Abstract
The immunoglobulin binding protein A (SpA) of Staphylococcus aureus is synthesized as a precursor with a C-terminal sorting signal. The sortase A enzyme mediates covalent attachment to peptidoglycan so that SpA is displayed on the surface of the bacterium. Protein A is also found in the extracellular medium, but the processes involved in its release are not fully understood. Here, we show that a portion of SpA is released into the supernatant with an intact sorting signal, indicating that it has not been processed by sortase A. Release of SpA was reduced when the native sorting signal of SpA was replaced with the corresponding region of another sortase-anchored protein (SdrE). Similarly, a reporter protein fused to the sorting signal of SpA was released to a greater extent than the same polypeptide fused to the SdrE sorting signal. Released SpA protected bacteria from killing in human blood, indicating that it contributes to immune evasion.
INTRODUCTION
Staphylococcus aureus is an important opportunistic pathogen causing serious invasive infections in the community and health care setting (1). Almost all clinical isolates of S. aureus express the major virulence factor, staphylococcal protein A (SpA) (2). Protein A is located both on the surface of the bacterium and in the extracellular medium (3–6) and comprises four or five repeated immunoglobulin-binding domains (IgBDs) (7, 8). The IgBDs of SpA (Fig. 1) adopt a triple helical structure and can bind to the Fc region of IgG via helices I and II (9) and to the Fab region of human IgM of the subclass VH3 via helices II and III (10). The binding of SpA to Fc and Fab domains contributes to S. aureus virulence in a mouse model of systemic infection (11). The interaction of SpA with IgM Fab triggers the proliferation and depletion of B cells (12), suppressing the development of adaptive immune responses. Thus, infection with SpA-expressing bacteria does not provide protection against subsequent S. aureus infection (11). Protein A also inhibits phagocytic killing of S. aureus in human and mouse blood (11, 13). This process is likely to be dependent on the interaction of SpA with IgG Fc since S. aureus expressing a variant of SpA lacking the ability to recognize IgG Fc survives poorly in mouse blood, akin to an SpA-deficient mutant (11). The IgBDs of SpA also promote inflammation through their interaction with tumor necrosis factor receptor 1 (14). The hypervariable Xr region of SpA (Fig. 1) comprises variable numbers of octapeptide repeats that contribute to inflammation by activating interferon-β signaling in airway epithelial and immune cells (15).
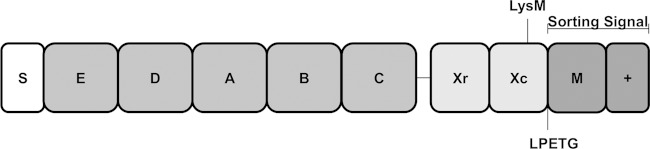
FIG 1.
Schematic representation of the domain organization of protein A. Protein A consists of an N-terminal signal sequence (S) followed by up to five IgG-binding domains (E to C), an antigenic variable region (Xr), and a cell wall-spanning region (Xc). The Xc region harbors an LysM domain which can mediate noncovalent binding of proteins to peptidoglycan. The sorting signal comprises an LPETG motif, a hydrophobic membrane-spanning region (M), and a positively charged tail region (+).
Protein A is synthesized as a precursor with an N-terminal signal sequence and C-terminal sorting signal (Fig. 1). The signal sequence is cleaved by signal peptidase during translocation of the precursor across the cytoplasmic membrane by the general secretory (Sec) pathway (16). The sorting signal comprises an LPETG motif, a hydrophobic membrane-spanning domain, and, at the extreme C terminus, a stretch of positively charged residues (Fig. 1). The last two elements delay secretion across the membrane and facilitate recognition and cleavage by sortase A (17). Sortase A cleaves between threonine and glycine of the LPETG motif, forming an acyl-enzyme intermediate capturing the C-terminal carboxyl group of the protein with its active-site cysteine thiol (18). Acyl intermediates are relieved by the nucleophilic attack of the amino group of the pentaglycine cross-bridge of lipid II (19). Following transglycosylation and transpeptidation, SpA becomes covalently anchored to peptidoglycan and is displayed on the surface of the bacterium (20).
