Abstract
Cytosolic SULT1A1 participates in the bioconversion of a plethora of endogenous and xenobiotic substances. Genetic variation in this important enzyme such as SNPs can vary by ethnicity and have functional consequences on its activity. Most SULT1A1 genetic variability studies have been centered on the SULT1A1*1/2 SNP. Highlighted here are not only this SNP, but other genetic variants associated with SULT1A1 that could modify drug efficacy and xenobiotic metabolism. Some studies have investigated how differential metabolism of xenobiotic substances influences susceptibility to or protection from cancer in multiple sites. This review will focus primarily on the impact of SULT1A1 genetic variation on the response to anticancer therapeutic agents and subsequently how it relates to environmental and dietary exposure to both cancer-causing and cancer-preventative compounds.
Keywords: cancer risk, chemoprevention, environmental exposures, pharmacogenetics, single nucleotide polymorphism, SULT1A1
Background
Human cytosolic sulfotransferases (SULTs) are Phase II biotransformation enzymes that catalyze the sulfation of a wide variety of structurally diverse endo- and xenobiotics [1]. They are members of the SULT gene superfamily and are divided into two major families that are well characterized, the phenol sulfotransferase family, SULT1, and the hydroxysteroid family, SULT2. The SULT1 family is further classified into four major subfamilies: phenol SULTs (SULT1A), thyroid hormone SULTs (SULT1B), hydroxyarylamine SULTs (SULT1C), and estrogen SULTs (SULT1E) [2]. At the time of this review, SULT pharmacogenetic studies have focused primarily on SULT phenol-preferring isoform 1A1 and the functional consequences of a common SNP, a non-synonymous G to A transition at nucleotide 638 in exon 7. This SNP results in an amino acid change at codon 213 from arginine (designated SULT1A1*1) to histidine (SULT1A1*2). The variant allozyme confers lower enzymatic activity and lower thermostability of the expressed protein [3,4]. There is, however, another coding region SULT1A1 SNP that is has not been commonly examined in pharmacogenetic studies until just recently. It is defined by an A to G conversion at nucleotide 667, resulting in a nonsynonymous change in amino acids from methionine to valine at codon 223 (SULT1A1 Met223Val) and it is given the designation SULT1A1*3. This allozyme is fairly common among African–Americans as opposed to Caucasians and Chinese, with an allele frequency of 0.229, 0.012 and 0.006, respectively [5]. This SNP is not included in many studies likely due to its low prevalence in the Caucasian populace and the under-representation of African–American subjects in pharmacogenetic studies. Several SNPs that are in linkage disequilibrium with each other and with SULT1A1*1 have also been identified in both the distal and proximal promoter region of SULT1A1, and are associated with platelet enzymatic activity [6,7]. Haplotype analysis also showed that there was considerable variability in the frequency of the haplotypes among different populations. Additionally, in 2007, copy number variation (CNV) was described in SULT1A1 [8]. Copy number was associated with enzymatic activity, and African–American subjects were significantly more likely to have higher CNV than Caucasians, providing a potential biological basis for the observation that African–Americans tend to have higher basal platelet SULT1A1 activity than Caucasians [9]. Other SNPs have been identified in the 3′-flanking region of the gene. These are in linkage with each other and in strong linkage with SULT1A1*1 (D′ = 0.85). When the collective effects of 3′-UTR SNPs, SULT1A1*1/2, and CNV on SULT1A1 activity were examined in 498 Caucasian and 127 African–American subjects, SULT1A1*1/2 did not significantly contribute to the variation in SULT1A1 enzymatic activity when the 3′-UTR SNPs were included in the statistical model. Two major haplotypes (ACG and GTA) were significantly associated with SULT1A1 activity, and when stratified by copy number, the SULT1A1 3′-UTR SNPs remain significantly associated with SULT1A1 enzymatic activity, explaining 21% of the variation seen in Caucasians. These models, however, accounted for little (12%) of the interindividual variability in activity observed in African–American subjects [10]. A recent genotype/phenotype association study in a Japanese population demonstrated that CNV was the strongest predictor of SULT1A1 platelet activity, and that all variants combined predicted only 14% of the observed variability in this population [11].
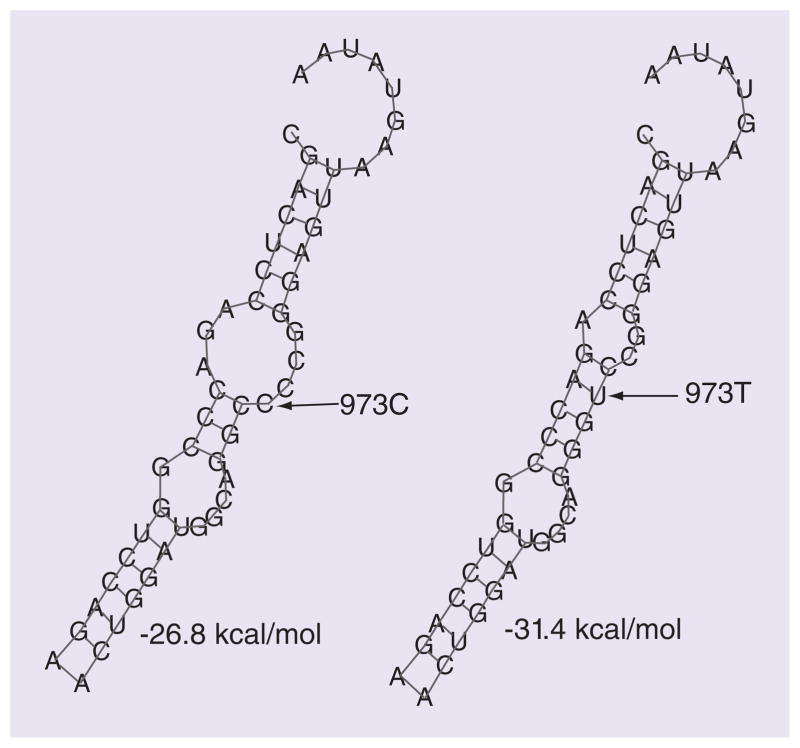
Nagar et al. discovered that the *2 allozyme demonstrated an almost sixfold lower half-life in both Sf-9 insect cells and stably transfected MCF-7 cells when compared with the *1 and *3 allozymes. They also suggest that *2 is ubiquitinated in the Sf-9 lysates while *1 is not [12]. Ubiquitination signifies a more rapid protein degradation and may explain the lower half-life and why *2 exhibits lower activity. Moreover, the 3′-UTR SNPs, which are linked to the *1/2 allele, confer differences in mRNA half-life, and could also explain activity differences. Functional characterization revealed that the 3′-UTR SNPs disrupted a binding site for the miR-631, thus also regulating SULT1A1 expression in a genotype-specific manner (Figure 1) [10].
Figure 1. miR-631 binding site by in silico analysis.
miR-631 differentially regulates SULT1A1 in allele-specific manner at 973C>T. In silico analysis of the pairing of miR-631 to the binding site in the 3′-UTR of SULT1A1. The C to T allele change decreased free energy from −26.8 to −31.4 kcal/mol.
Reproduced from [10] with permission from Oxford University.
SULT1A1 genetic variation & therapeutic agents
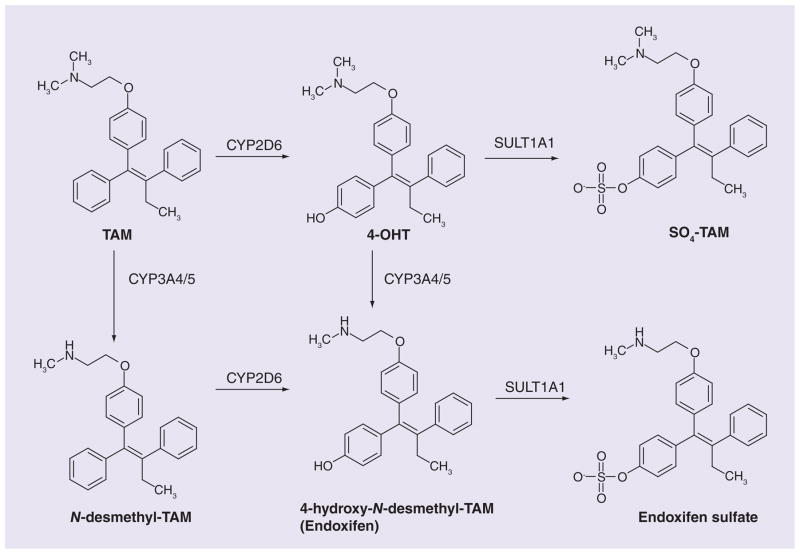
The efficacy of tamoxifen (TAM), the gold standard of treatment for estrogen receptor-positive breast cancer, varies widely between individuals. TAM is a prodrug that must be converted to its active metabolites by phase I and II drug-metabolizing enzymes, including SULT1A1, in order to exert its anti-estrogenic effects (Figure 2). Thus, SULT1A1 pharmacogenetic studies involving TAM have been conducted to interrogate patient outcome (Table 1). An initial report in 2002 by Nowell et al. involving 337 breast cancer cases in Arkansas, USA, of which 160 received TAM and 177 did not, indicated that the SULT1A1*1 high activity allele was significantly associated with improved overall survival in breast cancer patients receiving TAM. In fact, those individuals who were homozygous for the *2 allele had almost three-times the risk of death compared with either homozygous or heterozygous carriers of the *1 allele (hazard ratio [HR]: 2.9; 95% CI: 1.1–7.6) [13]. The same group conducted another study in TAM-treated breast cancer patients and found that carriers of the UGT2B15*2 allele along with homozygous SULT1A1*2 had significantly increased risk of recurrence and reduced 5-year survival [14]. In another early study the SULT1A1*1 homozygous patients treated with TAM in Sweden were found to have a decreased risk of recurrence (relative risk: 0.48; 95% CI: 0.21–1.12; p = 0.074). The association achieved significance when combined with carriers of the CYP2D6*4 SNP (relative risk: 0.38; 95% CI: 0.19–0.74; p = 0.0041) [15]. A subsequent study in the context of a clinical trial in the Stockholm Breast Cancer Group reported an improved recurrence-free survival for homozygous carriers of the SULT1A1*1 allele with 2 years of TAM treatment (HR: 0.33; 95% CI: 0.12–0.96; p = 0.04), but the same result was not evident in patients receiving TAM for 5 years [16]. No associations were found between the SNP and disease-free or overall survival for 216 lymph-node-positive breast cancer patients in Austria receiving TAM, hormonal, anthracycline or nonanthracycline therapy [17]. SULT1A1 CNV has also been explored in TAM pharmacogenetics with no significant associations reported either with disease-free survival or with recurrence after a 14 year median follow-up of 190 postmenopausal women in the North Central Cancer Treatment Group receiving TAM monotherapy [18]. A contradictory report came from a study of Finnish breast cancer patients. Those homozygous for the *2 low activity allele demonstrated a significantly improved overall survival (HR: 0.50; 95% CI: 0.29–0.88; p = 0.015). There was a similar yet statistically insignificant improvement in breast cancer specific survival (HR: 0.53; 95% CI: 0.26–1.05; p = 0.069) and no statistical difference was detected in relapse-free survival (p = 0.091). Nevertheless, these findings were in a cohort comprised of 145 patients who received either TAM, cyclophosphamide-based chemotherapy, or both [19].
