Abstract
The active form of vitamin D (1α,25-dihydroxyvitamin D, 1,25(OH)2D) exerts its genomic effects via binding to a nuclear high-affinity vitamin D receptor (VDR). Recent deep sequencing analysis of VDR binding locations across the complete genome has significantly expanded our understanding of the actions of vitamin D and VDR on gene transcription. However, these studies have also promoted appreciation of the extra-transcriptional impact of vitamin D on gene expression. It is now clear that vitamin D interacts with the epigenome via effects on DNA methylation, histone acetylation, and microRNA generation to maintain normal biological functions. There is also increasing evidence that vitamin D can influence pre-mRNA constitutive splicing and alternative splicing, although the mechanism for this remains unclear. Pre-mRNA splicing has long been thought to be a post-transcription RNA processing event, but current data indicate that this occurs co-transcriptionally. Several steroid hormones have been recognized to coordinately control gene transcription and pre-mRNA splicing through the recruitment of nuclear receptor co-regulators that can both control gene transcription and splicing. The current review will discuss this concept with specific reference to vitamin D, and the potential role of heterogeneous nuclear ribonucleoprotein C (hnRNPC), a nuclear factor with an established function in RNA splicing. hnRNPC, has been shown to be involved in the VDR transcriptional complex as a vitamin D-response element-binding protein (VDRE-BP), and may act as a coupling factor linking VDR-directed gene transcription with RNA splicing. In this way hnRNPC may provide an additional mechanism for the fine-tuning of vitamin D-regulated target gene expression.
Keywords: Vitamin D, RNA, Transcription, Splicing, Heterogenous nuclear ribonucleoprotein C
1. Introduction
Alternative splicing provides an efficient mechanism by which the genome of any given organism can greatly expand its protein repertoire. The aim of this review is to describe our current understanding of the link between alternative splicing and vitamin D. The review will initially focus on the canonical transcriptional actions of vitamin D, but will then explore the interaction between vitamin D and a key component of the machinery associated with the maintenance of splicing fidelity, heterogeneous nuclear ribonucleoprotein C (hnRNPC). In this way, the review will highlight a potentially important new mechanism for further expansion of the genomic actions of vitamin D.
1.1. Genomic actions of the vitamin D receptor
1α,25-dihydroxyvitamin D [1,25(OH)2D], the biologically active metabolite of vitamin D, performs the majority of its biological functions by regulating gene transcription through a nuclear high-affinity vitamin D receptor (VDR), a member of the superfamily of nuclear receptors that bind steroid hormones and other lipophilic ligands [1,2]. Upon ligand (1,25(OH)2D) binding, VDR heterodimerization occurs with the nuclear retinoid X receptor (RXR) [3]. The resulting VDR–RXR complex can then bind to specific DNA sequences, termed vitamin D-response elements (VDREs) within proximal or distal promoter regions of target genes [4,5]. After binding to a VDRE, a variety of VDR-interacting nuclear proteins (co-regulators) are recruited to the pre-initiation transcriptional complex, which act to enhance or suppress the rate of gene transcription by the liganded VDR [6,7]. Other factors that add to the diversity of VDR-mediated regulation of gene expression include the presence of distal enhancer elements [8], as well as the potential for ligand-independent actions of VDR [9].
As the only high-affinity receptor for 1,25(OH)2D, the VDR mediates genomic responses to 1,25(OH)2D [10]. Recent developments in deep sequencing technologies for unbiased analysis of VDR binding loci have expanded our understanding of the genome-wide actions of 1,25(OH)2D-VDR. Chromatin immunoprecipitation (ChIP) followed by DNA sequencing of the immunoprecipitated products, ChIP-sequencing (ChIP-Seq), using a variety of cell types has shown that there are between 1000 and 13,000 VDR-specific genomic binding sites [11,12]. The majority of these VDR binding sites appear to be distal to the target gene transcriptional start site, being located in either intergenic regions or within introns. This has had a dramatic effect on our understanding of transcriptional regulation by the 1,25(OH)2D-VDR complex. Most VDR-binding DNA loci have yet to be validated by specific quantitative PCR analysis of ChIP products, but, recent work by Pike and co-workers has confirmed that 1,25(OH)2D regulation of the genes for 24-hydroxylase (CYP24A1) and RANK ligand (RANKL), involves both proximal promoter VDREs and a complex set of downstream or upstream distal VDREs [13–15].
Genomic binding of VDR is also highly cell-specific. Tuoresmaki et al. recently conducted a re-analysis of publically available VDR ChIP-seq datasets for six different cell types. These datasets revealed a total of 23,409 non-overlapping genomic VDR binding sites, with a majority of these VDR loci (17,700) being unique to each of the analyzed cell types, and only 43 binding siteswere shared by all six cell types, emphasizing the cell-specificity of VDREs [16]. However, the underlying mechanisms that define cell-specific patterns of VDR binding remain unclear, and may involve alternative accessory factors for VDR. Intriguingly more than 60% of genomic VDR binding sites do not contain a canonical hexameric VDRE sequence direct-repeat (DR) 3 with two viable six-base half-elements separated by three base pairs with consensus sequence RGGTCAnnnRGTTCA, (r = A or G), suggesting that VDR may use alternative mechanisms to interact with genomic DNA [16]. For example, liganded VDR may partner with currently undefined partner proteins or interface with other DNA-binding transcription factors, such as pioneer factors.
These alternative binding mechanisms may explain some of the cell-specific actions of VDR as well as its repressive functions on gene transcription [11]. However, there is now strong evidence supporting the contribution of other mechanisms that diversify the effects of vitamin D on the transcriptome and proteome. For example, epigenetic modifications are known to play a key role in the maintenance of VDR-directed gene expression and dysregulation of these mechanisms can lead to pathological conditions [17,18]. The impact of epigenetics on VDR signaling has been well defined for chromatin remodeling via DNA methylation and histone acetylation, with VDR co-activators and co-repressors interfacing with chromatin modifiers and remodelers [6,19,20]. In addition, recent studies have also implicated microRNAs (miRNAs) in mediating the fine-tuning of vitamin D-mediated responses [21–23]. The liganded VDR complex can either suppress or induce miRNAs by either direct transcriptional regulation of autonomous miRNA genesor indirect regulation of miRNAs via host gene promoter sequences [24]. Conversely, miRNAs may act to regulate 1,25(OH)2D synthesis, catabolism, or signaling to form dynamic feedback mechanisms (comprehensive review, see [24]).
