Abstract
The CLAVATA3 (CLV3)-CLAVATA1 (CLV1) ligand-receptor kinase pair negatively regulates shoot stem cell proliferation in plants. clv1 null mutants are weaker in phenotype than clv3 mutants, but the clv1 null phenotype is enhanced by mutations in the related receptor kinases BARELY ANY MERISTEM 1, 2 and 3 (BAM1, 2 and 3). The basis of this genetic redundancy is unknown. Here, we demonstrate that the apparent redundancy in the CLV1 clade is in fact due to the transcriptional repression of BAM genes by CLV1 signaling. CLV1 signaling in the rib meristem (RM) of the shoot apical meristem is necessary and sufficient for stem cell regulation. CLV3-CLV1 signaling in the RM represses BAM expression in wild-type Arabidopsis plants. In clv1 mutants, ectopic BAM expression in the RM partially complements the loss of CLV1. BAM regulation by CLV1 is distinct from CLV1 regulation of WUSCHEL, a proposed CLV1 target gene. In addition, quadruple receptor mutants are stronger in phenotype than clv3, pointing to the existence of additional CLV1/BAM ligands. These data provide an explanation for the genetic redundancy seen in the CLV1 clade and reveal a novel feedback operating in the control of plant stem cells.
Keywords: CLV1, Plant development, Receptor kinase, Stem cells, Arabidopsis thaliana
Highlighted article: Transcriptional cross regulation between CLAVATA1 family receptor kinases operates in the stem cell niche, thereby explaining the apparent genetic redundancy amongst these receptor kinases.
INTRODUCTION
Genetic redundancy in developmental systems poses a challenge for understanding how cells interpret complex signal inputs and quantitatively respond to execute specific developmental programs. At the biochemical level, several mechanisms can account for genetic redundancy, making it difficult to infer relationships between components that have additive genetic effects. In plants this situation is often compounded by the unequal contributions of different genes to additive phenotypes (Briggs et al., 2006). Stem cell maintenance in plants is governed by diverse overlapping and redundant signal inputs (Barton, 2009). Plant stem cells are located in discrete niche-like environments called meristems. The shoot apical meristem (SAM) is a dome-shaped structure that harbors the stem cells that give rise to all above-ground tissue. The SAM stem cell population is located in a central zone (CZ), a cone-shaped cell population at the apex of the SAM. Cell division in the CZ replenishes the stem cell population, but daughter cells may also be displaced into the peripheral zone (PZ) on the lateral flanks of the SAM, where they can become incorporated into nascent lateral organs such as leaves or flowers.
The rate of exit and renewal of stem cells in the CZ is precisely balanced by cell-to-cell signaling networks controlled by the transmembrane receptor kinase (RK) CLAVATA1 (CLV1) and its peptide ligand CLAVATA3 (CLV3) (Clark et al., 1997; Fletcher et al., 1999; Ogawa et al., 2008). CLV3 is synthesized in stem cells of the CZ (Fletcher et al., 1999), where it is secreted and processed to a 13 amino acid glycopeptide (CLV3p) (Kondo et al., 2006; Ohyama et al., 2009). CLV3p diffuses from the CZ to the rib meristem (RM), an underlying organizing region where CLV1 is expressed (Nimchuk et al., 2011; Rojo et al., 2002). There, it promotes trafficking of CLV1 from the plasma membrane to the lytic vacuole (Nimchuk et al., 2011). It is thought that CLV3 promotes CLV1 signaling, which dampens stem cell production by negatively regulating the expression of WUSCHEL (WUS), which encodes a homeodomain transcription factor required for stem cell maintenance (Brand et al., 2000; Mayer et al., 1998; Schoof et al., 2000). WUS, in turn, promotes CLV3 expression, forming a feedback loop that maintains a constant stem cell pool size (Brand et al., 2002; Yadav et al., 2013).
Phenotypes for null mutations in clv3 and strong dominant-negative alleles of clv1 are roughly equivalent, both causing increases in SAM size and stem cell number (Clark et al., 1993, 1995). In floral meristems (FMs), these mutations result in increased numbers of floral organs, allowing a quantitative measure of allele strength. By contrast, null mutants in clv1 show considerably weaker phenotypes than clv3 (Dievart et al., 2003), but the phenotype can be enhanced by mutations in the CLV1-related RKs BARELY ANY MERISTEM 1 (BAM1) and BAM2 (DeYoung et al., 2006; DeYoung and Clark, 2008). These results suggest that BAM1 and BAM2 act as redundant receptors for CLV3p. Consistent, broad, high-level expression of BAM1 can complement clv1 null mutations (DeYoung et al., 2006). Triple mutants in bam1, bam2 and the related bam3 have the opposite effect on stem cell production, displaying reductions in meristem size (DeYoung et al., 2006). Thus, a complex set of interactions among related RKs controls stem cell production in the SAM. The molecular basis of these genetic interactions remains unknown. Transient transformation and overexpression studies in leaf tissue suggest that BAM1 and CLV1 receptors might interact; however, it is not clear if this potential interaction is relevant in the cells of the SAM (Guo et al., 2010).
Here, we present a new model explaining the genetic redundancy among BAM RKs and CLV1. CLV1 functions exclusively in the RM and specifically represses BAM gene transcription in response to CLV3p. Ectopic BAM expression in the RM partially compensates for the loss of CLV1. In addition, the negative regulation of BAM expression by CLV1 differs from the CLV1-dependent negative regulation of WUS, which may be subject to distinct signal inputs. These data clarify the role of specific RKs in stem cell maintenance and provide insights into the signaling outputs of these pathways.
