Abstract
Among 25 serogroup B Neisseria meningitidis clinical isolates, we identified four (16%) with high factor H binding protein (FHbp) expression that were resistant to complement-mediated bactericidal activity of sera from mice immunized with recombinant FHbp vaccines. Two of the four isolates had evidence of human FH-dependent complement downregulation independent of FHbp. Since alternative complement pathway recruitment is critical for anti-FHbp bactericidal activity, we hypothesized that in these two isolates binding of FH to ligands other than FHbp contributes to anti-FHbp bactericidal resistance. Knocking out NspA, a known meningococcal FH ligand, converted both resistant isolates to anti-FHbp susceptible isolates. The addition of a nonbactericidal anti-NspA monoclonal antibody to the bactericidal reaction also increased anti-FHbp bactericidal activity. To identify a role for FH ligands other than NspA or FHbp in resistance, we created double NspA/FHbp knockout mutants. Mutants from both resistant isolates bound 10-fold more recombinant human FH domains 6 and 7 fused to Fc than double knockout mutants prepared from two sensitive meningococcal isolates. In light of recent studies showing functional FH-PorB2 interactions, we hypothesized that PorB3 from the resistant isolates recruited FH. Allelic exchange of porB3 from a resistant isolate to a sensitive isolate increased resistance of the sensitive isolate to anti-FHbp bactericidal activity (and vice versa). Thus, some PorB3 variants functionally bind human FH, which in the presence of NspA enhances anti-FHbp resistance. Combining anti-NspA antibodies with anti-FHbp antibodies can overcome resistance. Meningococcal vaccines that target both NspA and FHbp are likely to confer greater protection than either antigen alone.
INTRODUCTION
Two meningococcal serogroup B vaccines contain factor H binding protein (FHbp) (1, 2). One of the vaccines, Bexsero (Novartis Vaccines), is licensed in Europe, Australia, and Canada for use in infants, older children, and adults. This vaccine also contains three other antigens capable of eliciting bactericidal antibodies (2) and is referred to as “4CMenB” (four component meningococcal B). The second vaccine, Trumenba (Pfizer), was licensed in the United States on 29 October 2014 for use with the age group from 10 to 25 years. This vaccine contains FHbp (see below).
As of October 2014, there were 758 distinct amino acid sequence variants of FHbp, each assigned a unique “peptide” identifier (ID) as designated on a public database (http://pubmlst.org/neisseria/fHbp/). Based on amino acid sequence relatedness, FHbp sequence variants can be subdivided into two subfamilies, A and B (3, 4), or into three variant groups (5). There is general agreement that protection conferred by anti-FHbp antibody is subfamily (3) or variant group (5) specific. There is also general agreement that isolates with very low FHbp expression are resistant to anti-FHbp bactericidal activity and that isolates with relatively high expression are susceptible as long as there is an antigenic match between the FHbp sequence variant expressed by the isolate and that of the vaccine used to raise the antibody (6–8).
The Pfizer FHbp vaccine contains two FHbp sequence variants, one from each subfamily. The Novartis 4CMenB vaccine contains only a subfamily B FHbp sequence variant. Antibodies elicited by the three other antigens in 4CMenB provide coverage against the majority of meningococci with subfamily A FHbp sequence variants. To date, defining threshold levels of FHbp expression and/or cross-reactivity that are sufficient for predicting susceptibility of an isolate to anti-FHbp bactericidal activity has largely been done empirically by correlating FHbp expression of isolates from different strain collections with susceptibility to bactericidal activity of serum pools from vaccinated infants or adolescents (9–11).
In the present study, we identified four disease-causing serogroup B isolates with relatively high expression of FHbp that were resistant to complement-mediated bactericidal activity of sera from mice immunized with FHbp sequence variants that matched those of the isolates. As described below, two of the four isolates had evidence of human FH-dependent complement evasion independent of FHbp. We report the results of investigations of the basis for the anti-FHbp bactericidal resistance in these two isolates.
MATERIALS AND METHODS
Neisseria meningitidis.
The isolates were a “convenience sample” of 25 isolates from previously described strain collections from patients hospitalized in Maryland (12), Norway (13), or pediatric hospitals in the United States (12, 14). The only selection criterion was that the isolates had to have moderate to relatively high FHbp expression. We assumed that if FHbp expression were low, we had an explanation for anti-FHbp bactericidal resistance. Each of the resistant isolates (an anti-FHbp bactericidal titer of <1:10) was matched with an anti-FHbp susceptible isolate (a titer of ≥1:1,000) with an identical respective FHbp amino acid sequence ID and similar respective FHbp expression as measured by flow cytometry. Characteristics of the two resistant isolates, designated R1 and R2, and the respective control sensitive isolates (designated S1 and S2) are summarized in Table 1.
TABLE 1.
Neisseria meningitidis serogroup B strains
| Isolate no.a | Original strain designation | Geographic location | Clonal complex (MLSTb) | PorA VR (VR1, VR2)c | PorB3 variantd (GenBank accession no.) | FHbp amino acid sequence (ID)e | LOS immunotypef |
|---|---|---|---|---|---|---|---|
| R1 | N02/06 | Norway | 41-44 (5780) | 7-2, 4 | 3-90 (KJ946412) | 4 | 8-1 |
| S1 | MD1327 | Maryland | 41-44 (2973) | 18, 25 | 3-ND (KJ946413) | 4 | 8-1 |
| R2 | SK50 | Ohio | 35 (5744) | 15-1, 3 | 3-7 (KJ997945) | 13 | 3-7-9 |
| S2 | SK57 | Tennessee | 213 (213) | 22, 14 | 3-14 (KJ946414) | 13 | 3-7-9 |
R, anti-FHbp-resistant isolates (titers < 1:10); S, anti-FHbp sensitive isolates (titers ≥ 1:1,000 [see Results]).
MLST, multilocus sequence type. Data are presented at http://pubmlst.org/neisseria/.
Based on PorA variable region (VR) sequence types presented at http://pubmlst.org/neisseria/.
All strains had PorB3 alleles inferred from DNA sequencing and as designated at http://pubmlst.org/neisseria/. 3-ND, not designated in database, but the sequence is one amino acid different from “3-16.”
All strains had FHbp in subfamily B (variant group 1) and were assigned to amino acid sequence variant ID 4 or 13, as indicated at http://pubmlst.org/neisseria/.
Based on reactivity with specific anti-LOS MAbs anti-L3,7,9, anti-L1, or anti-L8 (see Materials and Methods). The isolates were grown in broth containing CMP-NANA; R2 and S2 expressed sialylated LOS based on lack of binding by MAb 3F11 (see Materials and Methods).
Bacterial culture conditions.
N. meningitidis was grown overnight on chocolate agar plates at 37°C in an atmosphere with 5% CO2. GC plates with chemically defined supplements (IsoVitaleX enrichment BD, catalog no. 211875) were prepared and used for antibiotic selection. To measure bactericidal activity and for flow cytometric assays, the bacteria were grown to mid-log phase in Frantz medium supplemented with 4 mM d,l-lactate (Sigma, catalog no. number L-1250). We also added 2 mM cytidine 5′-monophospho-n-acetylneuramic acid (CMP-NANA; Carbosynth, catalog no. MC04931) to enhance the sialylation of lipooligosaccharide (LOS) (15).
Creation of mutant strains with inactivated genes encoding FHbp and/or NspA.
FHbp-knockout (KO), neisserial surface protein A (NspA)-KO, and FHbp/NspA double-KO mutants were generated as previously described (16, 17). In brief, the FHbp gene was replaced by an erythromycin resistance cassette carried on pBSΔgna1870erm (5). The genes encoding NspA were inactivated by transforming the wild-type or isogenic FHbp-KO mutants with a plasmid that contained a truncated nspA gene interrupted by a gene that conferred spectinomycin resistance (16). The resulting nonfunctional NspA genes were confirmed by sequencing of DNA obtained by PCR and by loss of binding of the anti-NspA monoclonal antibody (MAb) 14C7 (18), as measured by flow cytometry.
Sequence analysis of porB genes.
The PorB genes were amplified by PCR using the previously described primers PBA1 and PBA2 (19). The identification of the porB alleles was established according to the Neisseria PubMLST sequence database site (http://pubmlst.org/neisseria/porB/). The allelic variants and serotypes were inferred as described by Sacchi et al. (20) and Abad et al. (19) based on a combination of the sequences of the variable regions.