The current manuscript will review another mechanism with the potential to influence vitamin D regulated gene expression – namely RNA splicing, and alternative splicing. In particular, this review will focus on the potential role of hnRNPC, a key nuclear factor in post-transcriptional RNA-processing, as a mediator of vitamin D receptor-directed transcription and RNA splicing.
2. Vitamin D and RNA splicing
2.1. Pre-mRNA splicing and alternative splicing overview
In humans and other complex metazoans, the vast majority of protein-coding genes contain several exons separated by introns that will not appear in the mature mRNA. Removal of introns and the ligation of exons that contain the protein-coding open reading frame and the 5′ and 3′ untranslated regions (UTRs) is accomplished by pre-mRNA splicing, a process which is facilitated by a complex of small nuclear RNAs (snRNAs), splicing factors, and numerous RNA-binding proteins that collectively form the spliceosome [25]. Nuclear pre-mRNA splicing entails two consecutive trans-esterification reactions. First, the 2′-hydroxyl of an adenosine of the branch point sequence in the intron carries out a nucleophilic attack on the phosphodiester bond at the 5′ splicing site (SS). This results in cleavage at this site and ligation of the 5′ end of the intron to the branch adenosine, forming a free 5′ exon and a lariat structure. Subsequently, the phosphodiester bond at the 3′ SS is attacked by the 3′-hydroxyl of the 5′ exon, leading to the ligation of the 5′ and 3′ exons and release of the lariat intron.
During splicing, the spliceosome dynamically, and in a stepwise fashion, assembles and disassembles across the pre-mRNA to direct the correct recognition and pairing of the splice sites [26]. Whilst some exons are constitutively spliced, in that they are included in every mRNA produced from a given pre-mRNA, many others are alternatively spliced to generate variable forms of mRNA from a single pre-mRNA transcript [27]. The capacity for alternative splicing in eukaryotes has greatly enhanced transcriptome and proteome diversity leading to a higher order of organismal complexity but without the need for expansion of the genome. Importantly, alternative splicing can also be regulated differently according to cell type, developmental stage, or signal-dependent patterns [25,28].
2.2. Alternative RNA splicing and the vitamin D system
Previous studies linking vitamin D and alternative splicing have focused on the metabolism of vitamin D (Table 1). Published reports have described splice variant mRNA transcripts for the vitamin D-activating enzyme 25-hydroxyvitamin D-1α-hydroxylase (CYP27B1) [29–35]. Data from these studies have underlined a potential role for CYP27B1 splice variants in regulating localized synthesis of 1,25(OH)2D [32], notably in the context of tissue-specific regulation of this active from of vitamin D [31]. Other studies have suggested that dysregulation of CYP27B1 splice variant expression may contribute to the development of neoplasias via effects on cell-specific synthesis of 1,25(OH)2D [29,30,36]. The underlying mechanisms for this have yet to be defined, as reported splice variants of CYP27B1 appear to be expressed in very low abundance, and do not appear to encode functional proteins [32]. Nevertheless, knockdown of some of these CYP27B1 splice variants in human kidney cells using RNA-interference resulted in enhanced conversion of 25-hydroxyvitamin D (25OHD) to 1,25(OH)2D [32], suggesting a functional role for RNA splicing with respect to vitamin D metabolism. In this context the most informative gene for vitamin D and alternative splicing appears to be the vitamin D-catabolic enzyme 24-hydroxylase (CYP24A1), which is potently induced in cells following exposure to 1,25(OH)2D [37,38].
Table 1.
Overview of splice variants of the CYP24A1 and CYP27B1 genes.
| Gene | Transcript variant |
Organism/cell | Protein coding |
Enzymatic activity |
Biological meaning | Reference | |
|---|---|---|---|---|---|---|---|
| CYP24A1 | CYP24A1- wild type |
Full length | All VDR-expressing cells |
Yes | Yes | 23- and 24-hydroxylation of 25(OH)D/1,25(OH)2D | [38] |
| – | CYP24A1- SV |
Deletion exons 1 and 2, insertion part of intron 2 |
Chick macrophage cell line, Human/kidney, placenta, skin, and macrophages |
Yes | No | Suppress synthesis of 1,25(OH)2D as a cytosolic decoy for CYP24A1/CYP27B1 substrates |
[39–41] |
| CYP24A1- SV2 |
Deletion exons 1 and 2 | Human/colon cancer cells |
Unknown | No | Unknown | ||
| CYP24A1- SV3 |
Deletion exon 10 | Human/colon cancer cells |
Unknown | No | Unknown | ||
| - | |||||||
| CYP27B1 | CYP27B1- wild type |
Full length | Human/renal or extra- renal cells |
Yes | Yes | 1α-Hydroxylation of 25(OH)D | [38] |
| SV1 | Contains part of intron 2, exons 6–9 |
Human/kidney cell lines |
Noncoding | – | Predicted function: coordinate localized synthesis of 1,25(OH)2D by limiting the availability of the wild type 1α-hydroxylase enzyme activity |
[32] | |
| SV2 | Contains part of intron 2, part of exon 6, exons 7–9 |
Human/kidney cell lines |
Noncoding | – | |||
| SV3 | Contains part of intron 2, exon 3, part of intron 3, part of exon 8 and exon 9 |
Human/kidney cell lines |
Noncoding | – | |||
| SV4 | Contains part of intron 2, exon 3, intron 3, part of exon 4 and exon 9 |
Human/kidney cell lines |
Noncoding | – | |||
| SV5 | Contains part of intron 2, exons 3–4 and exon 9 |
Human/kidney cell lines |
Noncoding | – | |||
| SV6 | Contains intron 2, exons 3–5, intron 5 and part of exon 6 |
Human/ myelomonocytic cell lines |
Noncoding | – | |||
| SV7 | Contains part of intron 2, exons 3–5, part of exon 6 and part of exon 7 |
Human/ myelomonocytic cell lines |
Noncoding | – | |||
| SV8 | Contains part of intron 2, part of exon 4, exon 5, intron 5, exon 6 and part of exon 7 |
Human/ myelomonocytic cell lines |
Noncoding | – | |||
| SV9 | Contains part of intron 2, part of exon 4, exons 5–6 and part of exon 7 |
Human/ myelomonocytic cell lines |
Noncoding | – | |||
| SV10 | Contains part of intron 2, part of intron 5, exon 6, and part of exon 7 |
Human/ myelomonocytic cell lines |