RESULTS
CLV1 functions exclusively in WUS-expressing cells of the SAM
As a first step in dissecting the mechanism of redundancy in the CLV1 clade, we asked where CLV1 functions in the SAM. Although CLV1 has been postulated to repress the transcription of WUS, the two share largely overlapping expression domains in the RM of both the SAM and in FMs (Brand et al., 2000; Clark et al., 1997; Mayer et al., 1998; Schoof et al., 2000). To test whether CLV1 functions outside of WUS-expressing cells, we transformed clv1 null plants with constructs expressing either wild-type CLV1, expressed from the native WUS promoter, or a G201E mutant of CLV1 that is equivalent to the strong clv1-4 dominant-negative allele (Clark et al., 1993). Expression of pWUS::CLV1 was sufficient to fully complement the clv1-11 null mutant in the La-er background or the clv1-101 allele in a Col-0 background (Fig. 1) (Dievart et al., 2003; Kinoshita et al., 2010). By contrast, clv1-11 pWUS::clv1 G201E plants displayed a phenotype similar to that of existing clv1-4 mutants, complete with increased carpel production relative to the clv1-11 null as well as fasciation of the main stem and SAM (Fig. 1A,B; data not shown). These data indicate that CLV1 function in WUS-expressing RM cells of the SAM is necessary and sufficient in the control of stem cell proliferation.
Fig. 1.
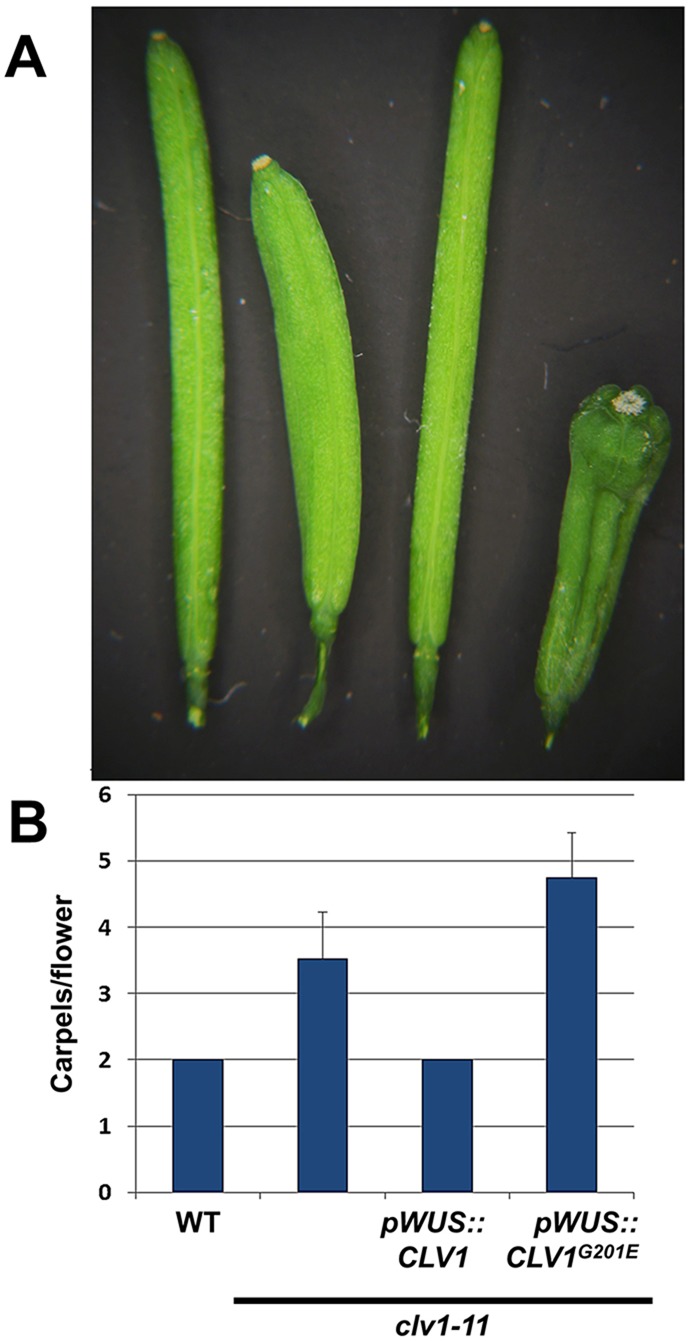
CLV1 functions exclusively in WUS-expressing cells of the rib meristem. (A) Carpel phenotypes of (left to right): wild-type Arabidopsis thaliana (La-er), clv1-11 null mutant, clv1-11 pWUS::CLV1 and clv1-11 pWUS::CLV1G201E. Ten lines of each transgenic line were selected and one typical line was chosen for analysis. (B) Quantification of carpel numbers. n=50, from five plants (ten siliques per plant). Error bars indicate s.d. A one-way ANOVA was carried out for the data set, and comparisons were performed with an SPSS Tukey HSD test at P<0.05 level. Only wild type (WT) and clv1-11 pWUS::CLV1 were not statistically different.
In order to dissect the genetic redundancy between CLV1 and BAM receptors, we next tested how CLV1 expression compares to the expression of BAM1 and BAM3 in the SAM and FM. We focused on BAM3, as its expression in the SAM has not been reported, and on BAM1, which is fully redundant with the nearly identical BAM2 (DeYoung et al., 2006). We generated native promoter binary vectors and used these to express a nuclear-targeted tandem Ypet fusion (2YN7). We transformed these binary vectors into wild-type plants to generate pCLV1::2YN7, pBAM1::2YN7 and pBAM3::2YN7 transgenic reporter lines. Imaging by confocal microscopy determined that the patterns of expression of CLV1, BAM1 or BAM3 reporters in roots were identical to those known from existing cell-specific transcriptomic profiling (Brady et al., 2007), indicating that our fluorescent reporter transgenes faithfully replicate cellular expression patterns (supplementary material Fig. S1A,B) (Depuydt et al., 2013). Consistent with this, expression of a wild-type genomic BAM1 coding sequence from the BAM1 promoter complemented the bam1 bam2 bam3 phenotype (supplementary material Fig. S1D). Furthermore, expression of a mutant BAM1 gene [G199E, equivalent to the clv1-4 dominant-negative allele (Shinohara et al., 2012)] that abolishes binding of the CLV3-related CLE9 peptide failed to restore rosette growth or fertility. Similarly, pBAM3:BAM3 complemented bam1 bam2 bam3 rosette size and leaf shape (not shown). Previous work demonstrated that genomic CLV1 expressed from the same CLV1 promoter used in these studies complements the clv1 phenotype (Nimchuk et al., 2011).
