrane (Bio-Rad, CA) for immunoprobing as previously described (22).
Real-time RT-PCR
Total RNA were extracted from colonic mucosa using Trizol (Invitrogen, CA), and reversely transcribed. The cDNA was used as templates for real-time RT-PCR with SYBR green qPCR mixture following manufacturer’s protocol (BioSciences Inc., CA). Gene specific primer sequences are listed in Supplementary Table S7.
ELISA
Colonic mucosa were minced with surgical scissors and homogenized in ice-cold PBS, followed by centrifugation at 4°C, 10,000g for 15 min. ELISA kits for interleukin-1β (IL-1β), interleukin-6 (IL-6) and interferon-γ (IFNγ) (R&D system, MN) were used to measure these protein levels following manufacturer’s instructions.
Reactive oxygen species
Reactive oxygen species in colonic mucosa were estimated using an antioxidant assay kit (Cayman Chemical Company, MI) following the instruction of the manufacturer.
Lipid peroxidation
Lipid peroxides were measured using a Lipid Peroxidation Microplate Assay Kit (Oxford Biomedical Research, MI), following the instruction of manufacturer.
Lipid synthesis
Radiochemicals 14C-butyrate (54mCi/mmol) was purchased from Moravek Biochemicals, Inc. CA. Colonocytes were isolated as previously described (27), and incubated with 5–10 µCi of 14C-butyrate in 5–10 ml medium for 2 hours. Cells were collected for lipid extraction and radioactivity tests using a scintillation counter (Beckman, CA), as previously described (22).
Next-generation sequencing
Genomic DNA was extracted with a standard method from mucosa and tumors. Genome-wide DNA mutations were analyzed by Next-generation sequencing (Otogenetics Corporation, GA) with a minimum of 30× average coverage and paired-end 100 base-pair (bp) reads. Exome coverage reports were provided for quality control (Table S1). DNA variants were identified using the platform provided by DNAnexus (https://dnanexus.com). Nucleotide-Level-Variations in AKR1B8 −/− and WT mouse genome were identified by variations from a reference genome.
Statistical analyses
Statistical analyses were carried out with Prism 4 (GraphPad Software, CA). Variance test, student t test or One-way ANOVA test, as appropriate, were used to compare the difference between AKR1B8 −/− and WT groups with P < 0.05 as statistical significance.
Results
AKR1B10 Expression is Diminished in Human UC and CAC
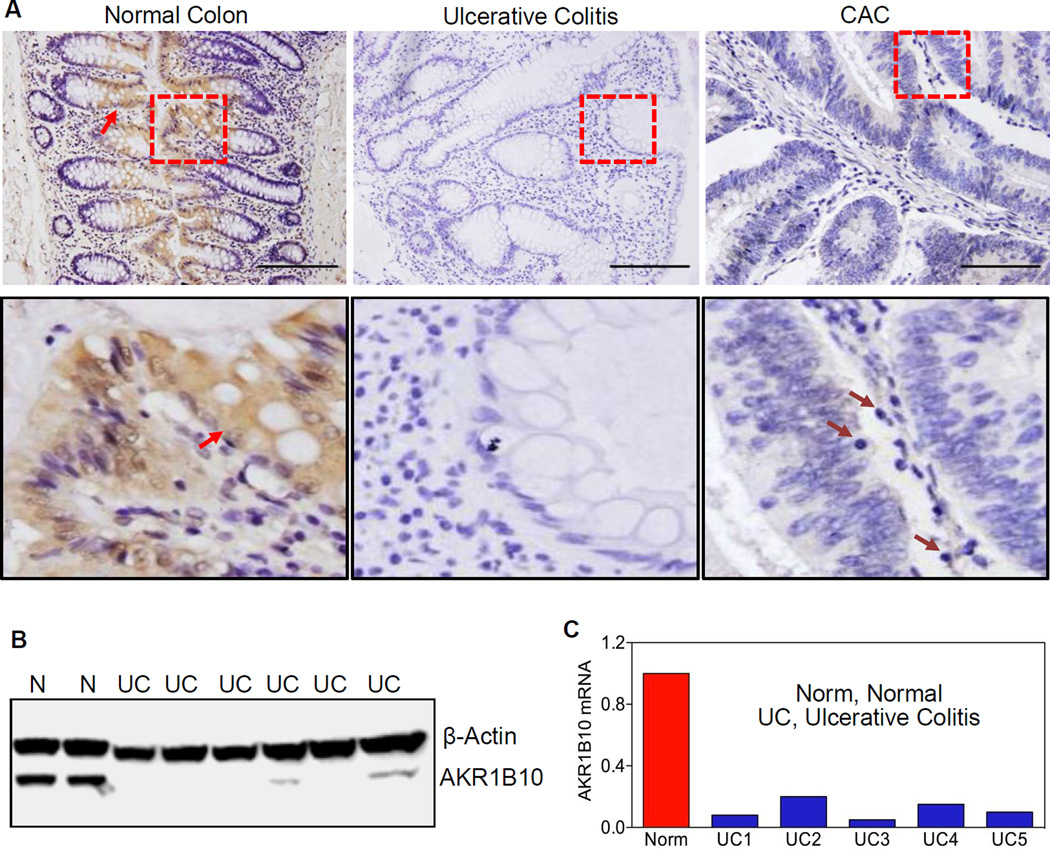
We examined the cellular distribution of AKR1B10 in the normal colon and found that it is specifically present in the mature epithelial cells (Fig. 1A, left). In contrast, AKR1B10 was significantly reduced in 45.6% (36/79) of UC tissues and undetectable in 50.6% (40/79) (Fig. 1A, middle). Furthermore, in 6 associated CAC in this collection, AKR1B10 was absent in 5 tumors (83.3%) (Fig. 1A, right) and was weakly positive in the other. We further examined AKR1B10 expression in frozen specimens and found that AKR1B10 protein (Fig. 1B) and mRNA (Fig. 1C) were decreased or undetectable in 77.3% (17/22) of UC tissues. Together these data suggest that AKR1B10 expression in UC and CAC is lost or significantly decreased.
Figure 1. AKR1B10 expression in normal, ulcerative colitis and cancerous colon tissues.
(A) Immunohistochemistry in paraffin-embedded sections of normal colon, ulcerative colitis and associated tumor tissues. AKR1B10 protein was detected in normal colonic epithelium (arrows), but not in ulcerative colitis and tumor tissues. Lower panel: amplification of the squared areas. Purple arrows indicate infiltrated mononuclear cells in CAC. Scale bar, 100µm. (B) Western blot. N, normal colon tissues; UC, ulcerative colitis. (C) Real-time RT-PCR. The AKR1B10 mRNA level in normal colon is set up at 1.0, and AKR1B10 mRNA levels in ulcerative colitis tissues are presented as fold over normal. GAPDH mRNA was detected as an internal control.
