Abstract
Generalized peeling skin syndrome (PSS) is an autosomal recessive genodermatosis characterized by lifelong, continuous shedding of the upper epidermis. Using whole-genome homozygozity mapping and whole-exome sequencing, we identified a novel homozygous missense mutation (c.229C>T, R77W) within the CHST8 gene, in a large consanguineous family with non-inflammatory PSS type A. CHST8 encodes a Golgi transmembrane N-acetylgalactosamine-4-O-sulfotransferase (GalNAc4-ST1), which we show by immunofluorescence staining to be expressed throughout normal epidermis. A colorimetric assay for total sulfated glycosaminoglycan (GAG) quantification, comparing human keratinocytes (CCD1106 KERTr) expressing wild type and mutant recombinant GalNAc4-ST1, revealed decreased levels of total sulfated GAGs in cells expressing mutant GalNAc4-ST1, suggesting loss of function. Western blotting revealed lower expression levels of mutant recombinant GalNAc4-ST1 compared to wild type, suggesting that accelerated degradation may result in loss of function, leading to PSS type A. This is the first report describing a mutation as the cause of PSS type A.
Keywords: Genodermatosis, Whole-exome, Mutation, Epidermis, Carbohydrate-sulfotransferase
1. Introduction
Peeling skin syndrome (PSS) is a group of rare autosomal recessive genodermatoses that can be divided into two main types: Acral PSS (APSS; OMIM 609796) and generalized PSS (OMIM 270300). A third form of facial peeling syndrome has also been reported [1]. Generalized PSS can be further divided into the non-inflammatory (type A) and the inflammatory (type B) forms [2].
PSS is described clinically and characterized histologically by asymptomatic lifelong and continuous shedding of the stratum corneum of the epidermis, due to the separation between keratinocyte layers at a subcorneal or intracorneal level [3,4]. This condition can present at birth or appear in early childhood [2,5]. PSS can be distinguished histologically from the different types of epidermolysis bullosa, in which blistering occurs within or below the basal layer of the epidermis [6,7], in contrast to the blistering high in the epidermis, at the stratum granulosum–corneum junction, seen in PSS.
APSS (localized form) involves the palmar, plantar and dorsal surfaces of hands and feet and is caused by mutations in the tranglutaminase 5 gene (TGM5) [8]. TGM5 is involved in cross-linking structural proteins for the formation of the cornified envelope, by catalyzing the formation of γ-glutamyl-ε-lysine isopeptide bonds between differentiation-specific proteins expressed mainly in corneocytes, including loricrin, involucrin and small proline-rich proteins [8].
Recently, it was found that recessive mutations in the corneodesmosin gene (CDSN) underlie PSS type B [9,10]. CDSN is a component of corneodesmosomes at the transition from the granular to cornified layers, and is involved in the late stages of epidermal differentiation, having a critical role in reinforcing cell–cell adhesion and maintaining the integrity of the stratum corneum. Patients with PSS type B share several clinical features with Netherton syndrome (NS, MIM 256500), presenting at birth with ichthyosiform erythroderma and white superficial exfoliation with recurrent flares of erythema, scaling and pruritis and increased predisposition to atopic diseases [9]. Interestingly, dominant mutations in CDSN are also associated with localized hypotrichosis simplex (MIM 146520) [11]. Homozygous loss-of-function mutations (a splice-site and a nonsense mutation) in the gene for protease inhibitor cystatin A (CSTA) have been recently identified as the underlying genetic cause of exfoliative ichthyosis. Patients with this condition present with dry, scaly skin over most of the body with coarse peeling of nonerythematous skin on the palms and soles. In contrast to PSS syndrome, electron microscopy of skin biopsies from affected individuals with exfoliative ichthyosis revealed that the level of detachment occurs in the basal and lower suprabasal layers [12].
We undertook this study to identify the gene responsible for non-inflammatory PSS type A, with the aim of defining a new genetic determinant of epidermal differentiation.
2. Results
2.1. Clinical findings
We studied a large consanguineous family from Pakistan showing features of non-inflammatory PSS, segregating as a recessive trait. Affected individuals reported symptoms starting during the second half of the first decade of life, consisting of generalized white scaling, most prominent over the upper and lower extremities (Fig. 1B), with pain-less and easy removal of the skin (Video S1). Moreover, they reported irritation when in contact with water, dust and sand. There was no history of erythema, pruritis or atopy and their hair was normal.
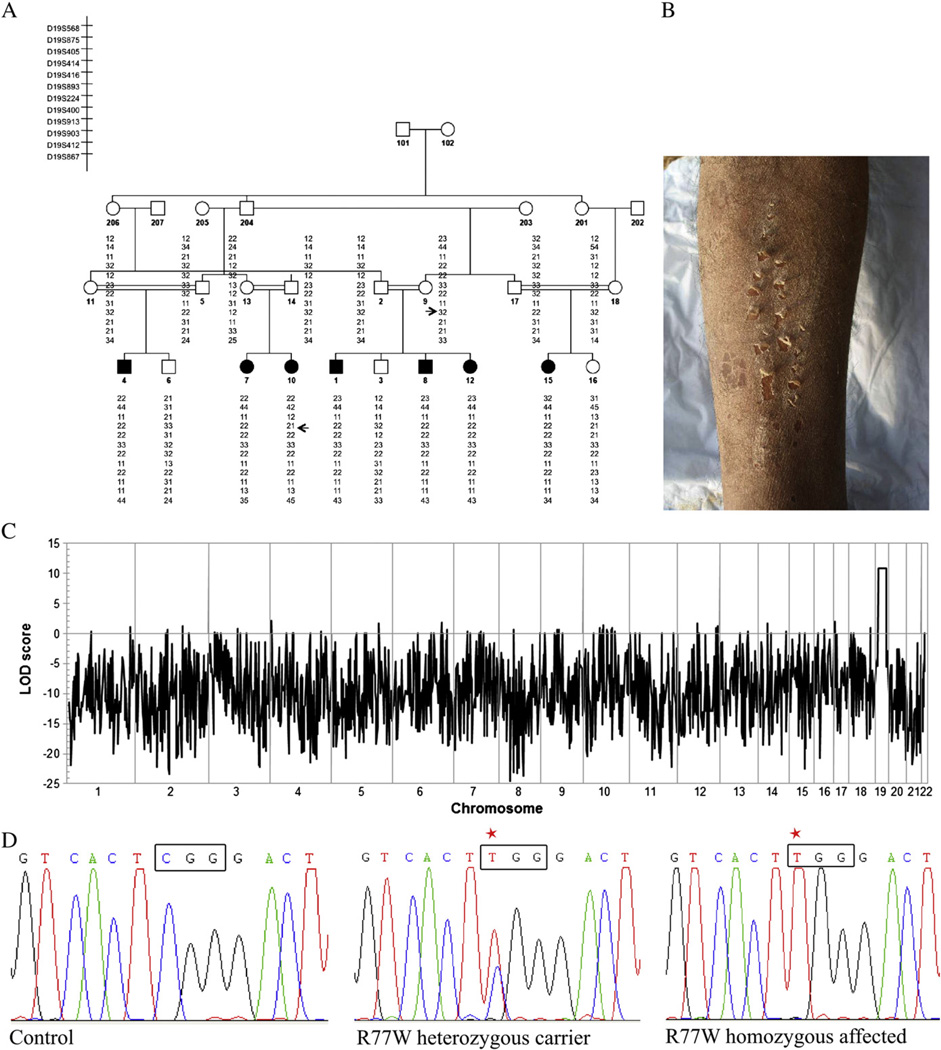
Fig. 1.