Generation of PorB3 allelic exchange mutants.
The 1,000-bp region downstream of porB3 was amplified by PCR from heat-killed bacterial cells of isolate R1 using the oligonucleotides 5′-GATAAATTGCCTTAGCATTGAATG-3′ and 5′-GGGAATGACTGAAACTCAAAAAAC-3′, which are referred to here as PBF1 and PBF2, respectively. The resulting 1-kb fragment was cloned into pGEM-T Easy vector (Promega), and a chloramphenicol resistance cassette (chloramphenicol acetyltransferase [CAT]) was cloned 500 bp from the 5′ end of the downstream fragment. The resulting construct (R1_Downstream_CAT) was used to transform both resistant and sensitive R1 and S1 isolates (Table 1) as previously described (21). Transformants were selected on GC agar plates containing 7 μg of chloramphenicol/ml. The porB3 gene (starting from the second codon) plus the region 3′ to porB3 that contained the newly inserted chloramphenicol-resistant gene was amplified by PCR from heat-killed cells of chloramphenicol-resistant R1 and S1 mutants using the primers PBA1 (19) and PBF2. The resulting ∼2.5-kb fragments were cloned separately into pGEM-T Easy vector and sequenced. For the allelic exchanges of porB3 between the R1 and S1 isolates, the plasmid carrying porB3 plus the region downstream of porB3 (with the cloned CAT cassette) from R1 isolate was used to transform the wild-type S1 isolate and vice versa. Transformants were selected on GC agar plates containing 7 μg of chloramphenicol/ml. Resistant colonies were tested by PCR and DNA sequencing for verification of the porB replacement.
LOS characterization.
Suspensions of heat-killed bacterial cells (∼108 cells in 50 μl) were serially diluted and applied to nitrocellulose membranes (Bio-Rad) using a vacuum manifold. LOS was immunotyped with MAbs to L3,7,9 (NIBSC, United Kingdom) or to L1 or L8 (kindly donated by Wendell Zollinger), followed by a 1:10,000 dilution of goat anti-mouse IgG-Alexa Fluor 488 (Abcam). The membranes were scanned at a wavelength of 800 nm using an infrared Odyssey scanner (Li-Cor, Lincoln, NE). The integrated intensities of the bands were calculated with software provided by the manufacturer (version 3.0.21). LOS sialylation can enhance binding of FH to C3 fragments deposited on bacteria (22). Sialylation of LOS was determined using MAb 3F11 (kindly donated by Michael A. Apicella), which recognizes the unsialylated lacto-N-neotetraose LOS; sialylation obscures the 3F11 epitope and decreases binding of the MAb (23). Controls in the assay included an LOS sialyltransferase (Lst) KO mutant of H44/76 that was unable to sialylate its LOS and a wild-type serogroup A strain that was positive for binding when grown in broth without CMP-NANA and negative for binding when grown with CMP NANA.
LOS samples also were prepared from bacteria by proteinase K digestion as previously described (24). Briefly, bacteria were grown in broth as described above, washed once, and resuspended in phosphate-buffered saline (PBS) to an optical density at 620 nm of 0.45. Next, 10 μl of the bacterial suspension was added to 50 μl of lysis buffer (2% sodium dodecyl sulfate [SDS], 4% 2-mercaptoethanol, 10% glycerol, 1 M Tris [pH 6.8], bromophenol blue), and the sample was heated at 100°C for 10 min. Then, 25 μg of proteinase K in 10 μl of lysis buffer was added to the samples, and the lysates were incubated at 60°C for 1 h. A total of 8 μl of the LOS preparation was electrophoresed on a bis-Tris 12% gel (100 V [15 mA] for 2 to 2.5 h). LOS bands were visualized by silver staining (Silver Staining Plus kit, catalog no. 161-0449; Bio-Rad, Hercules, CA).
Mouse antisera.
In initial experiments, we used mouse anti-FHbp antisera from a previous study (25). For additional antisera, we prepared recombinant vaccines from FHbp amino acid sequence variants ID 1, 4, and 55 as previously described (26). Groups of 6- to 8-week-old CD1 mice (Charles River Laboratories) were immunized intraperitoneally (i.p.) with three injections of each of the vaccines administered at 3-week intervals. The dose of vaccine was 10 μg of FHbp adsorbed with 600 μg of aluminum hydroxide. Control animals received aluminum hydroxide only. Blood was collected by cardiac puncture 3 weeks after the third dose.
Bactericidal assay.
Except where noted, the serum bactericidal activity was measured using 20% complement prepared by depleting human serum of IgG with a protein G column (HiTrap Protein G HP, 1 ml; GE Healthcare) (26), and survival of exponential growth-phase bacteria was measured as previously described (27). Where noted, in some experiments we used 20% intact serum that lacked endogenous bactericidal activity as a complement source. The final 40-μl bactericidal reaction mixture contained different dilutions of mouse sera that had been heated at 56°C for 30 min to inactivate endogenous complement, 20% (vol/vol) human complement, and ∼103 CFU of the test isolate. The CFU/ml were determined after 1 h of incubation at 37°C. Serum titers were assigned by the interpolated dilution resulting in 50% survival of the bacteria compared to the CFU/ml when samples were incubated for 60 min with negative-control sera and complement.
Human FH-dependent survival of bacteria in infant rat serum.
Pooled sera from 8- to 9-day-old wild-type Wistar rats, together with 0, 3, 10, 30, or 100 μg/ml of purified human FH (Complement Technologies, Inc.) and ∼400 CFU of bacteria, were added to the wells of microtiter plates. After incubation for 60 min with gentle agitation, the CFU/ml were determined.
Passive protection assay.
To measure the passive protective activity, human FH transgenic rats (28), ages 6 to 7 days, were administered 100 μl of diluted anti-FHbp sera i.p. Negative-control transgenic rats received diluted sera from mice immunized with aluminum hydroxide alone, and positive-control transgenic rats received 10 μg of a serogroup B anticapsular MAb (SEAM 12)/rat. At 2 h, the animals were challenged i.p. with 3,000 or 4,000 CFU of serogroup B isolate R1 or S1, respectively. Blood samples were obtained 12 h later. The CFU/ml of blood were measured by plating 1, 10, and 100 μl of blood onto chocolate agar, and these samples were incubated overnight at 37°C in 5% CO2.
Flow cytometric assays with live N. meningitidis bacteria.
We incubated ∼107 bacteria/ml for 1 h at room temperature with 10 μg/ml murine MAb to the serogroup B capsule (SEAM 12) (29), NspA (14C7) (30), or FHbp (JAR 41) (31). We also tested the binding of 50 μg/ml of previously described recombinant fragments of FH domain 6,7 fused to mouse IgG2a Fc (FH6,7/Fc) or domain 18-20 fused to Fc (FH18-20/Fc) (32). After washing, the bacteria were incubated for 1 h at room temperature with Alexa Fluor 488 goat anti-mouse IgG(H+L) (Invitrogen), diluted 1:500. The bacteria were washed twice with buffer (27) and fixed with 0.5% (vol/vol) formaldehyde in PBS, and binding was analyzed by flow cytometry.
For detection of binding of human FH to the bacterial surface, the bacteria were incubated for 1 h at room temperature with 100 μg/ml of purified human FH (Complement Technologies, Inc., TX) or different dilutions of heat-inactivated IgG-depleted human serum. After washing, bound FH was detected as previously described (26). The respective data testing purified FH or FH in heat-inactivated human serum gave similar results. For detection of C4b and C3b deposition the bacteria were grown as described above, resuspended in Dulbecco PBS-bovine serum albumin (BSA) buffer, mixed with different dilutions of antisera or MAbs and 5 or 20% IgG-depleted human complement. The reaction mixtures were incubated for 15 min at room temperature. Human C3b or C4b bound to bacteria was detected with a 1:100 dilution of fluorescein isothiocyanate-conjugated anti-human C3c or C4b antibody, respectively (Meridian Life Science), diluted in Dulbecco PBS containing 0.1 g of CaCl2 and 0.1 g of MgCl2·6H2O (Mediatech)/liter (pH 7.4) with 1% (wt/vol) BSA (Equitech-Bio).
Statistical analyses.
For calculation of geometric means, CFU/ml values below the limit of the detection were assigned half of the lower limit. A Student t test or, where appropriate, a Mann-Whitney test was used to compare the geometric mean CFU/ml between two independent test samples. All statistical tests were two-tailed; probability values of ≤0.05 were considered statistically significant.