AKR1B8 −/− Mice Have Defects in Crypt Development and Self-Renewal of the Colon
AKR1B8, the ortholog of human AKR1B10 in the mouse, is primarily expressed in the colon (24). To clarify the role of AKR1B10 in UC and CAC, we produced an AKR1B8 knockout (−/−) mouse strain by replacing 25 nucleotides (GCA GCA ACC ATG GCC ACG TTC GTG G) around the ATG translational start-site with a LacZ/Neo cassette (a selection marker) (Fig. S1A). This replacement leads to a 5 amino acid deletion and downstream frame-shifting. The AKR1B8 mutant was confirmed by Southern blot (Fig. S1B) and AKR1B8 expression at mRNA (Fig. S1C) and protein levels (Fig. S1D–E). AKR1B8 disruption in mice had not evident effect on general appearance, body weight, and reproductivity. The mutant AKR1B8 allele displayed a typical Mendelian dissemination, indicating that AKR1B8 is not an essential gene for embryo development and survival. The expression of other aldo-keto reductase isoforms in the colon of AKR1B8 −/− mice, such as AKR1B3, AKR1B7 and AKR1C12–14, was not significantly altered except for AKR1B7 in the DSS-treated mucosa (Fig.S2A–B).
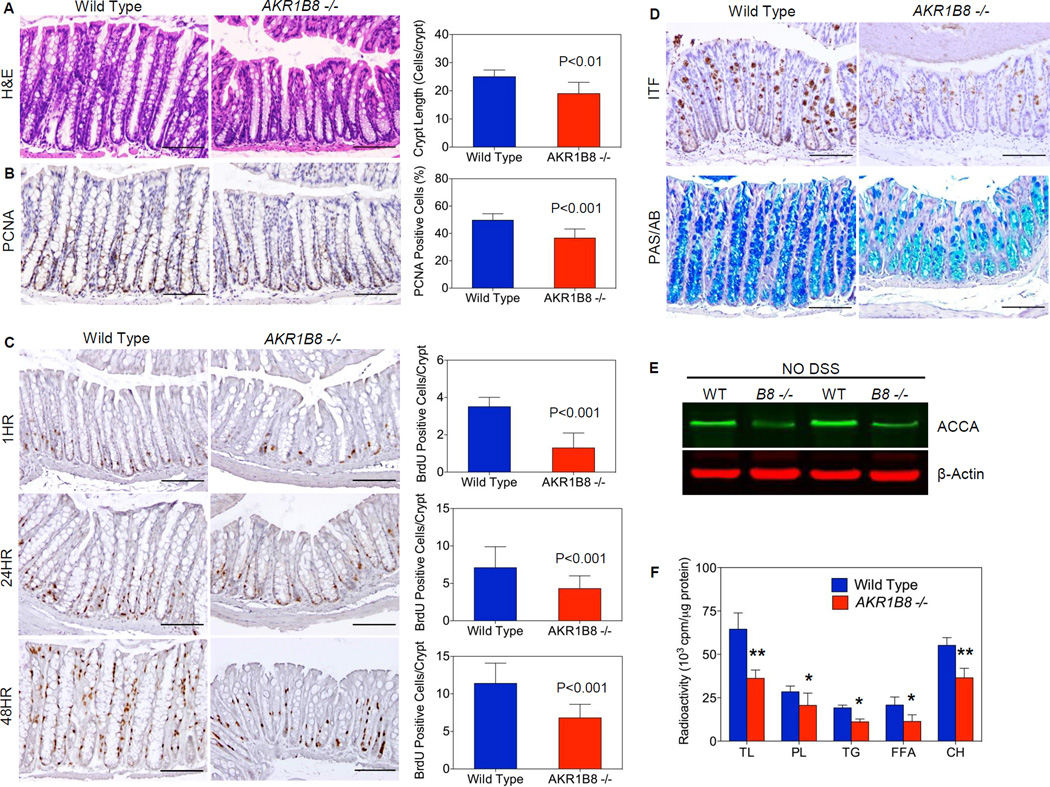
Histological assessments demonstrated that AKR1B8 abrogation significantly affected the colonic crypts. The colonic crypts were notably shorter in AKR1B8 −/− mice than in wild type mice (Fig. 2A, S3A). On average, the distal colon had 25±2.4 cells/crypt in wild type (WT) mice, but only 19±4.0 cells/crypt in age-matched AKR1B8 −/− animals (n=30, p<0.01). Proliferating cell nuclear antigen (PCNA)-positive cryptic cells were remarkably reduced to 36.6% in AKR1B8 −/− crypts vs. 49.7% in the WT (n=5, p < 0.001) (Fig. 2B, S3B). A time-course study of bromodeoxyuridine (BrdU)-labeling (i.e., 1, 24, and 48 hours) showed not only a decrease in the proliferating crypt cells, but also aberrations in the migration of crypt cells in the AKR1B8 −/− mice (Fig 2C, S3C). At 1 hour of BrdU labeling, the labeled cells were at approximately 3.5±0.5 cells/crypt in WT mice, but only 1.3±0.8 cells/crypt in AKR1B8 −/− (n=5, p < 0.001); at 24 hours post BrdU administration, BrdU-labeled cells migrated into the middle region of crypts were at approximately 7.1±2.8 cells/crypt in WT mice, but only 4.3±1.7 cells/crypt in AKR1B8 −/− (n=5, p < 0.001); and at 48 hours, the BrdU-labeled cells reached at the top of crypts in WT mice, but not in AKR1B8 −/− (n=5). We further evaluated the expression of mucins and intestinal trefoil factor (ITF) proteins, the differentiation markers expressed by mature goblet cells (28, 29), and our results demonstrated that both mucin and ITF levels were decreased (Fig. 2D, S3D–E) in the AKR1B8 −/− mouse colon when compared with WT. Together these data suggest that AKR1B8 deficiency impairs the proliferation, migration, and maturation of the crypt cells.
Figure 2. AKR1B8 deficiency interrupts growth and homeostasis of mouse colonic crypts.
(A) H&E histology (n=30 each); (B) PCNA expression (n=5 each); (C) BrdU labeling at 1, 24, and 48 hours (n=5 each); (D) Upper panel, ITF expression; and lower panel, Alcian blue and Periodic acid Schiff staining (n=5 each). Scale bar indicates 100 µm. (E) Western blot of acetyl-CoA carboxylase-α (ACCA) protein; (F) Incorporation of 14C-butyric acid into total lipids and various species: phospholipids (PL), triglycerides (TG), free fatty acids, and cholesterol (CH). n=5 each; *, P<0.05 and **, P<0.01 compared to wild type. Statistical significance was tested by One-way Anova test.