We examined expression of the CLV1, BAM1 and BAM3 transcriptional reporters in the SAM (Fig. 2; supplementary material Fig. S2). Consistent with previous accounts and our complementation data, CLV1 was highly expressed in the RM of the SAM (Clark et al., 1997). A minority of lines (2/20) showed weak expression in the L1, as also reported in a minority of lines in previous experiments (Nimchuk et al., 2011). In contrast to CLV1, the BAM1 reporter was highly expressed in the L1 cell layer of the SAM and was absent from the RM (Fig. 2). Expression of BAM1 extended into developing floral primordia and became enriched in the abaxial side during later stages of primordia emergence. In contrast to the BAM1 reporter, expression of the BAM3 reporter transgene is absent from the SAM and floral primordia proper. Further imaging analysis revealed that the BAM3 reporter is expressed exclusively in developing vascular strands below emerging primordia (Fig. 2). This expression pattern was maintained for BAM3 in developing FMs and is consistent with recent data that localized BAM3 expression to the vasculature in root tissue and with previous cell-specific transcriptomic analysis (supplementary material Fig. S1A-C) (Brady et al., 2007; Depuydt et al., 2013; Zhao et al., 2005). Taken together, these data demonstrate that BAM1 and BAM3 expression is absent in CLV1-expressing cells of the RM.
Fig. 2.
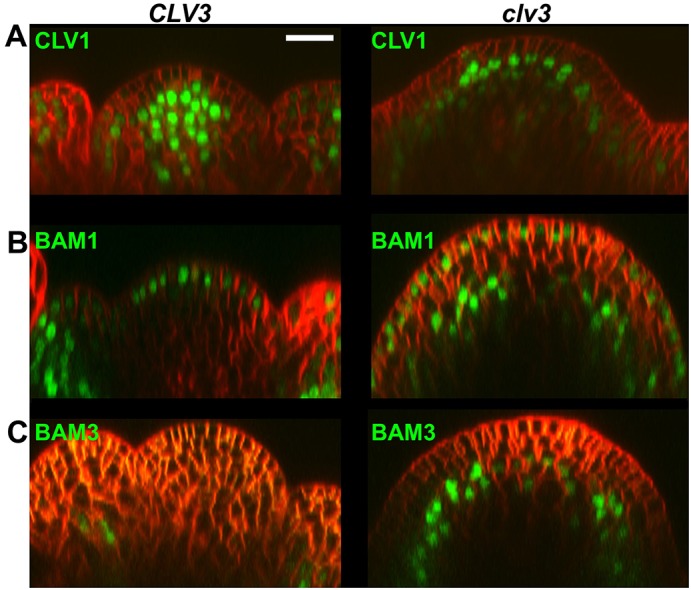
CLV3 signaling represses BAM1 and BAM3 expression from the rib meristem. Side view reconstruction through the center of FM4-64-stained inflorescence meristems. FM4-64 is in red, nuclear Ypet signal is in green. Each CLV3 and clv3 pair represents a single typical reporter line introgressed from wild type into the clv3 background and imaged using the same settings. CLV1 (A), BAM1 (B) and BAM3 (C) reporters in wild type (CLV3, left) and clv3 null (right). Scale bar: 20 µm.
We next examined CLV1, BAM1 and BAM3 reporter transgene expression in clv3-9 null plants and strong clv1-8 mutant plants (Fig. 2; supplementary material Fig. S2). Live imaging of these clv mutant SAMs revealed that both BAM1 and BAM3 are now expressed in the RM. The expression domain of the BAM reporters overlaps with the CLV1 expression domain in the SAM in both the clv3-9 and clv1-8 backgrounds, with the CLV1 expression domain being slightly broader than those of BAM1 and BAM3. BAM1 and BAM3 were repressed in L3 RM cells by CLV1 signaling (supplementary material Fig. S2), and the expression domains of BAM1 and BAM3 expanded laterally in the RM of clv SAMs, like WUS and CLV1 (Fig. 2; supplementary material Fig. S6) (Brand et al., 2000; Schoof et al., 2000). These data indicate that CLV3-CLV1 signaling is required to repress the transcription of BAM1 and BAM3 in the RM. CLV1 repression of BAM3 and BAM1 was also observed in FMs (supplementary material Fig. S2; data not shown). CLV1 expression levels appear unaffected in clv3 or clv1 mutant SAM tissue, despite the enlarged expression domain that contours to the overproliferated SAM tissue.
We next asked whether the BAM gene derepression seen in cells of the RM of clv1 mutants was due to the absence of CLV signaling or a consequence of the overproliferated SAM tissue. The SAM phenotype of the phb phv cna triple mutant superficially resembles that of clv1 mutants (Prigge et al., 2005), but these three homeodomain transcription factors are thought to act additively with the CLV1 pathway (Green et al., 2005; Williams et al., 2005). Live imaging of BAM1 and BAM3 reporter transgene expression in the phb phv cna triple mutant SAM revealed that BAM expression is not present in the RM. Therefore, expression of the BAM genes in RM cells of clv1 mutants is not an indirect consequence of the increase in SAM size (supplementary material Fig. S3B). Similarly, an L1-specific pATML1::mTFP-ER reporter (Chickarmane et al., 2012; Sessions et al., 1999), which is unrelated to the BAM1 transgene reporter, was not ectopically expressed in RM cells of clv3-2 or clv1-4 mutants. This indicates that derepression of the BAM1 reporter in the clv1 RM is not due to a general respecification of RM cells to an epidermal identity (supplementary material Fig. S3A; data not shown). Thus, BAM1 and BAM3 are specific transcriptional targets of CLV1 signaling in the RM.