A substantial amount of SpA is found in the extracellular medium (3–6). Released SpA can be detected in the skin lesions of mice infected with a USA300 strain of community-associated methicillin-resistant S. aureus (MRSA) and in fluids recovered from patients with S. aureus infection (21). However, the processes involved in SpA release are not completely understood. Becker et al. (4) described a mechanism whereby SpA is shed from the cell envelope of strain Newman into the culture medium following cleavage of the pentaglycine cross-bridge of peptidoglycan by the S. aureus glycyl-glycine endopeptidase LytM. The murein hydrolase LytN cleaves the amine bonds between N-acetylmuramic acid and the tetrapeptide side chains of peptidoglycan so that peptidoglycan fragments linked to SpA lack amino sugars (4). This is hypothesized to allow released SpA to avoid activating nucleotide-binding and oligomerization domain-containing protein 2 (NOD2) in vivo (4). The release of SpA is not completely inhibited in an lytM-deficient mutant of strain Newman, indicating that LytM activity is not the only factor involved in the release of SpA (4). Here, we identify a previously undescribed mode of SpA release by the USA300 strain LAC. We show that when this is interrupted, there is a significant reduction in the level of SpA release. We also investigate the biological significance of SpA release by studying the ability of extracellular SpA to promote S. aureus survival in human blood.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. aureus was grown on tryptic soy agar (TSA; Oxoid) or in brain heart infusion (BHI) broth (Difco) at 37°C. Escherichia coli was grown on Luria agar or broth (Difco). Cultures were supplemented with ampicillin (100 μg/ml; Melford Laboratories) or chloramphenicol (10 μg/ml), as required. Bacteria were diluted 1:200, washed in BHI broth, and allowed to grow to the optical density at 600 nm (OD600) required. Strains harboring the pRMC2 expression vector were grown to an OD600 of 0.3 and induced with anhydrotetracycline (ATc) until an OD600 of 1.2 was reached. Broth, where indicated, was supplemented with V8 protease (1 U/ml) and 3,4-dichloroisocoumarin (DCI; 200 μM). Unless otherwise stated, all reagents were obtained from Sigma.
Plasmid and strain construction.
All strains and plasmids are listed in Table 1. Strain LAC* spa was constructed by transduction of spa::Kanr by phage 85 into strain LAC*. LAC* spa sbi was constructed by transduction of sbi::Emr into strain LAC* spa::Kanr. Primer sequences are listed in Table S1 in the supplemental material. Cloning was carried out using the sequence- and ligation-independent cloning (SLIC) procedure as described by Li and Elledge (22). Primers for amplifying insert sequences contained 5′ extensions with homology to the target vector. The complete spa gene from strain Newman (100% amino acid sequence identity to the spa gene from LAC*) was amplified from genomic DNA by PCR using primers SpAF and SpAR (see Table S1). Plasmid pRMC2 (23) was used as the template for inverse PCR with primers pSLF and pSLR. Both amplimers were joined using SLIC to generate the plasmid pSpA, where spa is cloned between the SacI and EcoRI sites of pRMC2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus LAC* | Erythromycin-sensitive derivative of MRSA strain LAC; clonal complex 8 | 29 |
| LAC* spa | Derivative of LAC* deficient in protein A; spa::Kanr | This study |
| LAC* sbi | Derivative of LAC* deficient in Sbi; sbi::Emr | 13 |
| LAC* spa sbi | Derivative of LAC* deficient in protein A and Sbi; spa::Kanr sbi::Emr | This study |
| LAC* PD | Protease-deficient derivative of LAC*; Δaur ΔsspAB Δscp spl::Emr | 46 |
| LAC* sbi lytM | Derivative of LAC* sbi deficient in LytM; ΔlytM | This study |
| LAC* spa sbi lytM | Derivative of LAC* spa sbi deficient in LytM; ΔlytM | This study |
| LAC* sbi [SpAΩSdrESS] | Derivative of LAC* sbi where the SpA sorting signal has been replaced with the sorting signal of SdrE by allelic exchange | This study |
| LAC* sbi lytM [SpAΩSdrESS] | Derivative of LAC* sbi lytM where the SpA sorting signal has been replaced with the sorting signal of SdrE by allelic exchange | This study |
| Newman sbi | Derivative of S. aureus strain Newman, NCTC 8178, clonal complex 8, deficient in Sbi; sbi::Emr | 13 |
| Newman spa sbi | Derivative of Newman deficient in protein A and Sbi; spa::Kanr sbi::Emr | 13 |