AKR1B8 (AKR1B10 in humans) promotes de novo fatty acid synthesis by stabilizing ACCA (23, 24). In colonocytes, butyrate is a major carbon contributor to lipid synthesis (30, 31). Therefore, we evaluated the ACCA protein level in the mucosa and found that ACCA was markedly decreased in the AKR1B8 −/− mucosa (Fig 2E) and significantly affected incorporation of butyrate into complex lipids. The incorporation of butyrate into the phospholipids, cholesterols, and triglycerides was all decreased in the AKR1B8 −/− crypt cells compared to WT (Fig. 2F), suggesting that AKR1B8 deficiency disrupts the lipid biosynthesis from butyrate in the colonic mucosa.
AKR1B8 Deficiency Leads to Susceptibility and Severity of Colitis Induced by DSS
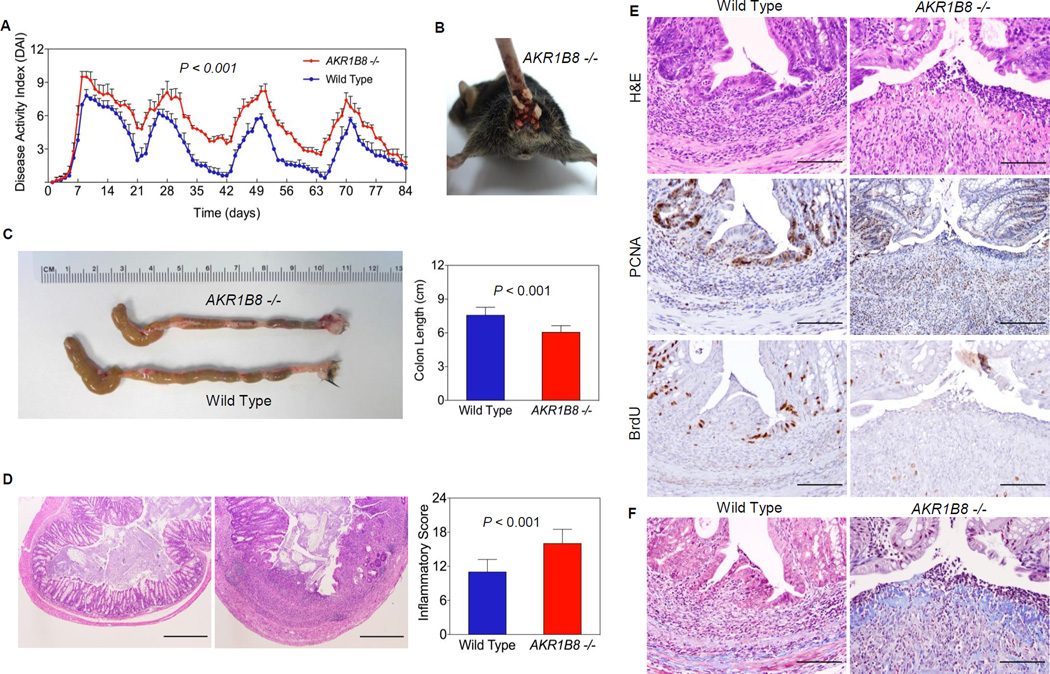
We further assessed susceptibility of AKR1B8 −/− mice to colitis induced by a low dose of 2% dextran sulfate sodium (DSS) (Fig. S4). Body weight, stool consistency, and gross and occult bleeding were monitored daily during DSS administration and recovery stages to evaluate disease activity index (32). During 4 cycles of DSS treatment, the overall DAI were significantly higher in AKR1 B8 −/− mice than in WT (Fig. 3A, n=15). Occult and gross bleeding (Fig. 3B) and liquid diarrhea occurred in all AKR1B8 −/− mice, but not in WT. On average, the colons of AKR1B8 −/− mice were over 1.5 cm shorter than those of WT animals (Fig. 3C), indicating more severe inflammation. In the AKR1B8 −/− mice the spleen was enlarged by two fold compared to that of WT (Fig. S5A). AKR1B8 −/− mice had large and continuous epithelial lesions, such as ulcerations, and intensive infiltration of inflammatory cells throughout mucosa, submucosa, and muscle layer of the colon (Fig. 3D, S5B); in contrast, WT mouse colons had minor epithelial damage. Furthermore, AKR1B8 −/− mice displayed impaired regeneration of the intestinal epithelium. DSS causes epithelial injury; after withdrawal of DSS the proliferation of colonic epithelial cells is increased (33). In WT mice, a layer of epithelial cells covered the ulcerated area within two days after DSS was withdrawn; however, AKR1B8 −/− mice failed to repair the ulcers. As a result, AKR1B8 −/− mice had contiguous focal epithelial cell loss and broader wound surface areas in the colonic epithelium (Fig. 3E, upper). The epithelial proliferation in ulcerated and adjacent areas was diminished in AKR1B8 −/− mice, as shown by PCNA expression and BrdU labeling (Fig. 3E, middle and lower). In contrast, increased collagen deposit was seen in the ulcerated areas of AKR1B8 −/− mice (Fig. 3F). These data suggest that AKR1B8 deficiency leads to susceptibility to and severity of colitis and impaired healing of the colonic mucosa.
Figure 3. DSS-induced colitis and mucosal lesion repair.
Wild type and AKR1B8 −/− mice (n=15 each) were administered with dextran sulfate sodium in drink water to induce colitis as described in Materials and Methods. (A) Disease activity index; (B) Rectal bleeding; (C) Colon length; and (D) Histological inflammatory lesions; (E) Upper panel, H&E histology; middle panel, PCNA expression; and lower panel, BrdU labeling (1 hour). (F) Masson Trichrome staining, showing collagen deposit at the ulcer. Scale bar, 100µm.
