Abstract
The Tat system can transport folded, signal peptide-containing proteins (Tat substrates) across energized membranes of prokaryotes and plant plastids. A twin-arginine motif in the signal peptide of Tat substrates is recognized by TatC-containing complexes, and TatA permits the membrane passage. Often, as in the model Tat systems of Escherichia coli and plant plastids, a third component – TatB – is involved that resembles TatA but has a higher affinity to TatC. It is not known why most TatA dissociates from TatBC complexes in vivo and distributes more evenly in the membrane. Here we show a TatBC-independent substrate-binding to TatA from Escherichia coli, and we provide evidence that this binding enhances Tat transport. First hints came from in vivo cross-linking data, which could be confirmed by affinity co-purification of TatA with the natural Tat substrates HiPIP and NrfC. Two positions on the surface of HiPIP could be identified that are important for the TatA interaction and transport efficiency, indicating physiological relevance of the interaction. Distributed TatA thus may serve to accompany membrane-interacting Tat substrates to the few TatBC spots in the cells.
Introduction
The twin-arginine translocation (Tat) system is a general translocation system that serves to translocate folded proteins with N-terminal signal peptides across energized membranes in prokaryotes and plant plastids [1]. Tat signal peptides contain a characteristic amino acid pattern that typically includes the two eponymous arginines [2]. Tat systems minimally consist of the two components TatA and TatC [3]. Often a third component that is sequence-related to TatA is found that has a higher affinity to TatC and that is termed TatB [2]. In E. coli as well as in plant plastids, TatBC-containing complexes recognize the twin-arginine motif and thereby bind the Tat substrates [4,5,6,7]. Some TatA associates with TatBC complexes already before substrate-binding and can be important for high-affinity binding of substrates at physiological conditions [8,9], but additional TatA is recruited or re-organized at the translocon upon substrate-binding [10] and plays an important role when the folded part is transferred through the membrane [10,11,12,13,14,15]. However, most TatA forms homo-oligomeric associations distributed in the membrane, whereas the active translocons are organized in few spots per cell [16,17,18,19]. TatA is a small (usually <10 kDa) membrane protein with a simple topology: a single N-terminal short membrane anchor, followed by an amphipathic helix and a highly charged and most likely largely unstructured C-terminus. The N-terminal membrane anchor and the amphipathic helix are connected by a hinge that contains a highly conserved FG motif. Near the end of the amphipathic helix, an equally conserved FK motif exists [2]. Deep in the lipid bilayer, a contact of the trans-membrane domain of TatA with Tat substrates has been demonstrated to depend on TatBC [20], suggesting that Tat substrates contact the membrane-embedded part of TatA late in transport. In agreement with this finding, the trans-membrane domain of TatA is proposed to allow the passage of the substrate by destabilizing the membrane [11,15]. Recent data showed that recombinant production of Tat substrates results in a recruitment of TatA to the active translocon sites, and the co-localization of TatA at active translocons depends on twin-arginine motif recognition [19]. It would be important to know whether individual membrane-interacting Tat substrates first associate with TatA before they encounter TatBC complexes, as such an interaction could attribute a role to the freely diffusing TatA oligomers. Such a function has been suggested for Streptomyces and Bacillus species [21,22,23], but since no TatBC-independent TatA/Tat substrate interactions were recognized in the model systems E. coli and plant plastids, these reports were so far not taken as evidence for a general TatA functionality.
Here we show in vivo and in vitro that E. coli TatA can bind Tat substrates. This interaction requires the signal peptide in conjunction with determinants of the mature domain surface and appears to be physiologically relevant for efficient transport of various Tat substrates, attributing a targeting function to the freely diffusing TatA oligomers.
Results
TatBC-independent TatA/Tat substrate interactions
We addressed the substrate-translocon interactions of the Tat system by a site-directed in vivo cross-linking method that has been developed by the group of Peter G. Schultz [24,25]. In this method, a specific amber stop codon suppressor tRNA is loaded exclusively with the artificial amino acid p-benzoyl-L-phenylalanine (pBpa), which is efficiently incorporated into engineered amber stop codons of recombinant proteins. Irradiation of the living cells with UV light of 365 nm then activates the pBpa residue that forms a covalent bond to molecules in its immediate environment. We introduced pBpa into selected positions of the model Tat substrate HiPIP from Allochromatium vinosum, which is exclusively Tat dependently translocated in E. coli [26], and used the low copy vector pRK-tatABC to express the tatABC operon ∼15-fold from its natural promoter [17]. After in vivo photo cross-linking, the membranes were prepared and SDS-solubilized, and HiPIP (that was produced with a C-terminal His6-tag) was affinity purified under denaturing conditions. Cross-links of the signal peptide to Tat system components resulted in shifts in SDS-PAGE analyses that were detected using specific antibodies after Western-blotting. A probe of the solubilized membrane sample prior to affinity chromatography was used to compare cross-link efficiencies. UV-induced cross-links eluted in the second and third elution fraction (S1 Fig.). As the method involves a purification step under denaturing conditions, any shifted bands are covalent cross-links that are due to photo-activated pBpa, and, as expected, minus UV-controls showed no cross-links (S1 Fig. shows minus UV-controls for the strongest cross-link position, M26pBpa, and the most intensively used position in this study, A13pBpa). Similarly, wt HiPIP did not cross-link to any wt Tat component in the presence of pBpa and the pEvol-system, as pBpa certainly cannot be incorporated into HiPIP or Tat components when the corresponding genes lack TAG stop codon positions (S1 Fig.).
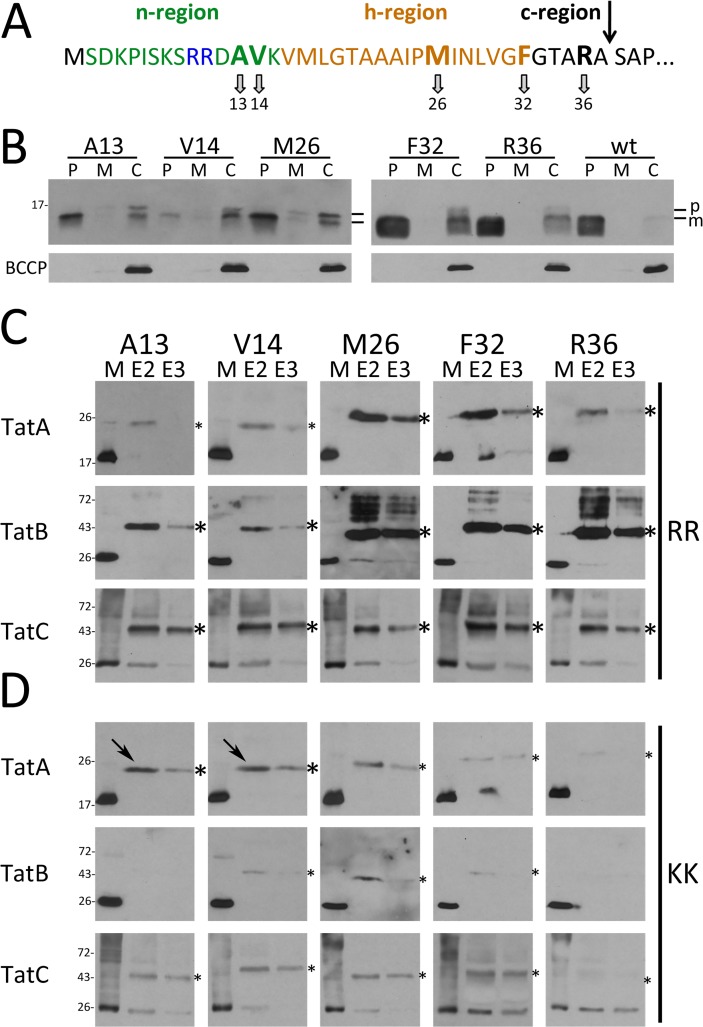
In an initial screen for Tat component interactions, we placed pBpa at selected positions in the twin-arginine motif (A13, V14), the h-region (M26, F32), and the c-region (R36) of the signal peptide (Fig. 1A). HiPIP variants with pBpa at these positions were still transported, as mature HiPIP was clearly detected in the periplasmic fractions of strains carrying the respective HiPIP variants (Fig. 1B). The V14pBpa exchange clearly reduced translocation efficiency. The in vivo cross-link data with signal peptides that contained unaltered twin-arginines demonstrated that all three Tat components are in close proximity to the signal peptide (Fig. 1C). TatA and TatB showed very pronounced cross-links to positions of the h- and c-regions (M26, F32, R36), indicating close substrate contacts. TatC, which is well-known to recognize the twin-arginine motif [4,5], cross-linked to all positions with high efficiency, indicating that the complete signal peptide is in direct contact with TatC under active translocation conditions in vivo, which is in agreement with structural and functional characteristics of TatC [7,27,28,29].
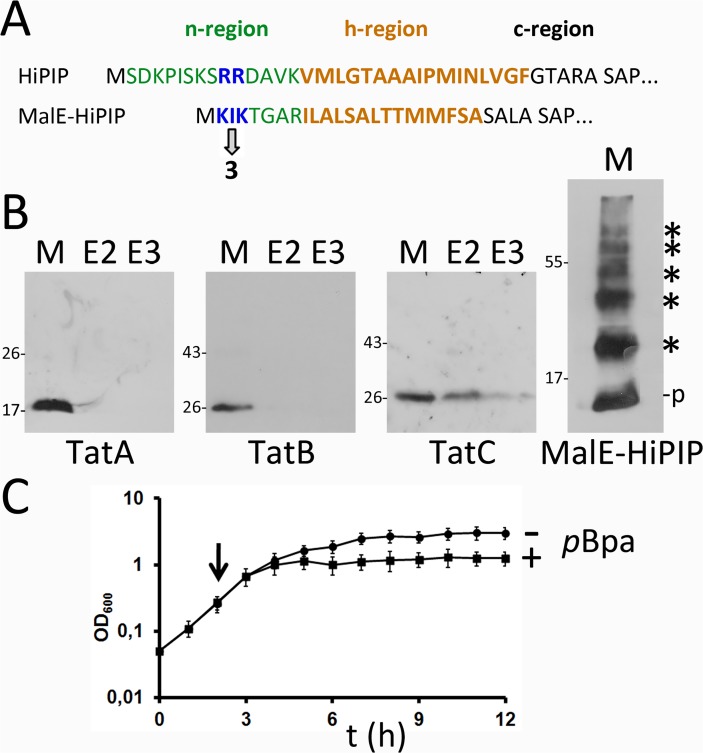
Fig 1. In vivo site-directed cross-linking of Tat signal peptides to the TatABC components.
(A) Amino acid sequence and positions of the pBpa cross-linker in the Tat signal peptide. The arrow indicates the signal peptide cleavage site. (B) Transport of native HiPIP and variants (produced from pEXH5-tac-H6 derivatives) with pBpa exchanges at indicated positions in pEvol-pBpF/pRK-tatABC-containing strains grown in the presence of 0.1 mM pBpa. Detection of HiPIP in subcellular fractions by SDS-PAGE/Western-blotting; the cytoplasmic biotin carboxyl carrier protein (BCCP; ∼22 kDa) was detected to control periplasmic purity. P: periplasm; M: membrane; C: cytoplasm; p: precursor HiPIP; m: mature HiPIP. (C) In vivo cross-links of twin-arginine signal peptides to Tat translocon components with pBpa at indicated positions. Specific cross-links are detected in affinity-chromatography elution fractions E2 and E3 by Western blotting using indicated Tat component-specific antibodies. Molecular weight standards are indicated at the left. (D) Effect of cross-links of RR>KK mutated signal peptides with pBpa exchanges; (strains and conditions in C) and D as in B)). Arrows indicate two TatA-cross-links with unaffected signal intensity. *, strong cross-links; *, weak cross-links; M, solubilized membrane fraction; E, elution fraction.