We confirmed this result using QRT-PCR analysis of BAM1 and BAM3 transcripts in clv3 plants that contain a conditional dexamethasone (DEX)-dependent CLV3 transgene (clv3 DEX>>CLV3). In these plants, clv3 activity is restored through inducible DEX application. We have previously shown that this system activates biologically relevant CLV1 endomembrane trafficking within 4 h post CLV3 induction (Nimchuk et al., 2011). Here, we find that activation of CLV1 signaling following induction of CLV3 caused rapid downregulation (30 min post DEX induction) of both BAM3 and WUS transcripts (Fig. 3). BAM1 expression is downregulated modestly at later time points (Fig. 3). This difference might reflect the fact that BAM1 expression in the L1 layer and primordia is CLV1 independent, potentially masking any repression of BAM1 transcripts in the RM. By contrast, CLV1 expression was upregulated in response to CLV3, consistent with previous data showing that WUS represses CLV1 expression (Busch et al., 2010). CLV1 expression in the L1, where BAM1 is expressed strongly, is very weak or absent, and we observed no CLV1 expression in the L1 of bam1; bam2; bam3 mutant SAMs (supplementary material Fig. S3). In addition, CLV1 expression overlapped with BAM1, BAM2 and BAM3 in a subset of root cells (supplementary material Fig. S1B; data not shown). Thus, although CLV1 signaling represses BAM1 there is no reciprocal repression of CLV1 expression by BAM1 signaling in the epidermis of the SAM.
Fig. 3.
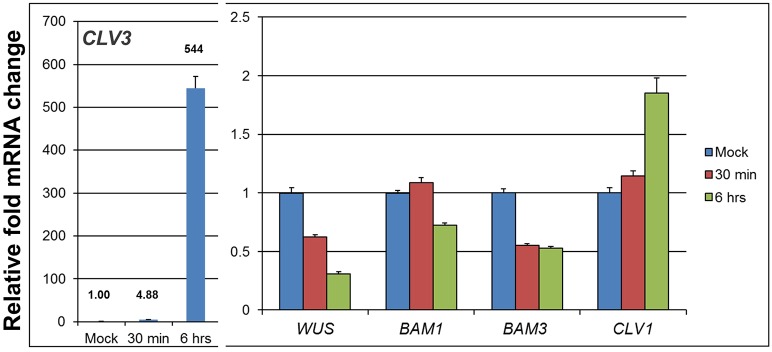
BAM3 and BAM1 are targets of the CLV3 signaling pathway. QRT-PCR was used to calculate fold changes for selected genes following CLV3 induction. RNA from apical shoot tissues was analyzed 30 min or 6 h post induction or from mock-treated plants at 6 h. See Materials and Methods for details. Error bars indicate s.d.
Our data demonstrate that CLV1 function is necessary and sufficient in the RM to repress stem cell proliferation. In wild-type plants, CLV1 signaling represses BAM1 and BAM3 expression in RM cells. Despite this, the clv1 null mutant phenotypes are enhanced by mutations in bam1 (DeYoung and Clark, 2008). Previous work, utilizing the ERECTA promoter, which is broadly expressed in the SAM, to drive expression of BAM1, demonstrated that BAM1 function can partially substitute for loss of CLV1 function (DeYoung et al., 2006). We therefore hypothesized that RM-specific derepression of BAM class receptors in clv1 null mutants functionally compensates for loss of CLV1 function. To test this we first examined whether RM cells are specifically sensitized to BAM1 levels. Given that BAM1 is expressed in both the L1 layer and RM of clv class mutants we used the L1-specific ATML1 promoter or the RM-specific WUS promoter to increase BAM1 levels in the clv1 null background. None of the pATML1::BAM1 lines complemented the clv1-11 mutant phenotype (0/60 T1 lines screened). By contrast, 17% of the pWUS::BAM1 lines complemented the clv1-11 phenotype (7/40 T1 lines screened), with some lines showing complete complementation. The proportion of complementing lines with the WUS promoter is consistent with previous studies using the ERECTA promoter (DeYoung et al., 2006). We also attempted to increase vascular expression levels of BAM3 in clv1 null plants by expressing additional copies of BAM3 from the S17 phloem-specific promoter, which overlaps with BAM3 expression in the root (supplementary material Fig. S1B) (Brady et al., 2007; Lee et al., 2006). None of the 70 T1 plant lines screened displayed any complementation of the clv1 phenotype. Thus, RM-specific, but not L1- or vascular-specific, expression of BAM can substitute for CLV1.
We next tested what phenotypic consequence the derepression of BAM1 and BAM3 expression has in RM cells by analyzing quadruple CLV1 clade receptor null mutants (bam1 bam2 bam3 clv1 plants, hereafter referred to as b1b2b3c1) in an isogenic Col-0 background. As a control, we used the b1b2b3 plants in an isogenic Col-0 background that displayed a mutant phenotype consistent with previous publications on b1b2b3 plants in a mixed background of La-er and Col-0 ecotypes (DeYoung et al., 2006). As reported, plants were very stunted, leaves were crinkled, higher order vascular complexity was reduced and flowers displayed male sterility. Despite these consistent phenotypes, we rarely noted the premature termination of shoot growth seen in the mixed La-er and Col-0 background. Apparently, the La-er background, which modifies clv1 phenotypes relative to Col-0 (Dievart et al., 2003), also modifies bam phenotypes. As a quantitative measurement of clv function we analyzed carpel number in Col-0, clv1, b1b2b3 and b1b2b3c1 plants (Fig. 4A,B). Loss of bam activity alone had no effect on CLV1 function, as Col-0 and b1b2b3 displayed wild-type carpel numbers indicating that BAM receptor function has no role in the negative regulation of stem cells when CLV1 is functional. However, the clv1 null phenotype was greatly enhanced in b1b2b3c1 plants (Fig. 4). This was also seen in the mixed La-er background, as b1b2b3c1 plants displayed a massive overproliferation of the vegetative SAM, something not seen in either b1b2b3 or clv1 nulls from the La-er background (supplementary material Fig. S4). The La-er quadruple mutant plants frequently died on soil before flowering, but rare single flowers displayed the enhanced carpel production seen in the b1b2b3c1 Col-0 plants. These results indicate that BAM receptor function can negatively regulate stem cell proliferation in the absence of CLV1 signaling. b1b2b3c1 quadruple mutant plants are phenotypically stronger than both the clv1-8 dominant-negative allele (supplementary material Fig. S7A) and null alleles of clv3 (supplementary material Fig. S7B).
