Summary
Background
There are regional variations in scalp hair miniaturization seen in androgenetic alopecia (AGA). Use of topical minoxidil can lead to reversal of miniaturization in the vertex scalp. However, its effects on other scalp regions are less well studied.
Methods
A placebo controlled double-blinded prospective pilot study of minoxidil topical foam 5% (MTF) vs placebo was conducted in sixteen healthy men ages 18-49 with Hamilton-Norwood type IV-V thinning. The subjects were asked to apply the treatment (active drug or placebo) to the scalp twice daily for eight weeks. Stereotactic scalp photographs were taken at the baseline and final visits to monitor global hair growth. Scalp biopsies were done at the leading edge of hair loss from the frontal and vertex scalp before and after treatment with MTF and placebo and microarray analysis was done using the Affymetrix GeneChip HG U133 Plus 2.0.
Results
Global stereotactic photographs showed that MTF induced hair growth in both the frontal and vertex scalp of AGA patients. Regional differences in gene expression profiles were observed before treatment. However, MTF treatment induced the expression of hair keratin associated genes and decreased the expression of epidermal differentiation complex (EDC) and inflammatory genes in both scalp regions.
Conclusions
These data suggest that MTF is effective in the treatment of both the frontal and vertex scalp of AGA patients.
Introduction
Androgenetic alopecia (AGA), also known as male pattern balding or hereditary thinning, is the most common type of hair loss. It has been reported that up to 50% of both men and women will manifest some degree of AGA by age 501. The patterned hair loss in AGA seen both in men and women can occur as early as the teens, twenties and thirties. The pathophysiology of AGA has been extensively studied and well characterized, particularly in men2,3. The scalp regions affected by the miniaturization process4 in men are the frontal hairline and the top and vertex scalp, while the temporo-occipital region is largely unaffected even in men with extensive balding5. These regional variations in patterns of scalp hair thinning may reflect differences in embryologic scalp patterning6, levels of hormonal receptors7 or other factors that may influence follicular growth.
Currently there are two medications approved by the US Food and Drug Administration (FDA) for hair regrowth and reversal of miniaturization in androgenetic alopecia: topical minoxidil and oral finasteride8,9. The mechanism of action of topical minoxidil on hair regrowth is not fully understood10. Minoxidil is a potent vasodilator that acts through nitric oxide pathways and as a potassium channel opener11, but its hair re-growth properties appear to be independent of its vasodilation properties. It has been suggested that human hair follicles contain two forms of ATP-sensitive potassium channels, only one of which is sensitive to minoxidil11. Since the clinical trials of topical minoxidil in men with AGA evaluated only hair growth properties of the vertex scalp12,13, it is unclear whether topical minoxidil might be effective in other scalp regions which are also susceptible to hair miniaturization.
In this study we sought to determine whether scalp biopsies from men with AGA show variable expression of genes before and after 8 weeks of treatment with minoxidil topical foam 5% (MTF) vs placebo. Our second aim was to determine whether microarray gene expression profiles in the frontal scalp would be the same as that seen in the vertex scalp.
Materials and Methods
A placebo controlled double-blinded prospective study was conducted. Institutional review board (IRB) approval was obtained and subjects were recruited by the Skin Study Center at Case Western Reserve University (CWRU), Cleveland OH. Sixteen healthy men ages 18-49 with Hamilton-Norwood type IV-V thinning were enrolled for this pilot study. The patients were advised to use Progaine shampoo for all hair cleansing purposes during the study and apply treatment products according to the instructions provided.
Global hair photographs of AGA patients were taken before and after treatment with minoxidil to monitor hair growth. Analysis of the photographs taken with a stereotactic device has emerged as a standard technique for monitoring hair growth and volume14. At the baseline visit, stereotactic scalp photographs were taken and scalp biopsies were done at the leading edge of hair loss from the frontal and vertex scalp. The subjects were instructed to apply the treatment (active drug or placebo) as per the manufacturer’s instructions “half a capful of 5% MTF or placebo topically to the affected area two times a day.” Since the onset of action for 5% Rogaine has been reported to be 8 weeks, we selected this time point to identify early changes in gene expression profiles after treatment with 5% MTF (Rogaine Extra Strength for Men package insert (Pharmacia & Upjohn Consumer Healthcare —US), Rec 2/98).
A blinded evaluation of the stereotactic photographs was done to monitor global hair regrowth on a “yes or no scale” and the subjects were categorized as either responders or non-responders. For the purpose of this study, a responder was defined as a subject who showed hair growth based on stereotactic photographs after 8 weeks treatment with 5% MTF. At the final visit, repeat stereotactic scalp photographs were taken and two scalp biopsies were done contra-lateral to where the first scalp biopsies were done at visit 0. Biopsies were obtained with written consent from patients in accordance with ethical standards of IRB and with the Helsinki Declaration of 1975, as revised in 1983.
Scalp biopsy samples were snap frozen with liquid nitrogen and stored at −80 °C until used. Once all of the scalp biopsies were completed, the specimens were batch processed. RNA was extracted using the Qiagen Rneasy Tissue Mini Kit (QIAGEN, Valencia, CA). cDNA was hybridized to GeneChip HG U133 Plus 2.0 Affymetrix human 3′ array chips using standard Affymetric protocols (Affymetrix, Santa Clara, CA). The data were analyzed using Affymetrix Microarray Suite 5.0 (MAS 5.0) algorithms found in their GeneChip Operating Software (GCOS). The stand alone “detection calls” and comparative “change calls” resulted from Wilcoxon signed rank statistical tests15. The differentially expressed genes in the active and placebo groups before and after treatment were compared using the multi-group Significance Analysis of Microarray (SAM) approach16 with a false discovery rate (FDR) of <0.003. The difference between pre- and post-treatment groups was considered significant for a 2-fold or greater difference in average expression with p<0.05 using a paired t-test.
