Abstract
Global eradication requires concerted efforts to combat emerging resistance to the potent antimalarial artemisinin.
Malaria kills more young children than any other infectious disease. The most pernicious causal agent, the protozoan parasite Plasmodium falciparum, is responsible for the death each year of more than half a million children, mostly in sub-Saharan Africa. Until recently, control efforts were thwarted by pyrimethamine-and chloroquine-resistant parasites, whose appearance in Africa was traced back to origins near the Thai-Cambodian border. Fortunately, the discovery of the potent antimalarial properties of artemisinin (1) has helped turn the tide against malaria. Artemisinin-based combination therapies (ACTs), which combine a potent but short-lived artemisinin derivative with a longer-lasting partner drug, have now been officially adopted across virtually the entire malaria-endemic world. Their deployment, along with efforts to distribute insecticide-treated bednets, is associated with recent substantial reductions in malaria burden. However, recent studies from Cambodia and now Thailand show that once again resistance is looming as a major threat to global control efforts (see the figure) (2, 3).
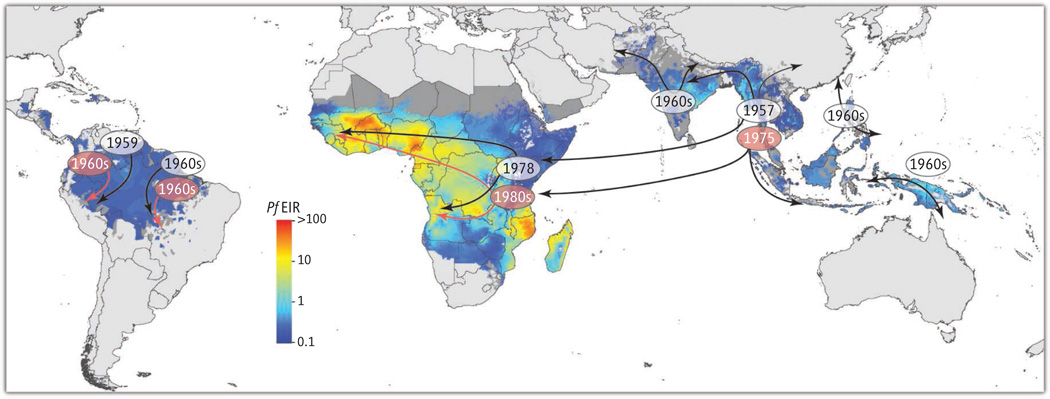
Spread of P. falciparum resistance.
Appearance and spread of resistance to former first-line antimalarials chloroquine and pyrimethamine (clear and orange ovals, respectively), showing migration from Southeast Asia to Africa. Emerging resistance to artemisinins is now documented in Southeast Asia, raising concerns about its future migration to Africa. Resistance patterns are overlaid onto a geostatistics map of P. falciparum entomological inoculation rates (EIR; numbers of infectious mosquito bites per individual per year) modeled for 2010 (16).
Emerging resistance to artemisinins is defined as a reduced rate of parasite clearance after administration of an artemisinin derivative such as artesunate or an ACT. In western Cambodia, the epicenter of resistance, artesunate treatment yielded a mean clearance half-life of 5.9 hours in Pursat, as opposed to 2 hours for a comparator drug-sensitive cohort in Wang Pha, western Thailand (4). Delayed clearance translates into an increased proportion of individuals having microscopically detectable blood-stage infections by the third day of treatment and raises the risk of parasite recrudescence (i.e., disease reappearance). Major efforts are under way in this region to eliminate malaria while ACTs remain clinically effective, with the theory that nothing short of elimination will prevent its global dissemination.
Mode-of-action studies, while controversial, mostly converge on the idea that artemisinins can be activated in the parasite via iron-mediated scission of the peroxide bridge following hemoglobin proteolysis and release of ferric heme iron (5). This can lead to the formation of carbon-centered radicals that trigger cell death. This mechanism can account for drug action during most of the 48-hour intraerythrocytic developmental cycle, although how artemisinins act against the very early “ring” stages that form shortly after host cell invasion remains enigmatic.
In vitro studies on P. falciparum cultures exposed to high concentrations of artemisinin show that parasites can acquire an initial state of tolerance (6) whereby early ring-stage parasites adopt a drug-induced quiescent or dormant state and then resume normal growth upon drug removal (7). This trait is distinct from mechanisms of resistance to other antimalarials, which typically permit robust growth despite drug treatment and often involve genetic changes that alter drug targets or efflux systems (e.g., PfCRT or PfMDR1). Further research is needed to define the molecular basis of in vitro tolerance or resistance, understand its relation to delayed clearance rates, and model how resistance, parasite fitness, transmission intensity, and treatment coverage collectively influence the spread of resistance.
Recent high-density genotyping studies with clinically defined Southeast Asian patient isolates showed that parasite clearance rates have a predominantly heritable component, which was associated with a 35-kb region on chromosome 13 (8). A separate genome-wide association study also associated delayed clearance with single-nucleotide polymorphisms on chromosomes 13 and 10 (9). The chromosome 13 candidate markers identified in these two studies did not overlap, suggesting intrinsic difficulties in obtaining sufficiently high resolution to identify the causal gene and/or the presence of multiple genetic determinants on that chromosome.
