Abstract
Complexity of multifactorial diseases as Parkinson’s disease (PD) often complicate identifying causal genetic factors by traditional approaches such as positional cloning and candidate gene analyses. PD is etiologically and genetically complex disease and second most common neurodegenerative disorder after Alzheimer’s disease. The most cases of PD are idiopathic and small growing subset of individuals have single gene defect as the cause. The main goal of this research was to identify the potential candidate genes for idiopathic PD by using biomedical discovery support system (BITOLA). For detecting the potential candidate genes for PD was used opened system of bioinformatics tool BITOLA. Data of chromosome location, tissue specific expression of potential candidate genes and their potential association with PD were obtained from Medline, Locus Link, Gene Cards and OMIM. By using BITOLA system is identified 17 genes as potential candidate genes for PD. The role of three genes (MAPT, PARK2, UCHL1) in PD were confirmed earlier. Discovering the novel candidate genes for multifactiorial diseases by using specially mentioned bioinformatics tool BITOLA could offer the new opportunity for researching genetics base of PD without using tissue samples of patients.
KEY WORDS: BITOLA, Parkinson’s disease, candidate genes
INTRODUCTION
Parkinson’s disease (PD) is complex multifactoral neurodegenerative disorder which affects people over the age of 50. It is estimated that most cases of PD are sporadic and have late age onset, yet small but growing subset of individuals has a single gene defect as the cause [1, 2]. The identification of the first gene(alpha synuclein) in familial PD only 10 years ago was a major step in the understanding of the molecular mechanisms in neurodegeneration [3]. Until today it have been identified at last 10 loci associated with PD: PARK1, PARK2, PARK3, PARK4, PARK5, PARK6, PARK7, PARK9, PARK9 and GBA [4]. Linkage analysis and gene expression studies indicate a very large number of candidate genes for PD [5, 6, 7, 8, 9]. Also, variation of genes which are associated with sporadic cases of PD in some cases increases or decreases risk of PD. Moreover, the concept of susceptibility genes allows the involvement of gene-gene and gene-environment interaction in sporadic PD [3]. Previous studies of genes identified as candidate genes for PD indicate that there are many interactions between mutated genes. Although they only occur in a small number of patients, the discovery of genetic forms of PD demonstrated conclusively that PD can occur through inheritance, which has opened a new and exciting area of research [10]. Traditional approaches such as positional cloning and candidate gene analyses, as well as modern methodologies such as gene expression profiling tend to fail to discover genes underlying diseases [11]. Also, DNA microarray analyses produce hundreds of differentially expressed genes which can not be distinguished from normally expressed genes. Genes that harbor no such mutations, but that play key roles in parts of the biological network that lead to disease, are systematically missed by forward genetics approach [12]. All those strategies fail to help researchers in reducing the target genes to a manageable number or to prioritize the disease specific causal genes for further analysis [11]. From this reason it is need to develop sophisticated techniques and tool to identify key candidates from gene list generated by disease gene discovery methods [11]. In terms of discovering new information from the literature, especially for candidate genes for various diseases, Peterlin and Hristovski have developed biomedical support discovery system (BITOLA) [13]. By using this system were identified candidate genes of interest for multiple sclerosis (MS) and bilateral polymicrogyria (BPP) [14, 15, 16]. Methods of integration data from the literature, using BITOLA tools can reveal new potential candidate genes for Parkinson’s disease. The main goal of this research was to identify candidate genes for PD which corresponds to the criteria nominated by authors in design of study.