Noncoding | – | |||
| Hyd-V1 | Insertion part of intron 2, deletion exons 4 and 5 |
Human/GBM cell lines | Yes | No | Predicted function: reduce 1α-hydroxylase enzyme activity; contribute to the development of neoplasias via effects on cell-specific synthesis of 1,25(OH)2D; potential as a diagnostic marker for cancer |
[29– 31,33,36] | |
| Hyd-V2 | Deletionexons 4 and 5 | Human/GBM, melanoma, cervix carcinoma and kidney cell lines |
Yes | No | |||
| Hyd-V3 | Deletion of part of exon 8 | Human/GBM, melanoma, cervix carcinoma and kidney cell lines |
Yes | No | |||
| Hyd-V4 | Insertion part of intron 2 | Human/GBM, melanoma, cervix carcinoma and kidney cell lines |
Yes | No | |||
| Hyd-V5 | Insertion part of intron 5 | Human/GBM, melanoma, cervix carcinoma and kidney cell lines |
Yes | No | |||
| Hyd-V6 | Insertion part of intron 1 | Human/GBM, melanoma, cervix carcinoma and kidney cell lines |
Yes | No | |||
| Hyd-V7 | Insertion part of intron 1, deletion exons 4 and 5 |
Human/GBM, melanoma, cervix carcinoma and kidney cell lines |
Yes | No | |||
| Hyd-V8 | Insertion introns 1 and 5 | Human/GBM, melanoma, cervix carcinoma and kidney cell lines |
Yes | No | |||
| Hyd-V9 | Insertion introns 2 and 6 | Human/GBM cell lines | Yes | No | |||
| Hyd-V10 | Insertion introns 1 and 2 | Human/GBM cell lines | Yes | No | |||
| Hyd-V11 | Insertion introns 1 and 3 | Human/GBM cell lines | Yes | No | |||
| Hyd-V12 | Insertion introns 1, 3 and 5 |
Human/GBM cell lines | Yes | No | |||
| Hyd-V13 | Insertion introns 1, 2 and 5 |
Human/GBM cell lines | Yes | No | |||
| Hyd-V14 | Insertion introns 1, 3 and 6, 7 |
Human/GBM cell lines | Yes | No | |||
| Hyd-V15 | Insertion intron s1, 2 and 3, 5 |
Human/GBM cell lines | Yes | No | |||
| Hyd-V16 | Insertion introns 2 and 5, deletion exons 6–9 |
Human/GBM cell lines | Yes | No | |||
| – | Insertion intron 1 | Human endometrial cancer cells/breast cancer cells |
Yes | No | Unknown | [34,35] | |
| Deletions exons 3–5 | Human endometrial cancer cells/breast cancer cells |
Yes | No |
GBM: glioblastoma multiforme.
Alternative splicing of CYP24A1 was first reported in the chick HD-11 macrophage cell line [39]. In this instance, exons 1 and 2 of the cyp24a1 gene are spliced out and replaced by a pseudo exon composing part of intron 2 [39]. Theresulting CYP24A1-splice variant (CYP24A1-SV) generates an in-frame shift in mRNA, using an alternative start codon that leads to a protein lacking the mitochondrial-targeting sequence for CYP24A1 which is therefore metabolically inactive. Despite this, CYP24A1-SV appears to play a pivotal role in vitamin D metabolism by acting as a decoy for substrate 25OHD and attenuating endogenous conversion of 25OHD to 1,25(OH)2D by CYP27B1 [39]. Thus, CYP24A1 may be particularly important in cells, such as macrophages, with significant intracrine CYP27B1 activity. Notably CYP24A1-SV has been detected in cancer cells, in particular colorectal and prostate cancer cells [40,41]. The precise impact of the CYP24A1 splice variant in this setting has yet to be fully defined. However, in common with its proposed role in macrophages, this may involve decoy actions on local vitamin D metabolism by these cells. In this way, expression of the variant protein form of CYP24A1 may play a crucial role in the efficacy of local 25OHD metabolism, with overabundance of CYP24A1-SV acting to diminish local concentrations of 1,25(OH)2D at the cell-specific level.
Other studies have shown that vitamin D itself may influence CYP24A splicing. Muindi et al. reported that 1,25(OH)2D can modulate CYP24A1 pre-mRNA splicing in prostate cancer cells in a dose- and time-dependent fashion, although the mechanism for this is unclear [40]. Another recent study showed that 1,25(OH)2Dcan also regulate CYP24A1 splicing in colon cancer cells [42]. In multiple colon cell lines, RNA splicing patterns regulated by 1,25(OH)2D were shown to be associated with cellular sensitivity to 1,25(OH)2D, with more significant induction of CYP24A1 splicing being observed in cells that were more sensitive to 1,25(OH)2D treatment. Regulation of RNA splicing by vitamin D also appears to be gene selective, with vitamin D target genes other than CYP24A1 showing no significant variations in splicing following treatment with 1,25(OH)2D [42]. The underlying mechanism for 1,25(OH)2D-mediated variations in CYP24A1 RNA splicing has yet to be clearly defined, and may involve multiple signaling pathway, such as PKAactivation and c-Jun terminal protein kinase inhibition [42]. Moreover, there is evidence that the role of 1,25(OH)2D in pre-mRNA splicing is neither confined to cancer cells nor to the CYP24A1 gene. Collectively these studies highlight the potential importance of RNA splicing as an alternative facet of vitamin D-mediated regulation of gene expression, and this will be discussed in greater detail in the remainder of the review.
2.3. Nuclear receptor co-transcriptional splicing
It is now clear that transcriptional regulation is physically and functionally integrated with pre-mRNA splicing [43–45]. Such co-transcriptional splicing has been demonstrated in a number of different organisms and has been shown to play a role in coordinating both constitutive and alternative splicing [46]. It has been reported that by recruiting receptor co-regulators that can both control gene transcription and splicing, steroid hormones (in this case progesterone and estrogen) may coordinately control gene transcription and splicing decisions leading to alternatively spliced transcripts [47]. This mechanism is controlled in a steroid nuclear receptor (NR) and promoter-dependent manner [47]. In particular a subset of hormonally recruited NR co-regulators including CoAA [48], CAPERα and CAPERβ [49], SKIP [50], and co-factor of BRCA 1 (COBRA1) [51], have been identified as coupling factors involved in coordinating steroid hormone-activated gene transcription and transcript splicing. This double-duty for the NR and co-regulators, in response to a steroid hormone signal,may act to ensure that the appropriate gene product is generated in the appropriate cell type [52].