We then used in vivo cross-linking to analyze the signal peptide interactions with RR>KK mutated Tat motifs (Fig. 1D). This exchange is known to lower dramatically the affinity to TatBC in vitro [4,29,30], resulting in a marked decrease or even block of translocation [26,31,32]. As expected, the KK-variants showed diminished overall cross-links to TatA, TatB or TatC at almost all positions, indicating that RR-motif-binding to the TatBC complex strongly enhances TatABC contacts to these positions. In control experiments, we ensured that RR and KK variants of HiPIP were all produced in comparable amounts (S1 Fig.). Residual contacts of KK-variants to Tat components are expected, as the RR>KK mutation does not fully abolish Tat transport in several test systems [6,33]. In line with this, we found that an optimization of the consensus hydrophobic residue by exchanges A13F as well as by A13pBpa resulted in partial compensation of the translocational block caused by the initial RR>KK exchange (S2 Fig.). An aromatic residue at this position, which corresponds to the F in the consensus motif [1], has been previously shown to strongly enhance TatBC complex binding [5] and our data suggest that this enhanced binding is highly supportive for Tat transport. We were surprised by the clear TatA cross-links and by the fact that some TatA cross-links were not negatively affected by the RR>KK exchange (A13pBpa and V14pBpa; Fig. 1D). Together, these initial data raised the possibility for “basal” signal peptide TatA contacts throughout the signal peptide that are TatBC-independent, while only TatA interactions with the h- and c-regions of the signal peptide are intensified in a TatBC-dependent way that is indicative for later contacts.
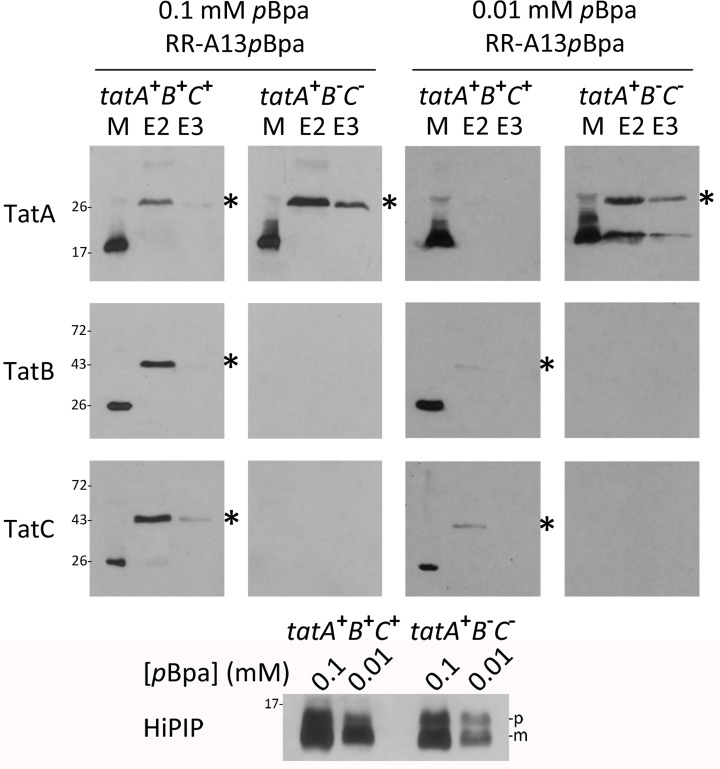
We thus analyzed the contacts in more detail by directly comparing cross-links to TatA at position A13 in the presence and absence of TatBC (Fig. 2). We chose this position, as its contacts to TatA were not enhanced by the RR-motif and as—in contrast to V14pBpa—the A13pBpa variant was nicely translocated (Fig. 1). Again, we included detections of TatB and TatC cross-links in these analyses. Tat substrate cross-links to TatA clearly accumulated in the absence of TatB and TatC, which demonstrates that the TatA interaction can in principle take place prior to TatBC interactions, and that the substrate is not degraded when TatBC interactions cannot take place. When the Tat substrate level was reduced by lowering the pBpa concentration, the TatA cross-links disappeared in the functional Tat background and still accumulated in the TatBC-deficient strain (Fig. 2, right panel). The decrease of substrate-level by lowering [pBpa] is accompanied by increased translational stop, which at this position results in a hydrophilic 12-residues peptide that did not influence the analyses.
Fig 2. The TatA/Tat signal peptide contact does not require the TatBC components.
Comparison of HiPIP-TatA cross-links in the presence/absence of TatBC using pBpa at position A13. Two pBpa concentrations (0.1 mM and 0.01 mM) in the growth medium were used to analyze effects of lowered HiPIP concentration. tatA + B + C +: BW25113/pRK-tatABC/pEvol-pBpF /pEXH5tac-H6-A13pBpa; tatA + B - C -: JBdBC/pRK-tatA/pEvol-pBpF /pEXH5tac-H6-A13pBpa. The bottom blot monitors the decrease of HiPIP levels by reduction of pBpa concentration in the medium. HiPIP detection in crude extracts. Significant HiPIP degradation to mature size is due to unspecific proteolysis of the signal peptide. See Fig. 1 for further details.
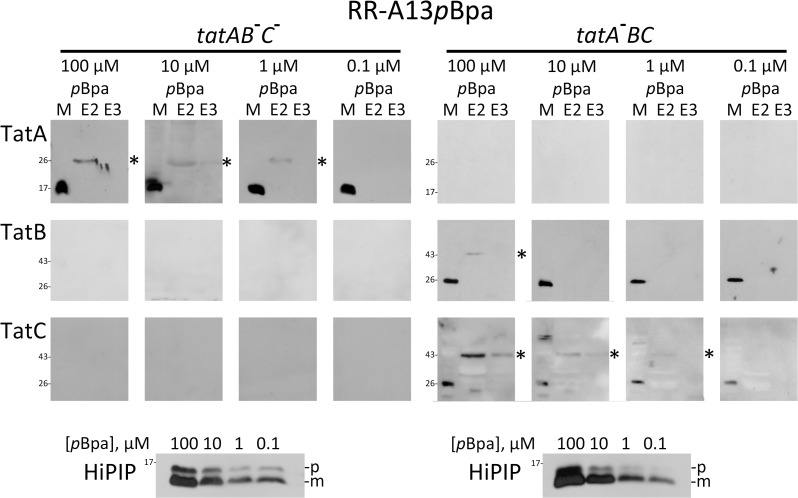
These data prompted an analysis of the lower limits of cross-link detectability, which showed that the observed TatA cross-links were also detectable with wild-type levels of the Tat components and very low substrate concentrations. Substrate cross-links in Tat-inactivated strains deficient in either TatA/E (strain JARV16) or TatB/C (strain JBdBC) were compared (Fig. 3). While the first strain monitors a TatA-independent TatBC interaction, the second strain monitors a TatBC-independent TatA interaction. Substrate levels were lowered by gradually lowering [pBpa] until cross-links were not detectable anymore. The data clearly demonstrated that—at wild-type Tat component levels—the TatBC-independent TatA cross-links were detected with the lowest substrate levels that could be used to detect TatC-cross-links (1 μM pBpa).
Fig 3. Substrate cross-links to TatA and TatC have comparable detection limits when substrate levels are gradually decreased.
Detection of cross-links of RR-HiPIP with pBpa at position A13 (RR-A13pBpa) either to wild-type level non-recombinant TatA in the absence of TatBC (left section: tatAB - C -, strain JBdBC) or to wild-type level non-recombinant TatBC in the absence of TatA (right section: tatA - BC; strain JARV16). Note that TatA and TatC cross-links are detectable at lowered substrate concentrations as achieved with 1 μM pBpa in the medium. TatB cross-links are depleted below detectability already with 10 μM pBpa, whereas TatA and TatC cross-links both become non-detectable with 0.1 μM pBpa. Bottom blots: SDS-PAGE-Western blot detection of HiPIP in extracts of cells grown in the presence of indicated pBpa concentrations. The bottom blots monitor the decrease of HiPIP levels by reduction of pBpa concentration in the medium, as described in Fig. 2. p, precursor of HiPIP; m, mature form of HiPIP. See Fig. 1 for further details.
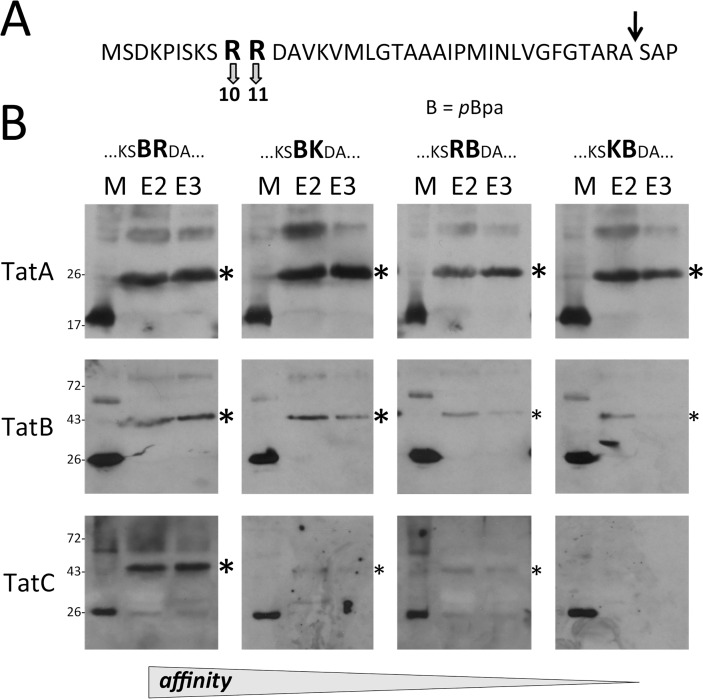
As only TatBC-containing complexes are known to recognize twin-arginine motifs, we analyzed the relevance of the twin-arginines for the TatA cross-links with four mutated motifs (pBpa abbreviated by B): BR, BK, RB, KB. Direct pBpa substitutions of the conserved arginine residues of the RR motif had not been examined in previous in vitro cross-linking analyses and most likely were not generated because they were expected to abolish Tat interactions (albeit very sensitive translocation assays could demonstrate residual Tat transport even with RR>KK mutations; [6,33]). The results showed highly efficient TatA-cross-linking to pBpa placed at both positions of the twin-arginines, whereas cross-links to TatB and TatC were gradually reduced in the order BR > BK > RB > KB, demonstrating that only TatBC components are involved in the recognition of the twin arginines (Fig. 4AB). Still remarkable TatBC cross-links in case of the BR motif reflect the high sensitivity of the cross-link detection. The KB-motif gave no detectable cross-links to TatC. The affinity-decrease in the order BR > BK > RB > KB highlights the preference of R over K as well as the outstanding importance of the positive charge at the second position for the motif recognition, which agrees with previous studies [6,31,33,34,35].
Fig 4. TatA does not recognize the RR motif.
(A) Positions of arginines in the twin-arginine motif that were mutated for experiments shown under B). (B) Analyses of cross-links to Tat components with pBpa (abbreviated “B”) at indicated twin-arginine motif positions (expression system and [pBpa] as in Fig. 1). Additional R>K exchanges are indicated. A decrease of TatBC cross-link intensities relates to decreasing affinities as indicative for the twin-arginine motif recognition (indicated at the bottom) that is confined to TatBC.