Figure 2. Tamoxifen metabolism.
TAM undergoes bioconversion to form its active metabolites by phase I drug-metabolizing enzymes. The active metabolites are substrates for SULT1A1.
4-OHT: 4-hydroxy-TAM; SO4-TAM: 4-sulfoxy-TAM; TAM: Tamoxifen.
Table 1.
SULT1A1 genetic variation in vivo and therapeutic agents.
| Study (year) | Cancer site | Population | Cases/controls (n) | Main findings | Ref. |
|---|---|---|---|---|---|
| Nowell et al. (2002) | Breast | African–American and Caucasian American | 337 cases (160 with TAM, 177 without TAM) | Increased risk of death with SULT1A1*2/*2 | [13] |
| Nowell et al. (2005) | Breast | African–American and Caucasian American | 162/175 | Significantly increased risk of recurrence and 5-year survival with UGT2B15*2 and SULT1A1*2/*2 | [14] |
| Wegman et al. (2005) | Breast | Swedish | 226 TAM treated cases | Significantly decreased risk of recurrence with CYP2D6*4 and SULT1A1*1/*1 | [15] |
| Wegman et al. (2007) | Breast | Swedish (Stockholm Breast Cancer Group) | 677 TAM treated cases | Significantly improved recurrence-free survival with treatment for 2 years and SULT1A1*1 | [16] |
| Tengström et al. (2012) | Breast | Finnish | 412 (76 treated with cyclophosphamide, 65 with TAM, four with both) | Significantly improved overall survival and moderately improved breast cancer specific survival associated with TAM or chemotherapy and SULT1A1*2/*2 NA with relapse-free survival | [19] |
| Grabinski et al. (2006) | Breast | Caucasian and Hispanic | 296 TAM treated cases | NA with age, tumor size, histological grade, stage of disease, menopausal status, HER2 overexpression, and ER or PR status | [20] |
| Serrano et al. (2011) | Breast | Caucasian (Italian Tamoxifen Prevention Trial) | 46 (20 cases treated with TAM, 26 cases received placebo)/135 (65 controls treated with TAM, 71 controls received placebo) | NA with breast cancer risk | [21] |
| Knechtel et al. (2010) | Breast | Austrian | 216 LN+ cases treated with TAM, hormone, anthracycline or nonanthracycline therapy | NA with either disease-free or overall survival | [17] |
| Moyer et al. (2011) | Breast | Mixed, but 95% Caucasian | 190 on TAM monotherapy | NA between either disease-free survival or recurrence and SULT1A1 copy number | [18] |
| Jin et al. (2005) | Breast | Mixed, but 91% Caucasian | 80 TAM treated cases | NA with TAM and metabolite serum concentrations Trend toward increase in 4-OHT and endoxifen with SULT1A1*2/*2 | [22] |
| Gjerde et al. (2008) | Breast | Caucasians in Norway | 151 TAM treated cases | NA with TAM and metabolite serum concentrations for either genotype or copy number Significant trend between increasing SULT1A1*1 alleles and serum metabolite ratios (+NDtam/TAM and -NDDtam/NDtam) |
[23] |
| Fernandez-Santander et al. (2013) | Breast | Spanish Caucasians | 135 TAM treated cases | NA with TAM and metabolite serum concentrations | [24] |
| Innocenti et al. (2013) | Multiple | Mixed | 47 ABT-751 treated cases | Increased clearance and metabolic ratios and decreased AUC with >2 copies | [25] |
AUC: Area under the curve; LN+: Lymph node positive; NA: No association; NDtam: N-demethyltamoxifen; TAM: Tamoxifen.
The role SULT1A1 genetic variation plays in the pharmacokinetics of TAM has also been evaluated. To determine whether genetic variation in TAM-metabolizing enzymes would influence TAM active metabolite bioavailability, Jin et al. conducted a small study in 80 patients across the USA, which would provide at least 93% power to detect a statistically significant increase or decrease in plasma drug concentration. They found that plasma concentrations of TAM and its metabolites were not significantly affected by SULT1A1*1/2 genotype. However, a trend toward higher concentrations of the active metabolites of TAM, 4-hydroxy-TAM (4-OHT) and endoxifen, could be seen among carriers of the *2/*2 genotype [22]. Additionally, in a study observing 151 Caucasian women in Norway, similar results were reported. Neither SULT1A1*1/2 genotype nor copy number was able to significantly influence TAM and TAM metabolite levels in patient serum. Yet, when ratios of metabolites were examined, increasing SULT1A1*1 alleles had significant trends in positive associations with N-demethyltamoxifen/TAM and negative associations with N-dedimethyltamoxifen/N-demethyltamoxifen [23]. More recently, Fernandez-Santander et al. conducted a small study in 135 Spanish breast cancer patients. SULT1A1*2 had a minor allele frequency of 30.08% in this population and there were no associations between SULT1A1 genotype and serum levels of TAM and TAM metabolites observed [24].
Based on available TAM pharmacogenetic data that was available at the time, Williams et al. in 2008 concluded that SULT1A1 pharmacogenetic screening was not warranted before administration of drug therapy, but that future studies involving novel drugs metabolized by SULT1A1 should take into consideration the contribution of genetic variation in the enzyme [26]. Moving forward, in a Phase 1 clinical trial, Innocenti et al. reported that SULT1A1 CNV plays a major role in the pharmacokinetics of a novel anticancer agent, ABT-751. When ABT-751 undergoes phase II metabolism by conjugation with a glucuronide (ABT-751G) or sulfate (ABT-751S), the antitubulin compound is rendered inactive. In cancer patients treated with ABT-751, those with greater than two copies of SULT1A1 experienced an average 34% increase in ABT-751 clearance (p = 0.044), an 18% reduction in ABT-751 area under the plasma concentration-time curve (p = 0.045), and a 50% increase in sulfation metabolic ratios (p = 0.025). They also observed a gene–dose effect of increasing metabolic ratios with increasing copies of SULT1A1 [25]. These data concordantly suggest a decrease in active ABT-751 bioavailability with higher SULT1A1 gene copies, implying that patients receiving this drug may have to have the dose adjusted based on individual genetic profiles of phase II enzymes.