In a similar fashion, the VDR co-regulator NcoA62/SKIP can promote RXR-VDR dependent gene transcriptional activation or repression in a cell-specific manner [53]. However, it has also been identified as a non-snRNP component of splicesome complexes, supporting a potential role for NCoA62/SKIP in pre-mRNA splicing [54,55]. Zhang et al. demonstrated that 1,25(OH)2D is able to influence target gene splicing through recruitment of NcoA62/SKIP. In this case the co-activator function of NcoA62/SKIP in the VDR-activated transcriptional process was demonstrated in HeLa cells by showing that NcoA62/SKIP interacts with VDR, and is associated with recruitment of VDR to CYP24A1 VDREs following treatment with 1,25(OH)2D [50]. However, using glutathione S-transferase (GST)-pull down analyses and HeLa cell nuclear extracts, NcoA62/SKIP was also shown to interact with components of the splicing machinery, such as the U5 small nuclear ribonucleoprotein (snRNP) [50]. Moreover, disruption of native NCoA62/SKIP through expression of a dominant-negative NCoA62/SKIP blocked conventional splicing of 1,25(OH)2D-induced RNA transcripts from a VDRE-driven reporter minigene cassette. This resulted in a 1,25(OH)2D-dependent accumulation of unspliced transcripts, suggesting that NcoA62/SKIP is required for correct splicing in VDR-activated gene expression. The mechanism by which NcoA62/SKIP affects RNA splicing has yet to be clarified, but these data suggest that NcoA62/SKIP, a common VDR co-factor, is involved in coupling VDR mediated transcription to RNA splicing [50].
There is currently no direct evidence showing that vitamin D affects nascent mRNA alternative splicing. However, a recent proteomic study identified a number of proteins functioning as splicing factors that were induced by 1,25(OH)2D in colon cancer cells [56]. As detailed above, VDR-interacting proteins with traditional roles as VDR transcriptional co-activators or co-repressors, may also be involved in the regulation of alternative splicing [57]. The remainder of this review will focus on an alternative view of this concept, in which a nuclear factor with an established function in RNA splicing, hnRNPC, has also been shown to be involved in the VDR transcriptional complex as a VDRE-binding protein [58].
3. HnRNPC1/C2, RNA splicing, and VDR-mediated transcription
The protein complex known as hnRNPC is a member of a subfamily of hnRNPs that act as RNA binding proteins by complexing with heterogeneous nuclear RNA. Transcript variants of the hnRNPC gene encode two major isoforms (C1 and C2) that form an hnRNPC1/C2 heterotetramer consisting of 3 hnRNPC1 and 1 hnRNPC2 subunits [59,60]. Although hnRNPC1/C2 was identified over 30 years ago as a core component of 40S hnRNP particles, whichform on all nascent RNA transcripts [61], conflicting reports still exist in regard to the sequence specificity and mode of hnRNPC1/C2 binding to RNA [62–65]. A recent study using in vivo crosslinking and immunoprecipitation followed by deep sequencing confirmed that hnRNPC1/C2 is predominantly positioned on pre-mRNAs via sequence-specific binding to neighboring uridine tracts of its RNA recognition motif (RRM) domains, with a defined spacing of 165 and 300 nucleotides [66]. In this study it was hypothesized that hnRNPC1/C2 act as an ‘RNA nucleosome’ that incorporates long regions of nuclear pre-mRNA [66]. This is consistent with the several previously reported functions of hnRNPC1/C2 in post-transcriptional RNA processing including RNA packaging [67,68], constitutive and alternative splicing [69–72] and RNA export [73].
3.1. HnRNPC1/C2 and RNA splicing
An essential role for hnRNPC1/C2 in pre-mRNA splicing was identified over 20 years ago [69,70]. However, more recent studies have described a versatile function for hnRNPC1/C2 in alternative splicing. Knockdown of hnRNPC1/C2 by small interfering RNAs (siRNAs) promoted exon skipping or the use of more internal 3′-splice sites, suggesting that hnRNPC1/C2 acts as a splicing enhancer by enforcing exon inclusion in a cell type-dependent manner [71]. Another study described an opposite role for hnRNPC1/C2 in regulating alternative pre-mRNA splicing to promote exon exclusion [74]. In this latter case cooperative interaction of hnRNPC1/C2 and human antigen R (HuR), an RNA binding protein which has been previously identified as an alternative splicing regulator in mammalian cells [74], promoted Fas cell surface death receptor (Fas) gene exon 6 skipping through binding to the exon splicing silencer (ESS) URE6. This, in turn, inhibited molecular events leading to exon definition, and subsequent exon skipping [72].
More recent approaches utilizing newly developed techniques such as individual-nucleotide resolution UV cross-linking and immunoprecipitation (iCLIP)[75], have been able to determine more precise locations of hnRNPC1/C2 binding to nascent transcripts and provide insights into the mechanism of hnRNPC1/C2 regulated alternative splicing [66,76]. Using iCLIP followed by deep sequencing, Konig et al. characterized a precise transcriptome-wide binding pattern of hnRNPC1/C2 at single-nucleotide resolution. These studies determined that hnRNPC1/C2 can promote either exon exclusion or inclusion, depending on the exact binding location of hnRNPC1/C2 on the nascent mRNA. Alternative exons were skipped by direct binding of the exon to the RNPC1/C2 tetramer, while binding of the hnRNPC1/C2 tetramer to the preceding intron enhanced the inclusion of alternative exons [66]. Another recent study by the same team found that hnRNPC1/C2 directly competes with the core splicing factor U2 auxillary factor 65 (U2AF65), with this mechanism acting to prevent a process known as Alu exonization [76]. Sequences of DNA known as Alu elements are amongst the most transposable of DNA elements in the human genome. However, Alu elements can introduce cryptic splice sites which, if used, lead to Alu exonization. There have been increasing numbers of reports of Alu exonization-associated disorders and the mechanisms that prevent spurious exonization of Alu elements are currently unknown [77]. In this setting hnRNPC1/C2 has been identified as a potential mediator of this protection against spurious exonization [76].