As the RR-motif was irrelevant for the TatA interaction, we assessed Tat specificity by replacing the Tat signal peptide of HiPIP by the Sec signal peptide of MalE (MalEsp-HiPIP; Fig. 5). A pBpa was positioned in between the two K residues of the n-region of the MalE signal peptide (Fig. 5A), which with respect to the distance to the h-region corresponds to the above tested RR-positions in Tat signal peptides that clearly cross-linked with TatA (see direct comparison in Fig. 5A). The MalEsp-HiPIP precursor was stable in vivo and many cross-links could be generated that proved the reactivity of the pBpa in the signal peptide, but no cross-link to TatA, suggesting that TatA differentiates between Tat and Sec signals by a distinct mechanism (Fig. 5B, right blot). Differences in other details could be involved, such as in the length of specific regions or in hydrophobicity, which are known to contribute to pathway preferences ([36,37,38]). An interesting side aspect is that MalEsp-HiPIP caused a severe growth inhibition, most likely due to jamming of the Sec translocon by the tightly folded HiPIP domain (Fig. 5C).
Fig 5. TatA does not cross-link to the signal peptide of the Sec substrate MalE.
(A) Position of the pBpa cross-linker in the MalE signal peptide relative to the position of the HiPIP signal peptide RR-motif. n-, h-, and c-regions are indicated (B) Cross-linking analysis of the MalE-signal peptide HiPIP fusion (expression systems and [pBpa] as in Fig. 1). As control, the fusion was detected in membranes by HiPIP-specific antibodies. It formed multiple cross-links with its N-terminal pBpa, indicating full-length of the signal peptide. See text for more details. (C) Growth defect upon production of MalE-HiPIP-I3pBpa, pointing to jamming of the Sec translocon by Sec-targeted folded HiPIP. + pBpa, Production of MalE-HiPIP-I3pBpa was induced by addition of 0.1 mM pBpa to the growth medium at the indicated time point. - pBpa, negative control without pBpa addition.
Preparation of soluble TatA and demonstration of substrate-binding in vivo and in vitro
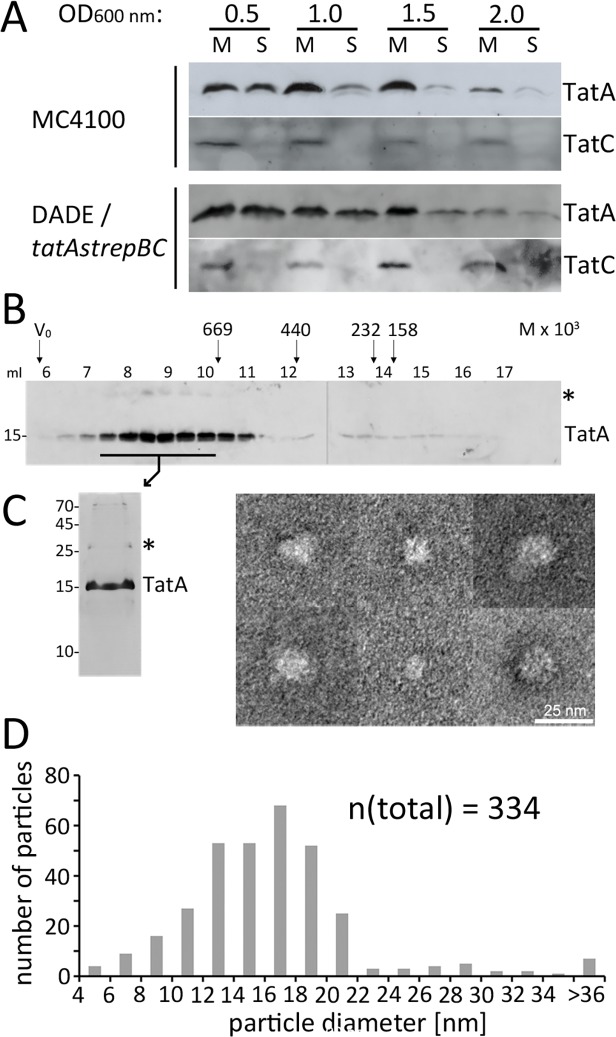
To address the TatBC-independent substrate/TatA interaction biochemically by a second, independent approach, we used detergent-free soluble TatA. Significant portions of TatA can be readily extracted from membranes without the use of detergents [39,40], which may be due to the very short membrane anchor or a population of soluble TatA that interacts with TatC at the membrane surface [21,41]. Indeed, soluble TatA has been observed in studies with archaea, bacteria, and plant plastids, strongly suggesting that TatA generally has the tendency to form soluble associations in biochemical preparations (Streptomyces lividans, Bacillus subtilis, Haloferax volcanii, as well as in the chloroplast stroma [23,39,42,43]). In Streptomyces lividans and in Bacillus subtilis, soluble TatA has been detected that had an intrinsic affinity to Tat substrates [22,23,41]. We also detected significant amounts of soluble TatA after cell disruptions, especially during early exponential growth (Fig. 6A). In the case of E. coli, this soluble TatA may result from a release from cell membranes during cell disruption, as the density of non-recombinant soluble TatA was experimentally determined to be ∼1.21 g/ml, which is higher than the density of cytoplasmic membranes (∼1.18 g/ml) and significantly lower than the density of pure protein (∼1.3 g/ml) (S3 Fig.). Like in the case of Bacillus or Streptomyces species, soluble TatA preparations from E. coli are large associations of many TatA protomers that broadly eluted from gel permeation chromatographies with a maximum size of about 800 kDa (Fig. 6B). Electron microscopy of purified Strep-tagged TatA revealed flattened, approx. round particles without characteristic features that would have been expected for a defined complex (Fig. 6C, see S4 Fig. for functionality confirmation of the construct and an overview micrograph). A statistical analysis of these TatA associations showed largely variable diameters with a most abundant size of ∼18 nm (Fig. 6D). Also few tube-like structures occurred. Together, the EM analyses suggested variable protein micelles rather than a defined complex. Protein micelles are expected to be formed by spontaneous interactions of the hydrophobic N-termini that must be shielded from the aqueous surrounding. Hydrophilic TatA regions in protein micelles are thus likely to be surface exposed just as in the case of TatA at the inner face of the cytoplasmic membrane.
Fig 6. Membrane-detached soluble TatA forms large micellar associations.
(A) Soluble TatA is abundant after cell disruption of bacteria at early exponential growth phase. Distribution of TatA in the membrane and cytoplasmic fraction in preparations from cultures of strains MC4100 and DADE tatA-strep-tatBC, harvested at indicated optical densities. Fractionation control was done by detection of the polytopic membrane protein TatC. Samples were normalized to the same amount of cells. (B) Size exclusion chromatography (SEC) of TatA-strep as purified from the cytoplasm after wild-type level production (DADE tatA-strep-BC) indicates formation of high molecular weight complexes. Western blot analysis of elution fractions; SEC molecular weight markers are indicated at the top (thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; aldolase, 158 kDa). (C) Silver-stained SDS-PAGE of purified TatA-strep and analysis of the TatA particles by electron microscopy. Six typical TatA assemblies are shown; see supplement S4 Fig. for an overview micrograph. (D) Statistical analysis of TatA particle diameters based on 334 measured particles. M, membrane fraction; S, soluble fraction; *, TatA dimer. Molecular weight marker positions (in kDa) are indicated on the left of SDS-PAGE blots.
Irrespective the debatable origin of soluble TatA, for this project most important was that the preparations of soluble TatA were stable over weeks and thus allowed for the assessment of substrate interactions in the absence of detergents. We initially tested interactions with HiPIP. Like in the cross-linking experiments, we included the RR>KK variant of HiPIP to test whether the RR motif in the signal peptide contributes to a possible Tat specificity of binding. We also included a mature variant of HiPIP that lacks the signal peptide. All HiPIP variants were C-terminally tagged to allow affinity chromatography. Strain E. coli MC4100/pEXH5tac-H6 was used that contains non-recombinant natural TatA and hexahistidine-tagged HiPIP. Cells were disrupted, soluble fractions were prepared and HiPIP was affinity-enriched. Soluble TatA clearly co-eluted with HiPIP and was detected by an increase of the TatA signal in the fractions where HiPIP eluted. The interaction was stable enough to be detectable even with soluble TatA at wild-type level (Fig. 7A, left side). As expected, the co-elution depended on the signal peptide but not on the twin-arginine motif. Since it was possible that soluble TatA can in principle interact with any kind of signal peptide, we again included the Sec-signal peptide of MalE in our analyses as negative control. Like in the case of in vivo cross-linking, MalEsp-HiPIP did not show any co-elution with TatA, indicating that indeed the Sec-signal peptide does not support the HiPIP interaction with TatA.
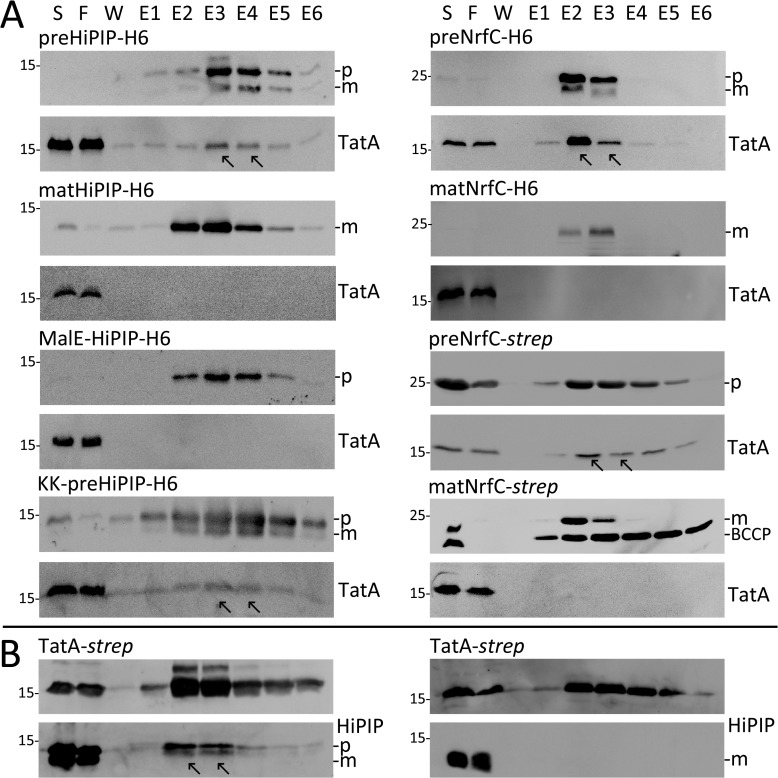
Fig 7. TatA/Tat substrate interactions in solution.
(A) Soluble non-recombinant TatA co-purifies with the precursor of HiPIP and NrfC and not with their mature forms. Precursor and mature forms of HiPIP were produced in E. coli strain MC4100 using pEXH5tac-H6 and pEXH5tac-mat-H6, respectively. The MalE-signal peptide-HiPIP fusion protein was produced using pEXH5tac-malE(sp)-H6, and the RR>KK mutated HiPIP variant was produced using pEXH5tac-H6-KK. Precursor and mature forms of NrfC were produced using pBW-nrfC-H6, pBW-nrfC-mat-H6, pBW-nrfC-strep or pBW-nrfC-mat-strep, as indicated. Soluble protein (S), flow-through (F), wash (W) and elution fractions (E1-6) were analyzed by SDS-PAGE Western blotting, using antibodies directed against HiPIP, the H6-Tag (in case of NrfC), or TatA as indicated. (B) In vitro folded pure HiPIP precursor associates with tagged soluble TatA. 5 μM of precursor (left panel) or mature (right panel) HiPIP were added to crude soluble extracts containing wild-type level TatA-strep (from strain DADE tatA-strep-tatBC), incubated at room temperature for 20 min and used for affinity chromatography. Further analyses were carried out as in A), using antibodies directed against TatA or HiPIP, as indicated. Arrows in A) and B) indicate co-elutions. p, precursor; m, mature form; BCCP, biotin carboxyl carrier protein.