MATERIALS AND METHODS
In order to include some gene as candidate genes in the list of candidate genes for PD, that gene had to meet following criteria nominated by authors: to show a specific pattern of expression in brain tissue, to be involved in processes of cell adhesion and cell death, and that so far in the literature have not been brought into relation with PD. The reasons because this criteria was nominated are that brain tissue is a target in PD which is characterized by a progressive loss of dopaminergic neurons of the substantia nigra. In the literature, oxidative stress and apoptotic cell death have been implicated in the dopaminergic cell loss. Cell adhesion molecules play a central role in neural development and are also critically involved in axon regeneration and plasticity of synapses in adult nerve system. Information about chromosome loci, tissue-specific expression and the function of potential candidate genes and their association with some genetics disorders were extracted from databases: Medline [17], Locus Link [18], Gene Cards [19] and Online Mendelian Inheritance in Man (OMIM) [20]. To find new potential biomedical relation between PD and pathogenetics mechanisms (cell adhesion and cell death) was used the BITOLA system [13]. The BITOLA system is an interactive literature-based biomedical discovery support system. The purpose of the system is to help the biomedical researchers make new discoveries by discovering potentially new relations between biomedical concepts [13]. The set of concepts currently contains Medical Subject Headings (MeSH), which is used to index Medline, and human genes from The Human Genome Organization (HUGO). The potential new relations are discovered by mining the Medline database [13]. The system is available in two versions: “closed discovery” and “opened discovery” [13]. Open discovery allows the input of a single concept, then categories for first-order relatives of that concept, then categories for relatives of those first order concepts [13]. The BITOLA system was used according to the authors proposed instruction [13]. Discovery algorithm for discovering new relations between medical concepts is described in Table 1 [14]. Discovery algorithm for finding new relations between the given concepts was adapted to PD. As the concept X we nominated Parkinson’s disease, then concept Y is cell function and concept Z is gene or gene products. The main goal was to first find all the concepts Y (cell function) related to the starting concepts X (PD). Then all the concepts Z (gene or gene product) are found. As the last step, we check if X (PD) and Z (gene or gene product) appears together in the medical literature, then we evaluated the proposed (X (PD), Z (gene or gene product)) pairs and select among them those that deserve further investigation. If the chromosomal region of PD matches the location of the related genes (Z) and if there are no MEDLINE documents mentioning both the PD and the genes Z, then the genes Z can be proposed as candidate genes for X (PD). Because in Medline each concept can be associated with many other concepts, the possible number of X→Z combinations can be extremely large [14]. In order to deal this combinatorial problem the author’s of BITOLA system integrated in discovery algorithm filtering (limiting) and ordering capabilities. The related concepts can be limited by the semantic type to which they belong and final possibility for limiting the number of related concepts or false related concepts is by setting thresholds on the support and confidence measures of the association rules [14]. The main goal of the ordering is to present best candidates first to make human review as easy as possible [14]. Our discovery algorithm integrated in opened BITOLA system Figure 1. As beginning concepts X was entered the name Parkinson’s disease. After limiting the related concepts Y by the semantic type Cell function, 53 concepts were obtained corresponding to MeSH descriptions. According to the research strategy, the cell adhesion and cell death were chosen as the most suited to PD. Using those concepts, all related concepts Z of the semantic type gene or gene products were searched and further limited to those matching chromosome location and discoveries only. The all chromosome loci which in literature associated with PD were examined.
TABLE 1.
The algorithm for discovering new relations between medical concepts [14]

FIGURE 1.
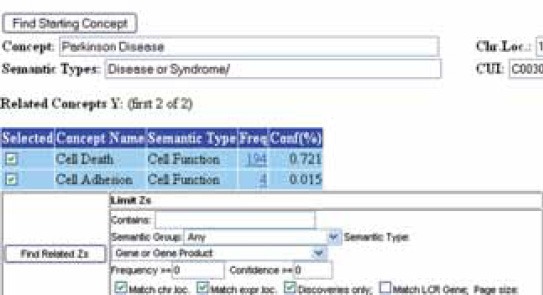
The user interface of open system BITOLA and integrated the discovery algorithm for finding new relation between medical concepts adapted to Parkinson’s disease
RESULTS
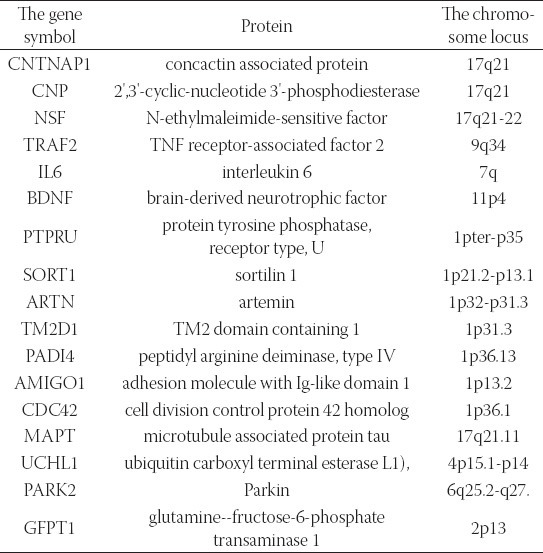