Autozygozity mapping and whole-exome sequencing identify homozygous missense mutation in CHST8 in a Pakistani family with PSS non-inflammatory type A. (A) Haplotype analysis in a large consanguineous family with PSS type A. Arrows indicate the key recombination events. (B) Clinical phenotype of an affected individual reveals superficial peeling of the skin. (C) Autozygozity mapping using the Affymetrix 10 K genotyping chip. A maximum LOD score of 10.9 was obtained for a 23 Mb region on chromosome 19. (D) Sanger sequencing of genomic DNA from patients 1–18 confirms presence of a homozygous c.229C>T mutation in affected individuals. Representative electropherograms for control, heterozygous carrier and homozygous affected individuals are shown (red star shows mutated nucleotide).
2.2. SNP genomic mapping and microsatellite linkage analysis
We undertook a genome-wide scan using DNA from ten family members on the low density Affymetrix 10K SNP array and identified a region of excess homozygosity shared among affected individuals on chromosome 19 (23 Mb), with a lod score of 10.9 (Fig. 1C). Micro-satellite markers spanning the region of autozygosity were then used to genotype all family members (Fig. 1A). Key recombination events were detected between markers D19S414 and D19S416, and D19S903 and D19S412 in the affected number 10 (Fig. 1A), which allowed the interval of linkage to be narrowed to 16 Mb flanked by markers D19S414 and D19S412. This region contained more than one hundred candidate genes.
2.3. Mutation detection and analysis
Because of our well-defined linkage region, we were able to perform whole-exome sequencing on a single affected family member. We identified a total of 1093 variants that were not present in any public databases or our internal database of ethnically-matched samples. However, only one of these variants, c.229C>T, located within exon 4 of the CHST8 gene, was homozygous and located within the autozygous region. This single base pair transition substitutes an arginine amino acid by a tryptophan (R77W). We validated the presence of the homozygous C to T transition mutation detected by exome sequencing, c.229C>T, using Sanger sequencing. We PCR amplified and sequenced genomic DNA from individuals 1–18 (Fig. 1A) using primers specific for exon 4 and found the c.229C>T mutation, homozygous in all seven affected family members and heterozygous in carrier individuals, showing complete co-segregation of the mutation with the phenotype. This mutation was excluded from 400 chromosomes from 200 unrelated healthy control individuals of Pakistani origin by direct sequencing.
2.4. Immunofluorescence analysis of skin proteins
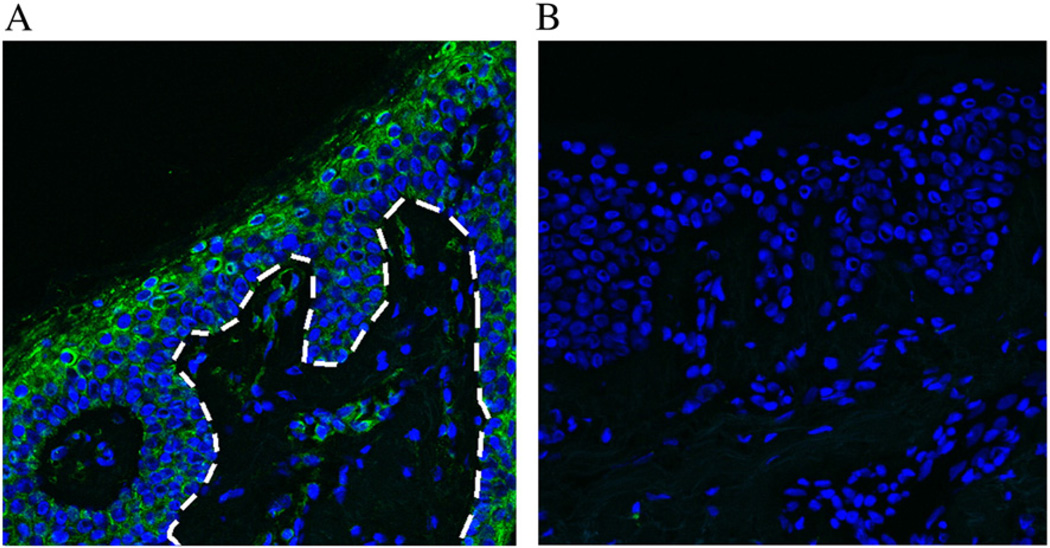
The CHST8 gene encodes a carbohydrate sulfotransferase, N-acetylgalactosamine-4-O-sulfotransferase 1 (GalNAc4-ST1). This enzyme is a type II transmembrane protein that functions almost exclusively in the Golgi apparatus and transfers sulfate groups to the C4 hydroxyl group of terminal β1,4-linked N-acetylgalactosamine moieties [13]. GalNAc4-ST1 is known to be highly expressed in the pituitary gland, cerebellum and brain and to be required for biosynthesis of the glycoprotein hormones lutropin and thyrotropin, by mediating sulfation of their carbohydrate structures. In order to assess whether GalNAc4-ST1 is expressed in normal human skin, we performed immunofluorescence staining of normal human frozen skin sections with a polyclonal antibody raised against GalNAc4-ST1 (Fig. 2). We observed expression of GalNAc4-ST1 throughout the epidermis, with highest expression levels in suprabasal layers (Fig. 2A).