Results
Enrollment and Sample Analysis
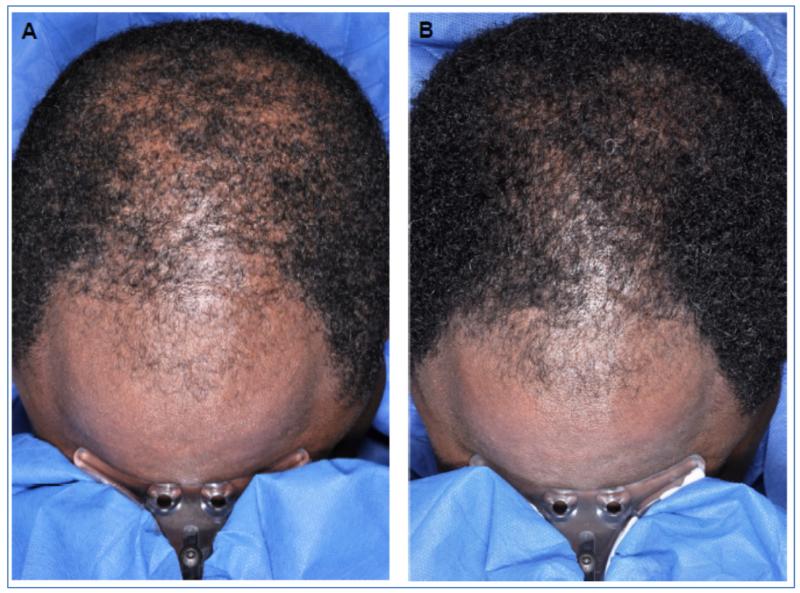
Sixteen men were enrolled for this pilot study; 10 used MTF and 6 used placebo. Three patients did not complete the study, this included 2 in the placebo group and one in the active drug group. RNA was extracted from all 13 patient samples (frontal and vertex, before and after treatment) and sent for microarray (a total of 52 samples). Of the 52 samples (36 active and 16 placebo), 5 were identified as outliers. Extreme expression values that lie outside the mean level of variation observed in the study were referred to as outliers. We used the principal component analysis (PCA) outlier detection method17 for identifying outliers in this study. Of the five samples identified as outliers, 3 were from active and 2 from placebo patients. The outliers were eliminated from the data analysis, thus the final results were based on 33 active and 14 placebo samples. Four of the nine subjects who were on active drug were determined to be responders based on hair growth observed at 8 weeks using global stereotactic photographs, and the five others were deemed non-responders. The four responders had hair growth noted in both the frontal and vertex scalp. Figure 1 shows stereotactic photographs of a responder at baseline and post-treatment with MTF. Increase in hair growth and hair volume was seen in both the vertex and frontal scalp suggesting that MTF induces hair regrowth in both scalp regions.
Figure 1.
A) Base-line and B) post-treatment global photographs of the scalp of an AGA patient taken with a stereotactic device. Hair growth and increase in volume was observed after 8 weeks of 5% MTF treatment and the patient was deemed a responder.
Vertex and Frontal Scalp before treatment
A comparison of vertex and frontal scalp before treatment identified 38 differentially expressed transcripts. As shown in Table 1, thirty three transcripts were down regulated and included both coding (DUSP1, FOS, FOSB, CYR61, HBB, EGR1, ZFP36, MS4A1, IGLI3, ATF3, PSG3, EFCAB4B, KRTAP19-1, 19-3 and 8-1) and non-coding RNAs (SNORDs, SNORAs, RNUs and Vault RNA). The five up regulated genes included MSL3L2, CD209, MUC7, SLC6A14 and ANKRD20B.
Table 1. Differentially expressed genes in vertex vs frontal scalp.
The Affymetrix probe IDs, Gene Symbols, Gene Description and fold-changes of the differentially expressed are shown. The down regulated genes include those that encode stress response proteins (pink highlight), keratin-associated proteins (yellow highlight) and non-coding RNA transcripts (green highlight). Five genes including non-coding RNAs were up regulated in the vertex compared to the frontal scalp.
| Probe id | Gene symbol Down regulated |
Fold change avg | Gene description |
|---|---|---|---|
| 8115831 | ATF3 | −1.954440184 | “Homo sapiens activating transcription factor 3 (ATF3), transcript variant 4, mRNA. ” |
| 7975779 | CYR61 | −1.998588958 | “Homo sapiens cysteine-rich, angiogenic inducer, 61 (CYR61), mRNA. ” |
| 8029693 | DUSP1 | −2.431645062 | “Homo sapiens dual specificity phosphatase 1 (DUSP1), mRNA. ” |
| 7902687 | EGR1 | −1.895999069 | “Homo sapiens early growth response 1 (EGR1), mRNA. ” |
| 7946033 | EFCAB4B | −1.906824031 | “Homo sapiens EF-hand calcium binding domain 4B (EFCAB4B), transcript variant 1, mRNA. ” |
| 8108627 | FOSB | −2.327675169 | “Homo sapiens FBJ murine osteosarcoma viral oncogene homolog B (FOSB), transcript variant 1, mRNA. ” |
| 8005547 | HBB | −1.862261909 | “Homo sapiens hemoglobin, beta (HBB), mRNA. ” |
| 7984259 | KRTAP19-1 | −1.854118595 | “Homo sapiens keratin associated protein 19-1 (KRTAP19-1), mRNA. ” |
| 7981730 | KRTAP19-3 | −2.205875048 | “Homo sapiens keratin associated protein 19-3 (KRTAP19-3), mRNA. ” |
| 7967028 | KRTAP8-1 | −2.076531532 | “Homo sapiens keratin associated protein 8-1 (KRTAP8-1), mRNA. ” |
| 8005553 | MS4A1 | −2.24590692 | “Homo sapiens membrane-spanning 4-domains, subfamily A, member 1 (MS4A1), transcript variant 1, mRNA. ” |
| 7948894 | IGLJ3 | −2.121862356 | “Homo sapiens mRNA for scFv collagenase IV antibody, complete cds. ” |
| 8013329 | PSG3 | −1.783784365 | “Homo sapiens pregnancy specific beta-1-glycoprotein 3 (PSG3), mRNA. ” |
| 7942594 | RNU2-1 | −1.991355015 | “Homo sapiens RNA, U2 small nuclear 1 (RNU2-1), non-coding RNA. ” |
| 7940287 | RNU4-2 | −2.280746608 | “Homo sapiens RNA, U4 small nuclear 2 (RNU4-2), non-coding RNA. ” |
| 7920873 | RNU5B-1 | −1.843214714 | “Homo sapiens RNA, U5B small nuclear 1 (RNU5B-1), non-coding RNA. ” |
| 7909610 | RNU5E | −1.902622286 | “Homo sapiens RNA, U5E small nuclear (RNU5E), non-coding RNA. ” |