SULT1A1 variation has been studied in vitro for its effects on other therapeutic agents it metabolizes. Aminoflavone (5-amino-2-[4-amino-3-fluorophenyl]-6,8-difluoro-7-methylchromen-4-one; AF) is a pro-drug that must be metabolized by phase I and II metabolic enzymes to exert its oxidative cytotoxic effects on breast cancer cells, inducing oxidative DNA damage and reactive oxidative species-mediated apoptosis [27]. Zheng et al. were able to stably express either human SULT1A1*1, *2 or *3 along with CYP1A1*1 and found that AF has an allele-specific sensitivity to SULT1A1 *3 (IC50, 0.01 μmol/l) > *1 (IC50, 0.01 μmol/l) > *2 (IC50, 0.01 μmol/l; Table 2). This suggests that patients receiving AF could benefit from the high metabolic activity alleles whereas carriers of the low-activity SULT1A1*2 allele may experience a poor therapeutic response [28]. Other in vitro studies in human liver cytosols have also been conducted to investigate the role of SULT1A1 3′-UTR SNPs and CNV. Fulvestrant is a pure antiestrogen that is approved to treat hormone receptor-positive metastatic breast cancer in postmenopausal women and its sulfation was correlated with both SULT1A1*1/2 genotype (p = 0.023) and copy number (p < 0.0001) [29]. Toremifene (TOR) is a selective estrogen receptor modulator given as adjuvant therapy for breast cancer and is being evaluated for prostate cancer prevention. Its active metabolite, 4-hydroxy-TOR, is sulfated by SULT1A1. SULT1A1 copy number (pANOVA = <0.0001) was significantly associated with 4-hydroxy-TOR sulfation demonstrating increasing activity with increasing copies. The SULT1A1*1/2 genotype was significant in influencing activity as well (pANOVA = 0.024). Even when haplo-types were constructed with 3′-UTR SNPs, significant associations were evident (ptrend = 0.008); however, these associations were not affected by the addition of SULT1A1*1/2 to the model [30]. To date, these variants have not been investigated in many disease-association studies. Seth et al. postulated that variations in enzymatic activity of sulfotransferases due to SNPs could influence breast cancer risk when they observed an increase in gene expression of the SULT1A subfamily of phenol SULTs in ZR75-1 human breast carcinoma cells by 4-OHT treatment. Although SULT1A1 SNP alone was not enough to modulate risk of developing breast cancer (p = 0.69), in this large study of 444 breast cancer patients and 227 controls they did report that carrying the SULT1A1*1 allele was associated with an earlier onset of disease and with presence of other tumors [31]. When Grabinski et al. sought to determine if associations exist between SULT1A1*1/2 genotype and prognostic clinical and biological markers in breast cancer patients that included Hispanics who received TAM, none were made [20]. TAM has also been approved for use in reducing the incidence of breast cancer in high-risk women. TAM efficacy in preventing breast cancer was evaluated in a nested case–control study from the Italian Tamoxifen Prevention Trial. Breast cancer risk was not altered by the SULT1A1*1/2 SNP across subjects participating in the placebo or TAM arms of the study [21]. Since the original hypothesis, genetic variation in SULT1A1 alone has been found to have varying effects on the risk of developing cancer in multiple sites [32–34]. It has been reported that approximately 80% of all human cancers can be attributed to lifestyle choices, such as diet, smoking and environmental chemical exposures, as a causal factor [35–38]. Those factors that are both controllable, such as environmental exposure to putative carcinogens and dietary lifestyle choices, and uncontrollable, that is, interindividual variation in xenobiotic-metabolizing enzymes, must be taken into consideration in order to perform pharmacogenetic analysis of disease risk and response.
Table 2.
SULT1A1 genetic variation in vitro and therapeutic agents.
| Study (year) | Drug | SULT1A1 source | Main findings | Ref. |
|---|---|---|---|---|
| Zheng et al. (2010) | Aminoflavone | Chinese hamster V79 cells stably coexpressing CYP1A1*1 with either SULT1A1*1, *2 or *3 | IC50*3 > *1 > *2 | [28] |
| Edavana et al. (2011) | Fulvestrant | 104 liver cytosols (Cooperative Human Tissue Network) | Significant correlation between fulvestrant sulfation and SULT1A1*1/2 genotype and copy number | [29] |
| Edavana et al. (2012) | 4-OH toremifene | 104 liver cytosols (Cooperative Human Tissue Network) | Significant trend with SULT1A1 3′-UTR haplotypes and activity Significant increase in activity with increasing copy number |
[30] |
SULT1A1 genetic variation, well-done meat consumption (heterocyclic amine exposure) & cancer risk
SULT1A1 utilizes the obligatory cosubstrate, 3′-phos phoadenosine-5′-phosphosulfate, to transfer a sulfonyl group to acceptor molecules including hydroxyl, sulfhydryl, amino and N-oxide groups of numerous substrates [39,40]. Sulfation tends to make molecules more water soluble and, therefore, more readily excreted, but in the case of certain pro mutagenic and procarcinogenic compounds, sulfation bioactivates to reactive metabolites capable of adducting to DNA, forming biological lesions that can initiate carcinogenesis if not repaired [41,42]. Heterocyclic amines (HCAs) are such compounds. They are present in meat cooked at high temperatures and N-hydroxy-HCAs, hydroxylated by phase I metabolic enzymes, are substrates for SULT1A1. The predominant HCA formed from cooking meat at high temperatures, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), is a potent prostate carcinogen in rats [43,44]. N-OH-PhIP is a substrate for and is bioactivated principally by SULT1A1 to form DNA adducts [45]. In platelet cytosols the SULT1A1*2 low-activity variant was shown to decrease its DNA-binding capability when compared with SULT1A1*1 [46]. Another dietary HCA, 2-amino-3-methylimidazo[4,4-f]quinoline, has the ability to form DNA adducts and become carcinogenic in both mice and rats [47]. One study revealed that a single human prostate cytosol with the SULT1A1*2/*2 low-activity genotype was able to form less DNA adducts than cytosols with the *1/*2 and *1/*1 genotypes in the presence of 2-amino-3-methylimidazo[4,4-f]quinoline [48]. These findings infer that this genetic variant is important in evaluating the carcinogenic potential of HCAs.
Studies examining the relationship between the consumption of well-done meat, SULT1A1 genetic variation and cancer risk have been performed (Table 3). A recent meta-analysis of available data revealed a slight increase in breast cancer risk among overall (odds ratio [OR]pooled overall = 1.12; 95% CI: 1.02–1.24) and postmenopausal (ORpooled post = 1.17; 95% CI: 1.03–1.32) carriers of the SULT1A1*2 low-activity variant allele. This finding was enhanced in Asian women (ORpooled Asian = 2.01; 95% CI: 1.24–3.26). This same group also performed a case–control study involving 400 cases and 400 controls that revealed no association between breast cancer risk and the SNP alone. However, upon further investigation, a positive interaction between high smoked meat intake and the SULT1A1*2 low-activity variant allele resulted [49]. They later reported that *2 was indeed associated with elevated breast cancer risk among premenopausal (OR: 3.31; 95% CI: 1.66–6.62) and postmenopausal women with high smoked meat intake (OR: 3.81; 95% CI: 1.79–8.10) [50].
Table 3.
SULT1A1 genetic variation, well-done meat consumption (heterocyclic amine exposure) and cancer risk.
| Study (year) | Cancer site | Population | Cases/controls (n) | Main findings | Ref. |
|---|---|---|---|---|---|
| Lee et al. (2012) | Breast | Chinese mixed (meta-analysis) | 400/400 | NA with SULT1A1*2 alone Positive interaction between high smoked meat intake and SULT1A1*2 | [49] |
| Tao et al. (2012) | Breast | Chinese | 400/400 | Significantly increased risk with SULT1A1*2 among both pre- and post-menopausal women and high smoked meat intake | [50] |
| Lilla et al. (2007) | Colorectal | German | 505/604 | Significantly increased risk with SULT1A1*2 and frequent consumption of red meat | [51] |
| Cotterchio et al. (2008) | Colorectal | Canadian (Ontario Familial Colorectal Cancer Registry) | 842/1251 | Increased risk with SULT1A1*2 and CYP1B1 variant and red meat doneness intake | [52] |
| Nowell et al. (2004) | Prostate | African–American and Caucasian American | 464/459 | NA between genotype and meat consumption. Significantly increased risk with increasing SULT1A1 activity and well-done meat consumption | [53] |
| Koutros et al. (2009) | Prostate | Mixed (Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial) | 1126/1127 | NA | [54] |
NA: No association.
Colorectal cancer risk associated with frequent consumption of red meat was found to be significantly elevated among carriers of the SULT1A1*2 allele but not increased among subjects with the SULT1A1*1/*1 genotype (OR: 2.1, 95% CI: 1.1–4.1; and OR; 1.0, 95% CI: 0.5–2.1, respectively) [51]. Another colorectal cancer study corroborated this finding, with colorectal cancer risk increasing when variants of SULT1A1 and a phase I metabolic enzyme, CYP1B1, were incorporated into the analysis along with red meat doneness intake [52]. High cooked meat intake can significantly increase the risk of both breast and colorectal cancer. The SULT1A1*2 allele can further modulate this association. The SULT1A1*1/2 SNP has also been interrogated in prostate cancer risk with the consumption of well-done meat and no significant associations were found in two studies [53,54]. Because SULT1A1 phenotype is dependent upon more than just SULT1A1*1/2, one of the studies also investigated the effects of SULT1A1 activity on prostate cancer risk. A significant association was then observed between increasing SULT1A1 activity, meat consumption and prostate cancer risk (ptrend = 0.02) [53].
SULT1A1 genetic variation, smoking (polycyclic aromatic hydrocarbon exposure) & cancer risk
Polycyclic aromatic hydrocarbons (PAHs) are environmental pollutants that are procarcinogenic, pro-mutagenic and/or proteratogenic. Benzo[a]Pyrene (B[a]P) has long been used to study the process of activation of PAHs to genotoxic compounds [55,56]. Environmental sources of B[a]P include smoke from automobile exhaust, wood burning and cigarettes, and, like HCAs, they can also be formed from the cooking of meats at high temperatures [57,58]. Benzo[a]pyrene-7,8-dione is a representative PAH O-quinone that is reduced to B[a]P-7,8-catechol in a futile redox cycle; this reduction product is electrophilic and genotoxic [59]. When metabolism by SULT isoforms 1A1, 1A3 and 1E1 was examined, Zhang, et al. found that SULT1A1 is the major isoform that can intercept B[a]P-7,8-catechol in human lung cells, thereby limiting its ability to redox cycle. They report that the catalytic efficiency of SULT1A1*3 for sulfation of B[a]P-7,8-catechol was about half that observed with SULT1A1*1 [60].