AKR1B8 Deficiency Aggravates Inflammatory Response, Oxidative Stress, and Carbonyl Lesions
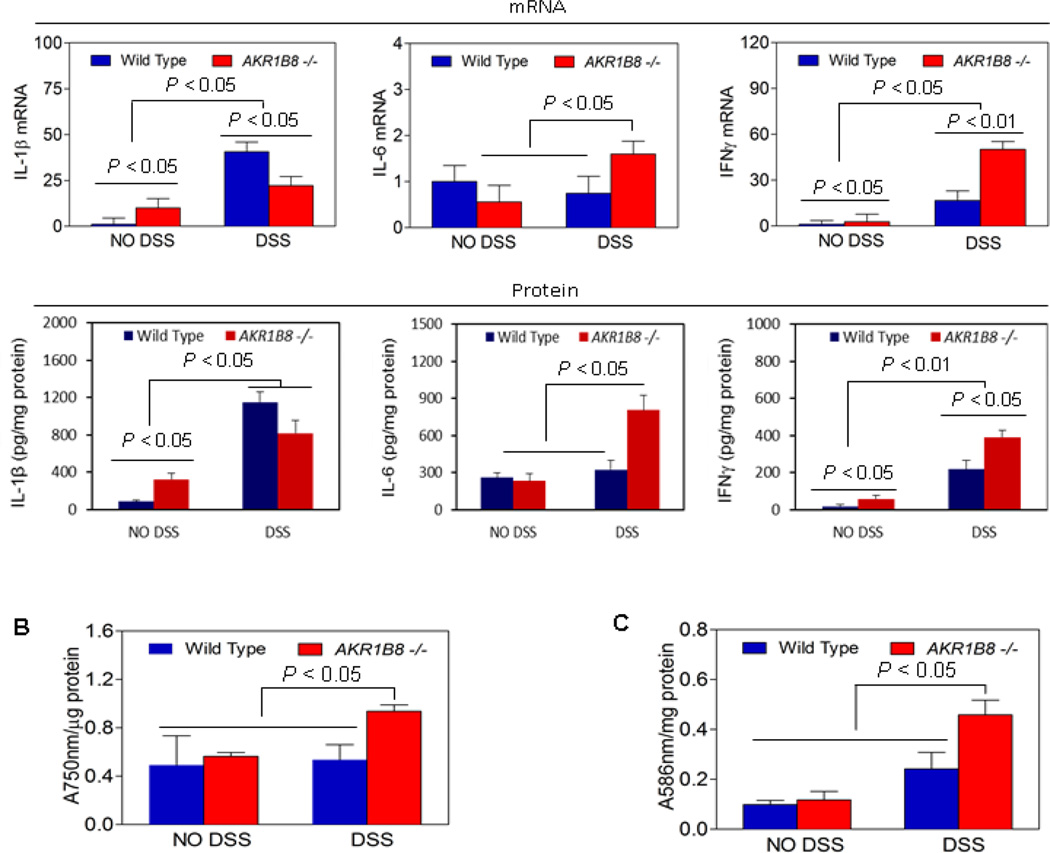
Cytokines regulates the inflammatory response (3, 34). We performed qRT-PCR for key proinflammatory cytokines (e.g., IL-1β, IL-6, and IFNγ) in the mucosa (Fig. 4A, upper panel). In the AKR1B8 −/− mice, IL-1β and IFNγ were upregulated at the baseline (p < 0.05, n=5). In response to DSS treatment, IL-6 and IFNγ were greatly induced in AKR1B8 −/− mice compared to WT, indicating an increased inflammatory response. In AKR1B8 −/− mucosa IL-1β was also induced by the DSS treatment, but to a lesser compared to that in WT, which may be due to its high basal level. We further measured the protein levels of these cytokines and found the protein levels were accordingly increased in the colonic mucosa (Figure 4A, lower panel).
Figure 4. Proinflammatory cytokines, oxidative stress, and carbomyl levels in AKR1B8 −/− mice.
(A) Expression of cytokines IL-1β, IL-6, and IFNγ. Upper panel: mRNA levels; lower panel: protein levels. Data indicate mean ± SD, n= 5. (B) Oxidative stress (n=5 each). (C) Lipid peroxides (n=5 each). Statistical significance was tested by Student’s t test.
Inflammatory oxidative stress is a driving force of cellular damage in UC (12). We measured reactive oxygen species in the colonic mucosa of AKR1B8 −/− and WT mice, and found that the radical levels were significantly higher in AKR1B8 −/− mucosa than in WT (Fig. 4B, n=5). In the AKR1B8 −/− mucosa, lipid peroxides were also higher than in WT (Fig. 4C, n=5), which may be due to increased lipid peroxidation and diminished elimination by AKR1B8. Together these data indicate that AKR1B8 deficiency exaggerates the inflammatory response and oxidative stress and thus exacerbates colitis induced by DSS.
AKR1B8 Deficiency Predisposes Mice to Colitis-Associated Tumorigenesis
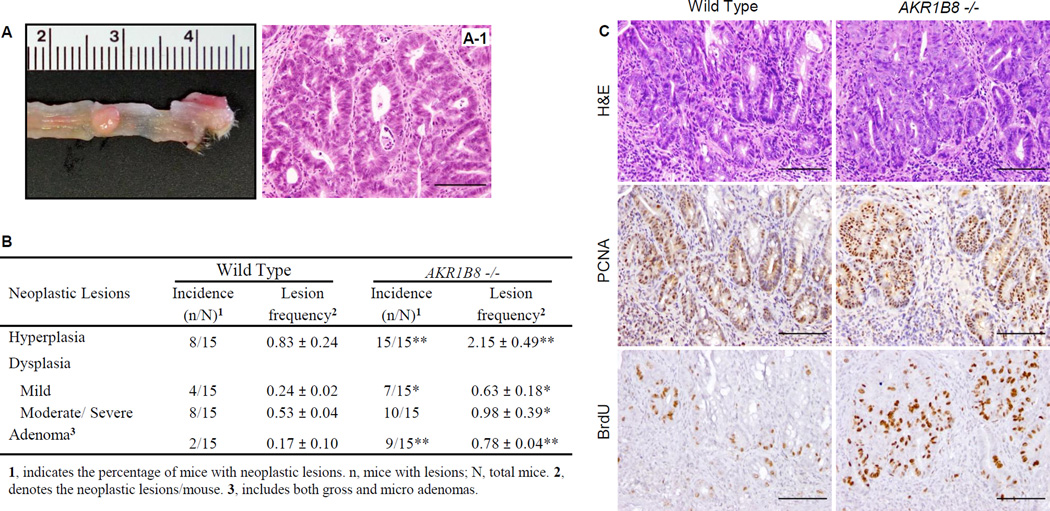
CAC correlates with extent and severity of inflammation (35, 36). In the AKR1B8 −/− mice, masses appeared (20.0%, n=5) after two cycles of DSS treatment, and after four cycles of DSS treatment, palpable tumors were larger and more frequent (30.0%, n=10) than in WT. Palpable tumors were not observed in any WT mice (n=15) after four cycles of DSS. All masses seen in AKR1B8 −/− mice were dome-shaped and present in the distal colon-rectum (Fig. 5A, S6A–B). We stratified the micro-neoplastic lesions in four classes, i.e., hyperplasia, mild dysplasia, moderate/severe dysplasia, and adenoma. As shown in Fig. 5B, the hyperplasia and dysplasia were 2–3 fold more frequent in AKR1B8 −/− mice than in WT (n=15, p < 0.05); in particular, the adenomas were 5 times more frequent at 0.78±0.04/mouse in AKR1B8 −/− vs. 0.17±0.10/mouse in WT (n=15, p < 0.01). Approximately 60% of AKR1B8 −/− mice had adenomas compared to 13.3% of WT (n=15, p < 0.01). The neoplasms in AKR1B8 −/− mice were of a higher grade and more aggressive, as illustrated by H&E histology (Fig. 5C upper, and S6C), PCNA expression (Fig. 5C, middle) and BrdU labeling (Fig. 5C, lower). These data indicate that the AKR1B8 deficiency leads to more severe colitis-associated tumorigenesis.