HiPIP is a heterologous Tat substrate, and the observed interaction was therefore likely reflecting a general property of TatA that should be observable also with E. coli Tat substrates. To test this, we examined the interaction with the E. coli Tat substrate NrfC [44]. Strikingly, non-recombinant TatA was even more pronounced co-eluting with NrfC and the interaction clearly depended on the signal peptide (Fig. 7A, right side).
While the above interactions have been established in vivo, it was unclear whether TatA / Tat substrate interactions can also be generated in vitro, i.e. outside the living cell. To test this, 5 μM purified in vitro folded HiPIP were added to cytoplasmic fractions containing Strep-tagged TatA at natural levels (from a strain with a single-copy chromosomally integrated tatA-strep-BC operon). HiPIP clearly co-eluted with soluble TatA-strep in Strep-tactin affinity chromatography in a signal sequence-dependent manner (Fig. 7B). TatA thus can recognize folded HiPIP precursor in vivo as well as in vitro.
The TatA/Tat substrate interaction involves determinants of the folded substrate domain and is important for efficient Tat transport in vivo
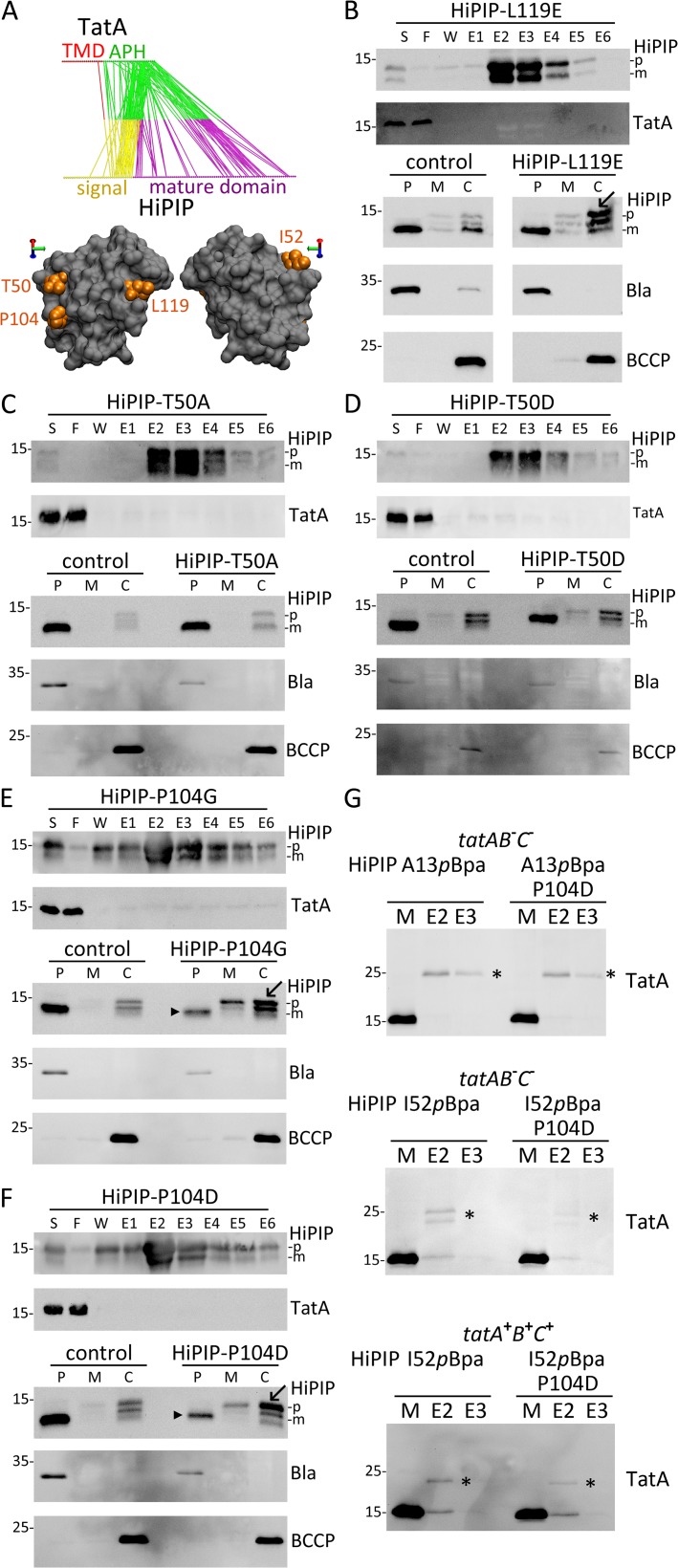
To address the physiological function of an alternative TatA-dependent targeting of substrates to the translocon, we searched for mutations that specifically affected TatBC-independent TatA/Tat substrate interactions to examine possible effects on transport. Candidate residues with possible importance for TatA/Tat substrate interactions were identified first by molecular dynamics (MD) simulations of HiPIP/TatA interactions in the membrane (Fig. 8A). The simulations predicted that the hydrophobic part of the HiPIP signal peptide can dip quite deeply into the membrane bilayer with its h-region kinked at a proline residue. Statistical analyses revealed possible TatA contact sites in the C-terminal half of the signal peptide and on the mature domain of HiPIP. As all Tat components contact the signal peptide (Fig. 1), and as TatBC complexes are not known to specifically recognize epitopes of mature domains and rather bind the RR-motif, the TatA contacts to the mature HiPIP domain raised the possibility that the mutation of mature domain surface residues could selectively weaken the TatBC-independent TatA recognition. The predicted contacts between TatA and the mature domain of HiPIP were all positioned on the same surface side of this small protein (Fig. 8A). Among the most prominent contacts in 50 independent simulations were contacts to T50, P104, and L119. To examine whether these contacts contributed to the affinity of TatA to HiPIP, we tested the system with T50D, T50A, P104D, P104G, and L119E exchanges (Fig. 8B-F). All mutations diminished the interactions to soluble TatA to an extent that compromised the co-elution. Co-purification requires strong interactions and a slow dissociation, and therefore an abolished co-elution can result already from minor changes that may not necessarily result in physiological effects in vivo. Nevertheless, the tested HiPIP mutations were promising candidates for mutations that might sufficiently affect the TatBC-independent TatA interaction to disclose a physiological role of the interaction. We therefore analyzed effects of these mutations on translocation efficiency. In control experiments, we ensured that the surface mutations did not compromise folding of HiPIP (S6 Fig.). While the T50A and T50D mutations did not affect transport, the other three mutations clearly caused an accumulation of precursor in the cytoplasm, and in the cases of P104D and P104G even in the membrane. With P104D of P104G mutations, the accumulation of precursor was clearly attributable to a lowered translocation efficiency, as the signal of transported mature HiPIP in the periplasmic fraction was strongly reduced (Fig. 8EF). To finally examine whether transport-compromising exchanges on the HiPIP surface did indeed reduce the TatA contacts in vivo, we carried out an in vivo cross-link experiment and monitored the TatA interactions with the signal peptide and the mature domain of HiPIP with or without the P104D exchange. We placed pBpa either at position A13 of the signal peptide or at position I52 of the mature domain, which was recently shown by us to contact TatA and TatB in vivo [45]. While the signal peptide interaction with TatA was unaffected by the P104D exchange, the I52pBpa cross-link to TatA was clearly reduced, and this reduction was independent of TatBC (Fig. 8G). The residual cross-link indicates that the exchange does not completely abolish the interaction, which is expected for contacts that are mediated by multiple interactions.
Fig 8. Identification of TatA-binding determinants on the surface of folded HiPIP.
(A) Most frequent interactions observed in 50 independent molecular dynamics (MD) simulations of TatA-HiPIP interactions (upper diagram). Color code: yellow: HiPIP signal peptide; magenta: HiPIP mature domain; red: TatA trans-membrane domain; green: TatA amphipathic helix. The three residues on the HiPIP surface with most contacts are positioned on one side of HiPIP and the pBpa exchange position I52 are highlighted (lower diagram). See S5 Fig. for a snapshot of a molecular dynamics simulation of TatA-HiPIP interaction. (B) – (F) Analysis of affinity purification, TatA co-elution (see Fig. 7 for experimental details) and in vivo translocation of HiPIP-L119E (B), -T50A (C), -T50D (D), -P104G (E), and -P104D (F). In vivo translocation was examined as described in Fig. 1B. For all subcellular fractionations, the periplasm control (β-lactamase, Bla) and the cytoplasm control (BCCP) are shown. Arrowheads indicate strongly reduced translocation and arrows indicate accumulating precursor HiPIP. p, precursor; m, mature; P, periplasm; M, membrane; C, cytoplasm; markers are indicated on the left of the blots. (G) Cross-linking of position I52 of the HiPIP mature domain to TatA is affected by the HiPIP-P104D mutation, whereas the interaction with the signal peptide A13 position remains unaffected. Analysis of HiPIP cross-links to wild-type level TatA in the absence of tatBC (tatAB − C −, strain: JBdBC) or with slightly increased levels of TatABC (pRK-tatABC), (tatA + B + C +, strain: MC4100). pBpa was positioned in the signal peptide (A13, upper blot) or in the mature domain (I52, lower two blots), and the effect of the TatA interaction-weakening HiPIP-P104D exchange on cross-link intensities was analyzed in parallel. *, cross-links.
Discussion
In vivo cross-linking data indicate a TatBC-independent TatA interaction
At the beginning of this study we used in vivo photo-activatable site directed cross-linking tools for the analysis of the protein translocating Tat system. So far, such Tat system analyses have been only carried out in vitro [4,5,20,46,47]. The in vivo system included an affinity chromatography-based enrichment of cross-links that increased the signal intensities of the Western-blot based assay to levels comparable to in vitro assays that can use additions of radiolabeled substrates [4]. The in vivo assay permitted the detection of weak cross-links of RR>KK mutated signal peptides to TatB and TatC (Fig. 1)—something that could not be detected so far in vitro, albeit a residual translocation activity with RR>KK mutated signal peptides has been shown in vivo by highly sensitive transport assays [6,33], and we similarly observed with HiPIP that RR>KK mutated signal peptides can mediate residual transport if only the Tat motif is optimized by a A13F or A13pBpa mutation (S2 Fig.). The in vivo cross-links with functional Tat substrates in the presence of active TatABC components monitor any interactions of the complete translocation path and it was thus possible to detect TatC cross-links to positions in the h- and c-region of the twin-arginine signal peptide (Fig. 1), which agrees with a deep contact of the signal peptide with TatC as supported by the TatC structure, protease protection studies, and the signal peptide insertase activity of TatC [27,28,29]. This agrees also with a recent report of a PMF-dependent TatC cross-link of a F32pBpa exchange in the HiPIP signal peptide [45].