Individuals exposed to a smoking environment are exposed to PAHs as well. The link between this environmental exposure and SULT1A1 genetic variation and cancer risk has been explored (Table 4). A meta-analysis examining lung cancer risk in a mixed population of 1669 cases and 1890 controls revealed an increase in risk for those with at least one SULT1A1*2 allele (OR: 1.66; 95% CI: 1.06–2.62; p = 0.03) in the dominant model. No significant association between this SNP and smokers versus nonsmokers in lung cancer risk was found [61]. Likewise, in a small Turkish study, carriers of the *2 low-activity allele had significantly increased risk for lung cancer (p = 0.027), yet when stratified by smoking status, no significant relationship was found [62]. The same researchers conducted another small Turkish study to identify the SNP’s role in prostate cancer and found similar results. Although carriers of the *2 allele had increased risk of prostate cancer, it was not significant. Neither was there a significant difference observed between smokers and nonsmokers [63]. In another meta-analysis of 20 case–control studies with 5915 cases and 7900 controls evaluating environment-related cancer (lung, esophageal and colorectal) risk, ten of those studies involved 1867 case and 1785 control subjects who smoked. Within the smoking population there were only marginally significant increases in risk between carriers of the SULT1A1*2 allele and susceptibility to both lung and esophageal cancers in the random-effects model (OR: 1.49, 95% CI: 0.99–2.24, p = 0.058; OR: 1.46, 95% CI: 1.00–2.14, p = 0.052, respectively). When subgroup analysis by ethnicity was performed, a stronger association between *2 carriers and cancer risk was clear among Asian smokers compared with Caucasian smokers (OR Asians: 2.00, 95% CI: 1.55–2.57, p < 0.001; OR Caucasians: 1.22, 95% CI: 1.02–1.46, p = 0.031) [64]. Liang et al. observed a significant increase in lung cancer risk among smokers with the variant SULT1A1*2 allele (OR: 1.85; 95% CI: 1.44–2.37). A more pronounced association was seen among younger smokers (OR: 2.28; 95% CI: 1.66–3.13, phomogeneity = 0.000). More-over, risk increased with increases in smoking dose (OR: 1.66, 95% CI: 0.75–3.68; OR 2.28, 95% CI: 1.47–3.54; OR: 3.35, 95% CI: 1.71–6.57 for those who smoked <15 pack-years, 15–36 pack-years and >36 pack-years, respectively, ptrend = 0.000) [65].
Table 4.
SULT1A1 genetic variation, smoking (polycyclic aromatic hydrocarbon exposure) and cancer risk.
| Study (year) | Cancer site | Population | Cases/controls (n) | Main findings | Ref. |
|---|---|---|---|---|---|
| Liao et al. (2012) | Lung | Mixed (meta-analysis) | 1669/1890 | Significantly increased risk with SULT1A1*2, but NA between smokers and nonsmokers | [61] |
| Li et al. (2012) | Lung and esophageal | Mixed (meta-analysis) | 1867/1785 | Marginally increased risk with SULT1A1*2 and smoking. Asian smokers had a stronger association | [64] |
| Arslan et al. (2009) | Lung | Turkish | 106/271 | Significantly increased risk with SULT1A1*2, but NA between smokers and nonsmokers | [62] |
| Liang et al. (2004) | Lung | Han Chinese | 805/809 | Significantly increased risk with SULT1A1*2 and smoking. Risk increased with smoking dose | [65] |
| Arslan et al. (2011) | Prostate | Turkish | 104/151 | NA | [63] |
| Li et al. (2012) | Colorectal | Mixed (meta-analysis) | 1867/1785 | NA | [64] |
| Cleary et al. (2010) | Colorectal | Canadian (Ontario Familial Colorectal Cancer Registry) | 1174/1293 | Significantly increased risk with SULT1A1*1/*1 and smoking >15 years | [66] |
| Lilla et al. (2007) | Colorectal | German | 505/604 | Significantly increased risk with SULT1A1*2 and 30+ pack-years of active smoking | [51] |
| Figueroa et al. (2008) | Bladder | Spanish Bladder Cancer Study | 1150/1149 | NA | [67] |
| Fortuny et al. (2006) | Bladder | Spanish | 958/1029 | NA | [68] |
| Kellen et al. (2006) | Bladder | Belgian | 200/385 | NA | [69] |
| Wang et al. (2008) | Bladder | Taiwanese | 300/300 | Significantly increased risk with SULT1A1*1/*1 and NQO1 C/T and T/T genotypes among ever-smokers | [70] |
| Wang et al. (2008) | Bladder | Taiwanese | 300/300 | Significantly increased risk with SULT1A1*1/*1 and exposure to more than one item of hazardous chemicals. Greater significantly increased risk with SULT1A1*1/*1 and heavy smoking. Greatest significantly increased risk with SULT1A1*1/*1 and exposure to more than one item of hazardous chemicals among ever-smokers | [71] |
| Santos et al. (2012) | Oral | Brazilian | 202/196 | Significantly increased risk with SULT1A1*1/*1 and smoking | [72] |
| Suzuki et al. (2008) | Pancreatic | Gastrointestinal Center at University of Texas MD Anderson Cancer Center | 755/636 | NA | [73] |
NA: No association.
Associations have also been seen in colorectal cancer risk. The aforementioned meta-analysis conducted by Li et al. to understand the involvement of the SULT1A1*1/2 SNP in environment-related cancer risk found no significant association with colorectal carcinoma in smokers [64]. Conversely, Canadian smokers who were carriers of the SULT1A1*1/*1 genotype, who were either current or former smokers, and who had reported smoking >15 years were at statistically significant increased risk of colorectal cancer [66]. By contrast, an increased risk of the disease was found among German carriers of the SULT1A1*2 allele who reported 30+ pack-years of active smoking (OR: 1.7; 95% CI: 1.0–3.2) compared with those with the SULT1A1*1/*1 genotype (OR: 1.1; 95% CI: 0.6–2.1) [51].
Smoking has been implicated in bladder cancer as well, yet there have been conflicting results for risk and smoking status with the SULT1A1*1/2 SNP. Three different studies in Spanish and Belgian populations reported no significant association [67–69], whereas two others from the same Taiwanese population of 300 urothelial cancer cases and 300 controls reported an increased risk for ever/heavy smokers homozygous for the *1 allele. In one of these studies, there was a significant increase in risk among those who had ever smoked and who had the SULT1A1*1/*1 high-activity genotype (OR: 5.3) and the observed association was made even stronger with gene–gene interactions between SULT1A1*1/*1 and NQO1 SNPs (OR: 8.6; 95% CI: 2.5–29.7) [70]. In the second study, those individuals who were ever-smokers, carried the SULT1A1*1/*1 genotype and had been exposed to more than one item of hazardous chemicals had the highest significantly increased bladder cancer risk (OR: 16.1; 95% CI: 2.9–87.2) [71].
A Brazilian study with 202 oral cancer cases and 196 sex- and age-frequency matched controls also reported increases in the risk of oral cancer and cigarette smoking regardless of SULT1A1 genotype, but individuals with the high-activity *1/*1 genotype exhibited greater risk (OR: 10.19; 95% CI: 3.90–26.61) than those with at least one *2 allele (OR: 4.50; 95% CI: 2.09–9.69) [72]. By contrast, pancreatic cancer and heavy smoking had no significant interactions with SULT1A1 genotypes in a study involving 755 cases and 636 healthy frequency-matched controls from the Gastrointestinal Center at The University of Texas MD Anderson Cancer Center (TX, USA) [73].
In light of the evidence that SULT1A1 activity is positively correlated with CNV, Palli et al. investigated gene CNV in male breast cancer patients and found that 10 out of 72 (13.9%) had gene deletions. Of the ten patients with gene deletions, all had one copy of SULT1A1 in the tumor samples and two copies in the corresponding blood samples, whereas copy number in blood and normal breast tissue were concordant. A link was also realized between SULT1A1 gene deletion and BRCA2 mutation (p = 0.0005) in a multivariate analysis. The researchers surmised that BRCA2 male breast cancer patients are susceptible to an increased exposure to estrogens and environmental PAHs and that this is due in part to lowered SULT1A1 copy number implying decreased enzymatic activity [74].
SULT1A1 genetic variation & chemopreventives (flavonoids, isoflavonoids & other phenols)
Flavonoids and other dietary polyphenols found in coffee, tea, red wine, soy and various fruits and vegetables represent potent noncompetitive inhibitors of SULT activity [75–80]. Owing to the role that SULTs play in the bioactivation of promutagens and procarcinogens, it has been postulated that dietary exposure to these compounds can exert a chemoprotective effect via inhibition of SULT activity [81], and tissue-specific expression of SULTs can lead to differential effects when comparing target tissues [82]. SULTs have a widespread tissue distribution with SULT1A1 being the most highly expressed isoform in the liver [83,84]. Coughtrie et al. found that epicatechin gallate and epigallocatechin gallate (components of green tea) showed noncompetitive inhibition of SULT1A1 whereas with SULT1A2 and 1A3 they demonstrated mixed type inhibition, suggesting that the mechanism of inhibition varies depending on the individual isoform [81].