Figure 5. Colitis-associated dysplastic lesions in AKR1B8.
Wild type and AKR1B8 −/− mice (n=15 each) were administered with dextran sulfate sodium in drink water to induce colitis as described in Materials and Methods. (A) A palpable tumor (arrow) from an AKR1B8 −/− mouse. (A-1), H&E histology. (B) Summary of colitis-associated dysplastic lesions. *, P<0.05 and **, P<0.01 compared to wild type control. Statistical significance was tested by Student’s t test. (C) Grade and aggressiveness of dysplasia: Upper panel, H&E histology; middle panel, PCNA expression; and lower panel, BrdU labeling (1 hour). Scale bar, 100µm.
Large-Scale Exome Sequencing Identifies More Frequent DNA Damage in AKR1B8 −/− Mouse Colons
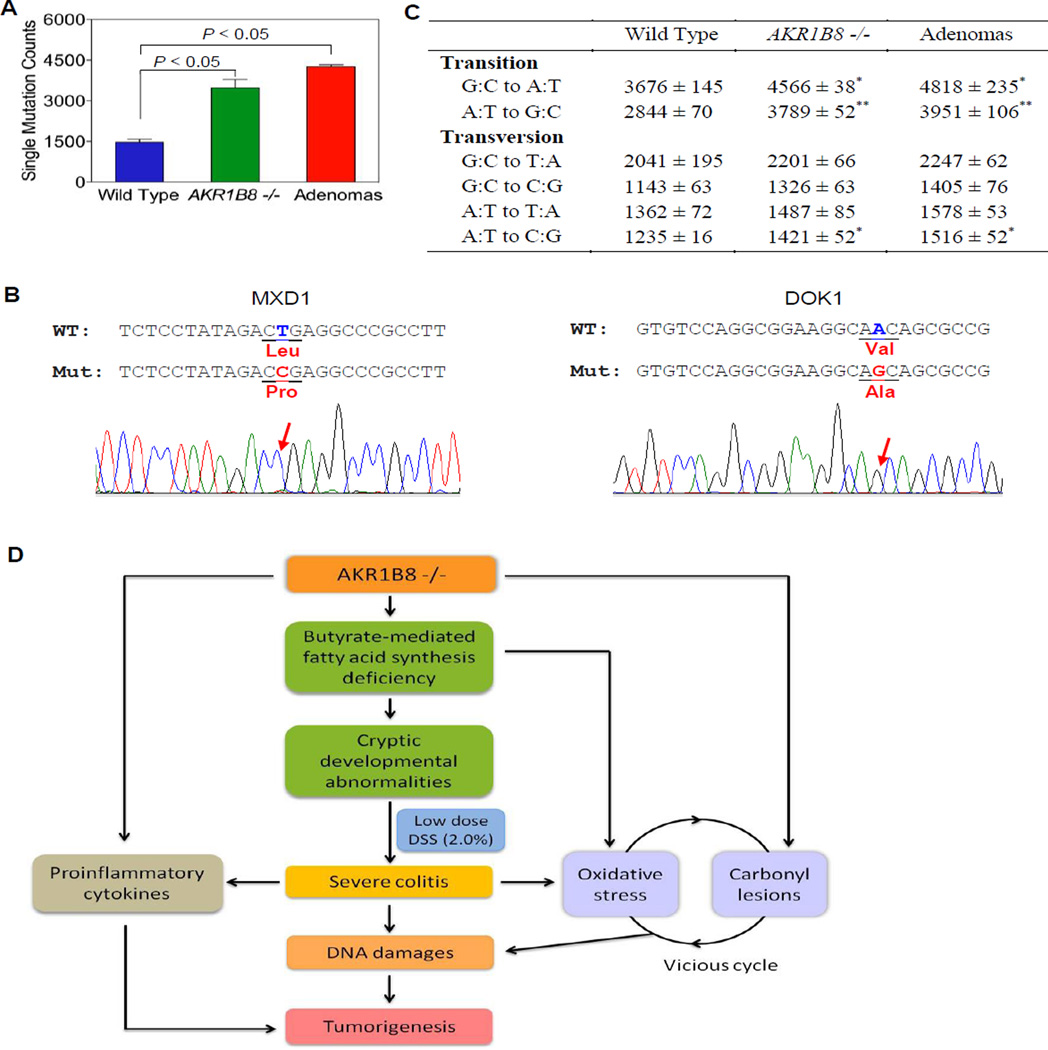
AKR1B8 −/− and WT mice received the same DSS treatment, but gross and micro tumors were seen earlier and more frequently in AKR1B8 −/− mice, suggesting differential DNA damage in these experimental animals. To understand the DNA damage profile that underscores the colitis-associated tumorigenesis, we conducted a genome-wide Exome sequencing analysis in the WT and AKR1B8 −/− colitis mucosa and tumors formed in the AKR1B8 −/− epithelium. The Exome coverage of this deep sequencing was satisfied (Table S1), and mutant analyses demonstrated that mutations were significantly higher in AKR1B8 −/− colitis and tumors than in WT (Fig. 6A, Table S2) (n=2, p<0.001). We further compared the gene mutants among WT colitis, AKR1B8 −/− colitis and tumors, and found that a total of 230 genes were mutated uniquely in AKR1B8 −/− colitis, tumors or both, but not in WT colitis. This includes non-synonymous point mutations, frameshifts, and stop gain and stop loss. Among the 230 genes, 104 genes were mutated in colitis only, 32 genes were mutated in tumors alone, and 94 genes were mutated in both AKR1B8 −/− colitis and tumors. In the 94 genes, 28 genes are oncogenes or tumor suppressors (Table S3); using conventional DNA sequencing, we confirmed the mutations of two tumor suppressor genes, DOK1 (37) and MXD1 (38), in AKR1B8 −/− colitis and tumors (Fig. 6B). These mutations of oncogenes and tumor suppressor genes may represent the DNA damage that drives the malignant transition of colitis. In addition, nucleotide transition of G:C to A:T (P < 0.05) or A:T to G:C (P < 0.01) and transversion of A:T to C:G (P < 0.05) were more prevalent in AKR1B8 −/− colitis and tumors compared to WT colits (Fig. 6C), consistent with the oxidative and carbonyl-induced DNA damage (10, 39).
Figure 6. DNA damage in AKR1B8 −/− mice.
Genome-wide Exome sequencing analyses of WT and AKR1B8 −/− colitis mucosa and tumors (n=2 each) were conducted as described in Materials and Methods. (A) Single homozygous nucleotide mutants. (B) Conventional DNA sequencing of DOX1 and MXD1 tumor suppressor genes, confirming the point mutations. (C) Transitional and transversional point mutations. *, P<0.05 and **, P<0.01 compared to wild type control. Statistical significance was tested by Student’s t test. (D) Hypothetic model of AKR1B8 in colitis and associated dysplastic lesions. AKR1B8 deficiency reduces long chain fatty acid/lipid synthesis from butyrate, which diminishes biomembrane assembly and cell proliferation. The AKR1B8 deficiency also leads to accumulation of toxic carbonyl compounds, which, together with oxidative stress, forms a vicious cycle, leading to DNA damage and subsequent dysplastic lesions. AKR1B8 may also affect proinflammatory cytokine expression via a mechanism unknown yet.