| 7960365 | SCARNA5 | −2.037973907 | “Homo sapiens small Cajal body-specific RNA 5 (SCARNA5), non-coding RNA. ” |
| 7897801 | SNORD15B | −1.843219815 | “Homo sapiens small nucleolar RNA, C/D box 15B (SNORD15B), non-coding RNA. ” |
| 8013325 | SNORD3A | −1.902175302 | “Homo sapiens small nucleolar RNA, C/D box 3A (SNORD3A), non-coding RNA. ” |
| 8133688 | SNORD3A | −1.902175302 | “Homo sapiens small nucleolar RNA, C/D box 3A (SNORD3A), non-coding RNA. ” |
| 8014755 | SNORD3A | −1.902175302 | “Homo sapiens small nucleolar RNA, C/D box 3A (SNORD3A), non-coding RNA. ” |
| 8028652 | SNORD3A | −1.902175302 | “Homo sapiens small nucleolar RNA, C/D box 3A (SNORD3A), non-coding RNA. ” |
| 8037231 | SNORD3A | −1.902175302 | “Homo sapiens small nucleolar RNA, C/D box 3A (SNORD3A), non-coding RNA. ” |
| 8049297 | SNORD82 | −1.856869421 | “Homo sapiens small nucleolar RNA, C/D box 82 (SNORD82), non-coding RNA. ” |
| 8059712 | SNORA14A | −1.7275943 | “Homo sapiens small nucleolar RNA, H/ACA box 14A (SNORA14A), non-coding RNA. ” |
| 8062490 | SNORA21 | −1.825872603 | “Homo sapiens small nucleolar RNA, H/ACA box 21 (SNORA21), non-coding RNA. ” |
| 8069822 | SNORA42 | −2.127392366 | “Homo sapiens small nucleolar RNA, H/ACA box 42 (SNORA42), non-coding RNA. ” |
| 8069831 | SNORA60 | −1.890606752 | “Homo sapiens small nucleolar RNA, H/ACA box 60 (SNORA6O), non-coding RNA. ” |
| 8069863 | SNORA74A | −1.867939307 | “Homo sapiens small nucleolar RNA, H/ACA box 74A (SNORA74A), non-coding RNA. ” |
| 8108370 | VTRNA1-1 | −2.631698263 | “Homo sapiens vault RNA 1-1 (VTRNA1-1), non-coding RNA. ” |
| 8108420 | FOS | −4.412605768 | “Homo sapiens v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS), mRNA. ” |
| 8013323 | ZFP36 | −1.746605695 | “Homo sapiens zinc finger protein 36, C3H type, homolog (mouse) (ZFP36), mRNA. ” |
| Up regulated | |||
| 8059852 | ANKRD20B | 1.832236142 | “Homo sapiens ankyrin repeat domain 20B (ANKRD20B), non-coding RNA. ” |
| 8020349 | CD209 | 1.620675753 | “Homo sapiens CD209 molecule (CD209), transcript variant 2, transcribed RNA. ” |
| 8169504 | MSL3L2 | 2.23003259 | “Homo sapiens male-specific lethal 3-like 2 (Drosophila) (MSL3L2), non-coding RNA. ” |
| 8095504 | MUC7 | 1.730630286 | “Homo sapiens mucin 7, secreted (MUC7), transcript variant 1, mRNA. ” |
| 8025301 | SLC6A14 | 1.780898386 | “Homo sapiens solute carrier family 6 (amino acid transporter), member 14 (SLC6A14), mRNA. ” |
Pre and Post Treatment with MTF – Vertex
A comparison of vertex scalp biopsy specimens after and before the use of MTF revealed the following up-regulated genes: keratin associated proteins (KRTAP7-1, KRTAP8-1, KRTAP19-1, KRTAP13-2, KRTAP19-3, KRTAP19-5) as well as small nucleolar non-coding RNAs (SNORD116-22, SNORD25, SNORA5, VTRNA1-1) (Table 2). The down-regulated genes included: the epidermal differentiation complex (EDC) genes including late cornified envelope precursors (LCE3D, LCE3E, LCE1C, LCE2A, LCE2C, LCE2D), small proline rich proteins (SPRR2B, SPRR2E, SPRR2G, SPRR2A), S100 protein (S100A7), loricrin (LOR), fillagrin (FLG2), and cornefelin (CNFN). Inflammatory genes (CCL18, IL1F7, CD177) were also down regulated. These changes were not seen in the placebo samples.
Table 2. Differentially expressed genes in vertex scalp after vs before treatment with minoxidil.
Epidermal differentiation complex (EDC) (yellow highlight) and inflammatory response genes (pink highlight) were down regulated after 5% MTF treatment for 8 weeks in the vertex scalp of AGA patients. Genes that were up regulated include those that encode keratin-associated proteins (yellow highlight) and non-coding RNAs (green highlight).
| Probe id | Gene symbol Down regulated |
Fold change avg | Gene Description |
|---|---|---|---|
| 7953775 | A2ML1 | −1.791380784 | “Homo sapiens alpha-2-macroglobulin-like 1 (A2ML1), mRNA. ” |
| 7983910 | AQP9 | −1.685857639 | “Homo sapiens aquaporin 9 (AQP9), mRNA. ” |
| 7905496 | C1orf46 | −2.061862602 | “Homo sapiens skin-specific protein (xp33) mRNA, partial cds. ” |
| 7905512 | C1orf68 | −2.309045267 | “Homo sapiens chromosome 1 open reading frame 68 (C1orf68), mRNA. ” |
| 7980828 | CCDC88C | −2.441701871 | “Homo sapiens coiled-coil domain containing 88C (CCDC88C), mRNA. ” |
| 8006594 | CCL18 | −2.076061673 | “Homo sapiens chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) (CCL18), mRNA. ” |
| 8037298 | CD177 | −2.10349745 | “Homo sapiens CD177 molecule (CD177), mRNA. ” |
| 8037179 | CNFN | −1.757184518 | “Homo sapiens cornifelin (CNFN), mRNA. ” |