Ethics statement.
Animal experiments were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the U.S. National Institutes of Health (NIH). The protocol (number 75) was approved by the Institutional Animal Care and Use Committee of UCSF Benioff Children's Hospital Oakland. Blood collection from the animals was performed under anesthesia, and all efforts were made to minimize suffering. The human complement source for measuring serum bactericidal activity was serum from an adult who participated in a protocol that was approved by the UCSF Benioff Children's Hospital Oakland Institutional Review Board (protocol 2004-039). Written informed consent was obtained from the subject.
RESULTS
Some meningococcal strains with high FHbp expression resist anti-FHbp complement-mediated bacteriolysis.
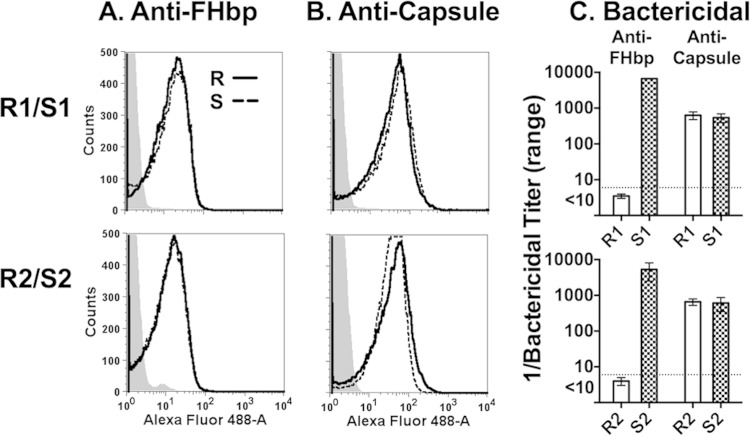
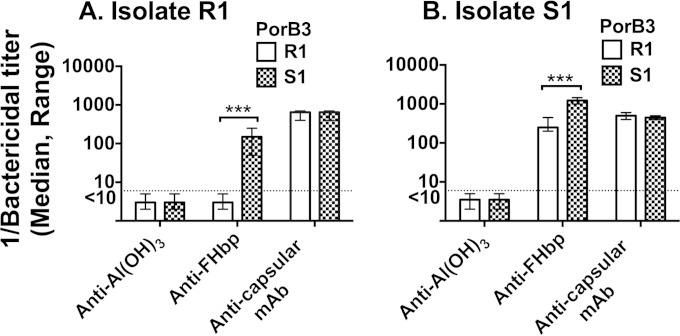
We measured susceptibility of 25 disease-causing serogroup B isolates to bactericidal activity elicited by mouse anti-FHbp antisera using 20% IgG-depleted human serum as a complement source. Four with relatively high FHbp expression as measured by flow cytometry were resistant (anti-FHbp bactericidal titers < 1:10). In two of the four resistant isolates we found evidence of human FH-dependent complement downregulation independent of FHbp (see below). These two isolates, designated R1 and R2, were also resistant to anti-FHbp bactericidal activity (titers < 1:10) when measured with intact human serum that lacked endogenous bactericidal activity as a complement source, and were selected for further study. R1 expressed FHbp amino acid sequence variant ID 4 and R2 had FHbp ID 13 as designated on a public database (http://pubmlst.org/neisseria/). Both were classified in variant group 1 (5) or subfamily B (4). Each of the resistant isolates was matched with an anti-FHbp-susceptible isolate (titer ≥ 1:1,000) with an identical respective FHbp amino acid sequence ID and similar respective FHbp expression as measured by flow cytometry (Fig. 1A). The respective susceptible control isolates were designated S1 and S2 (Table 1). These two isolates also had anti-FHbp bactericidal titers (≥1:1,000) when measured with intact human serum that lacked endogenous bactericidal activity as a complement source. The paired resistant and susceptible isolates showed similar binding with an anticapsular MAb (SEAM 12 [29]) (Fig. 1B). The anti-FHbp bactericidal titers for the two resistant and two susceptible isolates are shown in Fig. 1C. Both pairs showed similar susceptibility to complement-mediated bactericidal activity elicited by an anticapsular MAb (Fig. 1C, two bars on right).
FIG 1.
Anti-FHbp binding in relation to complement-mediated bactericidal activity. (A and B) Binding of MAb (10 μg/ml) to live bacteria as measured by flow cytometry. Solid black line, resistant (R) isolates; dashed black line, control sensitive (S) isolates. (A) Anti-FHbp MAb JAR 5; (B) anticapsular MAb SEAM 12. Representative data are shown. The results were replicated in two experiments. (C) Complement-mediated bactericidal activity. Anti-FHbp, antisera from mice immunized with an FHbp sequence variant that matched the FHbp amino acid sequence variant of the respective isolates. Anti-capsule, anticapsular MAb SEAM 12. The data are calculated from results of three assays. Error bars represent ranges.
The two anti-FHbp resistant isolates also were resistant to bactericidal activity elicited by mouse anti-FHbp ID 1 antibodies (the FHbp sequence variant in the Novartis 4CMenB vaccine), and mouse anti-FHbp ID 55 antibodies (the variant group 1) (subfamily B) antigen in the Pfizer meningococcal FHbp vaccine (see Fig. S1 in the supplemental material).
Among the surface components commonly used to characterize (or “type”) meningococci, no single factor appeared to differentiate between the anti-FHbp-resistant and -susceptible isolates (Table 1). As measured by dot blot analyses, the LOS immunotype of the R1 and S1 pair was L8 and L1, and that of the R2 and S2 pair was L3,7,9. By silver-stained SDS-PAGE, the respective LOS bands were similar for each pair (see Fig. S2 in the supplemental material). The bacterial broth used to grow the isolates included CMP-NANA (see Materials and Methods). Under these conditions, both the resistant and susceptible R2 and S2 isolates with an LOS phenotype of L3,7,9 expressed sialylated LOS based on lack of binding by MAb 3F11 by dot blotting (data not shown).
To determine whether resistance to anti-FHbp serum bactericidal activity correlated with in vivo resistance to passive protection by anti-FHbp antibody, we performed a bacterial challenge experiment with the R1 and S1 isolates. Human FH transgenic infant rats were treated i.p. with different dilutions of mouse serum pools and challenged i.p. 2 h later with 3,000 or 4,000 CFU. In two experiments, rats given a negative-control serum had similar respective levels of bacteremia with each of the isolates in blood obtained 12 h after the bacterial challenge (the endpoint of the experiments, Fig. 2A and D). All of the rats pretreated with 10 μg of a positive-control anticapsular MAb were protected (sterile blood cultures; data not shown on graph). In experiment 1, an anti-FHbp ID 1 serum pool (sequence variant in the Novartis 4CMenB vaccine [2]) conferred less protection against the R1 isolate than the S1 isolate (at a serum dilution of 1:40, P = 0.023 [Fig. 2B]; at a dilution of 1:160, P = 0.003 [Fig. 2C]). In experiment 2, a 1:40 dilution of an anti-FHbp serum pool from mice immunized with FHbp ID 4, which matched the FHbp sequence variant of the R1/S1 isolate pair, conferred protection against both isolates (Fig. 2E) but at a dilution of 1:160 there was less protection against the R1 isolate than the S1 isolate (P = 0.015, Fig. 2F). Thus, although only one of the two pairs of isolates was tested, in vitro anti-FHbp resistance of the R1/S1 pair correlated with in vivo resistance to passive protection against bacteremia by anti-FHbp antibodies.
FIG 2.