Cross-linking analysis with pBpa at RR or KK positions showed that TatC can distinguish arginines from lysines, with the second arginine position being much more important than the first, with a tolerance for even uncharged substitutions at the first position (Fig. 4). This fits to the analysis of Tat transport with a highly sensitive reporter system that could detect the same preferences [33]. In contrast to TatC, TatA did not show any recognition of the arginines and cross-linked to all pBpa substitutions at these positions with similar efficiencies. The interaction was nevertheless completely abolished when the Tat signal peptide was exchanged by the Sec signal peptide of MalE, suggesting a role of known Tat determinants other than the twin-arginine motif ([38] and Fig. 5). However, a strict differentiation between Sec and Tat signal peptides might even not be necessary in vivo: A targeting to the Sec translocon can occur either via SRP or SecA-mediated pathways, and for both pathways a signal peptide binding can already take place at the ribosome, which would prevent any TatA interaction [48,49,50]. TatB cross-links gave some intermediate result, which may be explained by the reported formation of TatAB complexes in vivo [19,51,52], that add to the well-known TatBC complexes [53].
Importantly, TatA cross-linked to Tat substrates independently of TatBC and at lowest substrate concentrations that were required to see TatC cross-links at wild-type Tat levels, which was suggestive for a delivery function of substrate-associated TatA in vivo (Figs. 2 and 3). This corresponds to the assembly of complete TatABC-Tat substrate translocons, as it is known that TatBC/Tat substrate recognition induces a recruitment and clustering of TatA at the translocon [10]. The resulting substrate-containing TatABC complex has been recently detected in E. coli by BN-PAGE analyses at ∼600 kDa [9]. The recognition of the RR motif by TatBC complexes is well-documented [4,5,6,7] and in full agreement with our cross-linking results (Fig. 1 and 4).
TatA binds the signal peptide as well as the mature domain of certain Tat substrates
As the cross-linking data can only demonstrate a close proximity between components, it was important to show that this corresponded to a binding of the proteins and that HiPIP is representative for other natural Tat substrates as well. This was achieved by co-purification and by including a second natural Tat substrate, E. coli NrfC. We found that soluble TatA is an ideal tool for studying the interactions: it is stable in solution, its purification does not require the use of detergents that could abolish interactions, and – most importantly – it binds Tat substrates (Figs. 6 and 7). With HiPIP as well as with NrfC, binding depended on the signal peptide. Since HiPIP is the structurally much better characterized Tat substrate, we used it to analyze the binding in more detail and we could show that a RR>KK exchange still allowed for an interaction whereas a MalE Sec signal peptide exchange diminished the affinity of HiPIP to TatA. These data agree with results reported in studies on the Streptomyces lividans Tat system [21]: In that study, recombinantly overproduced TatA from the soluble fraction bound Tat substrates with high affinity whereas a Sec signal peptide lowered the affinity by several orders of magnitude. The analysis of RR>KK exchanges was not reported in that study, which measured the affinities using surface plasmon resonance biosensors. A second report on a direct TatA/Tat substrate interaction comes from studies with Bacillus subtilis, where the Tat substrate PhoD interacts with a PhoD-specific TatAd [41]. In this case, positively charged residues in the RR-motif contributed to the affinity without being essential for the TatA interaction. This motif has the sequence DRRKFIQ in PhoD, and R>K exchanges had no effect and even a RRK>AAA exchange retained significant TatA affinity. These results are in full agreement with our findings and raised the question, what accounts for the specificity in the PhoD-TatAd interaction. Unexpectedly, in our experiments we found that mature domain can be highly important for the interaction. As the HiPIP and TatA structures are known, we could carry out MD simulations that identified three HiPIP surface residues that frequently contacted TatA, and single amino acid exchanges in these positions disabled the co-elution of TatA in affinity chromatographies. Such an effect on co-elution could already be due to a minor reduction in affinity, without abolishing a functional interaction in vivo. It was therefore important to assess, whether the mutations also affected Tat transport in vivo. Indeed, translocation assays showed that two of the three positions clearly affected translocation efficiency, strongly suggesting that the TatBC-independent TatA interaction is relevant for Tat transport (Fig. 8). With the P104D exchange, which affected transport most strongly, we showed that indeed the abolished TatA co-elution correlated with reduced TatA cross-links to the surface of the HiPIP mature domain (Fig. 8G). This was observed in the absence as well as in the presence of TatBC, confirming the TatBC-independence of this interaction. Interestingly, the signal peptide interaction with TatA was not affected by the exchange at the mature domain surface, suggesting that the two regions are both recognized by TatA, and only if both are bound, the interaction is strong enough to permit a co-purification. This identification of surface residues with importance of TatBC-independent TatA interactions and Tat transport indicates a physiological contribution to the transport process and it shows that it is most likely the interplay of signal peptide and mature domain characteristics that promotes TatA/Tat substrate interactions and thereby confers Tat substrate specificity. We believe that this is the key to understand the substrate-specific role of several TatA components, such as NosZ-transport-specific TatE in Pseudomonas stutzeri or PhoD transport-specific TatAd in Bacillus subtilis [23,54]. Future studies will have to focus on the exact determinants for mature domain interactions which are likely to play important roles for the recognition of specific Tat substrates, such as HiPIP, NrfC, PhoD or NosZ. The delivery of Tat substrates by TatA to the Tat translocon may explain the very distinct distribution of TatA and functional TatABC translocons. TatA associations are found distributed in the cytoplasmic membrane without a clear preference to specific subcellular regions [16,19,55]. In contrast, the RR-dependently formed TatC-containing translocation sites are located in few, often polar or sub-polar small foci [17,19]. TatA clearly is part of the active translocons, but the TatA/Tat substrate interaction and the improvement of transport suggest that the freely-diffusing population of TatA can contribute to targeting as outlined in Fig. 9, which shows our current working model for the sequence of events during Tat transport. This model includes the accepted current model and only extends it by the herein described TatA interaction. Tat substrates will first interact with membranes [56,57,58,59,60] and in E. coli, the first Tat component encountered by membrane-interacting Tat substrates is likely to be TatA. TatA exists in 50-fold molar excess [61] and the freely diffusing TatA is known to form tetramers, whereas TatC forms higher oligomers, further reducing the occurrence to a few spots in the cell [17,19]. Accordingly, TatA interactions are detectable already at very low substrate levels, at the same concentrations that are required to detect TatC interactions (Fig. 3). Early TatA interactions certainly do not substitute the later RR-motif recognition by TatC and are not expected to be essential for Tat transport, but nevertheless these interactions can positively affect Tat transport (Fig. 8). They may thus facilitate translocation by promoting TatBC interactions, accelerating the later TatA-recruitment step, and/or by protecting membrane-interacting signal peptides. Future studies will have to uncover the mode by which these early TatA/Tat substrate interactions contribute to Tat transport.
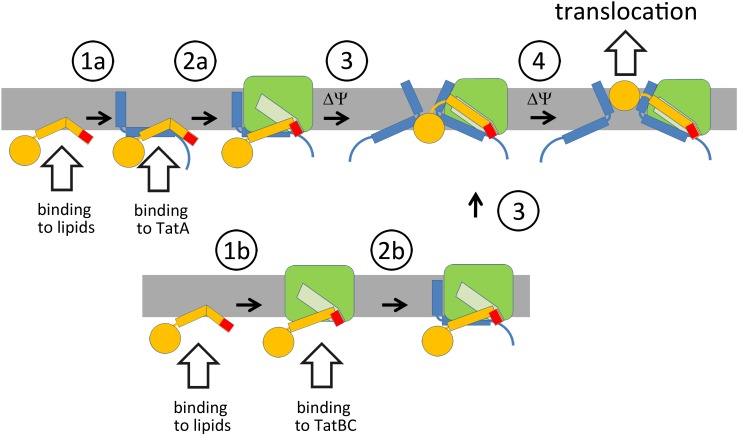
Fig 9. Interactions during Tat translocon assembly.
Tat substrate signal peptides such as from HiPIP or NrfC can spontaneously interact with membrane surfaces where they can either encounter free TatA (1a) or TatABC complexes (1b). TatA-bound substrates expose the RR-motif that is recognized by TatBC in the TatABC complex (2a). More TatA is recruited, possibly when TatBC imposes a force to the Tat substrate, which could influence TatA orientations and the local membrane curvature (3). Such a force could result from the known binding of larger signal peptide regions into the membrane-dunking binding-site regions at TatBC. Sufficient TatA recruitment finally permits translocation (4). If Tat substrates are first bound to TatBC, either after membrane interaction or directly (1b), the TatA assembly has to take place thereafter (2b). Such a binding may be more relevant for Tat substrates that do not readily interact with membranes, as the likelihood to first encounter TatA in membranes is high. “ΔΨ” designates steps that likely require the membrane potential. Color-code: Tat substrate, yellow; twin-arginine motif, red; TatA, blue; TatBC, green; binding-site for the signal peptide in TatBC, light green.
Materials and Methods
Strains and growth conditions
Escherichia coli strains MC4100 araR [16], its derivatives, BW25113 [62], and JBdBC (this study) were used for in vivo cross-linking and physiological studies. E. coli XL1-Blue Mrf’ Kan (Stratagene) and BW23473 [63] were used for cloning. The bacteria were grown aerobically at 37°C on LB-medium (1% (w/v) tryptone, 1% (w/v) NaCl, 0.5% (w/v) yeast extract) in the presence of the appropriate antibiotics (100 μg/ml ampicillin, 25 μg/ml chloramphenicol, 15 μg/ml Kanamycin 12.5 μg/ml tetracycline).
Plasmids and genetic methods
The single copy integration vector pAH120-Ptat-tatA-strep-tatBC was generated by amplification of the tat genes (from the 3’ region of tatA to tatC) using the primers tatA-Bam-s-ATG-B-F (5′-AGG TGG GAT CCT GGA GCC ACC CGC AGT TCG AAA AAT AAG CAG GTG TAA TCC ATG TTT GAT ATC GGT TTT AGC GAA C-3′) and pABS-tatC-BglII-R (5′-ATA TAG CGC GCT TAT TCT TCA GTT TTT TCG CTT TC-3′), followed by restriction with BamHI/BglII and ligation into the corresponding sites of a pAH120-Ptat-tatA-strep derivative [40] in which a second BamHI site downstream of tatA-strep had been removed by QuikChange (Stratagene) PCR. pRK-hip was used for constitutive low-level expression of the hip gene from its own promoter [26]. pEvol-pBpF (courteously provided by Peter G. Schultz) was used to incorporate pBpa at UAG stop codons [24,25,64]. When indicated, the plasmid pRK-tatABC [26] or its tatBC-deleted derivative pRK-tatA were used for constitutive moderate (15-fold) overproduction of TatABC or TatA, respectively. HiPIP was produced constitutively by plasmids pEXH5tac or pEXH5tac-KK, respectively [65], in which a C-terminal His6-tag-encoding sequence was fused to the hip gene by cloning the corresponding PvuI fragment from pEXH7 into the PvuI-digested pEXH5tac and pEXH5tac-KK [66], resulting in the vectors pEXH5tac-H6 and pEXH5tac-KK-H6, respectively. To construct HiPIP variants with amino acid exchanges, desired codons were introduced at specific sites by QuikChange mutagenesis (primers listed in S1 Table). The MalE-signal sequence-HiPIP-fusion constructs were created with and without the amber stop codon at position I3 of the MalE signal peptide (S2 Table). For construction of pEXH5tac-mat-H6, the mature domain-encoding sequence was amplified with pEXH5tac-H6 as template and used to substitute the precursor-encoding sequence in the template vector (see S2 Table). The L119E mutation was generated by amplification of the hip gene with mutational primers (see S2 Table). The pBW22 expression system was used for rhamnose-inducible expression of nrfC-H6 or nrfC-strep [67].
The strain JBdBC has been generated by deleting the tatBC genes in BW25113 by the method of Datsenko and Wanner [62].