Fig. 2.
GalNAc4-ST1 is expressed throughout normal human epidermis. (A) Immunofluorescence staining of normal human frozen skin sections with a polyclonal antibody raised against GalNAc4-ST1 reveals expression of this protein throughout the epidermis, most predominantly in the granular and cornified layers. The dotted white line depicts the boundary between the epidermis and the dermis. (B) Negative control.
2.5. The missense mutation R77W is predicted to alter amino acid hydrophilicity
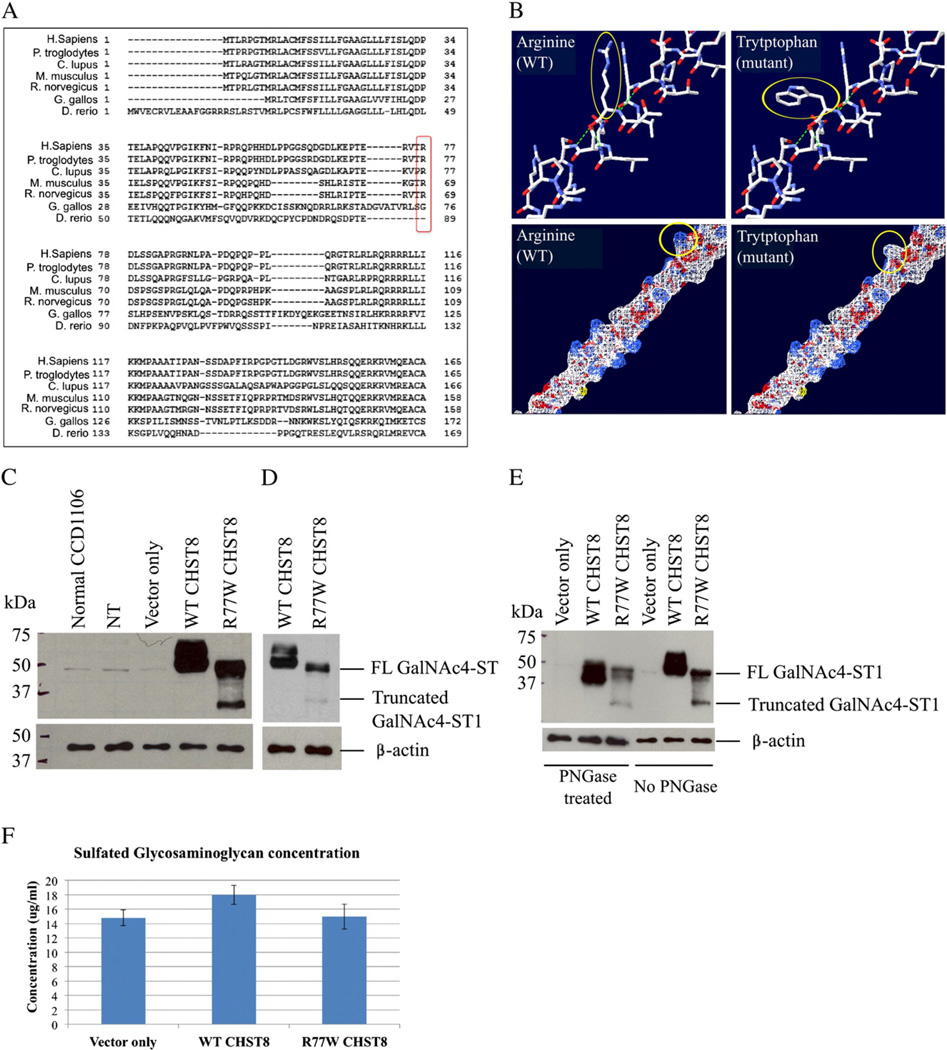
The c.229C>T substitution occurs in exon 4 of CHST8 and results in a single amino acid substitution, arginine to tryptophan (R77W), within the Golgi luminal domain of GalNAc4-ST1. The R77 amino acid is conserved across mammalian species, including human, chimpanzee, dog, mouse and rat, as depicted in Fig. 3A. Importantly, using the Swiss-PdbViewer 4.0, we show that the mutation is predicted to perturb amino acid hydrophilicity due to the substitution of the basic amino acid arginine to the non-polar amino acid tryptophan (Fig. 3B). This region of the protein is predicted to be highly hydrophilic as previously shown in a hydrophilicity plot for GalNAc4-ST1 [14]. These observations suggest that the mutation R77W is likely to have a detrimental effect on the protein.
Fig. 3.