Inhibition of SULT1A1-induced carcinogenesis has been the target of research with dietary flavonoids (Table 5). These substances are potent inhibitors of SULT1A1 activity, therefore the capacity of each SULT1A1 allozyme (*1, *2 and *3 recombinant proteins) to sulfate chrysin, genistein and quercetin was interrogated and significant differences were apparent. The highest rate of sulfation (Vmax) for each substrate was produced by *1 followed by *3, with the *2 allozyme producing the lowest sulfation rates [12]. The same kinetic measures were determined for three other compounds, apigenin, epicatechin and resveratrol, and again the *2 allozyme was less effective at conjugation than *1 and *3, following the same pattern for this set of substrates [85].
Table 5.
SULT1A1 genetic variation and chemopreventives (flavonoids, isoflavonoids and other phenols).
| Study (year) | Chemopreventive agent | Population | Cases/controls (n) | Main findings | Ref. |
|---|---|---|---|---|---|
| Nagar et al. (2006) | Chrysin, genistein, quercetin | Recombinant proteins | None | Vmax*1 > *3 > *2 | [12] |
| Ung et al. (2007) | Apigenin, epicatechin, resveratrol | Recombinant proteins | None | Vmax*1 > *3 > *2 | [85] |
| Kellen et al. (2006) | Fruit (antioxidants) | Belgian | 200/385 | NA between SULT1A1*1/2 SNP and ever-smokers in bladder cancer risk Decreased risk with high daily fruit consumption | [69] |
| Rybicki et al. (2011) | Red wine (resveratrol) | African–American and Caucasian American | 391 prostate cancer cases | 33% PhIP-DNA adducts explained by SULT1A1 and UGT1A10 SNPs, and African ancestry | [86] |
NA: No association; PhIP: 2-amino-1-methyl-6-phenylimidazo[4,5-f].
One study evaluated the role of fruit consumption in reducing the risk of developing bladder cancer among ever-smokers. They reported a higher risk for those with a low daily consumption of fruits (<188.7 g) than those with an increased daily consumption of fruits (>188.7 g; OR: 4.23, 95% CI: 1.91–9.0; OR: 2.15, 95% CI: 1.15–4.05, respectively). No significant associations were found between the SULT1A1*1/2 SNP and bladder cancer risk or fruit consumption in this population. The protective effect of fruit consumption was not modified by metabolic SNPs [69].
Resveratrol is a compound found in the skins of grapes and is therefore a component of red wine. Because PhIP must be bioactived by SULT1A1 to a genotoxic species and resveratrol can compete with PhIP as a substrate for SULT1A1, a study was conducted to measure PhIP–DNA adducts by beverage consumption and the genetic factors of SULT1A1*1/2 and UGT1A10 SNPs, and African ancestry. Red wine consumption could account for 13–16% of the variability in PhIP–DNA adduct formation in both African–Americans and white subjects. The genetic factors explained 33% of the PhIP–DNA adduct variation in African–American cases, while only 19% in white subjects. The authors conclude that red wine consumption does play a role in PhIP–DNA adduct variation but to a lesser extent than genetic variation [86].
Conclusion
Earlier pharmacogenetic studies observed correlations whereas many later investigations report no significant associations with SULT1A1*1/2 genotype or CNV with outcome of TAM-treated patients. Having higher SULT1A1 enzymatic activity does not result in a more rapid clearance of the active metabolites of TAM. These findings suggest that the bioavailability of active TAM metabolites due to variations in the most commonly studied coding region SNP, SULT1A1*1/2, and SULT1A1 copy number are not the major determining factors in TAM pharmacokinetics. Therefore any observed differences in outcome must be attributed to some other factor(s). The mechanistic basis for improved outcomes associated with the *1 high activity allele could be attributed to the observation that sulfated metabolites of TAM induced apoptosis in breast cancer cell lines [87]. In this case, rapid sulfation of the active metabolites of TAM in breast tumor cells could result in an increase in apoptosis and, hence, improved survival in individuals with the high activity SULT1A1 genotype. Because functional SNPs and CNVs cannot fully account for variable SULT1A1 activity, Yao-Borengasser et al. explored the role that transcription factor regulation plays in SULT1A1 gene expression. They found that the nuclear factor 1 (NF1) family members each have an impact on SULT1A1 gene expression in ZR75-1 cells. When NF1-A, -B, -C and -X were knocked down with siRNA, SULT1A1 expression was also decreased. While only some of these associations were significant (NF1-A and -C), the trend was still evident in others (NF1-B and -X) [88]. Taken all together, we are left with the knowledge that there is more to SULT1A1 variation than can be explained by functional SNPs, CNV and ethnic differences in allele frequency. More studies are needed to elucidate the pharmacogenetic implications of this dynamic protein. Like Innocenti et al. [25], data generated in preclinical trials can be used to prequalify candidate genes and therefore guide future pharmacogenetic studies.
HCAs are environmental procarcinogens found in meat cooked at high temperatures and they are bio-activated by SULT1A1. Individuals demonstrating high SULT1A1 activity would be expected to exhibit a greater risk of developing cancer owing to a supposed increase in exposure to carcinogenic and mutagenic compounds. Risk of prostate cancer was found to have no associations with genotype in two studies; but when phenotype was considered, one of these studies did find significant associations. Against the a priori hypothesis, high cooked meat consumption consistently increased the risk of both breast and colorectal cancer for carriers of the SULT1A1*2 allele. One explanation for this could be the fact that chemopreventive intake was not included in the analyses. Evaluating exposures to both cancer-causing and cancer-preventing agents in dietary assessments may allow for a more complete analysis. Intraindividual SULT1A1 expression is also tissue specific. Varying levels of SULT1A1 at each site of action could also explain differences in metabolic potential to initiate carcinogenesis.
Smoking exposes individuals to PAHs and SULT1A1 is able to intercept the formation of genotoxic metabolites. However, this exposure also yielded conflicting results with SULT1A1 genetic variation. Because SULT1A1 activity is thought to be protective against PAH damage, genetic variations that confer lower enzymatic activity are expected to have an increased risk of cancer susceptibility. Being a carrier of at least one SULT1A1*2 low-activity allele and a smoker appears to increase risk of prostate and esophageal cancer marginally. *2 alone was sufficient to increase risk of lung cancer, but when stratified by smoking status the significant association only persisted in one study which was a meta-analysis. There were associations between smokers, SULT1A1*1/*1 high-activity genotype, and increased risk for other cancers. Bladder cancer risk was only increased when *1/*1 and another factor was involved. Increased risk could also be seen among colorectal case–control studies, but not in a meta-analysis. Oral cancer subjects who smoked had increased risk regardless of SULT1A1 genotype; however, individuals homozygous for the *1 allele were at much greater risk. No association was observed in the single studies involving pancreatic and prostate cancer. More studies of SULT1A1 genetic variability and pancreatic and prostate cancer susceptibility are needed to understand how smoking can modulate this relationship. Current results vary greatly by ethnicity, another important factor in determining cancer pharmacogenetic risk. There are undoubtedly other gene–gene and gene–environment interactions as well as ethnic differences that can explain the inconsistent findings between the case–control studies and meta-analyses presented here.
Environmental exposure to promutagens, procarcinogens and chemopreventives along with variation in the SULT1A1 enzyme could exhibit a cumulative effect in modulating individual response to disease-related chemicals. Allele specific variation in the sulfation of chemopreventives has been demonstrated. The *1 allele has the greatest, *3 lower and *2 lowest activity toward chemopreventive compounds. The *2 low-activity SULT1A1 SNP may indeed decrease the formation of genotoxic species, thereby conferring lower susceptibility to those cancers that arise from SULT1A1-catalyzed HCA (meat cooked at high temperatures) bioactivation. Conversely, having lower SULT1A1 enzymatic activity could be detrimental in the case of detoxifying PAH (smoke) compounds. Further investigation into the role chemopreventive agents and SULT1A1 variation plays in modulating SULT1A1 activity and disease outcome is warranted, addressing cancer site differences, as well as ethnic differences in allele frequencies where an allele–dose effect may be observed.
Future perspective
Data presented in this review demonstrate the importance of evaluating genetic variation beyond the traditional SULT1A1*1/2 SNP in predicting response to chemicals and therapeutic agents. Future SULT1A1 pharmacogenetic studies should be more inclusive and take into consideration CNV, 3′-UTR SNPs and the SULT1A1*3 allozyme. The idea that SULT1A1 is not an inducible enzyme has been challenged. Discovering that 4-OHT may upregulate and NF1 knockdown can downregulate SULT1A1 expression affirms that these influences should not be ignored. Epigenetic evaluation is undoubtedly in the future for SULT1A1 genetic variability studies. Differential CNVs between tumor and matched blood samples warrants further exploration of the tumor microenvironment. Larger, more ethnically diverse studies of sufficient statistical power need to be conducted to more fully understand the contribution of SULT1A1 genetic variation and environmental and dietary contributors to cancer causation and prevention, as well as therapeutic efficacy.
Executive summary.
SULT1A1 genetic variation & therapeutic agents
-
Most pharmacogenetic studies with tamoxifen (TAM) report no significant associations with SULT1A1 genetic variation and either recurrence-free survival, prognostic clinical or biological markers, breast cancer prevention efficacy, disease-free survival, overall survival, recurrence or pharmacokinetics.