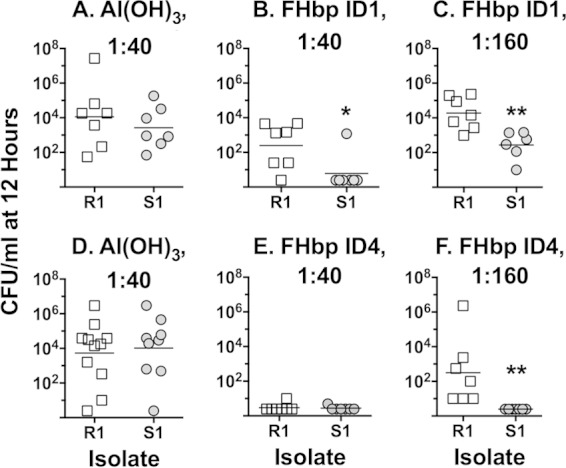
Anti-FHbp passive protective activity against bacteremia. Human FH transgenic infant rats were treated i.p. with sera from immunized mice and 2 h later challenged i.p. with 3,000 or 4,000 CFU of group B strain R1 or S1. Blood cultures were obtained 12 h after the bacterial challenge. Each symbol represents result of an individual animal. (A and D) Sera diluted 1:10 from negative-control mice immunized with aluminum hydroxide; (B and C) sera diluted 1:40 or 1:160, respectively, from mice immunized with FHbp ID 1 (antigenic variant in 4CMenB vaccine); (E and F) sera diluted 1:40 or 1:160, respectively, from mice immunized with FHbp ID 4 (matched the FHbp variant in the R1 and S1 isolates). Not shown are data from control animals treated with 10 μg of an anticapsular MAb, which resulted in sterile blood cultures. Panels with asterisks were statistically significant (panel B, P = 0.0239; panel C, P = 0.0032; panel F, P = 0.0146).
Anti-FHbp resistance requires expression of NspA.
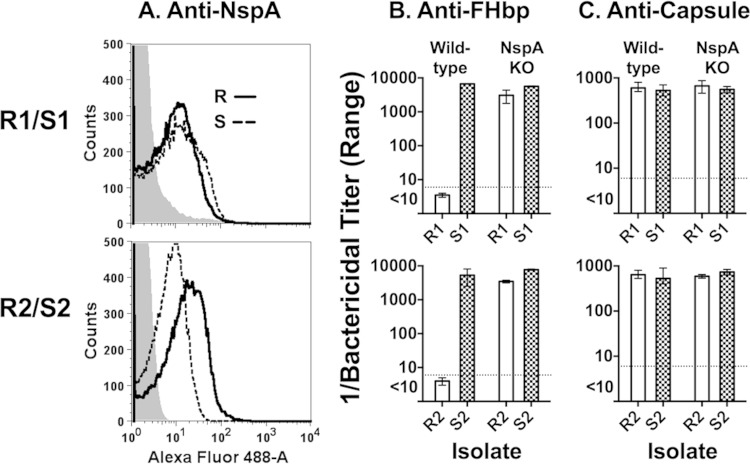
NspA and PorB2 have been identified as additional meningococcal FH ligands that can downregulate complement on the bacterial surface (17, 32). We hypothesized that binding of FH to ligands other than FHbp contributes to anti-FHbp resistance. Our initial studies focused on NspA because both of the resistant isolates had PorB3, which was reported not to functionally bind FH on meningococci (32). The respective NspA amino acid sequences of the R1, R2, and S1 isolates were identical, and that of S2 differed by only three amino acids (see Fig. S3 in the supplemental material); none of these amino acid polymorphisms was predicted to be surface exposed based on an NspA crystal structure (33). By flow cytometry, the R1 and S1 isolates showed similar binding by an anti-NspA MAb, while the R2 isolate bound more anti-NspA MAb than the control S2 isolate (Fig. 3A). When the NspA genes were inactivated, each of the resistant isolates became susceptible to complement-mediated anti-FHbp bactericidal activity (titer > 1:5,000, Fig. 3B). In contrast, there was no significant effect of knocking out NspA on the anti-FHbp titers of the two susceptible isolates (Fig. 3B) or on the anticapsular bactericidal activity of the resistant or sensitive isolates (Fig. 3C).
FIG 3.
Effect of knocking out NspA on Anti-FHbp bactericidal activity. (A) Binding of anti-NspA MAb to live bacteria. Anti-NspA MAb 14C7 was tested at 10 μg/ml. Black solid line, resistant (R) strains; dashed black lines, susceptible (S) strains; gray-filled histogram, mutants of R and S isolates with nspA inactivated. Typical results shown; data were replicated in two experiments. (B and C) Complement-mediated bactericidal activity. Mouse anti-FHbp antisera and anticapsular MAb are described in the legend for Fig. 1B and C, respectively. Error bars represent ranges of duplicate or triplicate measurements in three experiments.
We also tested the ability of a nonbactericidal anti-NspA MAb to augment anti-FHbp serum bactericidal titers. None of the resistant or susceptible isolates was killed by the anti-NspA MAb at 50 μg/ml (the highest concentration tested, data not shown). When 5 μg of the anti-NspA MAb/ml was added to the anti-FHbp bactericidal reaction mixtures, both of the anti-FHbp resistant strains became susceptible to complement-mediated bacteriolysis (Fig. 4A). There was no augmentation of the anti-FHbp titers by the anti-NspA MAb with either of the anti-FHbp susceptible isolates (Fig. 4B).
FIG 4.
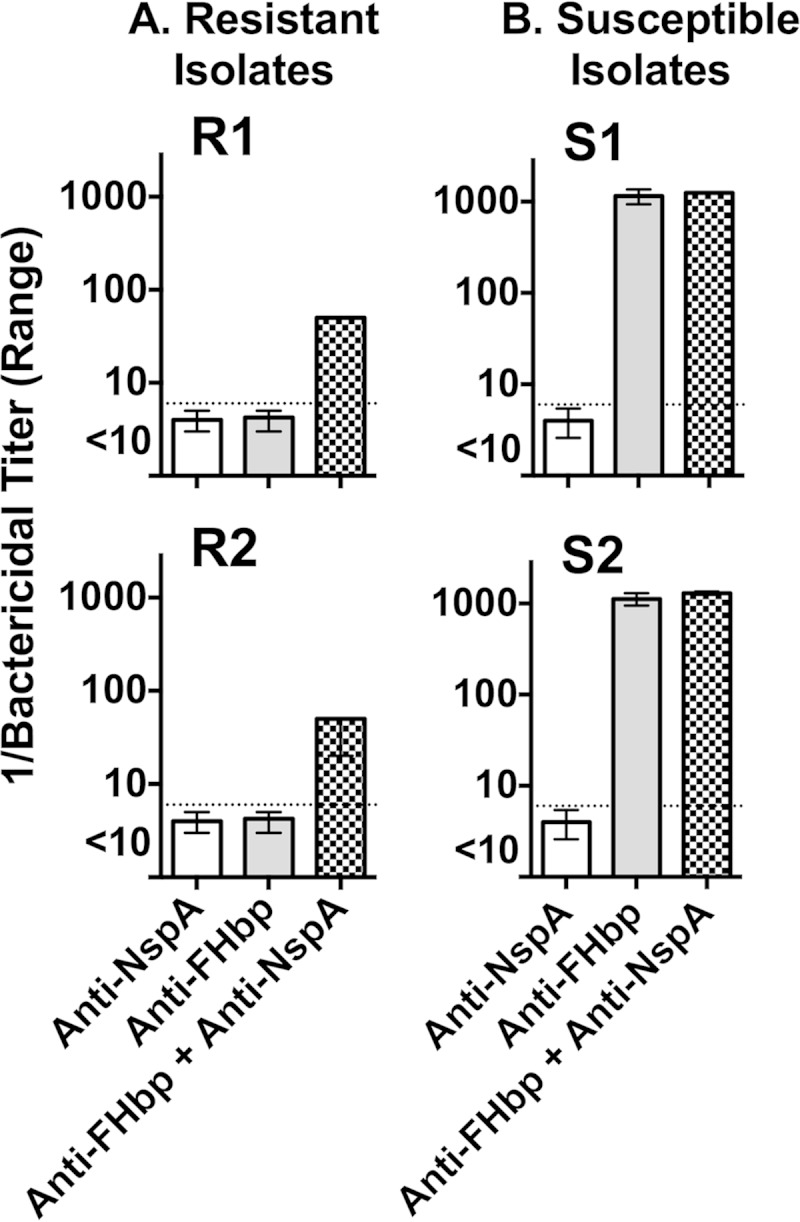
Effect of the addition of a nonbactericidal anti-NspA MAb on serum anti-FHbp bactericidal activity. The anti-NspA MAb (AL12) was tested at 5 μg/ml and 20% human complement. All isolates were resistant to anti-NspA bactericidal activity at MAb concentrations up to 50 μg/ml (the highest concentration tested). The anti-FHbp antisera matched the FHbp amino acid sequence of the test isolates. Ranges represent results from duplicate measurements. Representative results are shown. The data were replicated in two experiments.