| 7915896 | CYP4Z2P | −1.733354489 | “Homo sapiens cytochrome P450, family 4, subfamily Z, polypeptide 2 pseudogene (CYP4Z2P), non-coding RNA. ” |
| 7985555 | EFTUD1 | −1.851308237 | “Homo sapiens elongation factor Tu GTP binding domain containing 1 (EFTUD1), transcript variant 1, mRNA. ” |
| 7943562 | ELMOD1 | −1.876419323 | “Homo sapiens ELMO/CED-12 domain containing 1 (ELMOD1), transcript variant 1, mRNA. ” |
| 8034974 | EPHX3 | −1.673400532 | “Homo sapiens epoxide hydrolase 3 (EPHX3), transcript variant 1, mRNA. ” |
| 7920175 | FLG2 | −1.746052304 | “Homo sapiens filaggrin family member 2 (FLG2), mRNA. ” |
| 7952673 | FLJ45950 | −1.785333508 | “Homo sapiens cDNA FLJ45950 fis, clone PLACE7008136. ” |
| 8105331 | GZMK | −1.857252097 | “Homo sapiens granzyme K (granzyme 3; tryptase II) (GZMK), mRNA. ” |
| 8044532 | IL1F7 | −1.951182337 | “Homo sapiens interleukin 1 family, member 7 (zeta) (IL1F7), transcript variant 1, mRNA. ” |
| 8120378 | KIAA1586 | −1.580414979 | “Homo sapiens KIAA1586 (KIAA1586), mRNA. ” |
| 7905515 | KPRP | −2.395835622 | “Homo sapiens keratinocyte proline-rich protein (KPRP), mRNA. ” |
| 7963479 | KRT2 | −1.766520904 | “Homo sapiens keratin 2 (KRT2), mRNA. ” |
| 7905525 | LCE1B | −2.186144348 | “Homo sapiens late cornified envelope 1B (LCE1B), mRNA. ” |
| 7920193 | LCE1C | −1.711195411 | “Homo sapiens late cornified envelope 1C (LCE1C), mRNA. ” |
| 7905507 | LCE2A | −2.049191721 | “Homo sapiens late cornified envelope 2A (LCE2A), mRNA. ” |
| 7905505 | LCE2B | −2.499101451 | “Homo sapiens late cornified envelope 2B (LCE2B), mRNA. ” |
| 7905503 | LCE2C | −3.17507158 | “Homo sapiens late cornified envelope 2C (LCE2C), mRNA. ” |
| 7905500 | LCE2D | −2.132509503 | “Homo sapiens late cornified envelope 2D (LCE2D), mRNA. ” |
| 7920185 | LCE3D | −2.064755474 | “Homo sapiens late cornified envelope 3D (LCE3D), mRNA. ” |
| 7920182 | LCE3E | −1.686212956 | “Homo sapiens late cornified envelope 3E (LCE3E), mRNA. ” |
| 7928999 | LIPN | −1.579780344 | “Homo sapiens lipase, family member N (LIPN), mRNA. ” |
| 8115205 | LOC134466 | −1.845999017 | “Homo sapiens zinc finger protein 300 pseudogene (LOC134466), non-coding RNA. ” |
| 8119423 | LOC221442 | −1.688284707 | “Homo sapiens adenylate cyclase 10 pseudogene (LOC221442), non-coding RNA. ” |
| 8139796 | LOC441233 | −1.802369111 | “Homo sapiens cDNA FLJ46129 fis, clone TESTI2046188. ” |
| 7905563 | LOR | −1.681231089 | “Homo sapiens loricrin (LOR), mRNA. ” |
| 7957023 | LYZ | −1.766800783 | “Homo sapiens lysozyme (renal amyloidosis) (LYZ), mRNA. ” |
| 8059852 | MSL3L2 | −2.158304003 | “Homo sapiens male-specific lethal 3-like 2 (Drosophila) (MSL3L2), non-coding RNA. ” |
| 7937940 | OR52K3P | −1.860881202 | Homo sapiens clone IMAGE:110749 mRNA sequence. |
| 7939988 | OR5M3 | −2.029856502 | “Homo sapiens olfactory receptor, family 5, subfamily M, member 3 (OR5M3), mRNA. ” |
| 8062927 | PI3 | −1.988287725 | “Homo sapiens peptidase inhibitor 3, skin-derived (PI3), mRNA. ” |
| 7903404 | RNPC3 | −1.721533122 | “Homo sapiens RNA-binding region (RNP1, RRM) containing 3 (RNPC3), mRNA. ” |
| 7920252 | S100A7 | −1.945756393 | “Homo sapiens S100 calcium binding protein A7 (S100A7), mRNA. ” |
| 8023696 | SERPINB3 | −1.780693185 | “Homo sapiens serpin peptidase inhibitor, clade B (ovalbumin), member 3 (SERPINB3), mRNA. ” |
| 8023688 | SERPINB4 | −2.506533137 | “Homo sapiens serpin peptidase inhibitor, clade B (ovalbumin), member 4 (SERPINB4), mRNA. ” |
| 8021623 | SERPINB7 | −1.74912009 | “Homo sapiens serpin peptidase inhibitor, clade B (ovalbumin), member 7 (SERPINB7), transcript variant 1, mRNA. ” |
| 8169504 | SLC6A14 | −1.730808765 | “Homo sapiens solute carrier family 6 (amino acid transporter), member 14 (SLC6A14), mRNA. ” |
| 7981980 | SNORD116-16 | −1.673093454 | “Homo sapiens small nucleolar RNA, C/D box 116-16 (SNORD116-16), non-coding RNA. ” |
| 7951030 | SNORD6 | −2.356015722 | “Homo sapiens small nucleolar RNA, C/D box 6 (SNORD6), non-coding RNA. ” |
| 7920205 | SPRR2A | −1.990881958 | “Homo sapiens small proline-rich protein 2A (SPRR2A), mRNA. ” |
| 7920210 | SPRR2B | −2.112830411 | “Homo sapiens small proline-rich protein 2B (SPRR2B), mRNA. ” |
| 7920201 | SPRR2B | −2.116427818 | “Homo sapiens small proline-rich protein 2B (SPRR2B), mRNA. ” |
| 7920196 | SPRR2D | −1.676107252 | “Homo sapiens small proline-rich protein 2D (SPRR2D), mRNA. ” |
| 7920214 | SPRR2E | −1.944348255 | “Homo sapiens small proline-rich protein 2E (SPRR2E), mRNA. ” |
| 7920217 | SPRR2G | −2.494933605 | “Homo sapiens small proline-rich protein 2G (SPRR2G), mRNA. ” |
| 8066489 | WFDC12 | −3.050924897 | “Homo sapiens WAP four-disulfide core domain 12 (WFDC12), mRNA. ” |