3.2. HnRNPC1/C2 and vitamin D resistance
Despite their classical interaction with chromatin-associated single-stranded RNA, several studies have shown that hnRNPC1/C2 is a pluripotent protein complex with the potential for alternative modes of action [58,78–80]. In particular, previous studies from our group have demonstrated a role for hnRNPC1/C2 as an additional component of the VDR transcription complex, in which hnRNPC1/C2 functions as a trans-regulatory factor via interaction with double-stranded DNA cis-elements[79,81,82].
A role for hnRNPC1/C2 in vitamin D-mediated signaling was first reported following an outbreak of rachitic bone disease in the New World Primate (NWP) emperor tamarin (Saguinas imperator) colony at the Los Angeles Zoo [83]. Analysis of epidermal cells from these animal showed that the NWPs were protected against potential adverse effects of sustained exposure to high circulating levels of 1,25(OH)2D by elevated cellular expression of a protein that binds to VDR target gene promoter VDREs [79,81]. Overabundance of this so-called “VDRE-binding protein” (VDRE-BP) appears to attenuate vitamin D receptor-mediated gene expression by blocking the binding of 1,25(OH)2D-liganded VDR to target gene promoter VDREs. This occurs via direct binding of the VDRE-BP to the VDRE DNA half-sites, and has been proposed as the underlying cause of 1,25(OH)2D-insensitivity in NWPs [81,84]. Subsequent studies showed that a VDRE-BP was identical to hnRNPC1/C2 that was over-expressed in a human subject with a form of hereditary vitamin D resistant rickets (HVDRR) [79,82,85]. In this case, ChIP analyses confirmed the ability of hnRNPC1/C2 to compete in trans with VDR for occupation of the VDRE in human cells. In normal 1,25(OH)2D responsive human cells, the hnRNPC1/C2 occupying the CYP24A1 VDRE at baseline was then displaced from the promoter VDRE by the 1,25(OH)2D liganded VDR–RXR complex [79]. This reciprocal relationship between VDR and hnRNPC1/C2 occurs in a time-dependent cyclical fashion following exposure to 1,25(OH)2D, suggesting that hnRNPC1/C2 may be a key determinant of the temporal patterns of VDRE occupancy [79].In hnRNPC1/C2 over-expressing fibroblasts and B cells from the HVDRR patient, the reciprocal association of VDR and hnRNPC1/C2 with the VDRE was distorted relative to control vitamin D-responsive cells [79]. In a similar fashion, overexpression of hnRNPC1/C2 in vitro has been shown to disrupt the relationship between VDR and hnRNPC1/2, leading to suppression of 1,25(OH)2D-induced gene expression [79]. These data suggest that vitamin D resistance in the case of the HVDRR patient was similar to that previously described for NWPs [58,79].
3.3. hnRNPC1/C2 – a link between VDR-mediated transcription and alternative splicing?
Although there is evidence for effects of 1,25(OH)2D on pre-mRNA splicing, the mechanism for this is unclear. Because of its capacity to interact with nascent pre-mRNA as well as double-strand DNA, we hypothesize that hnRNPC1/C2 may play a key role in linking 1,25(OH)2D-induced transcription with RNA splicing, thereby providing an additional mechanism for fine-tuning of gene expression (see Fig. 1). This hypothesis is supported by the increasing evidence for functional integration between transcription and splicing machinery in mammalian cells [86]. This dual role for hnRNPC1/C2 may help to ensure more precise regulation of gene expression in response to vitamin D, in a manner that is more efficient for cellular energy. It is well recognized that the biosynthetic and metabolic energy cost of RNA splicing and the surveillance required to eliminate imprecisely spliced mRNAs is very high, as is the potential harm from defects in these processes [25].
Fig. 1.
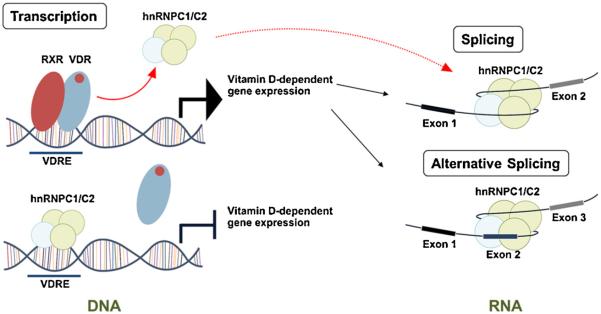
DNA and RNA binding functions of hnRNPC1/C2 and the action of 1,25(OH)2D. Schematic representation of the ability of hnRNPC1/C2 to act as: (1) a vitamin D-response element-binding protein (VDRE-BP) in the absence of liganded (1,25(OH)2D-bound) vitamin D receptor (VDR). In the presence of VDR-1,25(OH)2D, hnRNPC1/C2 is displaced from the VDRE; (2) a component of the RNA spliceosome, facilitating either exon exclusion or exon-inclusion. As yet it is unclear whether these two facets of hnRNPC1/C2 function are linked.
A key question to be answered in future studies is whether the hnRNPC1/C2 displaced from VDRE by liganded VDR is involved in facilitating RNA splicing. If this is the case, then another question arises as to how hnRNPC1/C2 migrates from gene promoter regions to a specific RNA splice site to direct pre-mRNA splicing or alternative splicing. Recent studies have shown that a ‘mediator complex’ may function to “hand over” a splicing regulator, heterogeneous nuclear ribonucleoprotein L (hnRNPL), from gene promoters to the elongating RNA polymerase II complex, thereby mediating specific splice site selection during co-transcription splicing [87]. Moreover, as outlined above, VDR co-activators or co-repressors may also function as splicing factors and, as such,these may also act as coupling factors linking transcription and RNA splicing responses to vitamin D. Further analysis of vitamin D signaling, hnRNPC expression and alternative splicing of RNA will broaden our perspective on how vitamin D is able to influence gene expression during normal physiology. However, alternative splicing may also be a contributor to human disease, and it in future studies it will also be important to assess the impact of vitamin D status on hnRNPC function and normal RNA splicing.
Acknowledgements
This work was supported by NIH grants AR037399 and AR063910, and by a scholarship from the China Scholarship Council (CSC).