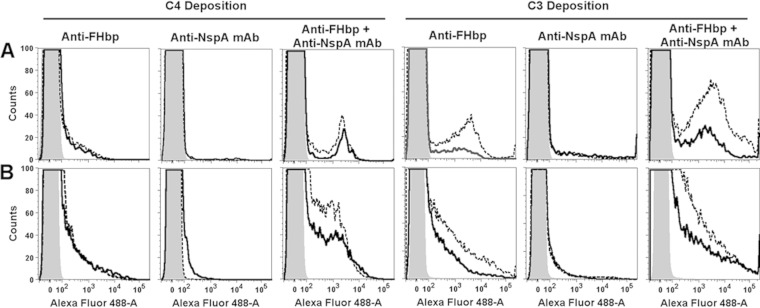
The ability of antibodies to elicit C4b deposition is a marker of classical complement pathway activation. The anti-FHbp antisera alone elicited similar respective C4b deposition with the R1/S1 isolates or R2/S2 isolates (Fig. 5A). As expected (based on differences in the bactericidal data), there was more C3b deposition elicited on the S1 and S2 isolates than the respective R1 and R2 isolates (combined classical and alternative pathway activation, Fig. 5B). Thus, there appeared to be selective alternative pathway downregulation on the R1 and R2 isolates, which resulted in less C3b deposition and resistance to anti-FHbp bactericidal activity. The anti-NspA MAb alone elicited minimal C4b or C3b deposition (Fig. 5). For both pairs of isolates, when 5 μg of the anti-NspA MAb/ml was added to the anti-FHbp reaction mixture, there was augmentation of C4b and C3b deposition, which resulted in bacteriolysis of both resistant isolates.
FIG 5.
Effect of the addition of a nonbactericidal anti-NspA MAb on complement deposition on live bacteria. (A and B) Deposition of C4b and C3b, respectively, by 1:50 dilutions of anti-FHbp antisera, anti-NspA MAb (5 μg/ml), or a combination of anti-FHbp antisera (1:50) with anti-NspA MAb (5 μg/ml). (A) S1 and R1 isolates; (B) S2 and R2 isolates. Solid black line, resistant (R) isolates; dashed black line, sensitive (S) isolates; shaded areas, negative-control without antibody. The data were replicated in three experiments testing 5% IgG-depleted human serum as a source of complement. We also obtained similar respective results for the R1 and S2 isolates testing 20% IgG-depleted human serum as a source of complement.
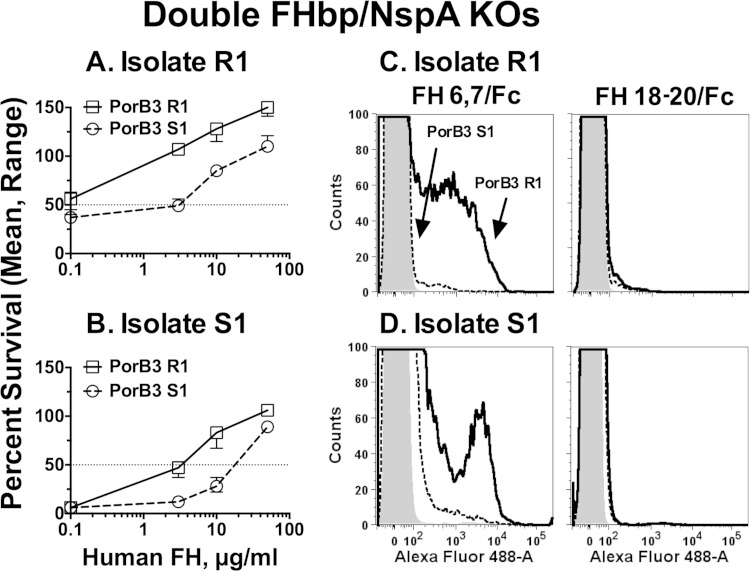
Double FHbp/NspA KO mutants from resistant isolates show greater human FH-dependent survival in infant rat serum than double-KO mutants from susceptible isolates.
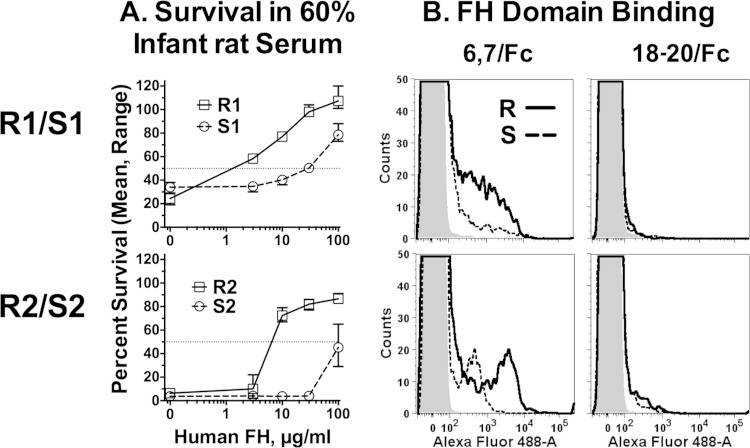
Resistance to anti-FHbp bactericidal activity required NspA. We hypothesized that binding of FH to an additional FH ligand present in the resistant isolates also contributed to anti-FHbp resistance. Since binding of FH to meningococcal FHbp and NspA is specific for human FH (17, 34), to identify an additional human FH ligand, we created double FHbp/NspA KO mutants for both isolate pairs and tested their ability to survive in infant rat serum when human FH was added. In the absence of added human FH, the resistant and susceptible double FHbp/NspA KO mutants were killed by 60% normal infant rat serum (<50% survival after 1 h of incubation, Fig. 6A). The addition of human FH enhanced survival of all four KO mutants in an FH dose-dependent manner. However, the doses of human FH required for increased survival of the resistant isolates were ca. 30- to 50-fold lower than those for the respective susceptible isolates. By flow cytometry, none of the FHbp/NspA double-KO mutants bound full-length human FH (data not shown). However, the functional data on human FH-dependent survival in infant rat serum suggested that the resistant isolates had an additional human FH ligand that was either absent or was less active in the susceptible isolates.
FIG 6.
Human FH enhances survival of FHbp/NspA double-KO mutants of N. meningitidis in infant rat serum. (A) Survival in 60% infant rat serum. Bacteria were incubated in pooled sera from wild-type infant rats, to which was added different concentrations of human FH. Open squares with solid black line, double-KO mutant of R1; open circles with dashed black line, double-KO mutant of S1. The data points represent median value (ranges) of triplicate measurements. The results were replicated in an independent experiment. (B) Binding of recombinant FH domain fragments fused to mouse Fc to FHbp/NspA double-KO mutants by flow cytometry. Solid black line, resistant (R) isolates; dashed black lines, susceptible (S) isolates; gray-filled histogram, bacteria without added recombinant fragment. Left, FH domain 6,7/Fc; right, FH domain 18-20/Fc. Representative data from one assay are shown. The results were replicated in a second experiment.
FH consists of 20 domains, called short consensus repeats. FH domain 6,7 binds to FHbp (35), to NspA (17), and to some PorB2 amino acid sequence variants (32). In previous studies, we found that recombinant fragments of FH domains 6 and 7 fused to mouse IgG2a or human IgG1 (FH6,7/Fc) bound with higher affinity than full-length human FH (32, 36). Therefore, we used a human FH6,7/mouse Fc fragment and flow cytometry to probe binding of the double FHbp/NspA KO mutants. For both pairs, there was greater binding of the fragment by the respective mutant derived from the anti-FHbp resistant member (Fig. 6B, left). As expected, binding was not detected with a negative-control human FH domain 18-20/Fc fragment (Fig. 6B, right), which, in the absence of deposited C3b (22), does not bind to N. meningitidis (32, 37).
PorB3 can mediate human FH-dependent survival of double FHbp/NspA KO meningococcal mutants in infant rat serum.
Each of the isolates in the R1/S1 and R2/S2 pairs had different PorB3 porin sequence variants (Table 1). Since PorB2 can functionally bind FH (32), we hypothesized that certain PorB3 sequence variants also might functionally bind human FH. To investigate this question, we created isogenic porB3 allelic exchange mutants from one of the FHbp/NspA double-KO mutant pairs, R1/S1. In the R1 double-KO mutant, porB3 (designated PorB3 R1) was replaced by either its own porB3 R1 or porB3 S1 derived from the S1 isolate. In the S1 double-KO mutant, porB3 S1 was replaced by either its own porB3 S1 or porB3 R1. Based on SDS-PAGE of detergent-extracted outer membrane vesicle preparations, the PorB3 expression by each of the allelic-exchange mutants was indistinguishable from the other (data not shown).