| Newman sbi lytM | Derivative of Newman sbi deficient in LytM; ΔlytM | This study |
| Newman sbi [SpAΩSdrESS] | Derivative of Newman sbi where the SpA sorting signal has been replaced with sorting signal of SdrE by allelic exchange | This study |
| E. coli DC10B | dam+ Δdcm ΔhsdRMS endA1 recA1 | 24 |
| Plasmids | ||
| pRMC2 | Anhydrotetracycline-inducible expression vector; Ampr Cmr | 23 |
| pSpA | Plasmid pRMC2 containing the full-length spa gene | This study |
| pSpAΔSS | Plasmid pSpA lacking DNA encoding the SpA sorting signal (residues 474 to 508) | This study |
| pSpAΩSdrESS | Plasmid pSpA where DNA encoding the SpA sorting signal (residues 474 to 508) has been replaced with DNA encoding the SdrE sorting signal (residues 1117 to 1154) | This study |
| pRMC2-sbi ΔD1D2 | Plasmid pRMC2 containing DNA encoding the Sbi signal sequence (residues 1 to 40) and the D3D4 domains (residues 153 to 253) | 13 |
| pD3D4-SpASS | Plasmid pRMC2 containing an in-frame fusion between DNA encoding Sbi D3D4 domains (residues 153 to 253) and DNA encoding the SpA sorting signal (residues 474 to 508) | This study |
| pD3D4-SdrESS | Plasmid pRMC2 containing an in-frame fusion between DNA encoding Sbi D3D4 domains (residues 153 to 253) and DNA encoding the SdrE sorting signal (residues 1117 to 1154) | This study |
| pSpAΩSdrESS-DS | Plasmid pSpAΩSdrESS containing 538 bp of sequence downstream of the spa gene | This study |
| pIMAY | Temperature-sensitive vector for allelic exchange; Cmr | 24 |
| pIMAY::ΔlytM | Plasmid for creating a lytM deletion mutant; carries 509 bp of DNA from upstream and 512 bp of DNA from downstream of the lytM gene amplified from LAC | This study |
| pIMAY::SpAΩSdrESS | Plasmid for replacing DNA encoding the sorting signal of SpA with DNA encoding the sorting signal of SdrE on the chromosome of LAC* | This study |
DNA encoding the sorting signal of SdrE was amplified by PCR using primers RESF and RESR using genomic DNA from strain Newman as the template. Plasmid pSpA was used as the template for inverse PCR with primers dSSF and dSSR. The amplimers were joined using SLIC to generate pSpAΩSdrESS.
Similarly, to generate plasmids pD3D4-SpASS and pD3D4-SdrESS, PCR was performed using primers dSSF and DWrR and plasmid pRMC2-sbi ΔD1D2 (13) as the template. DNA encoding the SpA sorting signal was amplified using primers DSpF and SpAR and plasmid pSpA as the template. DNA encoding the SdrE sorting signal was amplified using primers DSdF and RESR and plasmid pSpAΩSdrESS as the template. PCR products were joined by SLIC to generate pD3D4-SpASS and pD3D4-SdrESS.
Plasmid pSpAΩSdrESS-DS was generated by amplifying 538 bp of DNA located downstream of the spa gene using primers SpADSF and SpADSR and genomic DNA from S. aureus LAC* as the template and using primers pSLF and pSRER to amplify plasmid pSpAΩSdrESS. The PCR products were joined by SLIC so that the 538-bp fragment was incorporated into plasmid pSpAΩSdrESS directly downstream of the stop codon.
All plasmids were transformed into E. coli strain DC10B (24). Plasmids were isolated from DC10B, verified by DNA sequencing (Source Bioscience) using primers SEQF and SEQR, and transformed (5 μg) into S. aureus that was made electrocompetent as previously described (25).
Deletion of the lytM gene was achieved by allelic exchange using pIMAY (24). Primers lytM-A and lytM-B were designed to amplify 509 bp of DNA located upstream, and primers lytM-C and lytM-D amplified 512 bp of DNA located downstream of the lytM gene (see Table S1 in the supplemental material). The upstream and downstream PCR products were denatured and allowed to reanneal via the complementary sequences in primers lytM-B and lytM-C, and this was used as the template in a second PCR using primers lytM-A and lytM-D. The amplimer was cloned into pIMAY (24) between the KpnI and SacI restriction sites, and the resulting plasmid (pIMAY::ΔlytM) was transformed into E. coli DC10B (24) and verified by DNA sequencing. The plasmid was transformed into S. aureus strains LAC* spa sbi, LAC* sbi, and Newman sbi that were made electrocompetent, and deletion of the lytM gene was achieved by allelic exchange as previously described (24). The resulting LytM-deficient mutants were confirmed by DNA sequencing of a PCR amplimer. The mutants were phenotypically identical to the parent strains in terms of growth rate and hemolysis on sheep blood agar (data not shown).