The c.229C>T (R77W) missense mutation results in decreased expression levels of full-length protein, loss of protein glycosylation and decreased activity of GalNAc4-ST1. (A) GalNAc4-ST1 protein sequence alignment across species (using HomoloGene, NCBI) shows that R77 is conserved among human, chimpanzee, dog, mouse and rat mammal species, but not in chicken or zebra fish. (B) Amino acid structure and electrostatic potential of GalNAc4-ST1 shows that the mutation perturbs amino acid hydrophilicity. Top panel: arginine (wild type) and tryptophan (mutant) amino acids are indicated with a red arrow. Blue = N, red = O, white = C, Bottom panel: a change in hydrophilicity due to the substitution of the basic amino acid arginine (white) by the non-polar amino acid tryptophan (blue) is indicated by the red arrow. Blue = basic amino acid, gray = non-polar amino acid. Figure generated using the Swiss-PdbViewer 4.0 application of the ExPASy Bioinformatics Resource Portal. (C and D) Western blot of whole cell lysates from CCD1106 keratinocytes transfected with c.229C>T mutant CHST8-pCMVscript (R77W CHST8) shows reduced levels of full length GalNAc4-ST1 (~49 kDa) compared to cells transfected with wild type CHST8-pCMVscript (WT CHST8). In addition, a lower molecular weight band (~35 kDa) is observed in whole lysates from cells overexpressing mutant CHST8, which is not detected in cells expressing wild type CHST8 and suggests increased degradation of mutant proteins. Normal keratinocytes (normal CCD1106), non-transfected keratinocytes that were incubated with Fugene HD only (NT) and cells transfected with empty pCMVscript (vector only) show low levels of full length GalNAc4-ST1. β-actin was used as a control of equal loading. FL = full length. (D) Lower exposure of blot B clearly showing lower levels of full length GalNAc4-ST1 in cells expressing mutant construct compared to wild type. (E) Western blot of wild type and mutant recombinant GalNAc4-ST1 proteins before and after treatment with PNGase reveals that, in contrast with wild type proteins, the mutant proteins are not subject to glycosylation. After PNGase treatment, WT proteins have smaller molecular weight than the untreated ones, but mutant proteins have identical molecular weights before and after treatment. (F) Colorimetric assay for total sulfated GAG quantification and comparison between cells transfected with wild type and mutant CHST8 constructs. Decreased levels of total sulfated GAGs are observed in cells expressing mutant GalNAc4-ST1 compared to wild type (n = 6), suggesting loss of function of mutant GalNAc4-ST1 proteins.
2.6. The missense mutation R77W results in decreased expression levels of full-length protein and loss of protein glycosylation
To study the functional consequences of the R77W mutation at the protein level, we cloned both wild type and mutant CHST8 into the mammalian expression vector pCMV-script and transiently overexpressed these constructs in the human HPV16 immortalized keratinocyte cell line CCD1106 KERTr. Western blot analysis of whole cell lysates to test for recombinant protein expression revealed lower levels of full length GalNAc4-ST1 (~49 kDa), as well as a lower molecular weight (<37 kDa) immunoreactive band in cells transfected with the mutant CHST8 construct compared to wild type construct (Figs. 3C and D). These results suggest that mutant proteins are subject to increased degradation. GalNAc4-ST1 contains four potential glycosylation sites (Figure S2). Since protein glycosylation can protect proteins from degradation, we treated whole lysates from cells expressing both wild type and mutant recombinant GalNAc4-ST1 with 10% PGNase (an amidase that cleaves nearly all types of N-glycan chains from glycopeptides/glycoproteins) and performed western blots. We observed that in cells transfected with wild type CHST8, recombinant GalNAc4-ST1 is glycosylated, as indicated by the shift in the molecular weight of this protein upon treatment with PNGase. In contrast, in cells transfected with mutant CHST8, the mutant GalNAc4-ST protein was not glycosylated, as shown by the identical migration of these proteins before and after treatment with PNGase (Fig. 3E). These results demonstrate that, in contrast with wild type proteins, the mutant GalNAc4-ST proteins are not subject to glycosylation, which may result in increased degradation. Double immunofluorescence staining of 4% PFA fixed cells with the anti-CHST8 rabbit polyclonal antibody and a Golgi marker, anti-GM130 mouse monoclonal antibody (BD Biosciences), showed colocalization of GalNAc4-ST1 with the Golgi apparatus in both wild type and mutant cells (data not shown).
2.7. The missense mutation R77W results in decreased activity of GalNAc4-ST1
To test the function of mutant GalNAc4-ST proteins, we used a colorimetric assay (Biocolor assay) for the quantification of total sulfated GAGs, in keratinocytes transfected with both wild type and mutant CHST8 constructs. Decreased levels of total sulfated GAGs were observed in cells expressing mutant GalNAc4-ST1 compared to wild type proteins, suggesting loss of function of mutant GalNAc4-ST1.
3. Discussion
Mutations in two genes have been associated with PSS; TGM5 mutations are the underlying cause of the localized form (APPS) and CDSN mutations lead to type B (inflammatory) PSS. However, for the first time, we here describe a mutation in the CHST8 gene, encoding GalNAc4-ST1, as the underlying cause of type A non-inflammatory PSS.
The first case of PSS was described by Fox et al. [15] and named as “keratolysis exfoliativa congenital”. Since then, a number of cases of PSS have been described, particularly in India, most of which show characteristics of the type A non-inflammatory variant of PSS, which is otherwise asymptomatic with occasional pruritus. In contrast with the inflammatory type B PSS, type A PSS patients do not show the presence of vesicles or pustules, fever, sore throat, or involvement of mucous membrane and nails. Histologically, type A PSS is characterized by a slight hyperkeratosis, thinning of the granular layer and a separation of the stratum corneum from the underlying stratum granulosum or an intracorneal split [16]. Cytoplasmic, intracellular splitting in the lower stratum corneum has also been observed, as well as abnormal cribriform keratohyalin granules, detected by electron microscopy, which are indicative of disturbed keratinization. In some of these patients, reduced desmosomal plaques or intracellular electron-dense globular deposits within the stratum corneum have also been observed [4,17,18].