The R1 double FHbp/NspA KO mutant with PorB3 R1 had greater human FH-dependent survival in 60% infant rat serum than the corresponding R1 double FHbp/NspA KO mutant with PorB3 S1 (Fig. 7A). Similarly, the S1 double FHbp/NspA KO mutant with PorB3 R1 had greater human FH-dependent survival in 60% infant rat serum than the corresponding S1 double FHbp/NspA KO mutant with PorB3 S1 (Fig. 7B). Furthermore, the R1 and S1 double-KO mutants with PorB3 R1 showed greater binding of the FH domains 6,7/Fc than the corresponding mutants with PorB3 S1 (Fig. 7C and D). Collectively, the data indicated that certain PorB3 sequence variants can bind human FH and downregulate complement.
FIG 7.
Effect of allelic exchange of PorB3 on human FH-dependent susceptibility of FHbp and NspA double knockout mutants to killing by 60% infant rat serum. (A) Double-KO mutants of isolate R1 with its endogenous PorB3 replaced by PorB3 R1 or PorB3 S1. (B) Double-KO mutants of isolate S1 with its endogenous PorB replaced by PorB3 R1 or PorB3 S1. The data points represent median values from triplicate measurements. Error bars represent ranges. The results were replicated in an independent experiment. (C and D) Effect of PorB3 on binding of recombinant human FH domains by flow cytometry. Solid black lines, PorB3 R1; black dashed lines, PorB3 S1; gray-filled histogram, bacteria without added recombinant fragment. Panel C, double-KO isolate R1 with allelic replacement of PorB3 R1 or S1; panel D, double-KO isolate S1 with allelic replacement of PorB3 R1 or S1. The results were replicated in two independent experiments.
Certain PorB3 sequence variants can enhance resistance to human complement-mediated anti-FHbp bactericidal activity.
The data showing an effect of human FH and PorB3 on increasing survival of certain N. meningitidis isolates in infant rat serum were generated with double FHbp/NspA KO mutants. To investigate whether functional binding of FH to PorB3 contributes to resistance to anti-FHbp bactericidal activity, we created additional isogenic PorB3 allelic exchange mutants from wild-type R1 and S1 isolates in which their endogenous PorB3 variants were replaced by either PorB3 R1 or PorB3 S1. There was no effect of exchanging PorB3 S1 or R1 on susceptibility to bactericidal activity by a control anticapsular MAb (Fig. 8). In contrast, the R1 isolate with PorB3 S1 became susceptible to anti-FHbp bactericidal activity (geometric mean titer of <1:10 with PorB3 R1 to 1:95 with PorB3 S1, P = 0.002, Fig. 8A). Similarly, the S1 isolate with PorB3 R1 became 4.8-fold more resistant to anti-FHbp bactericidal activity than the S1 isolate with PorB3 S1 (geometric mean titer of 1:256 with PorB3 R1 compared to 1:1241 with PorB3 S1, P = 0.002, Fig. 8B). Since the S1 mutant with PorB3 R1 was not completely resistant to the anti-FHbp antiserum and the R1 mutant with PorB3 S1 was not as susceptible to anti-FHbp bactericidal activity as the wild-type S1 isolate, other factors intrinsic to the S1 or R1 isolates in addition to PorB3 contributed to their greater susceptibility or resistance to anti-FHbp bactericidal activity.
FIG 8.
Effect of allelic replacement of PorB3 variant on anti-FHbp bactericidal activity. The sera were from mice immunized with recombinant FHbp that matched the sequence variant of the isolates (FHbp ID 4). (A) Isolate R1 expressing wild-type FHbp and NspA with either allelic exchange of PorB3 R1 (open bars) or PorB3 S1 (hatched bars); (B) isolate S1 expressing wild-type FHbp and NspA with either allelic exchange of PorB3 R1 or PorB3 S1. The data were calculated from three assays. Error bars represent ranges. Asterisks indicate that a difference in bactericidal activity between respective PorB3 variants was significant (P = 0.002, Mann-Whitney).
DISCUSSION
Compared to other vaccine targets such as the polysaccharide capsule, FHbp is sparsely exposed on the bacterial surface (38). After the binding of anti-FHbp antibodies, the Fc density on the bacterial surface may be too low to permit sufficient C3b deposition by the classical complement pathway alone for the formation of a membrane attack complex without alternative pathway amplification (27, 39). Since FH downregulates the alternative pathway, the ability of anti-FHbp antibodies to block FH binding can be critical for eliciting anti-FHbp bacteriolysis (25, 27, 39, 40). We investigated whether resistance to anti-FHbp bactericidal activity might be explained by binding of FH to ligands other than FHbp (16).
N. meningitidis downregulates the alternative complement pathway by redundant mechanisms, including certain capsular polysaccharides (40), NspA (17), FHbp (41), PorB2 (32), and LOS sialylation (22). Our data indicate that the binding of FH to both NspA and certain PorB3 amino acid sequence variants can confer resistance of serogroup B isolates to anti-FHbp bactericidal activity. Thus, inactivation of nspA converted the resistant isolates to isolates susceptible to anti-FHbp bactericidal activity. Also, adding a nonbactericidal anti-NspA MAb to the bactericidal reaction mixture augmented anti-FHbp bactericidal activity. In previous studies, an anti-NspA MAb (14C7) inhibited the binding of FH to NspA (17). Thus, when NspA was absent or bound with an antibody that could inhibit binding of FH to NspA, functional binding of FH to PorB3 alone was not sufficient to confer anti-FHbp resistance.
When we inactivated both FHbp and NspA, the resistant double-KO mutants bound 10-fold more recombinant human FH 6,7/Fc than the respective sensitive mutants. Further, when we performed PorB3 allelic exchange between double FHbp/NspA KO mutants of one of the resistant and sensitive pairs (R1/S2), the R1 mutant with PorB3 R1 showed greater binding of FH domain 6,7 and had increased human FH-dependent survival in infant rat serum compared to the corresponding R1 double-KO mutant with PorB3 S1 (and vice versa after PorB3 allelic exchange in S1). Finally, with the wild-type S1/R1 pair, allelic exchange of PorB3 from a resistant or a sensitive isolate increased or decreased anti-FHbp bactericidal activity, respectively. Collectively, these results indicate that some PorB3 sequence variants can functionally bind human FH and, in the presence of NspA, confer anti-FHbp bactericidal resistance.
We compared the amino acid sequence alignments between PorB3 R1 and S1, and between PorB3 R2 and PorB3 S2 (see Fig. S4A and B, respectively, in the supplemental material). Most of the respective amino acid differences were located in exposed loops L1, L5, L6, L7, and L8, which are known to be variable. Further studies are needed to identify the specific amino acid residues responsible for PorB3 FH binding. We also visualized the locations of the respective amino acid differences onto the coordinates of a crystal structure of PorB (PDB ID 3W14) (42) (see Fig. S5 in the supplemental material). In the models, PorB is shown as a homotrimer, to reflect porin composition in the outer membrane formed by heteromers of PorA, PorB, and RmpM (43). Although we could not detect binding of full-length human FH to the double FHbp/NspA KO mutant of S1 or R1, we detected PorB3 R1-specific binding of recombinant FH domain 6,7/Fc. One possible explanation for detecting binding of the recombinant fragments, but not the full-length FH, is that the recombinant FH6,7/Fc contains two copies of FH domain 6,7 fused to Fc, which may permit cross-linking with PorB3 homotrimers or heterotrimers in the outer membrane, whereas the affinity of binding by the full-length FH with only one copy of domain 6,7 would be expected to be lower, yet functional. Note also that PorB homotrimers may exist in both PorA-positive and PorA-negative strains. Conceivably, only PorB homotrimers and not PorA/PorB heterotrimers can bind human FH. This question was beyond the scope of the present study and will require further investigation.
Our study focused on defining possible mechanisms for anti-FHbp resistance. An important limitation of our study was that we did not define the prevalence of resistant strains, which will need to be ascertained in future studies of serogroup B isolates from defined population-based studies. Identification of the four resistant isolates with moderately high FHbp expression in our study of 25 serogroup B clinical isolates, however, suggests a cautionary note about applying surrogate assays that only measure an isolate's FHbp expression and/or cross-reactivity with that of the vaccine antigen for predicting vaccine coverage without additional experimental data (7, 9–11, 44–46).