| Up regulated | |||
| 8014230 | AMAC1 | 1.585669668 | “Homo sapiens acyl-malonyl condensing enzyme 1 (AMAC1), mRNA. ” |
| 8176276 | ATRX | 2.298419926 | “Homo sapiens alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) (ATRX), transcript variant 1, mRNA. ” |
| 8175531 | CDR1 | 1.66741064 | “Homo sapiens cerebellar degeneration-related protein 1, 34kDa (CDR1), mRNA. ” |
| 7963845 | DCD | 2.186042377 | “Homo sapiens dermcidin (DCD), mRNA. ” |
| 8127396 | EYS | 2.220498184 | “Homo sapiens eyes shut homolog (Drosophila) (EYS), transcript variant 1, mRNA. ” |
| 7975779 | FOS | 5.135611804 | “Homo sapiens v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS), mRNA. ” |
| 7948354 | GLYATL2 | 1.942351062 | “Homo sapiens glycine-N-acyltransferase-like 2 (GLYATL2), mRNA. ” |
| 8043449 | IGK@ | 2.385066851 | “Homo sapiens immunoglobulin kappa locus, mRNA (cDNA clone MGC:22645 IMAGE:4700961), complete cds. ” |
| 8043436 | IGKC // IGKC | 2.63060007 | “Homo sapiens immunoglobulin kappa constant, mRNA (cDNA clone IMAGE:6282376). ” |
| 8069813 | KRTAP13-2 | 2.096151572 | “Homo sapiens keratin associated protein 13-2 (KRTAP13-2), nuclear gene encoding mitochondrial protein, mRNA. ” |
| 8069822 | KRTAP19-1 | 2.828617347 | “Homo sapiens keratin associated protein 19-1 (KRTAP19-1), mRNA. ” |
| 8069831 | KRTAP19-3 | 3.182862681 | “Homo sapiens keratin associated protein 19-3 (KRTAP19-3), mRNA. ” |
| 8069838 | KRTAP19-5 | 3.095847716 | “Homo sapiens keratin associated protein 19-5 (KRTAP19-5), mRNA. ” |
| 8069868 | KRTAP7-1 | 3.61335121 | “Homo sapiens keratin associated protein 7-1 (KRTAP7-1), mRNA. ” |
| 8069863 | KRTAP8-1 | 3.096976944 | “Homo sapiens keratin associated protein 8-1 (KRTAP8-1), mRNA. ” |
| 8013567 | LOC201229 | 1.751220303 | “Homo sapiens hypothetical protein LOC201229 (LOC201229), mRNA. ” |
| 7911276 | OR2T6 | 1.730463243 | “Homo sapiens olfactory receptor, family 2, subfamily T, member 6 (OR2T6), mRNA. ” |
| 7948148 | OR5M10 | 2.250498419 | “Homo sapiens olfactory receptor, family 5, subfamily M, member 10 (OR5M10), mRNA. ” |
| 7933561 | PARG | 1.609493237 | “Homo sapiens poly (ADP-ribose) glycohydrolase (PARG), mRNA. ” |
| 8146957 | PI15 | 1.551331779 | “Homo sapiens peptidase inhibitor 15 (PI15), mRNA. ” |
| 8136839 | PIP | 2.501813871 | “Homo sapiens prolactin-induced protein (PIP), mRNA. ” |
| 8088090 | RFT1 | 1.652418379 | “Homo sapiens RFT1 homolog (S. cerevisiae) (RFT1), mRNA. ” |
| 8049297 | SCARNA5 | 1.600678858 | “Homo sapiens small Cajal body-specific RNA 5 (SCARNA5), non-coding RNA. ” |
| 7940630 | SCGB1D2 | 1.817550739 | “Homo sapiens secretoglobin, family 1D, member 2 (SCGB1D2), mRNA. ” |
| 8021081 | SLC14A1 | 2.179329222 | “Homo sapiens solute carrier family 14 (urea transporter), member 1 (Kidd blood group) (SLC14A1), transcript variant 1, mRNA. ” |
| 7981992 | SNORD116-22 | 1.810373746 | “Homo sapiens small nucleolar RNA, C/D box 116-22 (SNORD116-22), non-coding RNA. ” |
| 7948910 | SNORD25 | 1.856333185 | “Homo sapiens small nucleolar RNA, C/D box 25 (SNORD25), non-coding RNA. ” |
| 8142685 | tcag7.977 | 2.029854813 | “Homo sapiens hypothetical protein LOC730130 (LOC730130), mRNA. ” |
| 7930631 | TDRD1 | 1.62149349 | “Homo sapiens tudor domain containing 1 (TDRD1), mRNA. ” |
| 7981732 | VSIG6 | 1.985693826 | Putative V-set and immunoglobulin domain-containing protein 6 gene:ENSG00000189039 |
| 8108627 | VTRNA1-1 | 2.295648077 | “Homo sapiens vault RNA 1-1 (VTRNA1-1), non-coding RNA. ” |
| 8089596 | WDR52 | 1.609278245 | WD repeat protein 52 gene:ENSG00000206530 |
| 7907156 | XCL1 | 2.275547194 | “Homo sapiens chemokine (C motif) ligand 1 (XCL1), mRNA. ” |
Pre and Post Treatment with MTF - Frontal
As seen with vertex scalp, the frontal scalp showed an up regulation of keratin associated proteins (KRTAP5-9, KRTAP7-1, KRTAP8-1, KRTAP10-5, KRTAP10-7, KRTAP10-11, KRTAP19-1, KRTAP19-3, KRTAP19-5) after MTF treatment. Intriguingly, the non-coding RNA, SNORD116-22, was also up regulated in the frontal scalp after MTF treatment. Similar to the vertex scalp, the EDC genes (LCE2A, LCE2B, LCE2C, LCE3D, S100A7, SPRR2A) were down regulated (Table 3).
Table 3. Differentially expressed genes in frontal scalp after vs before treatment with minoxidil.
As seen with the vertex scalp, the epidermal differentiation complex (EDC) (yellow highlight) and inflammatory response genes (pink highlight) were down regulated in the frontal scalp after 5% MTF treatment. Genes that were up regulated include those that encode keratin-associated proteins (yellow highlight) and non-coding RNAs (green highlight).