Using the adapted discovery algorithm to the PD and its integration into the opened BITOLA system, we searched all the related concepts Z (gene or gene product) and further limited them to those matching the chromosome loci described in Table 2. In this manner 181 genes were suggested by the opened BITOLA system. By further analysis of those genes we excluded 110 genes whose expression pattern did not preferentially included the brain tissue which is target in the PD. According to the information from Locus Link [18], Gene Cards [19] and OMIM [20] about tissue-specific pattern and the cell function (cell adhesion and cell death) of the remaining 71 genes we include 17 genes as potential candidate genes for PD (Table 3). In the next step, were tested the frequencies of those genes together with PD in the literature. The most frequencies have genes IL6, BDNF, MAPT, and PARK2. The gene IL6 appears with PD in 19 Medline records. The gene BDNF and PD in Medline records appears 88 times. The gene MAPT co-occurs in 157 Medline documents and PARK2 with PD is documented in 49 records. It could be explained by the facts that those genes were researched in population of patients with PD and the role of the genes MAPT and PARK2 in pathogenesis of PD is confirmed. The PTPRU gene with concept X (PD) appears in three Medline documents. However, none of the listed studies do confirm its role in PD, so we do not exclude it. The genes CNTNAP1, CNP, TRAF2, SORT1, ARTN, TM2D1, NSF, PADI4, CDC42, GFPT1 and AMIGO1 do not appear together with the concept X (PD) in Medline documents. One of the rules of discovery algorithm is that association between concept X (PD) and the concept Z (gene or gene product) must not exists. So we exclude from further analysis genes IL6, BDNF, MAPT and PARK2. However, their existence in list of offered genes in the BITOLA system is important because it is implicates on potential role of the BITOLA system in (re)identification disease candidate genes. According to the tissue-specific pattern of the remaining twelve genes and the facts that those genes are not researched into relation with PD, they could be proposed as interesting candidate genes for further analysis.
TABLE 2.
The chromosome regions examined in opened BITOLA system
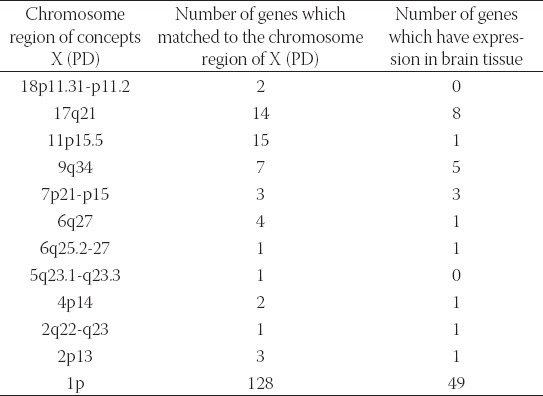
TABLE 3.
The genes extracted from the opened BITOLA system for the further analysis
DISCUSSION
By analyzing of results obtained by using bioinformatics tool BITOLA we compiled the list of potential candidate genes for PD. The twelve genes are chosen as potential candidate genes (CNTNAP1, NSF, CNP, TRAF2, PADI4, PTPRU, SORT1, ARTN, TM2D1, CDC42, GFPT1 and AMIGO1) whose role in PD should be further explored. The most interesting of those twelve genes are NSF, CDC42 and GFPT1 because their role in pathogenesis of PD has not been studied yet. N-ethylmaleimide sensitive factor (NSF) is an ATPases associated with various cellular activities protein (AAA), broadly required for intracellular membrane fusion [21, 22]. It does seem to interact with other proteins, such as the AMPA receptor subunit, GluR2, and beta2-AR and is thought to affect their trafficking patterns. Recently, it has been shown that NSF can be regulated by hydrogen peroxide. H2O2 is thought to inactivate NSF through oxidation of the Cys264 in NSF-D1 [21]. Consistently, mutation of Cys264 to threonine eliminates the sensitivity of NSF to H2O2 [23]. While this might suggest that NSF could be a redox sensor in the cell, whose activity is decreased when the oxidation state of the cytosol increases [21]. The interesting fact is that, NSF gene is located nearby to the MAPT gene. Mutations in the tau gene, MAPT, cause familial frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17), and common variation in MAPT is strongly associated with the risk of progressive supranuclear palsy (PSP), corticobasal degeneration and, to a lesser extent, AD and Parkinson’s disease (PD), implicating the involvement of tau in common neurodegenerative pathway(s) [24]. The genomic complexity around the MAPT locus is emphasized not only by complex arrangements of duplications close to the NSF gene but also in a recently identified de novo micro deletion of 500–600 kb of the locus in individuals with developmental delay and learning disabilities [24]. On the basis of previous facts it seems to be interesting to researching potential interaction between MAPT and NSF genes. Neuronal apoptosis or programmed cell death (PCD) is a crucial process occurring not only during normal development and tissue turnover but also in pathological situations such as stroke and neurodegenerative diseases [25]. Neuronal PCD involves the activation of a number of enzymes and genes and is regulated by specific growth factors, such as neurotrophins, which promote survival of particular neuronal populations by binding to specific cell surface receptors. Over expression of activated Rac1 or