References
- 1.Haussler MR, Whitfield GK, Kaneko I, Haussler CA, Hsieh D, Hsieh JC, Jurutka PW. Molecular mechanisms of vitamin D action. Calcif. Tissue Int. 2013;92:77–98. doi: 10.1007/s00223-012-9619-0. [DOI] [PubMed] [Google Scholar]
- 2.Haussler MR, Haussler CA, Whitfield GK, Hsieh JC, Thompson PD, Barthel TK, Bartik L, Egan JB, Wu Y, Kubicek JL, Lowmiller CL, Moffet EW, Forster RE, Jurutka PW. The nuclear vitamin D receptor controls the expression of genes encoding factors which feed the fountain of youth to mediate healthful aging. J Steroid Biochem. Mol. Biol. 2010;121:88–97. doi: 10.1016/j.jsbmb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 4.Jurutka PW, Bartik L, Whitfield GK, Mathern DR, Barthel TK, Gurevich M, Hsieh JC, Kaczmarska M, Haussler CA, Haussler MR. Vitamin D receptor: key roles in bone mineral pathophysiology, molecular mechanism of action, and novel nutritional ligands. J. Bone Miner. Res. 2007;22(Suppl. 2):V2–10. doi: 10.1359/jbmr.07s216. [DOI] [PubMed] [Google Scholar]
- 5.Pike JW, Meyer MB, Bishop KA. Regulation of target gene expression by the vitamin D receptor – an update on mechanisms. Rev. Endocr. Metab. Disord. 2012;13:45–55. doi: 10.1007/s11154-011-9198-9. [DOI] [PubMed] [Google Scholar]
- 6.Carlberg C, Campbell MJ. Vitamin D receptor signaling mechanisms: integrated actions of a well-defined transcription factor. Steroids. 2013;78:127–136. doi: 10.1016/j.steroids.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer MB, Pike JW. Corepressors (NCoR and SMRT) as well as coactivators are recruited to positively regulated 1alpha Pike, 25-dihydroxyvitamin D3-responsive genes. J. Steroid Biochem. Mol. Biol. 2013;136:120–124. doi: 10.1016/j.jsbmb.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martowicz ML, Meyer MB, Pike JW. The mouse RANKL gene locus is defined by a broad pattern of histone H4 acetylation and regulated through distinct distal enhancers. J. Cell. Biochem. 2011;112:2030–2045. doi: 10.1002/jcb.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skorija K, Cox M, Sisk JM, Dowd DR, MacDonald PN, Thompson CC, Demay MB. Ligand-independent actions of the vitamin D receptor maintain hair follicle homeostasis. Mol. Endocrinol. 2005;19:855–862. doi: 10.1210/me.2004-0415. [DOI] [PubMed] [Google Scholar]
- 10.Dusso AS, Brown AJ, Slatopolsky E. Vitamin D. Am. J. Physiol. Renal. Physiol. 2005;289:F8–28. doi: 10.1152/ajprenal.00336.2004. [DOI] [PubMed] [Google Scholar]
- 11.Carlberg C. Genome-wide (over) view on the actions of vitamin D. Front. Physiol. 2014;5:167. doi: 10.3389/fphys.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer MB, Goetsch PD, Pike JW. Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone. J. Steroid Biochem. Mol. Biol. 2010;121:136–141. doi: 10.1016/j.jsbmb.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1α, 25-dihydroxyvitamin D3. J. Biol. Chem. 2010;285:15599–15610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-κB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol. Cell. Biol. 2006;26:6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nerenz RD, Martowicz ML, Pike JW. An enhancer 20 kilobases upstream of the human receptor activator of nuclear factor-kappaB ligand gene mediates dominant activation by 1,25-dihydroxyvitamin D3. Mol. Endocrinol. 2008;22:1044–1056. doi: 10.1210/me.2007-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuoresmaki P, Vaisanen S, Neme A, Heikkinen S, Carlberg C. Patterns of genome-wide VDR locations Carlberg. PLoS One. 2014;9:e96105. doi: 10.1371/journal.pone.0096105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh PK, Doig CL, Dhiman VK, Turner BM, Smiraglia DJ, Campbell MJ. Epigenetic distortion to VDR transcriptional regulation in prostate cancer cells. J. Steroid Biochem. Mol. Biol. 2012 doi: 10.1016/j.jsbmb.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banwell CM, MacCartney DP, Guy M, Miles AE, Uskokovic MR, Mansi J, Stewart PM, O'Neill LP, Turner BM, Colston KW, Campbell MJ. Altered nuclear receptor corepressor expression attenuates vitamin D receptor signaling in breast cancer cells. Clin. Cancer Res. 2006;12:2004–2013. doi: 10.1158/1078-0432.CCR-05-1218. [DOI] [PubMed] [Google Scholar]
- 19.Pike JW, Meyer MB. Fundamentals of vitamin D hormone-regulated gene expression. J. steroid Biochem. Mol. Biol. 2013 doi: 10.1016/j.jsbmb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira F, Barbachano A, Singh PK, Campbell MJ, Munoz A, Larriba MJ. Vitamin D has wide regulatory effects on histone demethylase genes. Cell Cycle. 2012;11:1081–1089. doi: 10.4161/cc.11.6.19508. [DOI] [PubMed] [Google Scholar]
- 21.Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, Mehta MD, Vazirnia A, Rea TH, Sarno EN, Graeber TG, Modlin RL. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 2012;18:267–273. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang WL, Chatterjee N, Chittur SV, Welsh J, Tenniswood MP. Effects of 1α,25 dihydroxyvitamin D3 and testosterone on miRNA and mRNA expression in LNCaP cells. Mol. Cancer. 2011;10:58. doi: 10.1186/1476-4598-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisse TS, Chun RF, Rieger S, Adams JS, Hewison M. Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts. J. Bone Miner. Res. 2013;28:1478–1488. doi: 10.1002/jbmr.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lisse TS, Adams JS, Hewison M. Vitamin D and MicroRNAs in Bone. Crit. Rev. Eukaryot Gene Expr. 2013;23:195–214. doi: 10.1615/critreveukaryotgeneexpr.2013007147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Lu ZX, Jiang P, Xing Y. Genetic variation of pre-mRNA alternative splicing in human populations. Wiley Interdiscip. Rev. RNA. 2012;3:581–592. doi: 10.1002/wrna.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diesel B, Fischer U, Meese E. Gene amplification and splice variants of 25-hydroxyvitamin D3 1 Meese, alpha-hydroxylase (CYP27B1) in glioblastoma multiforme – a possible role in tumor progression? Recent Results Cancer Res. 2003;164:151–155. doi: 10.1007/978-3-642-55580-0_11. [DOI] [PubMed] [Google Scholar]