To construct strain DADE tatA-strep-tatBC, the tatA-strep-tatBC operon, expressed under control of the tatA promoter (pAH120-Ptat-tatA-strep-tatBC) was single copy integrated into the λatt-site of the tat deletion strain DADE [68], using the method of Haldiman and Wanner [63]. All constructs were confirmed by restriction analyses and sequencing.
Biochemical methods
For the site-specific in vivo incorporation of pBpa at the position of the amber stop codon by the pEvol System, pBpa (Bachem, dissolved in 0.5 M NaOH) was added with indicated concentrations simultaneously with 100 μM arabinose three hours prior to UV-cross-linking and harvest of the cells. For comparative analysis of HiPIP production 6 ml of the respective cell suspension was removed before cross-linking procedure. Cells were suspended in 1 ml Tris HCl, pH 8.0, disrupted by sonication and the supernatant was used for SDS-PAGE after sedimentation of cell debris. Cross-linking of pBpa was induced by irradiation of the cultures with UV light at 365 nm wavelength for 30 min at room temperature. Membranes were prepared from harvested cells and solubilised in buffer containing 3% SDS for 30 min and an adjacent dilution to 0.1% before affinity chromatography. Non-solubilised material was separated by ultracentrifugation (30 min, 130,000 x g, 4°C). Purification of His6-tagged proteins under denaturing conditions was carried out by Ni-NTA metal affinity chromatography as described elsewhere [69]. All purification buffers contained 0.1% SDS. Wash- and Elution-fractions were 10-fold concentrated by trichloroacetic acid precipitation and the presence of Tat-components and their cross-links was assessed by Western blotting as described below. Co-elution experiments were achieved by Ni2+-affinity chromatography [69] or Strep-Tactin Superflow chromatography (IBA).
The distribution of soluble and membrane-bound TatA at wild-type level was analyzed with cells harvested at different time points during growth (OD600nm = 0.5, 1.0, 1.5, 2.0) in cell-density normalized amounts. Cells were resuspended in 20 mM Tris HCl, pH 8.0 and disrupted by sonication. After removal of cell debris (15,000 x g, 10 min, 4°C), membranes were sedimented (130,000 x g, 30 min, 4°C) in 200 μl fractions and resuspended in the same amount of buffer.
Purification of soluble TatA was performed with cells harvested in the early exponential growth phase (OD600nm = 0.5). Cells were resuspended in 10 mM Hepes (4 ml/g cells), pH 8.0 and 150 mM NaCl, and passed twice through a French Press cell at 138 MPa. Cell debris and membranes were removed by centrifugations (20,000 x g, 15 min, 4°C and two times 130,000 x g, 1 h, 4°C). TatA-strep from the soluble fraction was purified by affinity chromatography on Strep-Tactin Superflow (IBA) columns according to the supplier′s protocol, but using the above mentioned buffer. Elution fractions containing TatA-strep were pooled, concentrated and further purified by size exclusion chromatography using a Superose 6 column. Inclusion bodies of precursor HiPIP were produced and folded as described previously. Mature HiPIP was obtained from folded precursor HiPIP by thermolysin treatment (200 μg/ml) for 30 min on ice [26].
SDS-PAGE was carried out by the method of Laemmli [70]. Subcellular fractionations were obtained by an optimized osmotic shock procedure. Briefly, 50 ml exponentially growing cultures were sedimented (4,500 x g, 4°C), cells were resuspended in 20 ml 20% sucrose/10 mM Tris HCl, pH 8.0/1 mM EDTA, incubated for 10 min at room temperature, and again sedimented (4,500 x g, 4°C). The supernatant-free cell pellet was resuspended in ice-cold 1 ml 5 mM MgSO4 and incubated for 20 min on ice. Shocked cells were sedimented (9,500 x g, 4°C) and the periplasm (supernatant) was carefully collected. The pellet was resuspended and cytoplasm and membranes were further separated by disintegration and centrifugation steps as described previously [26]. Western blotting was arranged as described previously using polyclonal rabbit serum against purified HiPIP, against synthetic TatA, TatB or TatC peptides (C-terminal 17, 17, 16 residues, respectively), against β-Lactamase (Acris), YidC (donated by Andreas Kuhn, Hohenheim) and His-tags (Qiagen). The biotin carboxyl carrier protein was detected by a Strep-Tactin-HRP-conjugate (IBA). Cells were visualized by differential interference contrast (DIC) as described previously [16].
Isopycnic ultracentrifugation
The different migration behavior of membrane-integral and membrane-free TatA was analyzed in a cesium chloride density gradient. 24.5 ml of a 2 M CsCl solution was over-layered with 0.5 ml of a membrane or soluble fraction derived from MC4100 culture grown to OD of 0.5. The samples were centrifuged at 360,000 x g for 20 h afterwards separated in 1 ml fractions and refractive indices measurements were achieved with a refractometer (RM40, Mettler Toledo). Densities were calculated as described [71].
Electron microscopy
Purified cytoplasmic TatA preparations were used for electron microscopical analyses. TatA particles were adsorbed to carbon foil and negatively stained with 1% uranyl acetate, as is described elsewhere [72]. Microscope settings were applied as described previously [73].
Molecular dynamics simulations
The molecular dynamics simulations were performed with the GROMACS simulation package [74], version 4.5.5. We used the MARTINI coarse-grained model [75,76] to simulate the lipids, amino acids and solvent. In all simulations, the system was coupled to a constant temperature bath [77] with a relaxation time of 1.0 ps. We performed our simulations at a temperature of 310 K. Periodic boundary conditions where applied to simulate bulk behavior. The time step used in the simulation was 20 fs. The dielectric constant in the simulations was εr = 15. The neighbor-list was updated every 10 simulation steps. The pressure was weakly coupled [77] to 1 bar with a relaxation time of 0.5 ps.
TatA was modeled using the MARTINI model for proteins, which qualitatively captures the chemical nature of each individual amino acid and implicitly includes the secondary structure. The secondary structure was modeled by both restraining proper dihedrals between four neighboring backbone beads with an harmonic potential and by altering the non-bonded interactions according to the imposed secondary structure (free in solution, or in a coil or bend the backbone has a more polar character than in a helix or -strand). Further details concerning this methodology can be found in the original publication [76]. In these simulations the secondary structure of TatA was obtained from a recently obtained NMR-structure [15] and translated according to the DSSP definition. The secondary structure of the HIPIP signal peptide was based on local hydropathy (http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm) and the prediction that the signal peptide may have a trans-membrane helical region between LEU17 to ALA36 (http://www.cbs.dtu.dk/services/TMHMM). In here, we allowed additional flexibility of PRO17 by modeling PRO17 as a coil, and ILE23 and MET25 as a band. Residues SER1 to MET16 were modeled as a random coil. This structure of the signal peptide should be regarded as a first approximation. The signal peptide was fused with the X-ray structure of the mature domain (PDB 1hip) of HIPIP. For sake of optical convenience, the 4 resolved Fe-S complexes were modeled by 4 polar beads (P4) and kept into the binding pocket by a harmonic elastic network (force constant of 500 kJ nm−1mol−1) formed with all nearby residues (< 0.8 nm cutoff). To additionally conserve the structure/secondary structure of the mature domain an additional elastic network was constructed between the backbone beads of residues ALA40-GLY121 (0.5 > cutoff < 0.8 nm).
A 6x6 nm piece of the bacterial membrane was modeled by symmetrically placing mimics of DPPE (102), DPPG(26) and Cardiolipin (8) in a bilayer configuration. The model of cardiolipin is described elsewhere [78]. Sodium counter-ions were added to neutralize the system.
The clustering between TatA and HIPIP was studied by performing 50 independent simulations of up to 4 microseconds [75] starting from different random initial velocities. An interaction between HIPIP and TatA is defined by a distance less than 0.5 nm between two atoms. For each simulation we listed the 3 most frequent observed interactions.
Supporting Information
(A) During affinity chromatography, specific cross-links elute in fractions E2 and E3. Shown is the immunoblot-detection of cross-links of RR-HiPIP with a M26pBpa substitution to TatB, using antibodies recognizing TatB. M: solubilized membranes, W: last wash fraction, E1-E4: elution fractions. (B) Without UV irradiation or pBpa, no shifted bands can be detected in the elution fractions. For this important control, we chose HiPIP M26pBpa, which gives strong cross-links to all Tat components after UV irradiation (see Fig. 1A). No cross-links could be detected without UV irradiation or pBpa (+ pBpa/-UV, −pBpa/-UV, left blots). We also analyzed the UV-dependence of the cross-links for the HiPIP A13pBpa variant that was the most important variant in our studies. As expected for the UV-activatable cross-linker pBpa, the cross-links were absent without UV-irradiation. The pBpa-dependence of cross-links to this position is already shown in Fig. 3. As additional control, we demonstrated that wild-type HiPIP per se does not give any shifts in the presence of pBpa and UV (right blots). The presence of TatA, TatB, and TatC (pRK-tatABC) in the membranes and elution fractions was assessed as shown in Figs. 1 and 2. (C) HiPIP RR/KK variants with pBpa (*) at indicated positions were produced in similar concentrations in the experiments shown in Fig. 1. Note significant precursor-accumulation in case of the KK-variants and more mature HiPIP in case of the RR-variants, indicating RR-dependent transport. Processing of KK-variants to mature size is mainly due to unspecific proteolytic degradation of the signal peptide. Detection in crude extracts with HiPIP specific antibodies.
(TIF)
Detection of HiPIP in subcellular fractions by SDS-PAGE/Western blotting, using HiPIP-specific antibodies. In control blots, purity of periplasmic and cytoplasmic fractions was confirmed by detection of periplasmic b-lactamase (bla) and cytoplasmic biotin carboxyl carrier protein (BCCP). Note some detectable transport of KK-HiPIP variants in which the Tat motif is optimized by an A13F or A13pBpa substitution (*: mature periplasmic HiPIP). This position corresponds to the consensus “F” position in the motif, and F as well as pBpa attribute a large, hydrophobic, aromatic side chain to the motif, which can promote the Tat translocon interaction and thus partially compensates for the transport-inactivating RR>KK exchange. Strains and conditions as in Fig. 1.
(TIF)
(A) Experimentally determined densities of the fractions analyzed in B, showing identity of the two density profiles. (B) Detection of TatA and the polytopic membrane protein YidC (membrane marker) after isopycnic CsCl gradient centrifugation of soluble or membrane fractions from strain MC4100. No YidC is detectable in the soluble fraction, indicating that this fraction is virtually membrane-free. Note that soluble TatA micelles sediment to a density of ∼1.21 g/ml, which suggests a tight association with lipids but not with vesicles. Membrane-associated TatA sediments with membrane vesicles to a density of ∼1.18 g/ml, as confirmed by the detection of the membrane marker YidC (lower two blots).
(TIF)
(A) Complementation of the chain formation phenotype of the Tat-deficient DADE strain by a single-copy chromosomally integrated tatA-strep-tatBC operon (strain DADE tatA-strep-tatBC). As controls, the Tat-system-containing parental strain MC4100 and the Tat-deficient strain DADE have been analyzed in parallel. (B) Tat transport of HiPIP as produced from pRK-hip in strain DADE tatA-strep-tatBC shown by Western-blot analysis of subcellular fractions. P: periplasm, M: membranes, C: cytoplasm. The periplasm contains transported mature (m) HiPIP, whereas only the cytoplasm contains unprocessed precursor (p) and some to mature size degraded HiPIP. The control blot on the right side detects the biotin carboxyl carrier protein (BCCP, cytoplasmic marker). (C) TatA micelles. Overview EM micrograph of purified TatA micelles from the cytoplasm of strain DADE tatA-strep-tatBC. The size bar indicates 100 nm.