Cdc42 in SCG neurons maintained in the presence of NGF induced apoptosis, whereas expression of dominant negative mutants of Cdc42 or Rac1 blocked apoptosis following NGF withdrawal. Furthermore, Cdc42-induced death was prevented by co expressing the c-Jun dominant negative FLAGΔ169 [26]. Taking into account the fact that CDC42 is a key component of the cell death machinery in sympathetic neurons [26], its potential the role in PD should be further considered. Glutamine-fructose-6-phosphate transaminase 1 (GFPAT1) is the rate-limiting enzyme of the hexosamine pathway that has been implicated in the pathogenesis of diabetic nephropathy [27]. Glucosamine 6-phosphate is subsequently converted to uridine diphosphate N-acetylglucosamine, which is used for the O-glycosylation of intracellular proteins. Although this gene is associated with diabetic nephropathy, it is differentially regulated in PD and may play a role in sporadic cases of PD and role in sporadic PD and represent candidate for as yet unidentified disease-causing genes [28]. Taking into account the fact that genes of NSF, CDC42 and GFPT1 have not been brought into correlation with the PD, as opposed to gene PARK2, MAPT and UCHL1, their significance for the PD may represent a potential target for further research.
CONCLUSION
On the basis of above mentioned, the role of NSF, CDC42 and GFPT1 genes, as well as the role of 11 genes that are selected in the list of candidate genes for PD in opened BITOLA system, could be readily tested by mutation screening of PD patients for mutation in those genes.
Discovering the novel candidate genes for multifactiorial diseases by using specially mentioned bioinformatics tool BITOLA could offer the new opportunity for researching genetics base of PD without using tissue samples of patients.
DECLARATION OF INTEREST
Th e autors state that there is no confl ict of interest.
REFERENCES
- [1].Grimes DA, Han F, Panniset M, Racacho L, Xiao F, Zou R, et al. Translated Mutation in the Nurr1 Gene as a Cause for Parkinson's Disease. Movement Disorders. 2006;21:906–909. doi: 10.1002/mds.20820. [DOI] [PubMed] [Google Scholar]
- [2].Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, Kruger R, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson's disease. JAMA. 2006;9(296):661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- [3].Schiesling C, Kieper N, Seidel K, Krüger R. Review: Familial Parkinson's disease –genetics, clinical phenotype andneuropathology in relation to the common sporadic form of the disease. Neuropathology and Neurobiology. 2008;34:225–271. doi: 10.1111/j.1365-2990.2008.00952.x. [DOI] [PubMed] [Google Scholar]
- [4].Feany MB. New Genetic Insights into Parkinson's disease. N Engl J Med. 2004;351:1937–40. doi: 10.1056/NEJMp048263. [DOI] [PubMed] [Google Scholar]
- [5].Pankratz N, Nichols WC, Uniacke SK. Genome-wide linkage analysis and evidence of gene-by-gene interaction in a sample of 362 multiplex Parkinson's disease families. Hum Mol Genet. 2003;12:2599–2608. doi: 10.1093/hmg/ddg270. [DOI] [PubMed] [Google Scholar]
- [6].Scott WK, Nance MA, Watts RL. Complete genomic screen in Parkinson's disease/evidence for multiple genes. JAMA. 2002;286:2239–2244. doi: 10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- [7].Martinez M, Brice A, Vaughan JR. Genome-wide scan for Parkinson's disease/the Gene PD Study. Neurology. 2004;41:900–907. doi: 10.1136/jmg.2004.022632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Elbaz A, Nelson ML, Payami H, Ioannidis JP, Fiske BK, Annesi G, et al. Lack of replication of thirteen single-nucleotide polymorphisms implicated in Parkinson's Disease: a large-scale international study. Lancet Neurology. 2006;5:917–923. doi: 10.1016/S1474-4422(06)70579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li YJ, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, et al. Age at onset in two common neurodegenerative diseases is geneti-cali controlled. Am J Hum Genet. 2007;70:985–983. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Collier KS, Maries E, Kordower JH. Etiology of Parkinson's disease: Genetic and environment revisited. PNAS. 2002;99:13972–13974. doi: 10.1073/pnas.242594999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gudivada RC, Qu XA, Jegga AG, Neumann EK, Aronow BJ. Banff, Alberta, Canada: 2007. A Genome – phenome integrated approach for mining disease-causal genes using semantic web. HCLS Workshop, 16th International World Wide Web Conference, May 8-12. [Google Scholar]
- [12].Chadt EE. Novel integrative genomics strategies to identify genes for complex traits. Animal Genetics. 2006;37:18–23. doi: 10.1111/j.1365-2052.2006.01473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Biomedical Discovery Support System (BITOLA) [accessed 17 May 2010]. Available at: http://ibmi.mfuni-lj.si/bitola .