| Probe id Down regulated |
Gene Symbol | Fold change avg | Gene description |
|---|---|---|---|
| 7905496 | C1orf46 | −1.786381293 | “Homo sapiens skin-specific protein (xp33) mRNA, partial cds. ” |
| 7905512 | C1orf68 | −1.901897993 | “Homo sapiens chromosome 1 open reading frame 68 (C1orf68), mRNA. ” |
| 8037298 | CD177 | −1.90208393 | “Homo sapiens CD177 molecule (CD177), mRNA. ” |
| 7917942 | FLJ35409 | −1.784675504 | “Homo sapiens cDNA FLJ35409 fis, clone SKNSH2009435. ” |
| 7918620 | FLJ36116 | −1.804717709 | “Homo sapiens cDNA FLJ36116 fis, clone TESTI2022338. ” |
| 7991762 | HBA1 | −2.08762655 | “Homo sapiens hemoglobin, alpha 1 (HBA1), mRNA. ” |
| 7991766 | HBA1 | −2.08762655 | “Homo sapiens hemoglobin, alpha 1 (HBA1), mRNA. ” |
| 7981737 | IGHA1 // IGHA1 |
−2.436976257 | “Homo sapiens SNC73 protein (SNC73) mRNA, complete cds. ” |
| 7981724 | IGHD | −2.140677262 | “Homo sapiens immunoglobulin heavy constant delta, mRNA (cDNA clone IMAGE:4855067). ” |
| 8043476 | IGKC | −1.865674065 | Ig kappa chain V-I region HK101 gene:ENSG00000211628 |
| 8043465 | IGKC // IGKC // IGKC // IGKC // IGKC // IGKC |
−1.796109167 | “Homo sapiens immunoglobulin kappa constant, mRNA (cDNA clone IMAGE:4692138). ” |
| 7905507 | LCE2A | −1.659016222 | “Homo sapiens late cornified envelope 2A (LCE2A), mRNA. ” |
| 7905505 | LCE2B | −2.030257255 | “Homo sapiens late cornified envelope 2B (LCE2B), mRNA. ” |
| 7905503 | LCE2C | −2.07719648 | “Homo sapiens late cornified envelope 2C (LCE2C), mRNA. ” |
| 7920185 | LCE3D | −1.873464294 | “Homo sapiens late cornified envelope 3D (LCE3D), mRNA. ” |
| 8045205 | LOC150527 | −1.792985994 | “Homo sapiens hypothetical LOC150527 (LOC150527), transcript variant 1, non-coding RNA. ” |
| 8055236 | LOC150527 | −1.792985994 | “Homo sapiens hypothetical LOC150527 (LOC150527), transcript variant 1, non-coding RNA. ” |
| 7940287 | MS4A1 | −1.891830913 | “Homo sapiens membrane-spanning 4-domains, subfamily A, member 1 (MS4A1), transcript variant 1, mRNA. ” |
| 8177130 | NHEDC1 | −1.622300855 | “Homo sapiens Na+/H+ exchanger domain containing 1 (NHEDC1), transcript variant 1, mRNA. ” |
| 7984259 | RNU5B-1 | −1.939784594 | “Homo sapiens RNA, U5B small nuclear 1 (RNU5B-1), non-coding RNA. ” |
| 8173176 | RP11-167P23.2 | −1.653311406 | Homo sapiens partial mRNA for XAGE-4 protein. |
| 7920252 | S100A7 | −1.775381601 | “Homo sapiens S100 calcium binding protein A7 (S100A7), mRNA. ” |
| 7920875 | SCARNA4 | −2.049700515 | “Homo sapiens small Cajal body-specific RNA 4 (SCARNA4), non-coding RNA. ” |
| 7938329 | SNORA23 | −2.148712212 | “Homo sapiens small nucleolar RNA, H/ACA box 23 (SNORA23), non-coding mRNA. ” |
| 8047780 | SNORA41 | −1.805948249 |
“Homo sapiens small nucleolar RNA, H/ACA box 41 (SNORA41), non-coding RNA. ” |
| 8096301 | SPP1 | −1.995863744 |
“Homo sapiens secreted phosphoprotein 1 (SPP1), transcript variant 1, RNA. ” |
| 7920205 | SPRR2A | −1.936677624 | “Homo sapiens small proline-rich protein 2A (SPRR2A), mRNA. ” |
| 8108627 | VTRNA1-1 | −2.631671674 | “Homo sapiens vault RNA 1-1 (VTRNA1-1), non-coding RNA. ” |
| 8066489 | WFDC12 | −2.638905738 | “Homo sapiens WAP four-disulfide core domain 12 (WFDC12), mRNA. ” |
| Up regulated | |||
| 7987315 | ACTC1 | 1.85182807 | “Homo sapiens actin, alpha, cardiac muscle 1 (ACTC1), mRNA. ” |
| 8042788 | ACTG2 | 1.657203708 | “Homo sapiens actin, gamma 2, smooth muscle, enteric (ACTG2), mRNA. ” |
| 8173349 | AWAT2 | 2.147338381 | “Homo sapiens acyl-CoA wax alcohol acyltransferase 2 (AWAT2), mRNA. ” |
| 7961075 | CD69 | 1.794297146 | “Homo sapiens CD69 molecule (CD69), transcript variant 1, mRNA. ” |
| 7902687 | CYR61 | 2.014127367 | “Homo sapiens cysteine-rich, angiogenic inducer, 61 (CYR61), mRNA. ” |
| 7975779 | FOS | 3.399003819 | “Homo sapiens v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS), mRNA. ” |
| 8029693 | FOSB | 2.209993813 | “Homo sapiens FBJ murine osteosarcoma viral oncogene homolog B (FOSB), transcript variant 1, mRNA. ” |
| 8157727 | GPR21 | 1.540559918 | “Homo sapiens G protein-coupled receptor 21 (GPR21), mRNA. ” |
| 8132843 | HAUS6 | 1.70772238 | “Homo sapiens HAUS augmin-like complex, subunit 6 (HAUS6), mRNA. ” |
| 8091550 | KIAA1328 | 1.74148094 | “Homo sapiens KIAA1328 (KIAA1328), mRNA. ” |
| 8069156 | KRTAP10-11 | 1.891391247 | “Homo sapiens keratin associated protein 10-11 (KRTAP10-11), mRNA. ” |
| 8070782 | KRTAP10-5 | 1.905336495 | “Homo sapiens keratin associated protein 10-5 (KRTAP10-5), mRNA. ” |
| 8069146 | KRTAP10-7 | 1.68653121 | “Homo sapiens keratin associated protein 10-7 (KRTAP10-7), mRNA. ” |
| 8069822 | KRTAP19-1 | 1.951151136 | “Homo sapiens keratin associated protein 19-1 (KRTAP19-1), mRNA. ” |
| 8069831 | KRTAP19-3 | 2.979072598 | “Homo sapiens keratin associated protein 19-3 (KRTAP19-3), mRNA. ” |
| 8069838 | KRTAP19-5 | 3.201872526 | “Homo sapiens keratin associated protein 19-5 (KRTAP19-5), mRNA. ” |
| 7942261 | KRTAP5-9 | 1.783844649 | “Homo sapiens keratin associated protein 5-9 (KRTAP5-9), mRNA. ” |
| 8069868 | KRTAP7-1 | 3.527598251 | “Homo sapiens keratin associated protein 7-1 (KRTAP7-1), mRNA. ” |
| 8069863 | KRTAP8-1 | 2.554485467 | “Homo sapiens keratin associated protein 8-1 (KRTAP8-1), mRNA. ” |
| 8113369 | SLCO4C1 | 1.866374928 | “Homo sapiens solute carrier organic anion transporter family, member 4C1 (SLCO4C1), mRNA. ” |
| 7981992 | SNORD116-22 | 1.790164025 | “Homo sapiens small nucleolar RNA, C/D box 116-22 (SNORD116-22), non- coding RNA. ” |
| 7920141 | TCHH | 1.796011989 | “Homo sapiens trichohyalin (TCHH), mRNA. ” |
| 8035838 | ZNF724P | 1.62142221 | “Homo sapiens cDNA FLJ56866 complete cds, moderately similar to Zinc finger protein 43. ” |
Discussion
Our data showed regional differences in gene expression profiles of vertex and frontal scalp of AGA patients before treatment with minoxidil indicating baseline molecular variations in these two scalp regions. Thus, miniaturized hairs from the vertex and frontal scalp of AGA patients could be considered to bear a distinct molecular signature despite appearing identical clinically and histologically. Whether this difference is a result of embryologic patterning of the scalp or variations in extent of hair thinning or both is unclear. Early stress response genes that participate in cell proliferation and differentiation were significantly decreased in vertex compared to frontal scalp. Hair keratin associated proteins were also significantly decreased. These changes likely reflect differences in the extent of hair loss between the two scalp regions. The other significant changes were in the expression of non-coding RNAs referred to as small nucleolar RNAs (snoRNAs). SnoRNAs were discovered as the regulators of ribosomes and the protein synthesis machinery18,19. Recent studies suggest that snoRNAs may have additional roles in the alternative splicing of mRNA and as microRNAs (miRNAs) that regulate gene expression transcriptionally or post-transcriptionally.20,21 The significance of our observation that snoRNAs are differentially expressed between vertex and frontal, although intriguing, is not immediately clear as we are not aware of previous reports of snoRNAs affecting follicular physiology. It is tempting to speculate that snoRNAs play a role in the differential regulation of gene expression in different regions of the scalp and in hair growth. Indeed, cross-talk between miRNA and mRNA gene networks are reported to participate in epidermal differentiation and tissue remodeling during hair growth22.