(TIF)
Color code: yellow: HiPIP signal peptide; magenta: HiPIP mature domain (FeS-cofactor in orange); red: TatA trans-membrane domain; green: TatA amphipathic helix.
(TIF)
Electronic absorption spectra of folded purified cofactor-containing HiPIP (wt) in comparison to the analyzed HiPIP variants with mutations in the mature domain (T50A, T50D, P104G, P104D, L119E). The spectra all show the typical [4Fe-4S]-cofactor absorption that indicates complete cofactor insertion and thus stable folding of HiPIP.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
We thank Jana Behrendt for strain JBdBC, Sybille Traupe for excellent technical assistance, Peter G. Schultz for donation of the pEvol system, Andreas Kuhn for YidC antibodies, and Hauke Lilie for helpful discussion. This work was funded by the Deutsche Forschungsgemeinschaft (GRK 1026: „Conformational transitions during macromolecular interactions“).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Deutsche Forschungsgemeinschaft (GRK 1026). The authors acknowledge support by the Open Access Publishing Fund of the Leibniz Universität Hannover and the Deutsche Forschungsgemeinschaft. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Palmer T, Berks BC (2012) The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol 10: 483–496. 10.1038/nrmicro2814 [DOI] [PubMed] [Google Scholar]
- 2. Hou B, Brüser T (2011) The Tat-dependent protein translocation pathway. Biomol Concepts 2: 507–523. [DOI] [PubMed] [Google Scholar]
- 3. Jongbloed JD, van der Ploeg R, van Dijl JM (2006) Bifunctional TatA subunits in minimal Tat protein translocases. Trends Microbiol 14: 2–4. [DOI] [PubMed] [Google Scholar]
- 4. Alami M, Lüke I, Deitermann S, Eisner G, Koch HG, Brunner J, et al. (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli . Mol Cell 12: 937–946. [DOI] [PubMed] [Google Scholar]
- 5. Gerard F, Cline K (2006) Efficient twin arginine translocation (Tat) Pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J Biol Chem 281: 6130–6135. [DOI] [PubMed] [Google Scholar]
- 6. Kreutzenbeck P, Kroger C, Lausberg F, Blaudeck N, Sprenger GA, et al. (2007) Escherichia coli twin arginine (Tat) mutant translocases possessing relaxed signal peptide recognition specificities. J Biol Chem 282: 7903–7911. [DOI] [PubMed] [Google Scholar]
- 7. Lausberg F, Fleckenstein S, Kreutzenbeck P, Fröbel J, Rose P, Müller M, et al. (2012) Genetic evidence for a tight cooperation of TatB and TatC during productive recognition of twin-arginine (Tat) signal peptides in Escherichia coli . PLoS One 7: e39867 10.1371/journal.pone.0039867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aldridge C, Ma X, Gerard F, Cline K (2014) Substrate-gated docking of pore subunit Tha4 in the TatC cavity initiates Tat translocase assembly. J Cell Biol 205: 51–65. 10.1083/jcb.201311057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Behrendt J, Brüser T (2014) The TatBC complex of the Tat protein translocase in Escherichia coli and its transition to the substrate-bound TatABC complex. Biochemistry 53: 2344–2354. 10.1021/bi500169s [DOI] [PubMed] [Google Scholar]
- 10.Dabney-Smith C, Cline K (2009) Clustering of C-Terminal Stromal Domains of Tha4 Homo-Oligomers during Translocation by the Tat Protein Transport System. Mol Biol Cell. [DOI] [PMC free article] [PubMed]
- 11. Brüser T, Sanders C (2003) An alternative model of the twin arginine translocation system. Microbiol Res 158: 7–17. [DOI] [PubMed] [Google Scholar]
- 12. Dabney-Smith C, Mori H, Cline K (2006) Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J Biol Chem 281: 5476–5483. [DOI] [PubMed] [Google Scholar]
- 13.Pal D, Fite K, Dabney-Smith C (2012) Direct Interaction between Precursor Mature Domain and Transport Component Tha4 during Twin Arginine Transport (Tat) of Chloroplasts. Plant Physiol. [DOI] [PMC free article] [PubMed]
- 14. Panahandeh S, Maurer C, Moser M, DeLisa MP, Müller M (2008) Following the path of a twin-arginine precursor along the TatABC translocase of Escherichia coli . J Biol Chem 283: 33267–33275. 10.1074/jbc.M804225200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodriguez F, Rouse SL, Tait CE, Harmer J, De Riso A, Timmel CR, et al. (2013) Structural model for the protein-translocating element of the twin-arginine transport system. Proceedings of the National Academy of Sciences of the United States of America 110: E1092–1101. 10.1073/pnas.1219486110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berthelmann F, Brüser T (2004) Localization of the Tat translocon components in Escherichia coli . FEBS Lett 569: 82–88. [DOI] [PubMed] [Google Scholar]
- 17. Berthelmann F, Mehner D, Richter S, Lindenstrauß U, Lünsdorf H, Hause G, et al. (2008) Recombinant expression of tatABC and tatAC results in the formation of interacting cytoplasmic TatA tubes in Escherichia coli . J Biol Chem 283: 25281–25289. 10.1074/jbc.M707757200 [DOI] [PubMed] [Google Scholar]
- 18. Leake MC, Greene NP, Godun RM, Granjon T, Buchanan G, Chen S, et al. (2008) Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc Natl Acad Sci U S A 105: 15376–15381. 10.1073/pnas.0806338105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rose P, Fröbel J, Graumann PL, Müller M (2013) Substrate-dependent assembly of the Tat translocase as observed in live Escherichia coli cells. PLoS One 8: e69488 10.1371/journal.pone.0069488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fröbel J, Rose P, Müller M (2011) Early Contacts between Substrate Proteins and TatA Translocase Component in Twin-arginine Translocation. J Biol Chem 286: 43679–43689. 10.1074/jbc.M111.292565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Keersmaeker S, Van Mellaert L, Lammertyn E, Vrancken K, Anne J, Geukens N. (2005) Functional analysis of TatA and TatB in Streptomyces lividans . Biochem Biophys Res Commun 335: 973–982. [DOI] [PubMed] [Google Scholar]
- 22. De Keersmaeker S, Vrancken K, Van Mellaert L, Anne J, Geukens N (2007) The Tat pathway in Streptomyces lividans: interaction of Tat subunits and their role in translocation. Microbiology 153: 1087–1094. [DOI] [PubMed] [Google Scholar]
- 23. Pop OI, Westermann M, Volkmer-Engert R, Schulz D, Lemke C, Schreiber S, et al. (2003) Sequence-specific binding of prePhoD to soluble TatAd indicates protein-mediated targeting of the Tat export in Bacillus subtilis . J Biol Chem 278: 38428–38436. [DOI] [PubMed] [Google Scholar]
- 24. Chin JW, Martin AB, King DS, Wang L, Schultz PG (2002) Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli . Proc Natl Acad Sci U S A 99: 11020–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Young TS, Ahmad I, Yin JA, Schultz PG (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli . J Mol Biol 395: 361–374. 10.1016/j.jmb.2009.10.030 [DOI] [PubMed] [Google Scholar]
- 26. Brüser T, Yano T, Brune DC, Daldal F (2003) Membrane targeting of a folded and cofactor-containing protein. Eur J Biochem 270: 1211–1221. [DOI] [PubMed] [Google Scholar]
- 27. Fröbel J, Rose P, Lausberg F, Blummel AS, Freudl R, Müller M, (2012) Transmembrane insertion of twin-arginine signal peptides is driven by TatC and regulated by TatB. Nat Commun 3: 1311 10.1038/ncomms2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerard F, Cline K (2007) The thylakoid proton gradient promotes an advanced stage of signal peptide binding deep within the Tat pathway receptor complex. J Biol Chem 282: 5263–5272. [DOI] [PubMed] [Google Scholar]
- 29. Rollauer SE, Tarry MJ, Graham JE, Jääskeläinen M, Jäger F, Johnson F, et al. (2012) Structure of the TatC core of the twin-arginine protein transport system. Nature 492: 210–214. 10.1038/nature11683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cline K, Mori H (2001) Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J Cell Biol 154: 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Halbig D, Wiegert T, Blaudeck N, Freudl R, Sprenger GA (1999) The efficient export of NADP-containing glucose-fructose oxidoreductase to the periplasm of Zymomonas mobilis depends both on an intact twin-arginine motif in the signal peptide and on the generation of a structural export signal induced by cofactor binding. Eur J Biochem 263: 543–551. [DOI] [PubMed] [Google Scholar]
- 32. Stanley NR, Palmer T, Berks BC (2000) The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli . J Biol Chem 275: 11591–11596. [DOI] [PubMed] [Google Scholar]
- 33. Ize B, Gerard F, Zhang M, Chanal A, Voulhoux R, Palmer T, et al. (2002) In vivo dissection of the Tat translocation pathway in Escherichia coli . J Mol Biol 317: 327–335. [DOI] [PubMed] [Google Scholar]
- 34. Hinsley AP, Stanley NR, Palmer T, Berks BC (2001) A naturally occurring bacterial Tat signal peptide lacking one of the 'invariant' arginine residues of the consensus targeting motif. FEBS Lett 497: 45–49. [DOI] [PubMed] [Google Scholar]
- 35. Molik S, Karnauchov I, Weidlich C, Herrmann RG, Klösgen RB (2001) The Rieske Fe/S protein of the cytochrome b6/f complex in chloroplasts: missing link in the evolution of protein transport pathways in chloroplasts? J Biol Chem 276: 42761–42766. [DOI] [PubMed] [Google Scholar]
- 36. Blaudeck N, Kreutzenbeck P, Freudl R, Sprenger GA (2003) Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J Bacteriol 185: 2811–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bogsch E, Brink S, Robinson C (1997) Pathway specificity for a ΔpH-dependent precursor thylakoid lumen protein is governed by a 'Sec-avoidance' motif in the transfer peptide and a 'Sec-incompatible' mature protein. EMBO J 16: 3851–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cristóbal S, de Gier JW, Nielsen H, von Heijne G (1999) Competition between Sec- and TAT-dependent protein translocation in Escherichia coli . EMBO J 18: 2982–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Keersmaeker S, Van Mellaert L, Schaerlaekens K, Van Dessel W, Vrancken K, Lammertyn E, et al. (2005) Structural organization of the twin-arginine translocation system in Streptomyces lividans . FEBS Lett 579: 797–802. [DOI] [PubMed] [Google Scholar]
- 40.Mehner D, Osadnik H, Lünsdorf H, Brüser T (2012) The Tat system for membrane translocation of folded proteins recruits the membrane-stabilizing Psp machinery in Escherichia coli. J Biol Chem. [DOI] [PMC free article] [PubMed]
- 41. Schreiber S, Stengel R, Westermann M, Volkmer-Engert R, Pop OI, Müller J (2006) Affinity of TatCd for TatAd elucidates its receptor function in the Bacillus subtilis Tat translocase system. J Biol Chem 281: 19977–19984. [DOI] [PubMed] [Google Scholar]
- 42. Dilks K, Gimenez MI, Pohlschröder M (2005) Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J Bacteriol 187: 8104–8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frielingsdorf S, Jakob M, Klösgen RB (2008) A stromal pool of TatA promotes Tat-dependent protein transport across the thylakoid membrane. J Biol Chem 283: 33838–33845. 10.1074/jbc.M806334200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tullman-Ercek D, Delisa MP, Kawarasaki Y, Iranpour P, Ribnicky B, et al. (2007) Export pathway selectivity of Escherichia coli twin-arginine translocation signal peptides. J Biol Chem. [DOI] [PMC free article] [PubMed]