- 30.Diesel B, Radermacher J, Bureik M, Bernhardt R, Seifert M, Reichrath J, Fischer U, Meese E. Vitamin D(3) metabolism in human glioblastoma multiforme: functionality of CYP27B1 splice variants, metabolism of calcidiol, and effect of calcitriol. Clin. Cancer Res. 2005;11:5370–5380. doi: 10.1158/1078-0432.CCR-04-1968. [DOI] [PubMed] [Google Scholar]
- 31.Flanagan JN, Wang L, Tangpricha V, Reichrath J, Chen TC, Holick MF. Regulation of the 25-hydroxyvitamin D-1α-hydroxylase gene and its splice variant. Recent Results Cancer Res. 2003;164:157–167. doi: 10.1007/978-3-642-55580-0_12. [DOI] [PubMed] [Google Scholar]
- 32.Wu S, Ren S, Nguyen L, Adams JS, Hewison M. Splice variants of the CYP27b1 gene and the regulation of 1 Hewison, 25-dihydroxyvitamin D3 production. Endocrinology. 2007;148:3410–3418. doi: 10.1210/en.2006-1388. [DOI] [PubMed] [Google Scholar]
- 33.Mitschele T, Diesel B, Friedrich M, Meineke V, Maas RM, Gartner BC, Kamradt J, Meese E, Tilgen W, Reichrath J. Analysis of the vitamin D system in basal cell carcinomas (BCCs) Lab. Invest. J. Techn. Method Pathol. 2004;84:693–702. doi: 10.1038/labinvest.3700096. [DOI] [PubMed] [Google Scholar]
- 34.Fischer D, Thome M, Becker S, Cordes T, Diedrich K, Friedrich M, Thill M. Expression of 25-hydroxyvitamin D3–24-hydroxylase in benign and malignant ovarian cell lines and tissue. Anticancer Res. 2009;29:3635–3639. [PubMed] [Google Scholar]
- 35.Cordes T, Diesing D, Becker S, Fischer D, Diedrich K, Friedrich M. Expression of splice variants of 1alpha-hydroxylase in mcf-7 breast cancer cells. J. Steroid Biochem. Mol. Biol. 2007;103:326–329. doi: 10.1016/j.jsbmb.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 36.Radermacher J, Diesel B, Seifert M, Tilgen W, Reichrath J, Fischer U, Meese E. Expression analysis of CYP27B1 in tumor biopsies and cell cultures. Anticancer Res. 2006;26:2683–2686. [PubMed] [Google Scholar]
- 37.Omdahl MB. The 25-Hydroxyvitamin D-24-Hydroxylase. Elsevier; New York: 2005. [Google Scholar]
- 38.Jones G, Prosser DE, Kaufmann M. Cytochrome P450-mediated metabolism of vitamin D. J. Lipid Res. 2014;55:13–31. doi: 10.1194/jlr.R031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren S, Nguyen L, Wu S, Encinas C, Adams JS, Hewison M. Alternative splicing of vitamin D-24-hydroxylase: a novel mechanism for the regulation of extrarenal 1,25-dihydroxyvitamin D synthesis. J. Biol. Chem. 2005;280:20604–20611. doi: 10.1074/jbc.M414522200. [DOI] [PubMed] [Google Scholar]
- 40.Muindi JR, Nganga A, Engler KL, Coignet LJ, Johnson CS, Trump DL. CYP24 splicing variants are associated with different patterns of constitutive and calcitriol-inducible CYP24 activity in human prostate cancer cell lines. J. Steroid. Biochem. Mol. Biol. 2007;103:334–337. doi: 10.1016/j.jsbmb.2006.12.060. [DOI] [PubMed] [Google Scholar]
- 41.Horvath HC, Khabir Z, Nittke T, Gruber S, Speer G, Manhardt T, Bonner E, Kallay E. CYP24A1 splice variants – implications for the antitumorigenic actions of 1,25-(OH) 2D3 in colorectal cancer. J. Steroid Biochem. Mol. Biol. 2010;121:76–79. doi: 10.1016/j.jsbmb.2010.03.080. [DOI] [PubMed] [Google Scholar]
- 42.Peng X, Tiwari N, Roy S, Yuan L, Murillo G, Mehta RR, Benya RV, Mehta RG. Regulation of CYP24 splicing by 1,25-dihydroxyvitamin D(3) in human colon cancer cells. J. Endocrinol. 2012;212:207–215. doi: 10.1530/JOE-11-0305. [DOI] [PubMed] [Google Scholar]
- 43.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 44.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. U. S. A. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merkhofer EC, Hu P, Johnson TL. Introduction to cotranscriptional RNA splicing. Method Mol. Biol. 2014;1126:83–96. doi: 10.1007/978-1-62703-980-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auboeuf D, Honig A, Berget SM, O’Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 48.Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, O’Malley BW. CoAA a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 2004;24:442–453. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O’Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 50.Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, Stein GS, MacDonald PN. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 2003;278:35325–35336. doi: 10.1074/jbc.M305191200. [DOI] [PubMed] [Google Scholar]
- 51.Sun J, Blair AL, Aiyar SE, Li R. Cofactor of BRCA1 modulates androgen-dependent transcription and alternative splicing. J. Steroid Biochem. Mol. Biol. 2007;107:131–139. doi: 10.1016/j.jsbmb.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auboeuf D, Dowhan DH, Dutertre M, Martin N, Berget SM, O'Malley BW. A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol. Cell. Biol. 2005;25:5307–5316. doi: 10.1128/MCB.25.13.5307-5316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leong GM, Subramaniam N, Issa LL, Barry JB, Kino T, Driggers PH, Hayman MJ, Eisman JA, Gardiner EM. Ski-interacting protein, a bifunctional nuclear receptor coregulator that interacts with N-CoR/SMRT and p300. Biochem. Biophys. Res. Commun. 2004;315:1070–1076. doi: 10.1016/j.bbrc.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 55.Ambrozkova M, Puta F, Fukova I, Skruzny M, Brabek J, Folk P. The fission yeast ortholog of the coregulator SKIP interacts with the small subunit of U2AF. Biochem. Biophys. Res. Commun. 2001;284:1148–1154. doi: 10.1006/bbrc.2001.5108. [DOI] [PubMed] [Google Scholar]