Fig. 4.
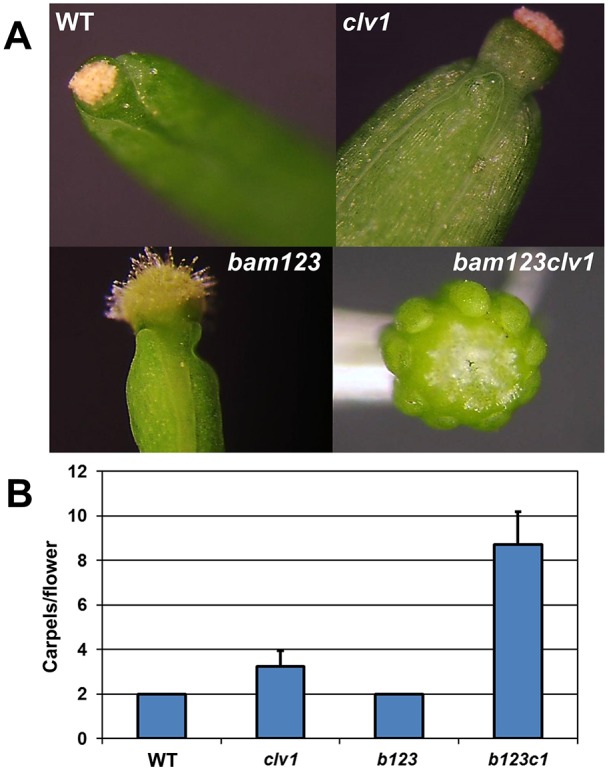
Loss of CLV1 is compensated for by BAM receptor genes. (A) Representative siliques from Col-0 wild type (WT) and null mutant combinations in bam1, bam2, bam3 or clv1. (B) Quantification of carpel number. x-axis, average carpel number per flower from plants in A. Error bars indicate s.d. n=60, from six plants (ten siliques per plant). Note that this number was chosen as bam1 bam2 bam3 clv1 flower production is strongly reduced due to disorganized shoot formation. A one-way ANOVA was carried out for the data set, and comparisons were performed with an SPSS Tukey HSD test at P<0.05 level. Only wild-type (WT) and bam1,bam2,bam3 (b123) carpel counts were not statistically different.
We then tested whether RM expression of CLV1 was sufficient to restore stem cell function to the b1b2b3c1 quadruple mutant by expressing wild-type CLV1 from the WUS promoter. Importantly, the WUS promoter is expressed in a wild-type pattern and at wild-type levels in the b1b2b3 mutant SAM (supplementary material Fig. S5). pWUS::CLV1 completely restored the regulation of shoot stem cell pool size back to a b1b2b3 phenotype (Fig. 5). This demonstrates that CLV1 function in the RM is necessary and sufficient for negative regulation of stem cell proliferation, independent of BAM receptor function. Collectively, these data demonstrate that RM cells are the sole domain from which the CLV receptor clade members act to functionally suppress the proliferation of stem cells within the SAM. The apparent genetic redundancy in this clade is due to compensation from the ectopic expression of BAM genes in the RM in the absence of CLV1 signaling.
Fig. 5.
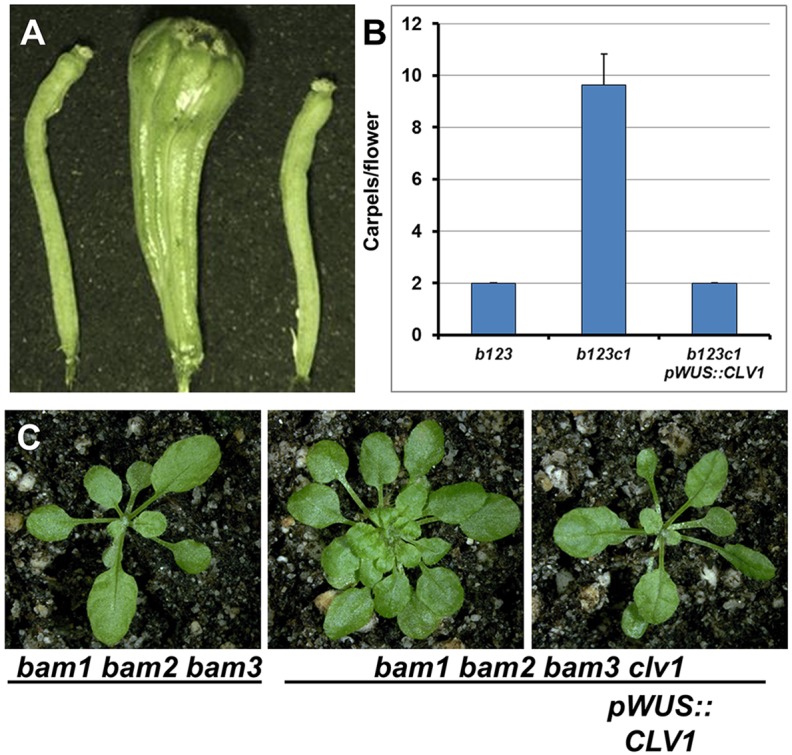
Expression of CLV1 in the RM is necessary and sufficient for all stem cell regulation. Expression of CLV1 from the WUS promoter fully complements the unregulated stem cell proliferation defects in bam1 bam2 bam3 clv1 plants back to that of bam1 bam2 bam3. (A) Silique phenotypes of bam1 bam2 bam3 (left), bam1 bam2 bam3 clv1 (middle) and bam1 bam2 bam3 clv1 pWUS::CLV1 (right). (B) Quantification of carpel numbers for plants displayed in A. Sample size as in Figure 4. Error bars indicate s.d. A one-way ANOVA was carried out for the data set, and comparisons were performed with an SPSS Tukey HSD test at P<0.05 level. b123c1 plants had significantly more carpel numbers than the other genotypes. (C) Rosette vegetative phenotypes for plants in A.