Discussion
AKR1B8 (AKR1B10 in humans) is primarily expressed in epithelial cells of the colon and intestine, but its biological function in the intestine is unknown. Here we found that targeted disruption of AKR1B8 gene resulted in diminished proliferation of the epithelial cells in the colon. In the DSS model of colitis, mice deficient in AKR1B8 had diminished epithelial repair and suffered more severe inflammation and disease activity and more severe dysplastic lesions. AKR1B8 is a critical protein in the self-renewal and barrier function of the colonic epithelium and in the pathogenesis of UC and CAC.
The two known functions of AKR1B8, i.e., control of fatty acid and lipid synthesis and detoxification of carbonyl compounds (24), may both relate to the decreased colonic epithelial proliferation, more severe inflammation, and the increased dysplasia in the DSS model of colitis seen in AKR1B8 deficient mice. Phospholipids are essential building blocks of biomembranes; constant renewal of colonic epithelial cells creates a need for phospholipids for cell growth and proliferation. The colonic epithelium is unique in that it is exposed to high concentrations of short chain fatty acids (SCFA) produced by commensal bacteria in the lumen. Butyric acid from commensal bacteria in the lumen is the major carbon source for lipid synthesis in the colonic epithelium (30). When radiolabeled butyric acid was given to mice deficient in AKR1B8, its incorporation into phospholipids and other complex lipids in the colonic mucosa was diminished when compared to wild type mice. It is possible that decreased incorporation of short chain fatty acids into phospholipids contributes to the decreased epithelial proliferation seen in AKR1B8 −/− mice.
The intestinal ecosystem consists of epithelium, immune cells and luminal microflora and contents. Intestinal epithelial cells are maintained in a dynamic balance between cell proliferation, differentiation, and apoptosis to serve as a robust barrier (40). In AKR1B8 −/− mice, the defects in epithelial proliferation and maturation may impair the barrier function, leading to susceptibility to DSS-induced colitis. Therefore, DSS at a low dose of 2.0% triggered much more severe inflammation in the AKR1B8 −/− mice, leading to serious diarrhea, bleeding, and rigid colonic tubes. Histologically, the AKR1B8 deficient mouse colons showed heavy inflammatory cell infiltration, larger area ulceration, and collagen fiber deposits. Inflammatory cytokines, such as IL-1β, IL-6, and INFγ, were also increased (Figure 4A).
Wound healing is the key of UC remission (40, 41). The epithelial cells surrounding the wound bed rapidly migrate to the denuded area and promptly proliferate to restore the pool of colonocytes (41). The DSS model is marked by epithelial injury during DSS exposure followed by epithelial repair and increased epithelial proliferation when DSS is withdrawn (42, 43). In the DSS treatment, AKR1B8 −/− mice experienced severe chronic epithelial damage, but lack of the proper restitution whereas the ulcerative area in WT mouse was covered by a layer of epithelial cells within two days, accompanied with active epithelial proliferation in adjacent areas. The impaired wound healing in the AKR1B8 deficient mice may explain the severity of inflammation triggered by DSS. The impaired proliferative response after DSS withdrawal in the AKR1B8 deficient mice also raises the possibility that their impaired ability to synthesize complex lipids may limit epithelial proliferation in the face of injury.CAC correlates with extent and severity of inflammation (35, 36). Oxidative stress plays a key role in inflammatory cellular and DNA damage in UC. Reactive oxygen species interact with DNA and form 8-oxo-deoxyguanosine (8-oxodG) adducts which leads to DNA mutations and breaks (13). In the oxidative stress associated with inflammation, lipid peroxidation mediates the destruction of biomembranes. The electrophilic lipid peroxides also induce cellular and DNA damage. The interaction of peroxide compounds with proteins results in covalent modifications and protein dysfunction (14, 16, 44). The interaction of peroxide compounds with DNA results in DNA mutations and breaks (17, 18). Lipid peroxidation and lipid peroxide-induced injury may aggravate oxidative stress by increasing mitochondrial permeability, forming a vicious cycle. AKR1B8 mediates the detoxification of carbonyl compounds to alcohols. We found evidence for increased oxidative stress and increased lipid peroxidation in the colonic mucosa of AKR1B8 −/− mice treated with DSS when compared to mucosa from wild type mice treated with DSS. Exome sequencing of colonic mucosal DNA from wild type and AKR1B8 −/− mice treated with DSS revealed a much higher rate of mutations in the AKR1B8 deficient mucosa. Moreover, the nucleotide transition of G:C to A:T or A:T to G:C and transversion of A:T to C:G were more prevalent in the AKR1B8 deficient mucosa. These changes are consistent with oxidative and carbonyl-induced DNA damage (10, 39). Therefore, the increased inflammatory cytokines and cellular and DNA damage induced by oxidative stress and lipid peroxides provide a mechanism for many of the events that drive the severity of UC and the progression of UC to cancer in this mouse model. It is noteworthy to note that same as literature report (45), p53 and Rb mutations were not evidenced in the colitis and associated tumors in the AKR1B8 −/− mice. However, we indeed observed APC mutations in one of two tested tumors (data not shown), but not in colitis mucosa, which is consistent with literature that APC mutation is less frequent and occurs later in the development of colitis-associated colorectal cancer (46).
AKR1B18 is an effective detoxicant of lipid peroxides (24). In the absence of AKR1B8, the colonic mucosa may be unable to efficiently eliminate oxidative lipid peroxides. In the mouse AKR1B protein family, AKR1B7 also has a substrate preference for 4-hydroxynonenal (HNE), a cytotoxic and genotoxic lipid peroxide (47). In the DSS-treated AKR1B8 −/− colonic mucosa, AKR1B7 was induced, which may represent a feedback mechanism responding to the carbonyl stress from oxidative lipid peroxidation (e.g., HNE) in the absence of AKR1B8.
The expression of AKR1B10 is known to be increased in many human cancers including breast cancer and lung cancer (48, 49), but is diminished in colorectal cancer (50). We found that AKR1B10 expression is also diminished in ulcerative colitis. We do not know if the diminished expression of AKR1B10 occurs before or after the development of colon cancer and UC and therefore, we cannot say if it may have a causal effect in these conditions. Our studies with the DSS model of colitis in AKR1B8 deficient mice would suggest that in the human colon, the diminished expression of AKR1B10 would result in increased susceptibility to chronic inflammation and dysplasia.