Novel insights into the genetic basis of artemisinin resistance were recently provided by whole-genome sequencing of 825 parasite strains from Asia and Africa, which identified a highly unusual population structure in parasites from western Cambodia (10). This region harbors four genetically highly differentiated but nonetheless sympatric parasite subpopulations termed KH1 to KH4. Three (KH2, 3, and 4) showed a significant prolongation of parasite clearance half-life relative to the KH1 subpopulation that was commonly observed in largely artemisinin-sensitive neighboring regions. One explanation might be the presence of multiple genetic loci that collectively confer upon each subpopulation the ability to survive artemisinin exposure; independent segregation of these genes during sexual recombination would thus be selected against during treatment. Alternatively, the lack of recombination may reflect relatively recent founder events that separately acquired a primary resistance determinant and that have not yet had time to recombine frequently and disrupt linkage disequilibrium. In KH2 parasites, chromosome 13 showed extensive linkage disequilibrium, with a single haplotype extending across half the 4-Mb chromosome; this makes the identification of specific changes associated with delayed clearance rates particularly challenging. Among the genome-wide candidates were several transporter genes that have highly differentiated sequences in the founder subpopulations, including the ABC transporters PfMDR1 and PF13_0271 that might restrict drug access to its site of action.
This study also identified substantial changes in the DNA mismatch repair pathway, including the repair heterodimer MutLα (consisting of PMS1 and MLH1) and its physical partner UvrD (10). Resistance-associated mutations in DNA repair pathways were separately observed for Rad5 (9), which is involved in the DNA damage tolerance pathway of postreplication repair (11). Mutations in this pathway have been implicated in cell cycle arrest in yeast and might play a similar role in P. falciparum. The presence of DNA repair variants in the KH1 subpopulation also suggests that some changes may predate artemisinin resistance— for example, by creating initial hypermutator strains that accelerate the frequency of resistance. This finding recalls the earlier report of an ARMD (accelerated resistance to multiple drugs) phenotype in some Southeast Asian parasites (12). Interestingly, some MLH1 variants in yeast can suppress meiotic crossovers (11), raising the possibility that the changes in P. falciparum might be used to preserve linkage disequilibrium and polygenetic traits.
Molecular tools are urgently needed to monitor artemisinin resistance and to identify therapeutic approaches to contain it. Forward genetic methods, which entail crossing drug-resistant and drug-sensitive parasites and which previously localized the genetic determinants mediating resistance to chloroquine and pyrimethamine, would be useful if the trait followed a pattern of Mendelian inheritance. This trait could be quantified using an in vitro assay recently used to study parasite ring-stage susceptibility to artemisinins (7). Such crosses, which until now have required nonhuman primates, may be achievable using a new human hepatocyte- and erythrocyte-engrafted FRG NOD mouse model that permits mosquito-delivered P. falciparum parasites to progress through the liver and be recovered as blood-stage forms (13). Reverse genetic approaches, where candidate genes are modified via allelic exchange and phenotypic changes assessed, also benefit from the recent development of customized zinc finger nuclease–mediated gene editing in P. falciparum (14). This makes it feasible to assess a panel of candidates, with the goal of defining DNA sequence markers to inexpensively and rapidly screen for the spread of resistance, and begin to delineate its molecular basis. Finally, taking inspiration from approaches in Mycobacterium tuberculosis (15), new high-throughput screening approaches are required to identify antimalarial compounds that effectively eliminate artemisinin-tolerant quiescent parasites.
Emerging resistance to artemisinins has not yet compromised the outstanding clinical efficacy of ACTs across most of the malaria-endemic regions of the world; even in western Cambodia most infections are reported to clear after ACT treatment. Malaria elimination remains an achievable goal, one that is critically dependent on an expanded and unified vision coordinated among funders, governments, health care providers, scientists, and the pharmaceutical industry. The issue of emerging artemisinin resistance highlights the necessity to intervene at all levels to prevent a looming disaster, and an opportunity to bring about a truly global accomplishment that directly or indirectly benefits us all.
References
- 1.White NJ. Science. 2008;320:330. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- 2.Dondorp AM, et al. N. Engl. J. Med. 2011;365:1073. doi: 10.1056/NEJMp1108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phyo AP, et al. Lancet. 2012;379:1960. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaratunga C, et al. Lancet Infect. Dis. 2012;12:851. doi: 10.1016/S1473-3099(12)70181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Neill PM, Barton VE, Ward SA. Molecules. 2010;15:1705. doi: 10.3390/molecules15031705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Q, Kyle DE, Gatton ML. Int. J. Parasitol. Drugs Drug Resist. 2012;2:249. doi: 10.1016/j.ijpddr.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witkowski B, et al. Antimicrob. Agents Chemother. 2013;57:914. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheeseman IH, et al. Science. 2012;336:79. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takala-Harrison S, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;110:240. [Google Scholar]
- 10.Miotto O, et al. Nat. Genet. 2013;45:648. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boiteux S, Jinks-Robertson S. Genetics. 2013;193:1025. doi: 10.1534/genetics.112.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rathod PK, McErlean T, Lee PC. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9389. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughan AM, et al. J. Clin. Invest. 2012;122:3618. doi: 10.1172/JCI62684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straimer J, et al. Nat. Methods. 2012;9:993. doi: 10.1038/nmeth.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gold B, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16004. [Google Scholar]
- 16.Gething PW, et al. Malar. J. 2011;10:378. doi: 10.1186/1475-2875-10-378. [DOI] [PMC free article] [PubMed] [Google Scholar]