Clinical trials have shown the efficacy of minoxidil in inducing hair growth in the vertex scalp13. However, the effects of minoxidil on other scalp regions and the mechanisms by which it induces hair growth are poorly understood. Although, regional differences in gene expression profiles were observed before treatment, the frontal and vertex scalp regions responded similarly to minoxidil treatment. This pilot study serves as a primer for future clinical trials that will have to be conducted to fully evaluate the effects of minoxidil on the frontal scalp. Genes encoding hair keratin-associated proteins were significantly up regulated after treatment in both the vertex and frontal scalp. Keratin-associated proteins are essential for the formation of a rigid and resistant hair shaft through their extensive disulfide bond cross-linking with abundant cysteine residues of hair keratins23-26. Thus, it is likely that minoxidil stimulates hair growth by inducing the expression of keratin associated proteins. The genes that were down regulated in both the frontal and vertex scalp after treatment with minoxidil included the epidermal differentiation complex (EDC) and inflammatory pathways. These pathways may potentially be involved in follicular miniaturization in AGA. The EDC genes map to chromosome 1q21 and encode the calcium-binding proteins of the S100 family, the small proline rich proteins (SPRRs) and the late cornified envelope (LCE) proteins27,28 that participate in keratinocyte differentiation. EDC gene expression is regulated by the AP1 family of transcription factors (jun/fos) which are major regulators of epidermal differentiation29. In addition to EDC genes, a number of inflammatory genes were also down regulated in both the vertex and frontal scalp of AGA patients after minoxidil treatment. Previous studies30-32 have implicated micro-inflammation in the pathogenesis of AGA. More recently, Garza et al33 have shown a role for prostaglandin D2 in the pathogenesis of AGA. The mechanism by which minoxidil down regulates the expression of inflammatory genes is not known. However, the Jun/AP-1 transcription factors regulate inflammation in the skin34 and the decreased expression of EDC and inflammatory genes may be due to the effects of minoxidil on AP1 transcription factors.
Our data suggests that despite regional baseline differences in the scalp of AGA patients, frontal and vertex scalp are responsive to MTF with a molecular pattern that is similar. After treatment with MTF, both regions of the scalp showed increased production of hair keratin-associated proteins and decreased keratinization of the epidermis and inflammatory signals which are known to be altered in AGA. Larger cohort studies are needed to correlate molecular alterations of the scalp with degree of clinical response to MTF which may potentially lead to development of response biomarkers. Additionally, further studies are needed to explain the novel finding of variable expression of non-coding RNAs of the frontal compared to the vertex scalp and to understand their potential role in follicular physiology.
What’s already known about this topic? There are regional variations in scalp hair miniaturization seen in AGA. Use of topical minoxidil can lead to reversal of miniaturization in the vertex scalp.
What does this study add? Frontal scalp of AGA patients is responsive to minoxidil treatment in a manner similar to vertex scalp.
Hair growth properties of MTF may be mediated through increased production of hair keratin-associated proteins and decreased epidermal differentiation complex and inflammatory gene expression.
Acknowledgements
This work was supported by an Independent Investigator grant to PM & PK from Johnson & Johnson Consumer Companies, Inc. and in part by NIH grant R01 AR056245 to PK.
Footnotes
Conflict of Interest: Supported by an Independent Investigator grant from Johnson & Johnson Consumer Companies, Inc.