The data present here demonstrate that AKR1B8 plays an essential role in regulating lipid synthesis in the colonic epithelium and in protecting the colonic epithelium from oxidative and carbonyl damage. Deficiency in AKR1B8 leads to diminished lipid synthesis and increased oxidative stress in the DSS model, resulting in impaired epithelial cell proliferation and increased DNA mutations and dysplasia (Fig. 6D). These data suggest a central role for AKR1B10 in modulating the development of human UC and CAC.
Supplementary Material
Translational relevance.
Ulcerative colitis (UC) occurs at 10–12 cases per 100,000 people affecting approximately 500,000 Americans; a serious scenario of UC is the increased risk of developing colorectal cancer in patients. However, etiopathological factors of UC and its tumorigenic transformation is not fully understood. This report demonstrated that aldo-keto reductase 1B8 (AKR1B8), the ortholog of human AKR1B10 in the mouse, is a critical protein that modulates the proliferation and injury repair of the colonic epithelium. ARK1B8 deficiency led to more severe inflammation and associated dysplastic lesions in the colonic mucosa. This study characterized a new risky factor of colitis and associated tumorigenesis and produced a new animal model for approaches of novel preventive and therapeutic strategies, having high translational potential.
Acknowledgements
Authors thank Dr. Martinez (Génétique, Reproduction & Développement, Clermont Université, 24, Avenue Des Landais 63171 Aubière Cedex) for a kind gift of AKR1B7 antibody. This work was supported in part by National Cancer Institute (CA122622) for DC and NIH (DK33165) for WS.
Footnotes
Author Contributions:
Y.S. and D. C. were involved in all aspect of this study, conceived the hypothesis, and performed experiments and data analysis; J.M. and R.Y. performed the experiments in human UC and CAC samples; H. L. did the real time qPCR; X. L. performed mouse Exome sequencing data analysis; W. F. S. reviewed data and revised the manuscript; W.Y., J.G., M. H., and D.F.L. provided critical pathology / gastroenterology support. The rest of author panel assisted in animal welfare, experimental preparation, and data analysis, or advices in the project design.
Conflicts of Interest: Authors declare no conflicts of financial interests.
References
- 1.Danese S, Fiocchi C. Ulcerative colitis. N Engl J Med. 2011;365:1713–1725. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- 2.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 3.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 4.Danese S, Malesci A, Vetrano S. Colitis-associated cancer: the dark side of inflammatory bowel disease. Gut. 2011;60:1609–1610. doi: 10.1136/gutjnl-2011-300953. [DOI] [PubMed] [Google Scholar]
- 5.Meira LB, Bugni JM, Green SL, Lee CW, Pang B, Borenshtein D, et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. The Journal of clinical investigation. 2008;118:2516–2525. doi: 10.1172/JCI35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton SR. Origin of colorectal cancers in hyperplastic polyps and serrated adenomas: another truism bites the dust. J Natl Cancer Inst. 2001;93:1282–1283. doi: 10.1093/jnci/93.17.1282. [DOI] [PubMed] [Google Scholar]
- 7.Kukitsu T, Takayama T, Miyanishi K, Nobuoka A, Katsuki S, Sato Y, et al. Aberrant crypt foci as precursors of the dysplasia-carcinoma sequence in patients with ulcerative colitis. Clin Cancer Res. 2008;14:48–54. doi: 10.1158/1078-0432.CCR-07-1835. [DOI] [PubMed] [Google Scholar]
- 8.Hawkins NJ, Ward RL. Sporadic colorectal cancers with microsatellite instability and their possible origin in hyperplastic polyps and serrated adenomas. J Natl Cancer Inst. 2001;93:1307–1313. doi: 10.1093/jnci/93.17.1307. [DOI] [PubMed] [Google Scholar]
- 9.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nature reviews Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 10.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nature reviews. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 11.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenzie SJ, Baker MS, Buffinton GD, Doe WF. Evidence of oxidant-induced injury to epithelial cells during inflammatory bowel disease. The Journal of clinical investigation. 1996;98:136–141. doi: 10.1172/JCI118757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roszkowski K, Jozwicki W, Blaszczyk P, Mucha-Malecka A, Siomek A. Oxidative damage DNA: 8-oxoGua and 8-oxodG as molecular markers of cancer. Med Sci Monit. 2011;17:CR329–CR333. doi: 10.12659/MSM.881805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada K, Wangpoengtrakul C, Osawa T, Toyokuni S, Tanaka K, Uchida K. 4-Hydroxy-2-nonenal-mediated impairment of intracellular proteolysis during oxidative stress. Identification of proteasomes as target molecules. The Journal of biological chemistry. 1999;274:23787–23793. doi: 10.1074/jbc.274.34.23787. [DOI] [PubMed] [Google Scholar]
- 15.Niki E, Yoshida Y, Saito Y, Noguchi N. Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochemical and biophysical research communications. 2005;338:668–676. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto M, Sibata T, Wasada H, Toyokuni S, Uchida K. Structural basis of protein-bound endogenous aldehydes. Chemical and immunochemical characterizations of configurational isomers of a 4-hydroxy-2-nonenal-histidine adduct. The Journal of biological chemistry. 2003;278:5044–5051. doi: 10.1074/jbc.M210129200. [DOI] [PubMed] [Google Scholar]
- 17.Eckl PM. Genotoxicity of HNE. Mol Aspects Med. 2003;24:161–165. doi: 10.1016/s0098-2997(03)00010-4. [DOI] [PubMed] [Google Scholar]
- 18.Yang IY, Chan G, Miller H, Huang Y, Torres MC, Johnson F, et al. Mutagenesis by acrolein-derived propanodeoxyguanosine adducts in human cells. Biochemistry. 2002;41:13826–13832. doi: 10.1021/bi0264723. [DOI] [PubMed] [Google Scholar]
- 19.Cao D, Fan ST, Chung SS. Identification and characterization of a novel human aldose reductase-like gene. The Journal of biological chemistry. 1998;273:11429–11435. doi: 10.1074/jbc.273.19.11429. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Zhong L, Johnson S, Cao D. Human aldo-keto reductases 1B1 and 1B10: A comparative study on their enzyme activity toward electrophilic carbonyl compounds. Chem Biol Interact. 2011;191:192–198. doi: 10.1016/j.cbi.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong L, Liu Z, Yan R, Johnson S, Zhao Y, Fang X, et al. Aldo-keto reductase family 1 B10 protein detoxifies dietary and lipid-derived alpha, beta-unsaturated carbonyls at physiological levels. Biochem Biophys Res Commun. 2009;387:245–250. doi: 10.1016/j.bbrc.2009.06.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Yan R, Luo D, Watabe K, Liao DF, Cao D. Aldo-keto reductase family 1 member B10 promotes cell survival by regulating lipid synthesis and eliminating carbonyls. The Journal of biological chemistry. 2009;284:26742–26748. doi: 10.1074/jbc.M109.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma J, Yan R, Zu X, Cheng JM, Rao K, Liao DF, et al. Aldo-keto reductase family 1 B10 affects fatty acid synthesis by regulating the stability of acetyl-CoA carboxylase-alpha in breast cancer cells. The Journal of biological chemistry. 2008;283:3418–3423. doi: 10.1074/jbc.M707650200. [DOI] [PubMed] [Google Scholar]