Conflicting significant associations with the *2/*2 genotype were found. One study reported improved overall survival and breast cancer specific survival, whereas another reported an almost threefold greater risk of death in *2/*2 carriers. Homozygous *2/*2 and UGT2B15*2 carriers were also associated with increased recurrence and decreased survival.
SULT1A1*1/*1 homozygotes were found to have a decreased risk of recurrence with the CYP2D6*4 variant allele and improved recurrence-free survival with 2 years of TAM treatment.
Pharmacokinetic analyses showed no significant associations between SULT1A1 genotype or copy number with plasma concentrations of TAM and TAM metabolites except when ratios of metabolites were considered in one study.
Results from a Phase 1 clinical trial investigating a novel anticancer therapeutic agent, ABT-751, reveals a clear indication for SULT1A1 copy number testing before prescribing this medication. Patients with high copy number variation of SULT1A1 receiving ABT-751 could potentially exhibit decreased benefit in terms of outcome owing to increased clearance of the active parent compound.
Aminoflavone has an allele specific sensitivity toward the SULT1A1 variants *3 > *1 > *2 suggesting that carriers of the low activity *2 allele may not respond to aminoflavone treatment as well as high activity carriers.
SULT1A1*1/2 and copy number could potentially influence the metabolism of fulvestrant and toremifene adjuvant therapies in that they were significantly associated with substrate sulfation in in vitro assays.
SULT1A1 genetic variation, well-done meat consumption (heterocyclic amine exposure) & cancer risk
N-hydroxylated heterocyclic amines are present in well-done meat and must undergo metabolism by SULT1A1 to form activated DNA damaging species that can initiate carcinogenesis.
Because the SULT1A1*2/*2 low activity genotype has been shown to produce less DNA adducts than the heterozygous or homozygous *1/*1 genotypes, one would expect this allele to be associated with lower risk of developing cancer.
No associations were observed for prostate cancer risk between well-done meat consumption and SULT1A1 genotype.
Surprisingly, the SULT1A1*2 low-activity variant allele was associated with increased risk of both breast and colorectal cancer among subjects who had high smoked meat intake.
SULT1A1 genetic variation, smoking (polycyclic aromatic hydrocarbon exposure) & cancer risk
Polycyclic aromatic hydrocarbons found in smoke are carcinogenic, but sulfation from SULT1A1 metabolism is able to intercept the carcinogenic redox cycle, which is a protective benefit.
The *2 variant allele is expected to exhibit a greater risk of cancer due to its lower enzymatic activity in the detoxication of polycyclic aromatic hydrocarbons; however, smoking and SULT1A1 SNP yielded conflicting results in cancer risk at multiple sites.
Increased risk for both lung and esophageal cancer were associated with smoking and SULT1A1*2 allele, albeit marginally, in a meta-analysis and more profoundly with lung cancer in another study. Otherwise there were no associations found with lung cancer.
Colorectal cancer risk with smoking had either no associations in a meta-analysis, increased risk with carrying the *1*1 genotype in Canadians or the *2 allele in Germans.
Bladder cancer and smoking had no associations with the SULT1A1 SNP except in two studies of the same population where other gene–gene and gene–environment factors were considered in those homozygous for *1/*1.
Oral cancer risk was increased among smokers with the *1/*1 genotype.
No associations were observed with prostate or pancreatic cancer risk.
Male breast cancer was associated with a decrease in SULT1A1 copy number and with the BRCA2 mutation.
SULT1A1 genetic variation & chemopreventives (flavonoids, isoflavonoids & other phenols)
Dietary flavonoids including epicatechin gallate, epigallocatechin gallate, chrysin, genistein, quercetin, apigenin, epicatechin and resveratrol are potent inhibitors of SULT1A1 and therefore may prevent sulfation-induced carcinogenesis.
The trend in recombinant SULT1A1 Vmax toward certain dietary flavonoids was *1 > *3 > *2, suggesting that *2 carriers might be at a greater disadvantage in preventing sulfation-induced carcinogenesis.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was funded in part by NIH grant number 5R01CA128897. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Jakoby WB, Ziegler DM. The enzymes of detoxication. J Biol Chem. 1990;265(34):20715–20718. [PubMed] [Google Scholar]
- 2.Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, Raftogianis RB. Sulfation and sulfotransferases 1: sulfotransferase molecular biology: cDNAs and genes. FASEB J. 1997;11(1):3–14. [PubMed] [Google Scholar]
- 3.Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun. 1997;239(1):298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- 4.Ozawa S, Tang YM, Yamazoe Y, Kato R, Lang NP, Kadlubar FF. Genetic polymorphisms in human liver phenol sulfotransferases involved in the bioactivation of N-hydroxy derivatives of carcinogenic arylamines and heterocyclic amines. Chem Biol Interact. 1998;109(1–3):237–248. doi: 10.1016/s0009-2797(97)00135-x. [DOI] [PubMed] [Google Scholar]
- 5.Carlini EJ, Raftogianis RB, Wood TC, et al. Sulfation pharmacogenetics: SULT1A1 and SULT1A2 allele frequencies in Caucasian, Chinese and African–American subjects. Pharmacogenetics. 2001;11(1):57–68. doi: 10.1097/00008571-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Lin ZN, Lin YC, Zhang X, et al. Differential promoter activities of functional haplotypes in the 5′-flanking region of human sulfotransferase 1A1. J Biochem Mol Toxicol. 2012;26(10):422–428. doi: 10.1002/jbt.21437. [DOI] [PubMed] [Google Scholar]
- 7.Ning B, Nowell S, Sweeney C, et al. Common genetic polymorphisms in the 5′-flanking region of the SULT1A1 gene: haplotypes and their association with platelet enzymatic activity. Pharmacogenet Genomics. 2005;15(7):465–473. doi: 10.1097/01.fpc.0000166823.74378.79. [DOI] [PubMed] [Google Scholar]
- 8.Hebbring SJ, Adjei AA, Baer JL, et al. Human SULT1A1 gene: copy number differences and functional implications. Human Mol Genet. 2007;16(5):463–470. doi: 10.1093/hmg/ddl468. [DOI] [PubMed] [Google Scholar]
- 9.Anderson RJ, Jackson BL. Human platelet phenol sulfotransferase: stability of two forms of the enzyme with time and presence of a racial difference. Clin Chim Acta. 1984;138(2):185–196. doi: 10.1016/0009-8981(84)90233-x. [DOI] [PubMed] [Google Scholar]
- 10••.Yu X, Dhakal IB, Beggs M, et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci. 2010;118(2):391–403. doi: 10.1093/toxsci/kfq296. Copy number and 3′-UTR variants most able to explain variable SULT1A1 activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X, Kubota T, Dhakal I, et al. Copy number variation in sulfotransferase isoform 1A1 (SULT1A1) is significantly associated with enzymatic activity in Japanese subjects. Pharmacogenomics Pers Med. 2013;6:19–24. doi: 10.2147/PGPM.S36579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagar S, Walther S, Blanchard RL. Sulfotransferase (SULT) 1A1 polymorphic variants *1, *2, and *3 are associated with altered enzymatic activity, cellular phenotype, and protein degradation. Mol Pharmacol. 2006;69(6):2084–2092. doi: 10.1124/mol.105.019240. [DOI] [PubMed] [Google Scholar]
- 13.Nowell S, Sweeney C, Winters M, et al. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst. 2002;94(21):1635–1640. doi: 10.1093/jnci/94.21.1635. [DOI] [PubMed] [Google Scholar]
- 14.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91(3):249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 15.Wegman P, Vainikka L, Stal O, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7(3):R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9(1):R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knechtel G, Hofmann G, Gerger A, et al. Analysis of common germline polymorphisms as prognostic factors in patients with lymph node-positive breast cancer. J Cancer Res Clin Oncol. 2010;136(12):1813–1819. doi: 10.1007/s00432-010-0839-2. [DOI] [PubMed] [Google Scholar]
- 18.Moyer AM, Suman VJ, Weinshilboum RM, et al. SULT1A1, CYP2C19 and disease-free survival in early breast cancer patients receiving tamoxifen. Pharmacogenomics. 2011;12(11):1535–1543. doi: 10.2217/pgs.11.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tengstrom M, Mannermaa A, Kosma VM, Hirvonen A, Kataja V. SULT1A1 rs9282861 polymorphism – a potential modifier of efficacy of the systemic adjuvant therapy in breast cancer? BMC Cancer. 2012;12:257. doi: 10.1186/1471-2407-12-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabinski JL, Smith LS, Chisholm GB, et al. Genotypic and allelic frequencies of SULT1A1 polymorphisms in women receiving adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2006;95(1):13–16. doi: 10.1007/s10549-005-9019-5. [DOI] [PubMed] [Google Scholar]