We next examined whether BAM1 and BAM3 repression by CLV1 signaling in RM cells is similar to the negative regulation of WUS expression, a target for repression by the CLV1 pathway in the RM. Unlike the observed BAM1 and BAM3 derepression, WUS expression is detected at comparable levels in wild type and clv3 mutants, although the expression domain is expanded (supplementary material Fig. S6). This apparent lack of any difference in WUS expression levels in the RM of clv3 mutants (e.g. CLV1-expressing cells) has also been noted in prior in situ expression analyses (Schoof et al., 2000).
DISCUSSION
Overlapping genetic function can complicate the phenotypic analysis of genetic traits in model systems. This is especially true in plants, where individual redundant genes often contribute unequally to phenotypic outcomes (Briggs et al., 2006). Here, we provide a new explanation for the apparent genetic redundancy of the CLV1 clade of RKs in stem cell regulation (Fig. 6). Our data demonstrate that BAM receptors are not crucial co-factors for CLV1 function at the genetic or biochemical levels and do not normally act within the CLV1 pathway in wild-type plants. Rather, the BAM genes appear genetically redundant with CLV1 function due to repression by CLV1 signaling and their ability to conditionally complement the clv1 phenotype due to ectopic expression in RM cells of clv mutant SAMs. In this model, clv1 null mutations are partially complemented due to depression of the BAM genes and BAM-specific signaling in RM cells of the clv SAM. In clv3 mutants, BAM receptors are derepressed but would fail to signal due to a lack of CLV3 ligand. In stronger, semi-dominant-negative clv1 alleles, the ectopic BAM receptors are presumably less competent to signal due to the presence of the interfering CLV1 proteins. Consistent with this, b1b2b3c1 quadruple mutant plants are measurably stronger in phenotype than clv1-8 dominant-negative alleles on their own. How dominant-negative CLV1 proteins act to dampen BAM function is unclear. Alternatively, it is possible that dominant-negative CLV1 proteins could interfere with common CLV1/BAM co-receptors or other receptors, such as RPK2 or CLV2, although both RPK2 and CLV2 are dispensable for wild-type CLV1 signaling (Kinoshita et al., 2010; Muller et al., 2008). b1b2b3c1 plants are phenotypically stronger than clv3 alleles in Col-0, suggesting that multiple CLV3-related ligands might contribute to CLV clade signaling and SAM function in Arabidopsis, as they do in rice (Suzaki et al., 2009). Consistent with this hypothesis, several CLV3-related CLE genes can substitute for loss of CLV3 when expressed from the CLV3 promoter (Ni and Clark, 2006). BAM receptor gene repression represents a specific transcriptional output as a consequence of the CLV1 signaling pathway in RM cells, which is not observed in the homeodomain transcription factor phb phv cna mutants. Despite this, phb phv cna triple mutants are completely resistant to CLV3-induced SAM termination when grown on CLV3 peptide-containing plates (supplementary material Fig. S8). This indicates that the commonly used peptide-based termination assays can select bypass mutants that do not affect CLV1 signaling per se, and so care should be used in the interpretation of these assays.
Fig. 6.
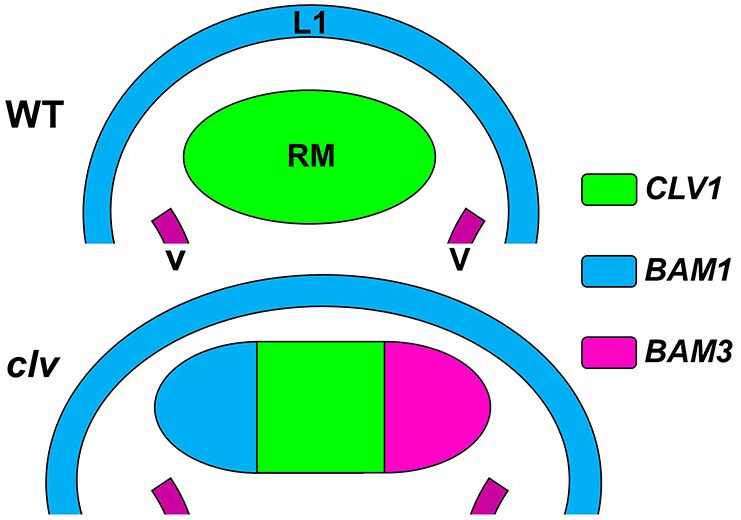
Model explaining CLV1-BAM genetic redundancy in stem cell suppression. The expression domains of CLV1, BAM1 and BAM3 are shown in wild-type SAMs (top) or in SAMs in clv clade mutants (bottom). L1, L1 epidermal layer; RM, rib meristem; V, vasculature. In wild-type (WT, top) SAMs, only CLV1 (green) is expressed in the crucial RM region necessary for stem cell regulation. In clv class SAMs (bottom), BAM1 (blue) and BAM3 (red) become ectopically expressed in the RM. In clv1 mutants, RM-specific ectopic expression of BAM genes partially compensates for loss of CLV1. See text for details.
The repression of BAM and of WUS are differentially sensitized to CLV1 signaling. In wild-type plants, WUS is robustly expressed in the RM, whereas BAM promoter activity is undetectable. Despite this, both genes are repressed by very high levels of CLV3. This suggests that a buffering mechanism might act preferentially on WUS to mask differences in WUS expression levels within a physiological range of CLV1 signaling. The WUS expression domain also expands when members of the PHB PHV CNA family are downregulated (Green et al., 2005; Williams et al., 2005), despite the continued BAM repression in RM cells in the phb phv cna triple mutant. As WUS relocalization also occurs in phb phv cna triple mutants, whereas BAM domain expansion does not, WUS upregulation in clv and likely in phb phv cna mutants might be due to expansion of the RM.
Conditional cross-complementation in the CLV1 clade is reminiscent of PIN family interactions in auxin-mediated development, where loss of specific PIN family members is compensated for by ectopic expression of other PIN genes (Vieten et al., 2005). CLV1 expression levels are directly or indirectly influenced by different stimuli (Busch et al., 2010; Chickarmane et al., 2012; Gordon et al., 2009; Yoshida et al., 2011). It is possible that cross-regulation of BAM expression reflects a mechanism to buffer fluctuations in CLV1 levels to ensure network robustness. The evolutionary maintenance of divergent CLV1 clade members might reflect selective pressures promoting both divergent functions and network robustness (Kafri et al., 2009). Deciphering the mechanism of BAM repression by CLV1 signaling should reveal novel downstream aspects of this key regulatory pathway in stem cell regulation.