Strains LAC* sbi [SpAΩSdrESS], LAC* sbi lytM [SpAΩSdrESS], and Newman sbi [SpAΩSdrESS] were constructed by allelic exchange using pIMAY. Plasmid pIMAY (24) was used as the template for PCR with primers pIMAYF and pIMAYR. Primers pIREF and pIRER were used to amplify DNA encoding the SdrE sorting signal and 521 bp of DNA upstream and 538 bp of DNA downstream using plasmid pSpAΩSdrESS-DS as the template. The PCR amplimers were joined by SLIC and transformed into E. coli DC10B (24). The resulting plasmid (pIMAY::SpAΩSdrESS) was transformed into S. aureus strains LAC* sbi, LAC* sbi lytM, and Newman sbi that were made electrocompetent, and replacement of DNA encoding the SpA sorting signal with DNA encoding the sorting signal of SdrE on the chromosome was achieved by allelic exchange as previously described (24). The resulting mutants were phenotypically identical to their respective parent strains in terms of growth rate and hemolysis on sheep blood agar (data not shown). The mutation was confirmed by DNA sequencing of a PCR amplimer.
Western immunoblotting.
To extract cell wall-associated proteins, cultures of S. aureus were harvested, washed in phosphate-buffered saline (PBS), and resuspended to an OD600 of 5 or 10 in lysis buffer (50 mM Tris-HCl, 20 mM MgCl2, pH 7.5) supplemented with raffinose (30%, wt/vol) and complete protease inhibitors (40 μl/ml; Roche). Cell wall proteins were solubilized by incubation with lysostaphin (100 μg/ml; Ambi, NY) for 8 min at 37°C. Protoplasts were removed by centrifugation at 16,000 × g for 5 min, and the supernatant containing solubilized cell wall proteins was aspirated and boiled for 10 min in final sample buffer. For supernatant fractions, bacteria were removed from cultures by centrifugation at 4,000 × g for 5 min, and the supernatant was passed through a 0.2-μm-pore-size filter. Following this, supernatant samples, where indicated in the figure legends, were concentrated using a 30,000-molecular-weight-cutoff spin column (Millipore).
Proteins were separated on 7.5%, 10%, or 12.5% (wt/vol) polyacrylamide gels, transferred onto polyvinylidene difluoride (PVDF) membranes (Roche), and blocked in 10% (wt/vol) skimmed milk proteins. Blots were probed with horseradish peroxidase (HRP)-conjugated rabbit anti-mouse IgG (1:2,000 or 1:500; Dako) to detect SpA. Alternatively, blots were probed with polyclonal rabbit anti-SdrE IgG (1:2,000) (26), rabbit anti-V8 serum (1:250) (a gift from Martin McGavin), or rabbit anti-D3D4 IgG (1:500) (13), and bound antibodies were detected using goat anti-rabbit IgG–HRP (1:2,000) or HRP-conjugated protein A (1:500). Biotin-labeled fibronectin (Fn) was used in ligand affinity blots. Human fibronectin (0.5 mg/ml; Calbiochem) was incubated with biotin (2 mg/ml) for 20 min at room temperature. The reaction was stopped by addition of NH4Cl (10 mM). Excess biotin was removed by dialysis against PBS overnight at 4°C. Blots were probed with biotin-labeled human fibronectin (15 μg/ml; Calbiochem) and HRP-conjugated streptavidin (0.5 μg/ml; Genscript). Reactive bands were visualized using LumiGLO reagent and a peroxide detection system (Cell Signaling Technology). Band quantification was performed using ImageQuant TL software (GE Healthcare).
Flow cytometry.
Cultures of S. aureus were washed once with PBS and once in a solution of bovine serum albumin ([BSA] 0.1%, wt/vol) and then adjusted to an OD600 of 0.3 in PBS. Bacteria were incubated with an equal volume of fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG (1:800 or 1:3,200; Dako) for 30 min. Unbound antibody was removed by one wash in PBS. Bacteria were resuspended in formaldehyde (2%, vol/vol), and bound IgG was detected using flow cytometry. Bacteria were gated on the basis of forward and side scatter. The fluorescence intensity of 20,000 bacteria was analyzed, and the mean fluorescence was calculated.