References
- 1.Price VH. Treatment of hair loss. N Engl J Med. 1999;341:964–73. doi: 10.1056/NEJM199909233411307. [DOI] [PubMed] [Google Scholar]
- 2.Trueb RM. Molecular mechanisms of androgenetic alopecia. Exp Gerontol. 2002;37:981–90. doi: 10.1016/s0531-5565(02)00093-1. [DOI] [PubMed] [Google Scholar]
- 3.Ellis JA, Sinclair R, Harrap SB. Androgenetic alopecia: pathogenesis and potential for therapy. Expert Rev Mol Med. 2002;4:1–11. doi: 10.1017/S1462399402005112. [DOI] [PubMed] [Google Scholar]
- 4.Whiting DA. Possible mechanisms of miniaturization during androgenetic alopecia or pattern hair loss. J Am Acad Dermatol. 2001;45:S81–6. doi: 10.1067/mjd.2001.117428. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton JB. Patterned loss of hair in man; types and incidence. Annals of the New York Academy of Sciences. 1951;53:708–28. doi: 10.1111/j.1749-6632.1951.tb31971.x. [DOI] [PubMed] [Google Scholar]
- 6.Dawber RP. The embryology and development of human scalp hair. Clinics in dermatology. 1988;6:1–6. doi: 10.1016/0738-081x(88)90059-4. [DOI] [PubMed] [Google Scholar]
- 7.Sawaya ME, Price VH. Different levels of 5alpha-reductase type I and II, aromatase, and androgen receptor in hair follicles of women and men with androgenetic alopecia. The Journal of investigative dermatology. 1997;109:296–300. doi: 10.1111/1523-1747.ep12335779. [DOI] [PubMed] [Google Scholar]
- 8.Leyden J, Dunlap F, Miller B, et al. Finasteride in the treatment of men with frontal male pattern hair loss. J Am Acad Dermatol. 1999;40:930–7. doi: 10.1016/s0190-9622(99)70081-2. [DOI] [PubMed] [Google Scholar]
- 9.Whiting DA. Advances in the treatment of male androgenetic alopecia: a brief review of finasteride studies. Eur J Dermatol. 2001;11:332–4. [PubMed] [Google Scholar]
- 10.Messenger AG, Rundegren J. Minoxidil: mechanisms of action on hair growth. Br J Dermatol. 2004;150:186–94. doi: 10.1111/j.1365-2133.2004.05785.x. [DOI] [PubMed] [Google Scholar]
- 11.Shorter K, Farjo NP, Picksley SM, et al. Human hair follicles contain two forms of ATP-sensitive potassium channels, only one of which is sensitive to minoxidil. FASEB J. 2008;22:1725–36. doi: 10.1096/fj.07-099424. [DOI] [PubMed] [Google Scholar]
- 12.Olsen EA, Dunlap FE, Funicella T, et al. A randomized clinical trial of 5% topical minoxidil versus 2% topical minoxidil and placebo in the treatment of androgenetic alopecia in men. J Am Acad Dermatol. 2002;47:377–85. doi: 10.1067/mjd.2002.124088. [DOI] [PubMed] [Google Scholar]
- 13.Olsen EA, Whiting D, Bergfeld W, et al. A multicenter, randomized, placebo-controlled, double-blind clinical trial of a novel formulation of 5% minoxidil topical foam versus placebo in the treatment of androgenetic alopecia in men. J Am Acad Dermatol. 2007;57:767–74. doi: 10.1016/j.jaad.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Olsen EA. Current and novel methods for assessing efficacy of hair growth promoters in pattern hair loss. J Am Acad Dermatol. 2003;48:253–62. doi: 10.1067/mjd.2003.81. [DOI] [PubMed] [Google Scholar]
- 15.Hochberg Y, Benjamini Y. More powerful procedures for multiple significancetesting. Stat Med. 1990;9:811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 16.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shieh AD, Hung YS. Detecting outlier samples in microarray data. Statistical applications in genetics and molecular biology. 2009;8 doi: 10.2202/1544-6115.1426. Article 13. [DOI] [PubMed] [Google Scholar]
- 18.Makarova JA, Ivanova SM, Tonevitsky AG, et al. New functions of smallnucleolar RNAs. Biochemistry. Biokhimiia. 2013;78:638–50. doi: 10.1134/S0006297913060096. [DOI] [PubMed] [Google Scholar]
- 19.Lui L, Lowe T. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays in biochemistry. 2013;54:53–77. doi: 10.1042/bse0540053. [DOI] [PubMed] [Google Scholar]
- 20.Ono M, Scott MS, Yamada K, et al. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–91. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falaleeva M, Stamm S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays. 2013;35:46–54. doi: 10.1002/bies.201200117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Botchkareva NV. MicroRNA/mRNA regulatory networks in the control of skin development and regeneration. Cell Cycle. 2012;11:468–74. doi: 10.4161/cc.11.3.19058. [DOI] [PubMed] [Google Scholar]
- 23.Shimomura Y, Ito M. Human hair keratin-associated proteins. J Investig Dermatol Symp Proc. 2005;10:230–3. doi: 10.1111/j.1087-0024.2005.10112.x. [DOI] [PubMed] [Google Scholar]
- 24.Rogers MA, Langbein L, Praetzel-Wunder S, et al. Human hair keratin-associated proteins (KAPs) Int Rev Cytol. 2006;251:209–63. doi: 10.1016/S0074-7696(06)51006-X. [DOI] [PubMed] [Google Scholar]
- 25.Rogers GE, Powell BC. Organization and expression of hair follicle genes. The Journal of investigative dermatology. 1993;101:50S–5S. doi: 10.1111/1523-1747.ep12362626. [DOI] [PubMed] [Google Scholar]
- 26.Gong H, Zhou H, McKenzie GW, et al. An updated nomenclature for keratin-associated proteins (KAPs) Int J Biol Sci. 2012;8:258–64. doi: 10.7150/ijbs.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kypriotou M, Huber M, Hohl D. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp Dermatol. 2012;21:643–9. doi: 10.1111/j.1600-0625.2012.01472.x. [DOI] [PubMed] [Google Scholar]
- 28.Henry J, Toulza E, Hsu CY, et al. Update on the epidermal differentiation complex. Front Biosci (Landmark Ed) 2012;17:1517–32. doi: 10.2741/4001. [DOI] [PubMed] [Google Scholar]
- 29.Eckert RL, Adhikary G, Young CA, et al. AP1 transcription factors in epidermal differentiation and skin cancer. J Skin Cancer. 2013:537028. doi: 10.1155/2013/537028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaworsky C, Kligman AM, Murphy GF. Characterization of inflammatory infiltrates in male pattern alopecia: implications for pathogenesis. Br J Dermatol. 1992;127:239–46. doi: 10.1111/j.1365-2133.1992.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 31.Whiting DA. Diagnostic and predictive value of horizontal sections of scalp biopsy specimens in male pattern androgenetic alopecia. J Am Acad Dermatol. 1993;28:755–63. doi: 10.1016/0190-9622(93)70106-4. [DOI] [PubMed] [Google Scholar]
- 32.Mahe YF, Michelet JF, Billoni N, et al. Androgenetic alopecia and microinflammation. International journal of dermatology. 2000;39:576–84. doi: 10.1046/j.1365-4362.2000.00612.x. [DOI] [PubMed] [Google Scholar]
- 33.Garza LA, Liu Y, Yang Z, et al. Prostaglandin D2 inhibits hair growth and is elevated in bald scalp of men with androgenetic alopecia. Science translational medicine. 2012;4:126ra34. doi: 10.1126/scitranslmed.3003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schonthaler HB, Guinea-Viniegra J, Wagner EF. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann Rheum Dis. 2011;70(Suppl 1):i109–12. doi: 10.1136/ard.2010.140533. [DOI] [PubMed] [Google Scholar]