In conclusion, we found that some serogroup B isolates can recruit FH independent of FHbp, which allowed the isolates to resist anti-FHbp complement-mediated bacteriolysis. FH recruitment sufficient for resistance required both a specific PorB3 sequence variant and NspA. Neither FH ligand alone was sufficient. Since a nonbactericidal MAb to NspA overcame anti-FHbp resistance, the addition of NspA to FHbp vaccines potentially could broaden protective immunity compared to the use of either vaccine antigen alone.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Public Health Service grants R01 AI 046464 and AI 082263 (to D.M.G.) and AI 054544 and AI 111728 (to S.R.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH). The work at Children's Hospital Oakland Research Institute was performed in a facility funded by Research Facilities Improvement Program grant C06 RR 016226 from the National Center for Research Resources, NIH.
D.M.G., S.G., R.P., and S.R. declare no conflicts of interest.
We thank Monica Konar and Peter Beernink, Children's Hospital Oakland Research Institute (CHORI), for providing mouse anti-FHbp antisera; David Vu, CHORI, for help measuring passive protection in the infant rat model; and Jutamas Shaughnessy, University of Massachusetts School of Medicine, for providing recombinant FH domains 6,7/Fc and 18-20/Fc. Lisa Lewis, University of Massachusetts School of Medicine, provided critical comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02984-14.
REFERENCES
- 1.McNeil LK, Zagursky RJ, Lin SL, Murphy E, Zlotnick GW, Hoiseth SK, Jansen KU, Anderson AS. 2013. Role of factor H binding protein in Neisseria meningitidis virulence and its potential as a vaccine candidate to broadly protect against meningococcal disease. Microbiol Mol Biol Rev 77:234–252. doi: 10.1128/MMBR.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R. 2012. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional, and structural characterization of the antigens. Vaccine 30(Suppl 2):B87–B97. doi: 10.1016/j.vaccine.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fletcher LD, Bernfield L, Barniak V, Farley JE, Howell A, Knauf M, Ooi P, Smith RP, Weise P, Wetherell M, Xie X, Zagursky R, Zhang Y, Zlotnick GW. 2004. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun 72:2088–2100. doi: 10.1128/IAI.72.4.2088-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy E, Andrew L, Lee KL, Dilts DA, Nunez L, Fink PS, Ambrose K, Borrow R, Findlow J, Taha MK, Deghmane AE, Kriz P, Musilek M, Kalmusova J, Caugant DA, Alvestad T, Mayer LW, Sacchi CT, Wang X, Martin D, von Gottberg A, du Plessis M, Klugman KP, Anderson AS, Jansen KU, Zlotnick GW, Hoiseth SK. 2009. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J Infect Dis 200:379–389. doi: 10.1086/600141. [DOI] [PubMed] [Google Scholar]
- 5.Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, Brunelli B, Pieri A, Santini L, Savino S, Serruto D, Litt D, Kroll S, Welsch JA, Granoff DM, Rappuoli R, Pizza M. 2003. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med 197:789–799. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnelly J, Medini D, Boccadifuoco G, Biolchi A, Ward J, Frasch C, Moxon ER, Stella M, Comanducci M, Bambini S, Muzzi A, Andrews W, Chen J, Santos G, Santini L, Boucher P, Serruto D, Pizza M, Rappuoli R, Giuliani MM. 2010. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc Natl Acad Sci U S A 107:19490–19495. doi: 10.1073/pnas.1013758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang HQ, Hoiseth SK, Harris SL, McNeil LK, Zhu D, Tan C, Scott AA, Alexander K, Mason K, Miller L, DaSilva I, Mack M, Zhao XJ, Pride MW, Andrew L, Murphy E, Hagen M, French R, Arora A, Jones TR, Jansen KU, Zlotnick GW, Anderson AS. 2010. Broad vaccine coverage predicted for a bivalent recombinant factor H binding protein based vaccine to prevent serogroup B meningococcal disease. Vaccine 28:6086–6093. doi: 10.1016/j.vaccine.2010.06.083. [DOI] [PubMed] [Google Scholar]
- 8.McNeil LK, Murphy E, Zhao XJ, Guttmann S, Harris S, Scott A, Tan C, Mack M, Dasilva I, Alexander K, Jiang HQ, Zhu D, Mininni T, Zlotnick GW, Hoiseth SK, Jones TR, Pride M, Jansen KU, Anderson A. 2009. Detection of LP2086 on the cell surface of Neisseria meningitidis and its accessibility in the presence of serogroup B capsular polysaccharide. Vaccine 27:3417–3421. doi: 10.1016/j.vaccine.2009.01.075. [DOI] [PubMed] [Google Scholar]
- 9.Frosi G, Biolchi A, Lo Sapio M, Rigat F, Gilchrist S, Lucidarme J, Findlow J, Borrow R, Pizza M, Giuliani MM, Medini D. 2013. Bactericidal antibody against a representative epidemiological meningococcal serogroup B panel confirms that MATS underestimates 4CMenB vaccine strain coverage. Vaccine 31:4968–4974. doi: 10.1016/j.vaccine.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Vogel U, Stefanelli P, Vazquez J, Taha MK, Claus H, Donnelly J. 2012. The use of vaccine antigen characterization, for example, by MATS, to guide the introduction of meningococcus B vaccines. Vaccine 30(Suppl 2):B73–B77. doi: 10.1016/j.vaccine.2011.12.061. [DOI] [PubMed] [Google Scholar]
- 11.Vogel U, Taha MK, Vazquez JA, Findlow J, Claus H, Stefanelli P, Caugant DA, Kriz P, Abad R, Bambini S, Carannante A, Deghmane AE, Fazio C, Frosch M, Frosi G, Gilchrist S, Giuliani MM, Hong E, Ledroit M, Lovaglio PG, Lucidarme J, Musilek M, Muzzi A, Oksnes J, Rigat F, Orlandi L, Stella M, Thompson D, Pizza M, Rappuoli R, Serruto D, Comanducci M, Boccadifuoco G, Donnelly JJ, Medini D, Borrow R. 2013. Predicted strain coverage of a meningococcal multicomponent vaccine (4CMenB) in Europe: a qualitative and quantitative assessment. Lancet Infect Dis 13:416–425. doi: 10.1016/S1473-3099(13)70006-9. [DOI] [PubMed] [Google Scholar]
- 12.Beernink PT, Welsch JA, Harrison LH, Leipus A, Kaplan SL, Granoff DM. 2007. Prevalence of factor H-binding protein variants and NadA among meningococcal group B isolates from the United States: implications for the development of a multicomponent group B vaccine. J Infect Dis 195:1472–1479. doi: 10.1086/514821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holst J, Comanducci M, Bambini S, Muzzi A, Comandi S, Oksnes J, DeTora L, Pizza M, Rappuoli R, Caugant DA. 2014. Variability of genes encoding surface proteins used as vaccine antigens in meningococcal endemic and epidemic strain panels from Norway. Vaccine 32:2722–2731. doi: 10.1016/j.vaccine.2014.02.068. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan SL, Schutze GE, Leake JA, Barson WJ, Halasa NB, Byington CL, Woods CR, Tan TQ, Hoffman JA, Wald ER, Edwards KM, Mason EO Jr. 2006. Multicenter surveillance of invasive meningococcal infections in children. Pediatrics 118:e979–984. doi: 10.1542/peds.2006-0281. [DOI] [PubMed] [Google Scholar]
- 15.Mandrell RE, Kim JJ, John CM, Gibson BW, Sugai JV, Apicella MA, Griffiss JM, Yamasaki R. 1991. Endogenous sialylation of the lipooligosaccharides of Neisseria meningitidis. J Bacteriol 173:2823–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giuntini S, Vu DM, Granoff DM. 2013. fH-dependent complement evasion by disease-causing meningococcal strains with absent fHbp genes or frameshift mutations. Vaccine 31:4192–4199. doi: 10.1016/j.vaccine.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis LA, Ngampasutadol J, Wallace R, Reid JE, Vogel U, Ram S. 2010. The meningococcal vaccine candidate neisserial surface protein A (NspA) binds to factor H and enhances meningococcal resistance to complement. PLoS Pathog 6:e1001027. doi: 10.1371/journal.ppat.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moe GR, Zuno-Mitchell P, Hammond SN, Granoff DM. 2002. Sequential immunization with vesicles prepared from heterologous Neisseria meningitidis strains elicits broadly protective serum antibodies to group B strains. Infect Immun 70:6021–6031. doi: 10.1128/IAI.70.11.6021-6031.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abad R, Alcala B, Salcedo C, Enriquez R, Uria MJ, Diez P, Vazquez JA. 2006. Sequencing of the porB gene: a step toward a true characterization of Neisseria meningitidis. Clin Vaccine Immunol 13:1087–1091. doi: 10.1128/CVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacchi CT, Lemos AP, Whitney AM, Solari CA, Brandt ME, Melles CE, Frasch CE, Mayer LW. 1998. Correlation between serological and sequencing analyses of the PorB outer membrane protein in the Neisseria meningitidis serotyping system. Clin Diagn Lab Immunol 5:348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koeberling O, Welsch JA, Granoff DM. 2007. Improved immunogenicity of a H44/76 group B outer membrane vesicle vaccine with overexpressed genome-derived neisserial antigen 1870. Vaccine 25:1912–1920. doi: 10.1016/j.vaccine.2006.03.092. [DOI] [PubMed] [Google Scholar]