GalNAc4-ST1 is a type II transmembrane protein with 424 amino acids that harbors four potential N-glycosylation sites and two putative 3′-phosphoadenosine, 5′-phosphosulfate (PAPS, a ubiquitous sulfate donor) binding domains [19] (Figure S2). This protein belongs to a family of sulphotransferases that includes HNK-1 sulphotransferase (HNK-1ST, encoded by CHST10), chondroitin-4-sulphotransferases-1–3 (C4ST1–3, encoded by CHST11, CHST12, and CHST13) and dermatan-4-sulphotransferase-1 (D4ST-1, encoded by CHST14) [13]. GalNAc4-ST1 exhibits 23% sequence homology to HNK-1ST, 28% to C4ST-1 and 26% to C4ST-2 [14]. Except for HNK-1ST, all the remaining three enzymes catalyze the transfer of a sulfate group from the PAPS sulfate donor to carbon-4 of either terminal or non-terminal β-1,4-linked GalNAc, but GalNAc4-ST1 is specific for oligosaccharides on glycoproteins bearing terminal β-1,4-linked GalNAc. Oligosaccharides terminating with β-1,4-linked GalNAc-4-SO4 have been described on a limited number of glycoproteins, including carbonic anhydrase VI (encoded by CA7), tenascin R (encoded by TNR), chondroitin, keratin sulfate, heparan sulfate and pro-opimelanocortin (POMC) [13], most of which can be expressed by human keratinocytes. These are potential substrates for GalNAc4-ST1, but also for some of the other sulphotransferases, suggesting some overlap between the different sulphotransferases.
Here, we identified a homozygous missense mutation (c.229C>T; R77W) in the CHST8 gene, encoding GalNAc4-ST1, in a family with autosomal recessive type A PSS, by using a combination of autozygozity mapping and whole-exome sequencing. Even though this amino acid substitution does not occur within a putative PAPS binding site or glycosylation site of GalNAc4-ST1, we show that it causes reduced expression levels of full-length GalNAc4-ST1 and that it alters its overall glycosylation state. In addition to these results, there is further evidence suggesting that this mutation may result in increased degradation of GalNAc4-ST1; (1) the R77 amino acid is conserved across mammalian species and resides within a stretch of amino acids that are highly hydrophilic, (2) the R77W mutation introduces a nonpolar (and hydrophobic) amino acid into this region of the protein, which is likely to alter the conformation and stability of the protein, (3) we observe a lower molecular weight peptide in cells transfected with the mutant CHST8 construct, in addition to lower levels of full-length protein, compared to cells transfected with wild type CHST8 construct, which suggests that the mutant protein is unstable and subject to degradation. Therefore, we postulate that the R77W mutation is likely to alter the conformation of the protein leading to reduction or absence of glycosylation, which may contribute to its increased turnover rate.
Sulfated groups in glycosaminoglycans and oligosaccharides have been shown to play crucial roles in conferring highly specific functions to glycoproteins, glycolipids and proteoglycans, including for cell–cell interactions, signal transduction and embryonic development [20]. Some of these functions include homing of lymphocytes via interaction of L-selectin with its ligands on the endothelial cells of high endothelial venule [21,22]; binding of the HNK-1 epitope to the sulfoglucuronyl sulfoglucuronyl carbohydrate-binding protein, which plays essential roles in higher brain functions [23,24]; rapid clearance of a pituitary glycoprotein hormone, lutropin, mediated by interaction with a hepatic reticulo-endothelial cell receptor [25,26]; myelin function and spermatogenesis mediated by sulfatide and seminolipid, respectively [27]. In addition, ablation of a specific sulfate group was shown to cause defects in both mouse and drosophila embryonic development [28,29]. Our observation of substantial GalNAc4-ST1 protein expression in normal human epidermis, with increasing levels detected in the upper spinous, granular and cornified layers, suggests that GalNAc4-ST1 plays a role in normal human epidermal differentiation. Supporting this hypothesis is the fact that we detected reduced levels of total GAG concentration in cells expressing mutant GalNAc4-ST1 compared to wild type, indicating loss of function of the mutant enzyme, which is likely to stem from its increased turnover rate. These results, coupled with the phenotype of our patients, which also suggests an abnormal epidermal differentiation program, lead us to postulate that GalNAc4-ST1 is important for epidermal homeostasis and that its loss of function results in increased and continuous desquamation of the stratum corneum.
The importance of sulphation of various substrates in the epidermis has been appreciated for a long time. For instance, cholesterol sulfate is an important molecule in the epidermis and plays a crucial role for the cohesion and desquamation of the stratum corneum [30]. The importance of cholesterol sulfate is underlined by the discovery that loss of steroid sulphatase activity underlies recessive X-linked ichthyosis [31,32]. In addition, the ability of cultured mouse and human normal and malignant keratinocytes to synthesize sulfated GAGs has been demonstrated, including hyaluronic acid, heparan sulfate and chondroitin sulfate [33,34]. However, the mechanisms of enzyme activity in these cells have not been elucidated.
In summary, we have identified a homozygous missense mutation in the CHST8 gene as the underlying cause of type A non-inflammatory PSS for the first time. These findings emphasize the importance of sulfation of epidermal molecules and, in particular, the importance of GalNAc4-ST1 for normal epidermal homeostasis. Further studies will be required to uncover the substrate(s) in the epidermis that are specific for GalNAc4-ST1.
4. Materials and methods
4.1. Patients
Informed consent was obtained from all subjects and approval for this study was provided by the Institutional Review Board of Columbia University in adherence to the Declaration of Helsinki Principles. Genomic DNA was isolated from peripheral blood samples according to standard techniques [35]. Control skin was obtained from discarded tissue resulting from routine surgical procedures and processed for immunefluorescence analysis.
4.2. Affymetrix SNP mapping array and linkage
We undertook a genome-wide scan on ten family members using the low density Affymetrix 10K SNP array. Genespring GT (Agilent Software) was used for quality control measures and to perform a number of analyses. SNPs that were not polymorphic or that displayed Mendelian inheritance errors were removed, so that the analyzed dataset contained 7861 SNPs. Haplotypes, or clusters of SNPs that tend to be inherited together, were inferred from the data by Genespring GT. Using haplotypes minimizes the effect of linkage disequilibrium on multipoint linkage analysis, reducing type I error. Initial analysis included genome-wide autozygosity mapping to identify regions identical by descent that were shared among affected individuals. Details of the methodology used by this test can be found on the Agilent website. We performed parametric linkage analysis twice, once using SNP genotypes and once using haplotypes. Microsatellite markers spanning the region of autozygosity were then used to genotype all family members (Fig. 1A).