- 56.Cristobo I, Larriba MJ, de los Rios V, Garcia F, Munoz A, Casal JI. Proteomic analysis of 1α, 25-dihydroxyvitamin D3 action on human colon cancer cells reveals a link to splicing regulation. J. Proteomics. 2011;75:384–397. doi: 10.1016/j.jprot.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 57.Campbell MJ. Vitamin D and the RNA transcriptome: more than mRNA regulation. Front. Physiol. 2014;5:181. doi: 10.3389/fphys.2014.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lisse, Hewison M, Adams JS. Hormone response element binding proteins: novel regulators of vitamin D and estrogen signaling. Steroids. 2011;76:331–339. doi: 10.1016/j.steroids.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merrill BM, Barnett SF, LeStourgeon WM, Williams KR. Primary structure differences between proteins C1 and C2 of HeLa 40S nuclear ribonucleoprotein particles. Nucleic Acids Res. 1989;17:8441–8449. doi: 10.1093/nar/17.21.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lisse TS, Vadivel K, Bajaj SP, Zhou R, Chun RF, Hewison M, Adams JS. The heterodimeric structure of heterogeneous nuclear ribonucleoprotein C1/C2 dictates 1,25-dihydroxyvitamin D-directed transcriptional events in osteoblasts. Bone Res. 2014;2 doi: 10.1038/boneres.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beyer AL, Christensen ME, Walker BW, LeStourgeon WM. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977;11:127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- 62.Huang M, Rech JE, Northington SJ, Flicker PF, Mayeda A, Krainer AR, LeStourgeon WM. The C-protein tetramer binds 230–240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol. Cell. Biol. 1994;14:518–533. doi: 10.1128/mcb.14.1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McAfee JG, Soltaninassab SR, Lindsay ME, LeStourgeon WM. Proteins C1 and C2 of heterogeneous nuclear ribonucleoprotein complexes bind RNA in a highly cooperative fashion: support for their contiguous deposition on pre-mRNA during transcription. Biochemistry. 1996;35:1212–1222. doi: 10.1021/bi951974k. [DOI] [PubMed] [Google Scholar]
- 64.Gorlach M, Wittekind M, Beckman RA, Mueller L, Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wan L, Kim JK, Pollard VW, Dreyfuss G. Mutational definition of RNA-binding and protein-protein interaction domains of heterogeneous nuclear RNP C1. J. Biol. Chem. 2001;276:7681–7688. doi: 10.1074/jbc.M010207200. [DOI] [PubMed] [Google Scholar]
- 66.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 68.Muller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 69.Choi YD, Grabowski PJ, Sharp PA, Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986;231:1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- 70.Sierakowska H, Szer W, Furdon PJ, Kole R. Antibodies to hnRNP core proteins inhibit in vitro splicing of human beta-globin pre-mRNA. Nucleic Acids Res. 1986;14:5241–5254. doi: 10.1093/nar/14.13.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, Lucier JF, Gervais-Bird J, Tremblay K, Prinos P, Klinck R, Elela SA, Chabot B. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol. Cell. Biol. 2008;28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Izquierdo JM. Heterogeneous ribonucleoprotein C displays a repressor activity mediated by T-cell intracellular antigen-1-related/like protein to modulate Fas exon 6 splicing through a mechanism involving Hu antigen R. Nucleic Acids Res. 2010;38:8001–8014. doi: 10.1093/nar/gkq698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335:1643–1646. doi: 10.1126/science.1218469. [DOI] [PubMed] [Google Scholar]
- 74.Izquierdo JM. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 2008;283:19077–19084. doi: 10.1074/jbc.M800017200. [DOI] [PubMed] [Google Scholar]
- 75.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP – transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J. Vis. Exp. 2011 doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vorechovsky I. Transposable elements in disease-associated cryptic exons. Hum. Genet. 2010;127:135–154. doi: 10.1007/s00439-009-0752-4. [DOI] [PubMed] [Google Scholar]
- 78.Anantha RW, Alcivar AL, Ma J, Cai H, Simhadri S, Ule J, Konig J, Xia B. Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination. PLoS One. 2013;8:e61368. doi: 10.1371/journal.pone.0061368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen H, Hewison M, Adams JS. Functional characterization of heterogeneous nuclear ribonuclear protein C1/C2 in vitamin D resistance: a novel response element-binding protein. J. Biol. Chem. 2006;281:39114–39120. doi: 10.1074/jbc.M608006200. [DOI] [PubMed] [Google Scholar]
- 80.Lisse TS, Liu T, Irmler M, Beckers J, Chen H, Adams JS, Hewison M. Gene targeting by the vitamin D response element binding protein reveals a role for vitamin D in osteoblast mTOR signaling. FASEB J. 2011;25:937–947. doi: 10.1096/fj.10-172577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen H, Hu B, Allegretto EA, Adams JS. The vitamin D response element-binding protein. A novel dominant-negative regulator of vitamin D-directed transactivation. J. Biol. Chem. 2000;275:35557–35564. doi: 10.1074/jbc.M007117200. [DOI] [PubMed] [Google Scholar]
- 82.Chen H, Hewison M, Hu B, Adams JS. Heterogeneous nuclear ribonucleoprotein (hnRNP) binding to hormone response elements: a cause of vitamin D resistance. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6109–6114. doi: 10.1073/pnas.1031395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adams JS, Gacad MA. Phenotypic diversity of the cellular 1,25-dihydroxyvitamin D3-receptor interaction among different genera of New World primates. J. Clin. Endocrinol. Metab. 1988;66:224–229. doi: 10.1210/jcem-66-1-224. [DOI] [PubMed] [Google Scholar]
- 84.Adams JS, Chen H, Chun RF, Nguyen L, Wu S, Ren SY, Barsony J, Gacad MA. Novel regulators of vitamin D action and metabolism: lessons learned at the Los Angeles zoo. J. Cell. Biochem. 2003;88:308–314. doi: 10.1002/jcb.10333. [DOI] [PubMed] [Google Scholar]
- 85.Hewison M, Rut AR, Kristjansson K, Walker RE, Dillon MJ, Hughes MR, O’Riordan JL. Tissue resistance to 1,25-dihydroxyvitamin D without a mutation of the vitamin D receptor gene. Clin. Endocrinol. (Oxf) 1993;39:663–670. doi: 10.1111/j.1365-2265.1993.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 86.Ji X, Fu XD. The mediator couples transcription and splicing. Mol. Cell. 2012;45:433–434. doi: 10.1016/j.molcel.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang Y, Li W, Yao X, Lin QJ, Yin JW, Liang Y, Heiner M, Tian B, Hui J, Wang G. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol. Cell. 2012;45:459–469. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