- 21.Serrano D, Lazzeroni M, Zambon CF, et al. Efficacy of tamoxifen based on cytochrome P450 2D6, CYP2C19 and SULT1A1 genotype in the Italian Tamoxifen Prevention Trial. Pharmacogenomics J. 2011;11(2):100–107. doi: 10.1038/tpj.2010.17. [DOI] [PubMed] [Google Scholar]
- 22.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97(1):30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 23.Gjerde J, Hauglid M, Breilid H, et al. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19(1):56–61. doi: 10.1093/annonc/mdm434. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Santander A, Gaibar M, Novillo A, et al. Relationship between genotypes Sult1a2 and Cyp2d6 and tamoxifen metabolism in breast cancer patients. PLoS ONE. 2013;8(7):e70183. doi: 10.1371/journal.pone.0070183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Innocenti F, Ramirez J, Obel J, et al. Preclinical discovery of candidate genes to guide pharmacogenetics during Phase I development: the example of the novel anticancer agent ABT-751. Pharmacogenet Genomics. 2013;23(7):374–381. doi: 10.1097/FPC.0b013e3283623e81. Used preclinical data to guide pharmacogenetic study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams JA, Andersson T, Andersson TB, et al. PhRMA white paper on ADME pharmacogenomics. J Clin Pharmacol. 2008;48(7):849–889. doi: 10.1177/0091270008319329. [DOI] [PubMed] [Google Scholar]
- 27.Mclean L, Soto U, Agama K, et al. Aminoflavone induces oxidative DNA damage and reactive oxidative species-mediated apoptosis in breast cancer cells. Int J Cancer. 2008;122(7):1665–1674. doi: 10.1002/ijc.23244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Q, Sha X, Liu J, Heath E, Lorusso P, Li J. Association of human cytochrome P450 1A1 (CYP1A1) and sulfotransferase 1A1 (SULT1A1) polymorphisms with differential metabolism and cytotoxicity of aminoflavone. Mol Cancer Ther. 2010;9(10):2803–2813. doi: 10.1158/1535-7163.MCT-10-0597. [DOI] [PubMed] [Google Scholar]
- 29.Edavana VK, Yu X, Dhakal IB, et al. Sulfation of fulvestrant by human liver cytosols and recombinant SULT1A1 and SULT1E1. Pharmacogenomics Pers Med. 2011;4:137–145. doi: 10.2147/PGPM.S25418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edavana VK, Dhakal IB, Yu X, Williams S, Kadlubar S. Sulfation of 4-hydroxy toremifene: individual variability, isoform specificity, and contribution to toremifene pharmacogenomics. Drug Metabol Dispos. 2012;40(6):1210–1215. doi: 10.1124/dmd.111.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seth P, Lunetta KL, Bell DW, et al. Phenol sulfotransferases: hormonal regulation, polymorphism, and age of onset of breast cancer. Cancer Res. 2000;60(24):6859–6863. [PubMed] [Google Scholar]
- 32•.Daniels J, Kadlubar S. Sulfotransferase genetic variation: from cancer risk to treatment response. Drug Metabol Rev. 2013;45(4):415–422. doi: 10.3109/03602532.2013.835621. Reviews the role of SULT1A1 genetic variation in cancer risk and treatment response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi JY, Lee KM, Park SK, et al. Genetic polymorphisms of SULT1A1 and SULT1E1 and the risk and survival of breast cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1090–1095. doi: 10.1158/1055-9965.EPI-04-0688. [DOI] [PubMed] [Google Scholar]
- 34.Glatt H, Meinl W. Pharmacogenetics of soluble sulfotransferases (SULTs) Naunyn Schmiedeberg’s Arch Pharmacol. 2004;369(1):55–68. doi: 10.1007/s00210-003-0826-0. [DOI] [PubMed] [Google Scholar]
- 35.Perera FP. Environment and cancer: who are susceptible? Science. 1997;278(5340):1068–1073. doi: 10.1126/science.278.5340.1068. [DOI] [PubMed] [Google Scholar]
- 36.Danaei G, Vander Hoorn S, Lopez AD, Murray CJ, Ezzati M Comparative Risk Assessment Collaborating Group (Cancers) Causes of cancer in the world: comparative risk assessment of nine behavioural and environmental risk factors. Lancet. 2005;366(9499):1784–1793. doi: 10.1016/S0140-6736(05)67725-2. [DOI] [PubMed] [Google Scholar]
- 37.Key TJ, Allen NE, Spencer EA, Travis RC. The effect of diet on risk of cancer. Lancet. 2002;360(9336):861–868. doi: 10.1016/S0140-6736(02)09958-0. [DOI] [PubMed] [Google Scholar]
- 38.Sankpal UT, Pius H, Khan M, et al. Environmental factors in causing human cancers: emphasis on tumorigenesis. Tumour Biol. 2012;33(5):1265–1274. doi: 10.1007/s13277-012-0413-4. [DOI] [PubMed] [Google Scholar]
- 39.Strott CA. Sulfonation and molecular action. Endocr Rev. 2002;23(5):703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- 40.Klaassen CD, Boles JW. Sulfation and sulfotransferases 5: the importance of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 1997;11(6):404–418. doi: 10.1096/fasebj.11.6.9194521. [DOI] [PubMed] [Google Scholar]
- 41.Chou CY, Shen MR, Wu SN. Volume-sensitive chloride channels associated with human cervical carcinogenesis. Cancer Res. 1995;55(24):6077–6083. [PubMed] [Google Scholar]
- 42.Minchin RF, Ilett KF, Teitel CH, Reeves PT, Kadlubar FF. Direct O-acetylation of N-hydroxy arylamines by acetylsalicylic acid to form carcinogen–DNA adducts. Carcinogenesis. 1992;13(4):663–667. doi: 10.1093/carcin/13.4.663. [DOI] [PubMed] [Google Scholar]
- 43.Shirai T, Sano M, Tamano S, et al. The prostate: a target for carcinogenicity of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) derived from cooked foods. Cancer Res. 1997;57(2):195–198. [PubMed] [Google Scholar]
- 44.Nakai Y, Nelson WG, De Marzo AM. The dietary charred meat carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine acts as both a tumor initiator and promoter in the rat ventral prostate. Cancer Res. 2007;67(3):1378–1384. doi: 10.1158/0008-5472.CAN-06-1336. [DOI] [PubMed] [Google Scholar]
- 45.Muckel E, Frandsen H, Glatt HR. Heterologous expression of human N-acetyltransferases 1 and 2 and sulfotransferase 1A1 in Salmonella typhimurium for mutagenicity testing of heterocyclic amines. Food Chem Toxicol. 2002;40(8):1063–1068. doi: 10.1016/s0278-6915(02)00032-7. [DOI] [PubMed] [Google Scholar]
- 46.Nowell S, Ambrosone CB, Ozawa S, et al. Relationship of phenol sulfotransferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenetics. 2000;10(9):789–797. doi: 10.1097/00008571-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 47.Ohgaki H, Hasegawa H, Kato T, et al. Carcinogenicity in mice and rats of heterocyclic amines in cooked foods. Environ Health Perspect. 1986;67:129–134. doi: 10.1289/ehp.8667129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Buheissi SZ, Patel HR, Meinl W, et al. N-acetyltransferase and sulfotransferase activity in human prostate: potential for carcinogen activation. Pharmacogenet Genomics. 2006;16(6):391–399. doi: 10.1097/01.fpc.0000204998.22301.09. [DOI] [PubMed] [Google Scholar]
- 49.Lee H, Wang Q, Yang F, et al. SULT1A1 Arg213His polymorphism, smoked meat, and breast cancer risk: a case–control study and meta-analysis. DNA Cell Biol. 2012;31(5):688–699. doi: 10.1089/dna.2011.1403. [DOI] [PubMed] [Google Scholar]
- 50.Tao P, Li H, Wang Q, et al. A case–control study on association of SULT1A1 polymorphism, smoked meat intake with breast cancer risk. Zhonghua Yu Fang Yi Xue Za Zhi. 2012;46(9):831–835. [PubMed] [Google Scholar]
- 51.Lilla C, Risch A, Verla-Tebit E, Hoffmeister M, Brenner H, Chang-Claude J. SULT1A1 genotype and susceptibility to colorectal cancer. Int J Cancer. 2007;120(1):201–206. doi: 10.1002/ijc.22156. [DOI] [PubMed] [Google Scholar]
- 52.Cotterchio M, Boucher BA, Manno M, Gallinger S, Okey AB, Harper PA. Red meat intake, doneness, polymorphisms in genes that encode carcinogen-metabolizing enzymes, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17(11):3098–3107. doi: 10.1158/1055-9965.EPI-08-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowell S, Ratnasinghe DL, Ambrosone CB, et al. Association of SULT1A1 phenotype and genotype with prostate cancer risk in African–Americans and Caucasians. Cancer Epidemiol Biomarkers Prev. 2004;13(2):270–276. doi: 10.1158/1055-9965.epi-03-0047. [DOI] [PubMed] [Google Scholar]
- 54.Koutros S, Berndt SI, Sinha R, et al. Xenobiotic metabolizing gene variants, dietary heterocyclic amine intake, and risk of prostate cancer. Cancer Res. 2009;69(5):1877–1884. doi: 10.1158/0008-5472.CAN-08-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gelboin HV. Benzo[alpha]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol Rev. 1980;60(4):1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- 56.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42(12):4875–4917. [PubMed] [Google Scholar]