4.3. Whole-exome sequencing
We performed whole exome sequencing on a single affected family member using the SureSelect Human All Exon Kit (Agilent) to capture the exomes of the affected member. This kit targets approximately 38 Mb of the genome in a single tube, covering 1.22% of human genomic regions corresponding to the coding exons. Captured exons were then subject to paired end sequencing on an Applied Biosystems SOLiD 4 next generation sequencing system, and reads were aligned using the Bioscope software for SOLiD.
4.4. Mutation screening
In order to validate our exome-sequencing findings, we PCR amplified exon 4 of the CHST8 gene using primers 5′GTGGTGAGAGCCTGACCTGTG and 3′GAGCTGTTGGCCGGGATGGTC. The PCR products were directly sequenced in an ABI Prism 310 Automated Sequencer, with the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems). In addition to patients and family members, DNA from 200 unrelated healthy control individuals of Pakistani origin was used to amplify exon 4 of the CHST8 gene, and the resulting DNA fragments were sequenced directly.
4.5. Immunofluorescence staining
Normal skin cryosections (8-µm thick) were fixed in 4% paraformaldehyde (PFA, Sigma) for 15 min and blocked in 5% goat serum for 40 min, followed by incubation overnight at 4 °C with an anti-CHST8 rabbit polyclonal antibody (1:200 dilution, HPA016004, Sigma Prestige). Incubation with Alexa Fluor 488- (green) conjugated goat anti-rabbit IgG secondary antibody (1:800 dilution, Invitrogen) was then performed for 40 min at room temperature. Sections were mounted with DAPI-containing VECTASHIELD Mounting Medium (Vector Laboratories). Primary antibody was omitted from the negative control. Immunofluorescent images were acquired with a Zeiss LSM 510 laser scanning confocal microscope (Carl Zeiss).
4.6. Recombinant GalNAc4-ST1 expression
We cloned both wild type and mutant CHST8 into the mammalian expression vector pCMV-script (Addgene). To accomplish this, human fetal brain cDNA (Clontech) was PCR amplified using the CHST8-specific primers 5′ CGCGAATTCGATGACCCTGCGACCTGGAA CAATG and 3′ CAGAAGCTTTCAGAGCCCTGTTGCTCCCAGGAT. The PCR cycling conditions were 94 °C for 2 min followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 68 °C for 1.5 min, with a final extension step of 5 min at 68 °C. The PCR products were directly cloned into the pCR™4-TOPO® TA vector, using the TOPO® TA Cloning® Kit (Invitrogen) and fully sequenced to confirm that the correct CHST8 sequence was cloned. To generate the c.229C>T mutation, site directed mutagenesis was performed using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies), and the primers used were 5′ CCACAGAGAGGGTCACTTGGGACTTATCCAGTGGG and 3′ CCCACTGGATAAGTCCCAAGTGACCCTCTCTGTGG. Following confirmation of the correct sequence by sequencing analysis, wild type and mutant cDNA clones were subcloned into the pCMVscript mammalian expression vector using the EcoRI and HindIII restriction enzymes (NEB). We transiently overexpressed these constructs, using Fugene HD (Promega) transfections, in the human HPV16 immortalized keratinocyte cell line CCD1106 KERTr, under culture conditions specified by the manufacturer (ATCC, CRL-2309).
4.7. Immunoblotting
Whole-cell protein extracts were prepared from cells lysed in 0.125 M Tris–HCl pH 6.8, 4% (w/v) SDS, 20% (v/v) glycerol, 0.001% (w/v) bromophenol and 1.44 M β-mercaptoethanol, boiled for 5 min and resolved on 12% Mini-PROTEAN® TGX™ Precast Gel (Biorad) according to the manufacturer's instructions. The primary antibody used was the anti-CHST8 rabbit polyclonal antibody (HPA016004, Sigma Prestige). To determine the glycosylation state of both wild type and mutant recombinant GalNAc4-ST1, whole lysates from cells expressing wild type and mutant constructs were treated with 10% PGNase according to the manufacturer's instructions (New England Biolabs), prior to western blotting.
4.8. Biocolor assay
A colorimetric assay for quantification of total sulfated GAGs was performed. Keratinocytes were first lysed in 400 µl of K2HPO4 100 mM pH 8.0, Triton X-100 0.5% buffer. A 100 µl-aliquotwas used to determine protein concentration, using the Pierce BCA Protein Assay Kit (Thermo Scientific), and the remaining 300 µl of protein lysate were digested with proteinase K (50 µg/ml final concentration) at 56 °C for 12 h. Heat inactivation of the enzyme was carried out at 90 °C for 10 min and the mixture was centrifuged (10,000×g, 10 min) through an ultra-free 0.22 µm filter (Millipore). Sulfated GAG quantification was performed according to the procedure previously published by Barbosa et al [36]. Briefly, 1 ml of DMMB solution was added to adjusted-100 µl-aliquots of digested samples and shaken for 30 min. Following centrifugation at 12,000×g for 10 min, the supernatant was discarded and 1 ml of decomplexation solution (4 M guanidine hydrochloride in 10% propan-1-ol and acetate tri-hydrate buffer 50 mM pH 6.8) was added to the precipitate. The mixture was shaken for 30 min and its absorbance at 655 nm was measured. The sulfated GAG concentration was determined by comparison with a chondroitin sulfate calibration curve.
4.9. Protein sequence alignment and in silico modeling
GalNAc4-ST1 protein sequence alignment across species was performed using HomoloGene, NCBI. In silico modeling of amino acid structure and electrostatic potential of GalNAc4-ST1 was performed using the Swiss-PdbViewer 4.0 application of the ExPASy Bioinformatics Resource Portal.
Supplementary Material
Acknowledgments
We are grateful to the family members for their participation in this study. We would like to thank Dr. Munenari Itoh for his valuable discussion, comments and ideas and H. Lam and M. Zhang for technical assistance. We appreciate the collaboration with Drs. C. Higgins, C. Luke and other members of the Christiano laboratory. Supported in part by USPHS/NIH grant RO1AR44924 from NIAMS (to A.M.C.).