- 22.Lewis LA, Carter M, Ram S. 2012. The relative roles of factor H binding protein, neisserial surface protein A, and lipooligosaccharide sialylation in regulation of the alternative pathway of complement on meningococci. J Immunol 188:5063–5072. doi: 10.4049/jimmunol.1103748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apicella MA, Bennett KM, Hermerath CA, Roberts DE. 1981. Monoclonal antibody analysis of lipopolysaccharide from Neisseria gonorrhoeae and Neisseria meningitidis. Infect Immun 34:751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apicella MA. 2008. Isolation and characterization of lipopolysaccharides, p 1–11. In DeLeo FR, Otto M (ed), Bacterial pathogenesis: methods and protocols, vol 431 Humana Press, Totowa, NJ. [Google Scholar]
- 25.Konar M, Granoff DM, Beernink PT. 2013. Importance of inhibition of binding of complement factor H for serum bactericidal antibody responses to meningococcal factor H-binding protein vaccines. J Infect Dis 208:627–636. doi: 10.1093/infdis/jit239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beernink PT, Shaughnessy J, Braga EM, Liu Q, Rice PA, Ram S, Granoff DM. 2011. A meningococcal factor H binding protein mutant that eliminates factor H binding enhances protective antibody responses to vaccination. J Immunol 186:3606–3614. doi: 10.4049/jimmunol.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giuntini S, Reason DC, Granoff DM. 2011. Complement-mediated bactericidal activity of anti-factor H binding protein monoclonal antibodies against the meningococcus relies upon blocking factor H binding. Infect Immun 79:3751–3759. doi: 10.1128/IAI.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vu DM, Shaughnessy J, Lewis LA, Ram S, Rice PA, Granoff DM. 2012. Enhanced bacteremia in human factor H transgenic rats infected by Neisseria meningitidis. Infect Immun 80:643–650. doi: 10.1128/IAI.05604-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granoff DM, Bartoloni A, Ricci S, Gallo E, Rosa D, Ravenscroft N, Guarnieri V, Seid RC, Shan A, Usinger WR, Tan S, McHugh YE, Moe GR. 1998. Bactericidal monoclonal antibodies that define unique meningococcal B polysaccharide epitopes that do not cross-react with human polysialic acid. J Immunol 160:5028–5036. [PubMed] [Google Scholar]
- 30.Moe GR, Zuno-Mitchell P, Lee SS, Lucas AH, Granoff DM. 2001. Functional activity of anti-Neisserial surface protein A monoclonal antibodies against strains of Neisseria meningitidis serogroup B. Infect Immun 69:3762–3771. doi: 10.1128/IAI.69.6.3762-3771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vu DM, Pajon R, Reason DC, Granoff DM. 2012. A broadly cross-reactive monoclonal antibody against an epitope on the N terminus of meningococcal FHbp. Sci Rep 2:341. doi: 10.1038/srep00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis LA, Vu DM, Vasudhev S, Shaughnessy J, Granoff DM, Ram S. 2013. Factor H-dependent alternative pathway inhibition mediated by porin B contributes to virulence of Neisseria meningitidis. mBio 4:e00339-13. doi: 10.1128/mBio.00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandeputte-Rutten L, Bos MP, Tommassen J, Gros P. 2003. Crystal structure of neisserial surface protein A (NspA), a conserved outer membrane protein with vaccine potential. J Biol Chem 278:24825–24830. doi: 10.1074/jbc.M302803200. [DOI] [PubMed] [Google Scholar]
- 34.Granoff DM, Welsch JA, Ram S. 2009. Binding of complement factor H (fH) to Neisseria meningitidis is specific for human fH and inhibits complement activation by rat and rabbit sera. Infect Immun 77:764–769. doi: 10.1128/IAI.01191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider MC, Prosser BE, Caesar JJ, Kugelberg E, Li S, Zhang Q, Quoraishi S, Lovett JE, Deane JE, Sim RB, Roversi P, Johnson S, Tang CM, Lea SM. 2009. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature 458:890–893. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaughnessy J, Vu DM, Punjabi R, Serra-Pladevall J, DeOliveira RB, Granoff DM, Ram S. 2014. Fusion protein comprising factor H domains 6 and 7 and human IgG1 Fc as an antibacterial immunotherapeutic. Clin Vaccine Immunol 21:1452–1459. doi: 10.1128/CVI.00444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis LA, Vu DM, Granoff DM, Ram S. 2014. Inhibition of the alternative pathway of non-human infant complement by porin B2 contributes to virulence of Neisseria meningitidis in the infant rat model. Infect Immun 82:2574–2584. doi: 10.1128/IAI.01517-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welsch JA, Ram S, Koeberling O, Granoff DM. 2008. Complement-dependent synergistic bactericidal activity of antibodies against factor H-binding protein, a sparsely distributed meningococcal vaccine antigen. J Infect Dis 197:1053–1061. doi: 10.1086/528994. [DOI] [PubMed] [Google Scholar]
- 39.Giuntini S, Reason DC, Granoff DM. 2012. Combined roles of human IgG subclass, alternative complement pathway activation, and epitope density in the bactericidal activity of antibodies to meningococcal factor H binding protein. Infect Immun 80:187–194. doi: 10.1128/IAI.05956-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal S, Vasudhev S, DeOliveira RB, Ram S. 2014. Inhibition of the classical pathway of complement by meningococcal capsular polysaccharides. J Immunol 193:1855–1863. doi: 10.4049/jimmunol.1303177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madico G, Welsch JA, Lewis LA, McNaughton A, Perlman DH, Costello CE, Ngampasutadol J, Vogel U, Granoff DM, Ram S. 2006. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol 177:501–510. doi: 10.4049/jimmunol.177.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kattner C, Toussi DN, Zaucha J, Wetzler LM, Ruppel N, Zachariae U, Massari P, Tanabe M. 2014. Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition. J Struct Biol 185:440–447. doi: 10.1016/j.jsb.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez S, Arenas J, Abel A, Criado MT, Ferreiros CM. 2005. Analysis of outer membrane protein complexes and heat-modifiable proteins in Neisseria strains using two-dimensional diagonal electrophoresis. J Proteome Res 4:91–95. doi: 10.1021/pr049846i. [DOI] [PubMed] [Google Scholar]
- 44.Bettinger JA, Scheifele DW, Halperin SA, Vaudry W, Findlow J, Borrow R, Medini D, Tsang R, members of the Canadian Immunization Monitoring Program . 2013. Diversity of Canadian meningococcal serogroup B isolates and estimated coverage by an investigational meningococcal serogroup B vaccine (4CMenB). Vaccine 32:124–130. doi: 10.1016/j.vaccine.2013.03.063. [DOI] [PubMed] [Google Scholar]
- 45.Hong E, Giuliani MM, Deghmane AE, Comanducci M, Brunelli B, Dull P, Pizza M, Taha MK. 2013. Could the multicomponent meningococcal serogroup B vaccine (4CMenB) control Neisseria meningitidis capsular group X outbreaks in Africa? Vaccine 31:1113–1116. doi: 10.1016/j.vaccine.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 46.Tzanakaki G, Hong E, Kesanopoulos K, Xirogianni A, Bambini S, Orlandi L, Comanducci M, Muzzi A, Taha MK. 2014. Diversity of Greek meningococcal serogroup B isolates and estimated coverage of the 4CMenB meningococcal vaccine. BMC Microbiol 14:111. doi: 10.1186/1471-2180-14-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.