- 45. Taubert J, Brüser T (2014) Twin-arginine translocation-arresting protein regions contact TatA and TatB. Biol Chem 395: 827–836. 10.1515/hsz-2014-0170 [DOI] [PubMed] [Google Scholar]
- 46. Holzapfel E, Eisner G, Alami M, Barrett CM, Buchanan G, Lüke I, et al. (2007) The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 46: 2892–2898. [DOI] [PubMed] [Google Scholar]
- 47. Maurer C, Panahandeh S, Jungkamp AC, Moser M, Müller M (2010) TatB Functions as an Oligomeric Binding Site for Folded Tat Precursor Proteins. Mol Biol Cell 21: 4151–4161. 10.1091/mbc.E10-07-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huber D, Rajagopalan N, Preissler S, Rocco MA, Merz F, Kramer G, et al. (2011) SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol Cell 41: 343–353. 10.1016/j.molcel.2010.12.028 [DOI] [PubMed] [Google Scholar]
- 49. Karamyshev AL, Johnson AE (2005) Selective SecA association with signal sequences in ribosome-bound nascent chains: a potential role for SecA in ribosome targeting to the bacterial membrane. J Biol Chem 280: 37930–37940. [DOI] [PubMed] [Google Scholar]
- 50. Xie K, Dalbey RE (2008) Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol 6: 234–244. 10.1038/nrmicro1845 [DOI] [PubMed] [Google Scholar]
- 51. Bolhuis A, Bogsch EG, Robinson C (2000) Subunit interactions in the twin-arginine translocase complex of Escherichia coli . FEBS Lett 472: 88–92. [DOI] [PubMed] [Google Scholar]
- 52. Sargent F, Gohlke U, De Leeuw E, Stanley NR, Palmer T, Saibil HR, et al. (2001) Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur J Biochem 268: 3361–3367. [DOI] [PubMed] [Google Scholar]
- 53. Bolhuis A, Mathers JE, Thomas JD, Barrett CM, Robinson C (2001) TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli . J Biol Chem 276: 20213–20219. [DOI] [PubMed] [Google Scholar]
- 54. Heikkilä MP, Honisch U, Wunsch P, Zumft WG (2001) Role of the Tat ransport system in nitrous oxide reductase translocation and cytochrome cd1 biosynthesis in Pseudomonas stutzeri . J Bacteriol 183: 1663–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ray N, Nenninger A, Mullineaux CW, Robinson C (2005) Location and mobility of twin arginine translocase subunits in the Escherichia coli plasma membrane. J Biol Chem 280: 17961–17968. [DOI] [PubMed] [Google Scholar]
- 56. Bageshwar UK, Whitaker N, Liang FC, Musser SM (2009) Interconvertibility of lipid- and translocon-bound forms of the bacterial Tat precursor pre-SufI. Mol Microbiol 74: 209–226. 10.1111/j.1365-2958.2009.06862.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brehmer T, Kerth A, Graubner W, Malesevic M, Hou B, Brüser T, et al. (2012) Negatively charged phospholipids trigger the interaction of a bacterial Tat substrate precursor protein with lipid monolayers. Langmuir 28: 3534–3541. 10.1021/la204473t [DOI] [PubMed] [Google Scholar]
- 58. Hou B, Frielingsdorf S, Klösgen RB (2006) Unassisted membrane insertion as the initial step in ΔpH/Tat-dependent protein transport. J Mol Biol 355: 957–967. [DOI] [PubMed] [Google Scholar]
- 59. Kerth A, Brehmer T, Meister A, Hanner P, Jakob M, Klösgen RB, et al. (2012) Interaction of a Tat substrate and a Tat signal peptide with thylakoid lipids at the air-water interface. Chembiochem 13: 231–239. 10.1002/cbic.201100458 [DOI] [PubMed] [Google Scholar]
- 60. Shanmugham A, Wong Fong Sang HW, Bollen YJ, Lill H (2006) Membrane binding of twin arginine preproteins as an early step in translocation. Biochemistry 45: 2243–2249. [DOI] [PubMed] [Google Scholar]
- 61. Jack RL, Sargent F, Berks BC, Sawers G, Palmer T (2001) Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J Bacteriol 183: 1801–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Haldimann A, Wanner BL (2001) Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183: 6384–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ryu Y, Schultz PG (2006) Efficient incorporation of unnatural amino acids into proteins in Escherichia coli . Nat Methods 3: 263–265. [DOI] [PubMed] [Google Scholar]
- 65. Richter S, Brüser T (2005) Targeting of unfolded PhoA to the TAT translocon of Escherichia coli . J Biol Chem 280: 42723–42730. [DOI] [PubMed] [Google Scholar]
- 66. Brüser T, Deutzmann R, Dahl C (1998) Evidence against the double-arginine motif as the only determinant for protein translocation by a novel Sec-independent pathway in Escherichia coli . FEMS Microbiol Lett 164: 329–336. [DOI] [PubMed] [Google Scholar]
- 67. Wilms B, Hauck A, Reuss M, Syldatk C, Mattes R, Siemann M, et al. (2001) High-cell-density fermentation for production of L-N-carbamoylase using an expression system based on the Escherichia coli rhaBAD promoter. Biotechnol Bioeng 73: 95–103. [DOI] [PubMed] [Google Scholar]
- 68. Wexler M, Sargent F, Jack RL, Stanley NR, Bogsch EG, Robinson C, et al. (2000) TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J Biol Chem 275: 16717–16722. [DOI] [PubMed] [Google Scholar]
- 69. Lindenstrauß U, Brüser T (2009) Tat transport of linker-containing proteins in Escherichia coli . FEMS Microbiol Lett 295: 135–140. 10.1111/j.1574-6968.2009.01600.x [DOI] [PubMed] [Google Scholar]
- 70. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 71. Scotti PD (1985) The estimation of virus density in isopycnic cesium chloride gradients. J Virol Methods 12: 149–160. [DOI] [PubMed] [Google Scholar]
- 72. Valentine RC, Shapiro BM, Stadtman ER (1968) Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli . Biochemistry 7: 2143–2152. [DOI] [PubMed] [Google Scholar]
- 73. Standar K, Mehner D, Osadnik H, Berthelmann F, Hause G, Lünsdorf H, et al. (2008) PspA can form large scaffolds in Escherichia coli . FEBS Lett 582: 3585–3589. 10.1016/j.febslet.2008.09.002 [DOI] [PubMed] [Google Scholar]
- 74. Hess B, Kutzner C, van der Spoel DEL (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4: 435–447. [DOI] [PubMed] [Google Scholar]
- 75. Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH (2007) The MARTINI forcefield: coarse grained model for biomolecular simulations. J Phys Chem B 111: 7812–7824. [DOI] [PubMed] [Google Scholar]
- 76. Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ (2008) The MARTINI coarse-grained force field: extension to proteins. J Chem Theory Comput 4: 819–834. [DOI] [PubMed] [Google Scholar]
- 77. Berendsen HJC, Postma JPM, van Gunsteren WF, Dinola AJRH (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81: 3684–3690 [Google Scholar]
- 78. Dahlberg M, Maliniak A (2010) Mechanical Properties of Coarse-Grained Bilayers Formed by Cardiolipin and Zwitterionic Lipids. J Chem Theory Comput 6: 1638–1649 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) During affinity chromatography, specific cross-links elute in fractions E2 and E3. Shown is the immunoblot-detection of cross-links of RR-HiPIP with a M26pBpa substitution to TatB, using antibodies recognizing TatB. M: solubilized membranes, W: last wash fraction, E1-E4: elution fractions. (B) Without UV irradiation or pBpa, no shifted bands can be detected in the elution fractions. For this important control, we chose HiPIP M26pBpa, which gives strong cross-links to all Tat components after UV irradiation (see Fig. 1A). No cross-links could be detected without UV irradiation or pBpa (+ pBpa/-UV, −pBpa/-UV, left blots). We also analyzed the UV-dependence of the cross-links for the HiPIP A13pBpa variant that was the most important variant in our studies. As expected for the UV-activatable cross-linker pBpa, the cross-links were absent without UV-irradiation. The pBpa-dependence of cross-links to this position is already shown in Fig. 3. As additional control, we demonstrated that wild-type HiPIP per se does not give any shifts in the presence of pBpa and UV (right blots). The presence of TatA, TatB, and TatC (pRK-tatABC) in the membranes and elution fractions was assessed as shown in Figs. 1 and 2. (C) HiPIP RR/KK variants with pBpa (*) at indicated positions were produced in similar concentrations in the experiments shown in Fig. 1. Note significant precursor-accumulation in case of the KK-variants and more mature HiPIP in case of the RR-variants, indicating RR-dependent transport. Processing of KK-variants to mature size is mainly due to unspecific proteolytic degradation of the signal peptide. Detection in crude extracts with HiPIP specific antibodies.
(TIF)
Detection of HiPIP in subcellular fractions by SDS-PAGE/Western blotting, using HiPIP-specific antibodies. In control blots, purity of periplasmic and cytoplasmic fractions was confirmed by detection of periplasmic b-lactamase (bla) and cytoplasmic biotin carboxyl carrier protein (BCCP). Note some detectable transport of KK-HiPIP variants in which the Tat motif is optimized by an A13F or A13pBpa substitution (*: mature periplasmic HiPIP). This position corresponds to the consensus “F” position in the motif, and F as well as pBpa attribute a large, hydrophobic, aromatic side chain to the motif, which can promote the Tat translocon interaction and thus partially compensates for the transport-inactivating RR>KK exchange. Strains and conditions as in Fig. 1.
(TIF)
(A) Experimentally determined densities of the fractions analyzed in B, showing identity of the two density profiles. (B) Detection of TatA and the polytopic membrane protein YidC (membrane marker) after isopycnic CsCl gradient centrifugation of soluble or membrane fractions from strain MC4100. No YidC is detectable in the soluble fraction, indicating that this fraction is virtually membrane-free. Note that soluble TatA micelles sediment to a density of ∼1.21 g/ml, which suggests a tight association with lipids but not with vesicles. Membrane-associated TatA sediments with membrane vesicles to a density of ∼1.18 g/ml, as confirmed by the detection of the membrane marker YidC (lower two blots).
(TIF)
(A) Complementation of the chain formation phenotype of the Tat-deficient DADE strain by a single-copy chromosomally integrated tatA-strep-tatBC operon (strain DADE tatA-strep-tatBC). As controls, the Tat-system-containing parental strain MC4100 and the Tat-deficient strain DADE have been analyzed in parallel. (B) Tat transport of HiPIP as produced from pRK-hip in strain DADE tatA-strep-tatBC shown by Western-blot analysis of subcellular fractions. P: periplasm, M: membranes, C: cytoplasm. The periplasm contains transported mature (m) HiPIP, whereas only the cytoplasm contains unprocessed precursor (p) and some to mature size degraded HiPIP. The control blot on the right side detects the biotin carboxyl carrier protein (BCCP, cytoplasmic marker). (C) TatA micelles. Overview EM micrograph of purified TatA micelles from the cytoplasm of strain DADE tatA-strep-tatBC. The size bar indicates 100 nm.
(TIF)
Color code: yellow: HiPIP signal peptide; magenta: HiPIP mature domain (FeS-cofactor in orange); red: TatA trans-membrane domain; green: TatA amphipathic helix.
(TIF)
Electronic absorption spectra of folded purified cofactor-containing HiPIP (wt) in comparison to the analyzed HiPIP variants with mutations in the mature domain (T50A, T50D, P104G, P104D, L119E). The spectra all show the typical [4Fe-4S]-cofactor absorption that indicates complete cofactor insertion and thus stable folding of HiPIP.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.