Abbreviations
- GalNAc4-ST
N-acetylgalactosamine-4-O-sulfotransferase
- PSS
peeling skin syndrome
- APSS
acral peeling skin syndrome
- GAG
glycosaminoglycan
- TGM5
tranglutaminase 5
- CDSN
corneodesmosin
- NS
Netherton syndrome
- CSTA
cystatin A
- DMMB
1,9-dimethylmethylene blue
- PAPS
3′-phosphoadenosine, 5′-phosphosulfate 3′-phosphoadenosine 5′-phosphosulfate
- C4ST
chondroitin-4-sulphotransferases
- D4ST
dermatan-4-sulphotransferase
- POMC
proopimelanocortin
Footnotes
Supplementary materials related to this article can be found online at doi:10.1016/j.ygeno.2012.01.005.
WEB RESOURCES
The URLs for data presented herein are as follows:
Agilent, http://www.chem.agilent.com/cag/bsp/products/gsgt/Downloads/pdf/autozygosity.pdf1000
1000 Genomes, http://www.1000genomes.org/
dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
Online Mendelian Inheritance in Man, http://www.ncbi.nlm.nih.gov/omim
UCSC Genome Browser database, http://genome.ucsc.edu/
Swiss-PdbViewer 4.0, http://spdbv.vital-it.ch/
References
- 1.Janjua SA, Hussain I, Khachemoune A. Facial peeling skin syndrome: a case report and a brief review. Int. J. Dermatol. 2007;46:287–289. doi: 10.1111/j.1365-4632.2006.03074.x. [DOI] [PubMed] [Google Scholar]
- 2.Köse O, Safali M, Koç E, Arca E, Açikgöz G, Ozmen I, Yeniay Y. Peeling skin diseases: 21 cases from Turkey and a review of the literature. J. Eur. Acad. Dermatol. Venereol. 2011 doi: 10.1111/j.1468-3083.2011.04166.x. [Ahead of print]. [DOI] [PubMed] [Google Scholar]
- 3.Bowden PE. Peeling skin syndrome: genetic defects in late terminal differentiation of the epidermis. J. Investig. Dermatol. 2011;3:561–564. doi: 10.1038/jid.2010.434. [DOI] [PubMed] [Google Scholar]
- 4.Ishida-Yamamoto A, Igawa S, Kishibe M. Order and disorder in corneocyte adhesion. J. Dermatol. 2011;7:645–654. doi: 10.1111/j.1346-8138.2011.01227.x. [DOI] [PubMed] [Google Scholar]
- 5.Telem DF, Israeli S, Sarig O, Sprecher E. Inflammatory peeling skin syndrome caused a novel mutation in CDSN. Arch. Dermatol. Res. 2011 doi: 10.1007/s00403-011-1195-z. [Ahead of print]. [DOI] [PubMed] [Google Scholar]
- 6.Lane EB, McLean WH. Keratins and skin disorders. J. Pathol. 2004;204:355–366. doi: 10.1002/path.1643. [DOI] [PubMed] [Google Scholar]
- 7.Uitto J, Richard G. Progress in epidermolysis bullosa: genetic classification and clinical implications. Am. J. Med. Genet. C. Semin. Med. Genet. 2004;15:61–74. doi: 10.1002/ajmg.c.30035. [DOI] [PubMed] [Google Scholar]
- 8.Cassidy AJ, van Steensel MA, Steijlen PM, van Geel M, van der Velden J, Morley SM, Terrinoni A, Melino G, Candi E, McLean WH. A homozygous missense mutation in TGM5 abolishes epidermal transglutaminase 5 activity and causes acral peeling skin syndrome. Am. J. Hum. Genet. 2005;77:909–917. doi: 10.1086/497707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oji V, Eckl KM, Aufenvenne K, Nätebus M, Tarinski T, Ackermann K, Seller N, Metze D, Nürnberg G, Fölster-Holst R, Schäfer-Korting M, Hausser I, Traupe H, Hennies HC. Loss of corneodesmosin leads to severe skin barrier defect, pruritus, and atopy: unraveling the peeling skin disease. Am. J. Hum. Genet. 2010;87:274–281. doi: 10.1016/j.ajhg.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Israeli S, Zamir H, Sarig O, Bergman R, Sprecher E. Inflammatory peeling skin syndrome caused by a mutation in CDSN encoding corneodesmosin. J. Invest. Dermatol. 2011;131:779–781. doi: 10.1038/jid.2010.363. [DOI] [PubMed] [Google Scholar]
- 11.Dávalos NO, García-Vargas A, Pforr J, Dávalos IP, Picos-Cárdenas VJ, García-Cruz D, Kruse R, Figuera LE, Nöthen MM, Betz RC. A non-sense mutation in the corneodesmosin gene in a Mexican family with hypotrichosis simplex of the scalp. Br. J. Dermatol. 2005;153:1216–1219. doi: 10.1111/j.1365-2133.2005.06958.x. [DOI] [PubMed] [Google Scholar]
- 12.Blaydon DC, Nitoiu D, Eckl K, Cabral RM, Bland P, Hausser I, van Heel DA, Rajpopat S, Fischer J, Oji V, Zvulunov A, Traupe H, Hennies HC, Kelsell DP. Mutations in CSTA, encoding cystatin A, underlie exfoliative ichthyosis and reveal a role for this protease inhibitor in cell–cell adhesion. Am. J. Hum. Genet. 2011 doi: 10.1016/j.ajhg.2011.09.001. doi: 10.1016/j.ajhg.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baenziger JU. Glycoprotein hormone GalNAc-4-sulphotransferase. Biochem. Soc. Trans. 2003;31:326–330. doi: 10.1042/bst0310326. [DOI] [PubMed] [Google Scholar]
- 14.Xia G, Evers MR, Kang HG, Schachner M, Baenziger JU. Molecular cloning and expression of the pituitary glycoprotein hormone N-acetylgalactosamine-4-Osulfotransferase. J. Biol. Chem. 2000;275:38402–38409. doi: 10.1074/jbc.M007821200. [DOI] [PubMed] [Google Scholar]
- 15.Fox H. Skin shedding (keratolysis exfoliativa congenital): report of a case. Arch. Dermatol. 1921;3:202. [Google Scholar]
- 16.Garg K, Singh D, Mishra D. Peeling skin syndrome: current status. Dermatol. Online J. 2010;16:10. [PubMed] [Google Scholar]