MATERIALS AND METHODS
Mutant seed stocks
All seed stock lines were ordered from the Arabidopsis Biological Resource Center (ABRC) via the TAIR database, except the clv3-9 allele, which was a gift from Rüdiger Simon (Heinrich-Heine-University, Düsseldorf, Germany), and phb phv can seeds, which were obtained from Steven Clark (University of Michigan, Ann Arbor, MI, USA). The null clv1-101 (in Col-0 background), null clv1-11 (in La-er background) and dominant-negative clv1-8 (in Col-0 background) alleles were described previously (Dievart et al., 2003; Kinoshita et al., 2010), as was the La-er/Col-0 bam1-1 bam2-1 bam3-2 triple mutant (DeYoung et al., 2006). clv3-9 Col-0 is an EMS allele resulting in a W62STOP mutation that deletes the critical CLE domain region. The bam1-4 (SALK_107290), bam2-4 (SAIL_1053_E09) and bam3-2 (SALK_044433) mutant seeds that were used to generate the bam1 bam2 bam3 triple mutant in the Col-0 background were identified from the SALK T-DNA expression databases and ordered from ABRC. Each T-DNA insertion mutant is located early in the first exon of each gene and is likely to lead to a null phenotype. bam3-2 has been characterized previously (DeYoung et al., 2006). clv2-101 was used as a null Col-0 allele (Kinoshita et al., 2010).
Reporter line construction and transgenic plant line generation
The pCLV1 binary vector construction has been described previously, with the exception that this version is kanamycin selectable in planta (Nimchuk et al., 2011). The pBAM1 binary vector was created by recombinant overlapping PCR to generate a 5.2 kb NotI fragment consisting of 3.5 kb of BAM1 upstream promoter and 1.6 kb of downstream promoter separated by a unique BamHI site. The NotI fragment was cloned into pMOA34 binary vector (Barrell and Conner, 2006). The pBAM3 binary vector was generated by a similar strategy using 3.5 kb and 1.5 kb of the upstream and downstream promoter. A tandem 2xYpet fusion gene containing the N7 nuclear targeting sequence (Cutler et al., 2000) at the C-terminus was cloned into each vector by standard cloning methods. Plants were transformed by floral dipping (Clough and Bent, 1998) and transgenic lines were selected on either hygromycin (pBAM1 and pBAM3) or kanamycin (pCLV1) plates. Approximately 10-20 lines per construct were examined by confocal microscopy and representative single insertion lines were selected for crossing and further analysis.
Genetic analysis
Standard emasculation and crossing techniques were used. F2 and F3 families were selected and confirmed using allele-specific PCR genotyping. Complementation lines of CLV1, BAM1 and BAM3 were transformed into either clv1 plants or bam1 bam3 bam2/+ plant lines. bam2 homozygous T1s and T2s of wild-type appearance were identified by PCR genotyping.
Confocal imaging
Confocal imaging and FM4-64 staining were performed as described (Nimchuk et al., 2011).
Quantitative PCR (QRT-PCR) analysis
clv3-2 (in La-er background) plants harboring a DEX-inducible CLV3 gene were grown under continuous light (Nimchuk et al., 2011). Once shoots were ∼1 cm in height they were treated with either 0.1% ethanol/0.02% Silwet-70 (mock vehicle treatment) or 20 µM DEX/0.02% Silwet-70 for 0 min, 30 min or 6 h. Twenty individual inflorescence meristems from each treatment group were dissected to remove all mature flowers and developing flowers (stage 4-5 and older). Total RNA was purified from the 20 pooled tissue samples using the Qiagen RNeasy RNA purification kit. One microgram of total RNA was reverse transcribed by random priming using SuperScript II (Invitrogen). For QRT-PCR analysis 1 µl cDNA from each treatment was used with SYBR Green Master Mix (Bioline). Samples were analyzed on a Roche Light Cycler 480 with a two-step PCR protocol for 40 cycles. Data were analyzed by the ΔΔCt method using the mock treatment as the control sample. Each data point represents average Ct values calculated from three technical replicates run on the same plate. The experiment was repeated twice.
Supplementary Material
Acknowledgements
We thank Arnavaz Garda for lab support; Dr Steven Clark for providing the La-er/Col-0 bam1-1 bam2-1 bam3-2 seed; and Dr Rüdiger Simon for providing the clv3-9 allele.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Z.L.N. and E.M.M. conceived the project. Z.L.N. created the vectors and plant lines and performed the confocal imaging and genetics. Y.Z. analyzed the T1 complementation assays in the clv1 background. P.T.T. performed all QRT-PCR analysis. B.A.P. analyzed some of the multiple mutants. Z.L.N. and E.M.M. analyzed the data. Z.L.N. wrote and revised the paper with P.T.T. and E.M.M.