- [14].Hristovski D, Peterlin B, Džeroski S, Stare J. Literature Based Discovery Support System and its Application to Disease Gene Identification. Proc AMIA Symp. 2001:928. [Google Scholar]
- [15].Hristovski D, Stare J, Peterlin B, Džeroski S. Supporting Discovery in Medicine by Association Rule Minining in Medline and UMLS. Stud Health Technol Inform. 2001;84(Pt 2):1344–1348. [PubMed] [Google Scholar]
- [16].Hristovski D, Peterlin B, Mitchell JA, Humphrey SM. Improving Literature Based Discovery Support by Genetic Knoweledge Integration. Stud Health Technol Inform. 2003;95:68–73. [PubMed] [Google Scholar]
- [17].U.S. National Library of Medicine-MEDLINE. [accessed 11 October 2010]. Available at: http://nlm.nih.gov/databases/databases_MEDLINE .
- [18].U.S.National Library of Medicine-MEDLINE. [accessed 11 October 2010]. Available at: http://nlm.nih.gov/sites/entrez/databases .
- [19].GeneCards. [accessed from 2005 to 2008]. Available at: http://genecards.org/
- [20].Online Mendelian Inheritance in Man (OMIM) [accessed 11 October 2010]. Available at: http://nlm.nih.gov/omim .
- [21].Zhao C, Slevin JT, Whiteheart SW. Cellular function of NSF: Not just SNAPs and SNAREs. FEBS Lett. 2007;581:2140–2149. doi: 10.1016/j.febslet.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Morrell CN, Matsushita K, Lowenstein CJ. A novel inhibitor of N-ethylmaleimide-sensitive factor decreases leukocyte trafficking and peritonitis. J Pharmacol Exp Ther. 2005;314:155–161. doi: 10.1124/jpet.104.082529. [DOI] [PubMed] [Google Scholar]
- [23].Matsushita K, Morrell CN, Mason RJ, Yamakuchi M, Khanday FA, Irani K, et al. Hydrogen peroxide regulation of endothelial exocytosis by inhibition of N-ethylmaleimide sensitive factor. J Cell Biol. 2005;170:73–79. doi: 10.1083/jcb.200502031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pittman AM, Fung HC, de Silva R. Untatling the tau gene association with neurodegenerative dissiorders. Hum Mol Genet. 2006;15:188–195. doi: 10.1093/hmg/ddl190. [DOI] [PubMed] [Google Scholar]
- [25].Oppenheim RW. Cell death during development of the nervous system. Annu Rew Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- [26].Bazenet CE, Mota M, Rubin LL. The small GTP-binding protein Cdc42 is required for nerve growth factor withdrawal-induced neuronal death. Proc Natl Acad Sci USA. 1998;95:3984–3989. doi: 10.1073/pnas.95.7.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ng DP, Walker WH, Chia KS, Choo S, Warram JH, et al. Scrutiny of the Glutamine-Fructose-6-Phosphate Transaminase 1 (GFPT1) Locus Reveals Conserved Nephropathy. Diabetes. 2004;53:865–869. doi: 10.2337/diabetes.53.3.865. [DOI] [PubMed] [Google Scholar]
- [28].Moran LB, Duke DC, Deprez M, Dexter DT, Pearce RK, Graeber MB, et al. Whole genome expression profiling of the medial and lateral substantia nigra in Parkinson's disease. Neurogenetics. 2006;7:1–11. doi: 10.1007/s10048-005-0020-2. [DOI] [PubMed] [Google Scholar]