- 17.Silverman AK, Ellis CN, Beals TF, Woo TY. Continual skin peeling syndrome: an electron microscopic study. Arch. Dermatol. 1986;122:71–75. [PubMed] [Google Scholar]
- 18.Mevorah B, Frenk E, Saurat JH, Siegenthaler G. Peeling skin syndrome: a clinical, ultra-structural and biochemical study. Br. J. Dermatol. 1987;116:117–125. doi: 10.1111/j.1365-2133.1987.tb05799.x. [DOI] [PubMed] [Google Scholar]
- 19.Okuda T, Mita S, Yamauchi S, Fukuta M, Nakano H, Sawada T, Habuchi O. Molecular cloning and characterization of GalNAc 4-sulfotransferase expressed in human pituitary gland. J. Biol. Chem. 2000;275:40605–40613. doi: 10.1074/jbc.M007983200. [DOI] [PubMed] [Google Scholar]
- 20.Hiraoka N, Misra A, Belot F, Hindsgaul O, Fukuda M. Molecular cloning and expression of two distinct human N-acetylgalactosamine 4-O-sulfotransferases that transfer sulfate to GalNAc beta 1→4GlcNAc beta 1→R in both N- and O-glycans. Glycobiology. 2001;11:495–504. doi: 10.1093/glycob/11.6.495. [DOI] [PubMed] [Google Scholar]
- 21.Hemmerich S, Rosen SD. Carbohydrate sulfotransferases in lymphocyte homing. Glycobiology. 2000;10:849–856. doi: 10.1093/glycob/10.9.849. [DOI] [PubMed] [Google Scholar]
- 22.Kannagi R. Regulatory roles of carbohydrate ligands for selectins in the homing of lymphocytes. Curr. Opin. Struct. Biol. 2002;12:599–608. doi: 10.1016/s0959-440x(02)00365-2. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto S, Oka S, Inoue M, Shimuta M, Manabe T, Takahashi H, Miyamoto M, Asano M, Sakagami J, Sudo K, Iwakura Y, Ono K, Kawasaki T. Mice deficient in nervous system-specific carbohydrate epitope HNK-1 exhibit impaired synaptic plasticity and spatial learning. J. Biol. Chem. 2002;277:27227–27231. doi: 10.1074/jbc.C200296200. [DOI] [PubMed] [Google Scholar]
- 24.Senn C, Kutsche M, Saghatelyan A, Bosl MR, Lohler J, Bartsch U, Morellini F, Schachner M. Mice deficient for the HNK-1 sulfotransferase show alterations in synaptic efficacy and spatial learning and memory. Mol. Cell. Neurosci. 2002;20:712–729. doi: 10.1006/mcne.2002.1142. [DOI] [PubMed] [Google Scholar]
- 25.Roseman DS, Baenziger JU. Molecular basis of lutropin recognition by the mannose/GalNAc-4-SO4 receptor. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9949–9954. doi: 10.1073/pnas.170184597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi Y, Shapiro SD, Baenziger JU. Regulation of lutropin circulatory half-life by the mannose/N-acetylgalactosamine-4-SO4 receptor is critical for implantation in vivo. J. Clin. Invest. 2002;109:269–276. doi: 10.1172/JCI13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honke K, Hirahara Y, Dupree J, Suzuki K, Popko B, Fukushima K, Fukushima J, Nagasawa T, Yoshida N, Wada Y, Taniguchi N. Paranodal junction formation and spermatogenesis require sulfoglycolipids. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4227–4232. doi: 10.1073/pnas.032068299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bullock SL, Fletcher JM, Beddington RS, Wilson VA. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 1998;12:1894–1906. doi: 10.1101/gad.12.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin X, Buff EM, Perrimon N, Michelson AM. Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development. 1999;126:3715–3723. doi: 10.1242/dev.126.17.3715. [DOI] [PubMed] [Google Scholar]
- 30.Epstein EH, Langston AW, Leung J. Sulfation reactions of the epidermis. Ann. N. Y. Acad. Sci. 1988;548:97–101. doi: 10.1111/j.1749-6632.1988.tb18795.x. [DOI] [PubMed] [Google Scholar]
- 31.Jobsis AC, van Duuren CY, de Vries GP, Koppe JG, Rijken Y, vanKempen GMJ, de Groot WP. Trophoblast sulphatase deficiency associated with X-chromosomal ichthyosis. Nederlandsch tijdschrift voor geneeskunde. 1976;120:1980. [Google Scholar]
- 32.Shapiro LJ, Weiss R, Buxman MM, Vidgoff J, Dimond RL, Roller JA, Wells RS. Enzymatic basis of typical X-linked icthyosis. Lancet. 1978;2:756–757. doi: 10.1016/s0140-6736(78)92646-6. [DOI] [PubMed] [Google Scholar]
- 33.King IA. Characterization of epidermal glycosaminoglycans synthesized in organ culture. Biochim. Biophys. Acta. 1981;674:87–95. doi: 10.1016/0304-4165(81)90350-0. [DOI] [PubMed] [Google Scholar]
- 34.Lamberg SI, Yuspa SH, Hascall VC. Synthesis of hyaluronic acid is decreased and synthesis of proteoglycans is increased when cultured mouse epidermal cells differentiate. J. Invest. Dermatol. 1986;86:659–667. doi: 10.1111/1523-1747.ep12275707. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Barbosa I, Garcia S, Barbier-Chassefiere V, Caruelle JP, Martelly I, Papy-Garcia D. Improved and simple micro assay for sulfated glycosaminoglycans quantification in biological extracts and its use in skin and muscle tissue studies. Glycobiology. 2003;13:647–653. doi: 10.1093/glycob/cwg082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.