Funding
This work was funded by a National Institutes of Health (NIH) grant [1R01GM086639] to E.M.M.; by funds from the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation [through grant GBMF3406]; and supported in part by start-up funds provided by Virginia Tech to Z.L.N. Deposited in PMC for release after 6 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.119677/-/DC1
References
- Barrell P. J. and Conner A. J. (2006). Minimal T-DNA vectors suitable for agricultural deployment of transgenic plants. Biotechniques 41, 708-710 10.2144/000112306 [DOI] [PubMed] [Google Scholar]
- Barton M. K. (2009). Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341, 95-113 10.1016/j.ydbio.2009.11.029 [DOI] [PubMed] [Google Scholar]
- Brady S. M., Orlando D. A., Lee J.-Y., Wang J. Y., Koch J., Dinneny J. R., Mace D., Ohler U. and Benfey P. N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801-806 10.1126/science.1146265 [DOI] [PubMed] [Google Scholar]
- Brand U., Fletcher J. C., Hobe M., Meyerowitz E. M. and Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617-619 10.1126/science.289.5479.617 [DOI] [PubMed] [Google Scholar]
- Brand U., Grünewald M., Hobe M. and Simon R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565-575 10.1104/pp.001867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs G. C., Osmont K. S., Shindo C., Sibout R. and Hardtke C. S. (2006). Unequal genetic redundancies in Arabidopsis--a neglected phenomenon? Trends Plant Sci. 11, 492-498 10.1016/j.tplants.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Busch W., Miotk A., Ariel F. D., Zhao Z., Forner J., Daum G., Suzaki T., Schuster C., Schultheiss S. J., Leibfried A. et al. (2010). Transcriptional control of a plant stem cell niche. Dev. Cell 18, 841-853 10.1016/j.devcel.2010.03.012 [DOI] [PubMed] [Google Scholar]
- Chickarmane V. S., Gordon S. P., Tarr P. T., Heisler M. G. and Meyerowitz E. M. (2012). Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 109, 4002-4007 10.1073/pnas.1200636109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. E., Running M. P. and Meyerowitz E. M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397-418. [DOI] [PubMed] [Google Scholar]
- Clark S. E., Running M. P. and Meyerowitz E. M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057-2067. [Google Scholar]
- Clark S. E., Williams R. W. and Meyerowitz E. M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575-585 10.1016/S0092-8674(00)80239-1 [DOI] [PubMed] [Google Scholar]
- Clough S. J. and Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cutler S. R., Ehrhardt D. W., Griffitts J. S. and Somerville C. R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97, 3718-3723 10.1073/pnas.97.7.3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S., Rodriguez-Villalon A., Santuari L., Wyser-Rmili C., Ragni L. and Hardtke C. S. (2013). Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc. Natl. Acad. Sci. USA 110, 7074-7079 10.1073/pnas.1222314110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung B. J. and Clark S. E. (2008). BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180, 895-904 10.1534/genetics.108.091108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung B. J., Bickle K. L., Schrage K. J., Muskett P., Patel K. and Clark S. E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45, 1-16 10.1111/j.1365-313X.2005.02592.x [DOI] [PubMed] [Google Scholar]
- Dievart A., Dalal M., Tax F. E., Lacey A. D., Huttly A., Li J. and Clark S. E. (2003). CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15, 1198-1211 10.1105/tpc.010504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. C., Brand U., Running M. P., Simon R. and Meyerowitz E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911-1914 10.1126/science.283.5409.1911 [DOI] [PubMed] [Google Scholar]
- Gordon S. P., Chickarmane V. S., Ohno C. and Meyerowitz E. M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 106, 16529-16534 10.1073/pnas.0908122106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K. A., Prigge M. J., Katzman R. B. and Clark S. E. (2005). CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17, 691-704 10.1105/tpc.104.026179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Han L., Hymes M., Denver R. and Clark S. E. (2010). CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 63, 889-900 10.1111/j.1365-313X.2010.04295.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R., Springer M. and Pilpel Y. (2009). Genetic redundancy: new tricks for old genes. Cell 136, 389-392 10.1016/j.cell.2009.01.027 [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H. and Sawa S. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911-3920 10.1242/dev.048199 [DOI] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H. and Sakagami Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845-848 10.1126/science.1128439 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Colinas J., Wang J. Y., Mace D., Ohler U. and Benfey P. N. (2006). Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 103, 6055-606 10.1073/pnas.0510607103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K. F. X., Schoof H., Haecker A., Lenhard M., Ju¨rgens G. and Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805-815 10.1016/S0092-8674(00)81703-1 [DOI] [PubMed] [Google Scholar]
- Muller R., Bleckmann A. and Simon R. (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934-946 10.1105/tpc.107.057547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. and Clark S. E. (2006). Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140, 726-733 10.1104/pp.105.072678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z. L., Tarr P. T., Ohno C., Qu X. and Meyerowitz E. M. (2011). Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21, 345-352 10.1016/j.cub.2011.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y. and Matsubayashi Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294 10.1126/science.1150083 [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M. and Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5, 578-580 10.1038/nchembio.182 [DOI] [PubMed] [Google Scholar]
- Prigge M. J., Otsuga D., Alonso J. M., Ecker J. R., Drews G. N. and Clark S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61-76 10.1105/tpc.104.026161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Sharma V. K., Kovaleva V., Raikhel N. V. and Fletcher J. C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14, 969-977 10.1105/tpc.002196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K. F. X., Ju¨rgens G. and Laux T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635-644 10.1016/S0092-8674(00)80700-X [DOI] [PubMed] [Google Scholar]
- Sessions A., Weigel D. and Yanofsky M. F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259-263 10.1046/j.1365-313x.1999.00594.x [DOI] [PubMed] [Google Scholar]
- Shinohara H., Moriyama Y., Ohyama K. and Matsubayashi Y. (2012). Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J. 70, 845-854 10.1111/j.1365-313X.2012.04934.x [DOI] [PubMed] [Google Scholar]
- Suzaki T., Ohneda M., Toriba T., Yoshida A. and Hirano H.-Y. (2009). FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice. PLoS Genet. 5, e1000693 10.1371/journal.pgen.1000693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A., Vanneste S., Wiśniewska J., Benková E., Benjamins R., Beeckman T., Luschnig C. and Friml J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132, 4521-4531 10.1242/dev.02027 [DOI] [PubMed] [Google Scholar]
- Williams L., Grigg S. P., Xie M., Christensen S. and Fletcher J. C. (2005). Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132, 3657-3668 10.1242/dev.01942 [DOI] [PubMed] [Google Scholar]
- Yadav R. K., Perales M., Gruel J., Ohno C., Heisler M., Girke T., Jonsson H. and Reddy G. V. (2013). Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 9, 654 10.1038/msb.2013.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Mandel T. and Kuhlemeier C. (2011). Stem cell activation by light guides plant organogenesis. Genes Dev. 25, 1439-1450 10.1101/gad.631211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Craig J. C., Petzold H. E., Dickerman A. W. and Beers E. P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138, 803-818 10.1104/pp.105.060202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