- 24.Joshi A, Rajput S, Wang C, Ma J, Cao D. Murine aldo-keto reductase family 1 subfamily B: identification of AKR1B8 as an ortholog of human AKR1B10. Biol Chem. 2010;391:1371–1378. doi: 10.1515/BC.2010.144. [DOI] [PubMed] [Google Scholar]
- 25.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 26.Okayasu I, Yamada M, Mikami T, Yoshida T, Kanno J, Ohkusa T. Dysplasia and carcinoma development in a repeated dextran sulfate sodium-induced colitis model. Journal of gastroenterology and hepatology. 2002;17:1078–1083. doi: 10.1046/j.1440-1746.2002.02853.x. [DOI] [PubMed] [Google Scholar]
- 27.Roediger WE, Truelove SC. Method of preparing isolated colonic epithelial cells (colonocytes) for metabolic studies. Gut. 1979;20:484–488. doi: 10.1136/gut.20.6.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 29.Mashimo H, Wu DC, Podolsky DK, Fishman MC. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science. 1996;274:262–265. doi: 10.1126/science.274.5285.262. [DOI] [PubMed] [Google Scholar]
- 30.Zambell KL, Fitch MD, Fleming SE. Acetate and butyrate are the major substrates for de novo lipogenesis in rat colonic epithelial cells. J Nutr. 2003;133:3509–3515. doi: 10.1093/jn/133.11.3509. [DOI] [PubMed] [Google Scholar]
- 31.Thibault R, Blachier F, Darcy-Vrillon B, de Coppet P, Bourreille A, Segain JP. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflammatory bowel diseases. 2010;16:684–695. doi: 10.1002/ibd.21108. [DOI] [PubMed] [Google Scholar]
- 32.Borradaile NM, de Dreu LE, Huff MW. Inhibition of net HepG2 cell apolipoprotein B secretion by the citrus flavonoid naringenin involves activation of phosphatidylinositol 3-kinase, independent of insulin receptor substrate-1 phosphorylation. Diabetes. 2003;52:2554–2561. doi: 10.2337/diabetes.52.10.2554. [DOI] [PubMed] [Google Scholar]
- 33.Ni J, Chen SF, Hollander D. Effects of dextran sulphate sodium on intestinal epithelial cells and intestinal lymphocytes. Gut. 1996;39:234–241. doi: 10.1136/gut.39.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito R, Shin-Ya M, Kishida T, Urano A, Takada R, Sakagami J, et al. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin Exp Immunol. 2006;146:330–338. doi: 10.1111/j.1365-2249.2006.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. American journal of physiology Gastrointestinal and liver physiology. 2004;287:G7–G17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 36.Shacter E, Weitzman SA. Chronic inflammation and cancer. Oncology (Williston Park) 2002;16:217–226. 229. discussion 30-2. [PubMed] [Google Scholar]
- 37.Mercier PL, Bachvarova M, Plante M, Gregoire J, Renaud MC, Ghani K, et al. Characterization of DOK1, a candidate tumor suppressor gene, in epithelial ovarian cancer. Mol Oncol. 2011;5:438–453. doi: 10.1016/j.molonc.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P, et al. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell death & disease. 2014;5:e1144. doi: 10.1038/cddis.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Oliveira RC, Ribeiro DT, Nigro RG, Di Mascio P, Menck CF. Singlet oxygen induced mutation spectrum in mammalian cells. Nucleic Acids Res. 1992;20:4319–4323. doi: 10.1093/nar/20.16.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blikslager AT, Moeser AJ, Gookin JL, Jones SL, Odle J. Restoration of barrier function in injured intestinal mucosa. Physiological reviews. 2007;87:545–564. doi: 10.1152/physrev.00012.2006. [DOI] [PubMed] [Google Scholar]
- 41.Rieder F, Brenmoehl J, Leeb S, Scholmerich J, Rogler G. Wound healing and fibrosis in intestinal disease. Gut. 2007;56:130–139. doi: 10.1136/gut.2006.090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nature protocols. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 43.Perse M, Cerar A. Dextran sodium sulphate colitis mouse model: traps and tricks. Journal of biomedicine & biotechnology. 2012:718617. doi: 10.1155/2012/718617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchida K, Stadtman ER. Modification of histidine residues in proteins by reaction with 4-hydroxynonenal. Proc Natl Acad Sci U S A. 1992;89:4544–4548. doi: 10.1073/pnas.89.10.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper HS, Murthy S, Kido K, Yoshitake H, Flanigan A. Dysplasia and cancer in the dextran sulfate sodium mouse colitis model. Relevance to colitis-associated neoplasia in the human: a study of histopathology, B-catenin and p53 expression and the role of inflammation. Carcinogenesis. 2000;21:757–768. doi: 10.1093/carcin/21.4.757. [DOI] [PubMed] [Google Scholar]
- 46.Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 47.Lefrancois-Martinez AM, Tournaire C, Martinez A, Berger M, Daoudal S, Tritsch D, et al. Product of side-chain cleavage of cholesterol, isocaproaldehyde, is an endogenous specific substrate of mouse vas deferens protein, an aldose reductase-like protein in adrenocortical cells. The Journal of biological chemistry. 1999;274:32875–32880. doi: 10.1074/jbc.274.46.32875. [DOI] [PubMed] [Google Scholar]
- 48.Ma J, Luo DX, Huang C, Shen Y, Bu Y, Markwell S, et al. AKR1B10 overexpression in breast cancer: Association with tumor size, lymph node metastasis and patient survival and its potential as a novel serum marker. Int J Cancer. 2012;131:E862–E871. doi: 10.1002/ijc.27618. [DOI] [PubMed] [Google Scholar]
- 49.Fukumoto S, Yamauchi N, Moriguchi H, Hippo Y, Watanabe A, Shibahara J, et al. Overexpression of the aldo-keto reductase family protein AKR1B10 is highly correlated with smokers' non-small cell lung carcinomas. Clin Cancer Res. 2005;11:1776–1785. doi: 10.1158/1078-0432.CCR-04-1238. [DOI] [PubMed] [Google Scholar]
- 50.Ohashi T, Idogawa M, Sasaki Y, Suzuki H, Tokino T. AKR1B10, a transcriptional target of p53, is downregulated in colorectal cancers associated with poor prognosis. Molecular cancer research : MCR. 2013;11:1554–1563. doi: 10.1158/1541-7786.MCR-13-0330-T. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.