- 57.Grimmer G, Bohnke H. Polycyclic aromatic hydrocarbon profile analysis of high-protein foods, oils, and fats by gas chromatography. J Assoc Off Anal Chem. 1975;58(4):725–733. [PubMed] [Google Scholar]
- 58.Burczynski ME, Lin HK, Penning TM. Isoform-specific induction of a human aldo-keto reductase by polycyclic aromatic hydrocarbons (PAHs), electrophiles, and oxidative stress: implications for the alternative pathway of PAH activation catalyzed by human dihydrodiol dehydrogenase. Cancer Res. 1999;59(3):607–614. [PubMed] [Google Scholar]
- 59.Shultz CA, Quinn AM, Park JH, et al. Specificity of human aldo-keto reductases, NAD(P)H:quinone oxidoreductase, and carbonyl reductases to redox-cycle polycyclic aromatic hydrocarbon diones and 4-hydroxyequilenin-O-quinone. Chem Res Toxicol. 2011;24(12):2153–2166. doi: 10.1021/tx200294c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L, Huang M, Blair IA, Penning TM. Detoxication of benzo[a]pyrene-7,8-dione by sulfotransferases (SULTs) in human lung cells. J Biol Chem. 2012;287(35):29909–29920. doi: 10.1074/jbc.M112.386052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao SG, Liu L, Zhang YY, Wang Y, Wang YJ. SULT1A1 Arg213His polymorphism and lung cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 2012;13(2):579–583. doi: 10.7314/apjcp.2012.13.2.579. [DOI] [PubMed] [Google Scholar]
- 62.Arslan S, Silig Y, Pinarbasi H. An investigation of the relationship between SULT1A1 Arg(213)His polymorphism and lung cancer susceptibility in a Turkish population. Cell Biochem Funct. 2009;27(4):211–215. doi: 10.1002/cbf.1558. [DOI] [PubMed] [Google Scholar]
- 63.Arslan S, Silig Y, Pinarbasi H. Sulfotransferase 1A1 Arg(213) His polymorphism and prostate cancer risk. Exp Ther Med. 2011;2(6):1159–1162. doi: 10.3892/etm.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li K, Ren YW, Wan Y, Yin ZH, Wu W, Zhou BS. SULT1A1 Arg213His polymorphism and susceptibility of environment-related cancers: a meta analysis of 5,915 cases and 7,900 controls. Mol Biol Rep. 2012;39(3):2597–2605. doi: 10.1007/s11033-011-1012-y. [DOI] [PubMed] [Google Scholar]
- 65.Liang G, Miao X, Zhou Y, Tan W, Lin D. A functional polymorphism in the SULT1A1 gene (G638A) is associated with risk of lung cancer in relation to tobacco smoking. Carcinogenesis. 2004;25(5):773–778. doi: 10.1093/carcin/bgh053. [DOI] [PubMed] [Google Scholar]
- 66.Cleary SP, Cotterchio M, Shi E, Gallinger S, Harper P. Cigarette smoking, genetic variants in carcinogen-metabolizing enzymes, and colorectal cancer risk. Am J Epidemiol. 2010;172(9):1000–1014. doi: 10.1093/aje/kwq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Figueroa JD, Malats N, Garcia-Closas M, et al. Bladder cancer risk and genetic variation in AKR1C3 and other metabolizing genes. Carcinogenesis. 2008;29(10):1955–1962. doi: 10.1093/carcin/bgn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fortuny J, Kogevinas M, Garcia-Closas M, et al. Use of analgesics and nonsteroidal anti-inflammatory drugs, genetic predisposition, and bladder cancer risk in Spain. Cancer Epidemiol Biomarkers Prev. 2006;15(9):1696–1702. doi: 10.1158/1055-9965.EPI-06-0038. [DOI] [PubMed] [Google Scholar]
- 69••.Kellen E, Zeegers M, Paulussen A, Van Dongen M, Buntinx F. Fruit consumption reduces the effect of smoking on bladder cancer risk. The Belgian case control study on bladder cancer. Int J Cancer. 2006;118(10):2572–2578. doi: 10.1002/ijc.21714. Examined the involvement of SULT1A1 genetic variation along with both carcinogenic and chemopreventive agents in the risk of bladder cancer. [DOI] [PubMed] [Google Scholar]
- 70.Wang YH, Lee YH, Tseng PT, Shen CH, Chiou HY. Human NAD(P)H:quinone oxidoreductase 1 (NQO1) and sulfotransferase 1A1 (SULT1A1) polymorphisms and urothelial cancer risk in Taiwan. J Cancer Res Clin Oncol. 2008;134(2):203–209. doi: 10.1007/s00432-007-0271-4. [DOI] [PubMed] [Google Scholar]
- 71.Wang YH, Juang GD, Hwang TI, Shen CH, Shao KY, Chiou HY. Genetic polymorphism of sulfotransferase 1A1, cigarette smoking, hazardous chemical exposure and urothelial cancer risk in a Taiwanese population. Int J Urol. 2008;15(12):1029–1034. doi: 10.1111/j.1442-2042.2008.02166.x. [DOI] [PubMed] [Google Scholar]
- 72.Santos SS, Koifman RJ, Ferreira RM, et al. SULT1A1 genetic polymorphisms and the association between smoking and oral cancer in a case–control study in Brazil. Front Oncol. 2012;2:183. doi: 10.3389/fonc.2012.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzuki H, Morris JS, Li Y, et al. Interaction of the cytochrome P4501A2, SULT1A1 and NAT gene polymorphisms with smoking and dietary mutagen intake in modification of the risk of pancreatic cancer. Carcinogenesis. 2008;29(6):1184–1191. doi: 10.1093/carcin/bgn085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palli D, Rizzolo P, Zanna I, et al. SULT1A1 gene deletion in BRCA2-associated male breast cancer: a link between genes and environmental exposures? J Cell Mol Med. 2013;17(5):605–607. doi: 10.1111/jcmm.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Littlewood JT, Glover V, Sandler M. Red wine contains a potent inhibitor of phenolsulphotransferase. Br J Clin Pharmacol. 1985;19(2):275–278. doi: 10.1111/j.1365-2125.1985.tb02644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gibb C, Glover V, Sandler M. In vitro inhibition of phenolsulphotransferase by food and drink constituents. Biochem Pharmacol. 1987;36(14):2325–2330. doi: 10.1016/0006-2952(87)90598-3. [DOI] [PubMed] [Google Scholar]
- 77.Bamforth KJ, Jones AL, Roberts RC, Coughtrie MW. Common food additives are potent inhibitors of human liver 17 alpha-ethinyloestradiol and dopamine sulphotransferases. Biochem Pharmacol. 1993;46(10):1713–1720. doi: 10.1016/0006-2952(93)90575-h. [DOI] [PubMed] [Google Scholar]
- 78.Jones AL, Hagen M, Coughtrie MW, Roberts RC, Glatt H. Human platelet phenolsulfotransferases: cDNA cloning, stable expression in V79 cells and identification of a novel allelic variant of the phenol-sulfating form. Biochem Biophys Res Commun. 1995;208(2):855–862. doi: 10.1006/bbrc.1995.1414. [DOI] [PubMed] [Google Scholar]
- 79.Coughtrie MW, Sharp S, Maxwell K, Innes NP. Biology and function of the reversible sulfation pathway catalysed by human sulfotransferases and sulfatases. Chem Biol Interact. 1998;109(1–3):3–27. doi: 10.1016/s0009-2797(97)00117-8. [DOI] [PubMed] [Google Scholar]
- 80.Eaton EA, Walle UK, Lewis AJ, Hudson T, Wilson AA, Walle T. Flavonoids, potent inhibitors of the human P-form phenolsulfotransferase. Potential role in drug metabolism and chemoprevention. Drug Metabol Dispos. 1996;24(2):232–237. [PubMed] [Google Scholar]
- 81.Coughtrie MW, Johnston LE. Interactions between dietary chemicals and human sulfotransferases-molecular mechanisms and clinical significance. Drug Metabol Dispos. 2001;29(4 Pt 2):522–528. [PubMed] [Google Scholar]
- 82.Moon YJ, Wang X, Morris ME. Dietary flavonoids: effects on xenobiotic and carcinogen metabolism. Toxicol In vitro. 2006;20(2):187–210. doi: 10.1016/j.tiv.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 83.Glatt H. Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact. 2000;129(1–2):141–170. doi: 10.1016/s0009-2797(00)00202-7. [DOI] [PubMed] [Google Scholar]
- 84.Glatt H, Boeing H, Engelke CE, et al. Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutat Res. 2001;482(1–2):27–40. doi: 10.1016/s0027-5107(01)00207-x. [DOI] [PubMed] [Google Scholar]
- 85.Ung D, Nagar S. Variable sulfation of dietary polyphenols by recombinant human sulfotransferase (SULT) 1A1 genetic variants and SULT1E1. Drug Metabol Dispos. 2007;35(5):740–746. doi: 10.1124/dmd.106.013987. [DOI] [PubMed] [Google Scholar]
- 86.Rybicki BA, Neslund-Dudas C, Bock CH, et al. Red wine consumption is inversely associated with 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-DNA adduct levels in prostate. Cancer Prev Res. 2011;4(10):1636–1644. doi: 10.1158/1940-6207.CAPR-11-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87••.Mercer KE, Apostolov EO, Da Costa GG, et al. Expression of sulfotransferase isoform 1A1 (SULT1A1) in breast cancer cells significantly increases 4-hydroxytamoxifen-induced apoptosis. Int J Mol Epidemiol Genet. 2010;1(2):92–103. Provides a mechanistic basis (inducing apoptosis) for outcome in tamoxifen treatment. [PMC free article] [PubMed] [Google Scholar]
- 88.Yao-Borengasser A, Rogers LJ, Edavana VK, et al. Sulfotransferase 1A1 (SULT1A1) gene expression is regulated by members of the NFI transcription factors in human breast cancer cells. BMC Clin Pathol. 2014;14(1):1. doi: 10.1186/1472-6890-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]