Purification of SpA from S. aureus culture supernatants by affinity chromatography.
S. aureus strain LAC* sbi was grown in BHI broth for 16 h to stationary phase. Bacteria were removed by centrifugation at 4,000 × g for 10 min, and the supernatant was passed through a 0.2-μm-pore-size filter. A total of 150 ml of filtered supernatant was allowed to pass through a gravity feed column packed with a 4-ml bed volume of IgG-Sepharose (GE Healthcare). Briefly, the column was equilibrated with 16 ml of elution buffer (0.5 M acetic acid [HAc]), followed by at least 20 ml of Tris-saline-Tween 20 (TST; 50 mM Tris buffer, pH 7.6, 150 mM NaCl, and 0.05% Tween 20) until the column eluate was at neutral pH. The column was washed with 40 ml of TST and 10 ml of ammonium acetate ([NH4Ac] 5 mM; pH 5.5), and SpA was eluted with 12 ml of elution buffer. The purified SpA was dialyzed against PBS at 4°C overnight. Protein purity was assessed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western immunoblotting, and the protein concentration was determined with a bicinchoninic acid (BCA) protein assay kit (Pierce). For N-terminal sequencing, samples were transferred to PVDF, and sequencing was carried out by Abingdon Health Laboratory Services, United Kingdom.
Trypsin digestion.
Purified SpA (2 μg) was resuspended in a solution containing urea (6 M) and dithiothreitol (4 mM). The solution was heated at 60°C for 60 min. The sample was allowed to cool before iodoacetamide (15 mM) was added and incubated for 30 min at 37°C. A solution (120 μl) containing NH4HCO3 (50 mM; pH 7.8) and CaCl2 (1 mM) was added to dilute the urea concentration to below 1 M. Sequencing-grade modified trypsin was added (1 μg; Promega), and the samples were incubated at 37°C overnight. Prior to mass spectrometry (MS) analysis, the samples were cleaned using ZipTips (Millipore).
Liquid chromatography and tandem mass spectrometry (LC-MS/MS).
The samples were run on a Thermo Scientific Q Exactive mass spectrometer connected to a Dionex Ultimate 3000 (RSLCnano) chromatography system. Peptides were resuspended in formic acid (0.1%). Each sample was loaded onto a Biobasic Picotip emitter (120-mm length; 75-μm inside diameter [i.d.]) packed with ReproSil-Pur C18 (1.9 μm) reverse-phase medium and was separated by an increasing acetonitrile gradient over 37 min at a flow rate of 250 nl/min. The mass spectrometer was operated in positive ion mode with a capillary temperature of 220°C and with a potential of 2,000 V applied to the frit. All data were acquired with the mass spectrometer operating in automatic data-dependent switching mode. A high-resolution (70,000) MS scan (300 to 2,000 m/z) was performed using the Q Exactive to select the 12 most intense ions prior to MS/MS analysis using higher-energy collisional dissociation with stepped normalized collision energy.
Database search.
The raw data were de novo sequenced and searched against the Homo sapiens subset of the UniProt Swiss-Prot database (to which the full-length amino acid sequence of SpA from strain LAC* was added) using the search engine Peaks Studio 7 (Bioinformatics Solutions) for peptides cleaved with trypsin. Each peptide used for protein identification met specific Peaks parameters; i.e., only peptide scores that corresponded to a false discovery rate (FDR) of ≤1% were accepted from the Peaks posttranslational modification (PTM) database search. The Peaks de novo results were filtered using an average local confidence (ALC) of ≥50% and a peptide score of [−10 log10(P value)] of ≥15.
ELISA.
Microtiter plates (Nunc MaxiSorp) were coated with chicken anti-protein A polyclonal IgY (1 μg/ml; GenScript) diluted in coating buffer (100 mM NaHCO3, 34 mM Na2CO3, pH 9.6) at 4°C overnight. Wells were washed five times with PBS and blocked with 100 μl of BSA (5%, wt/vol; Fisher Scientific) at 37°C for 2 h. Supernatant samples were diluted (1:8 or 1:50), and 100 μl of each was added into the appropriate well and incubated with shaking at room temperature for 1 h and at 37°C with no shaking for 1 h. Wells were washed five times with PBS; 100 μl of mouse monoclonal biotin-conjugated anti-protein A-IgG (1 μg/ml; GenScript) was added and incubated at 37°C for 1 h. Wells were washed five times with PBS, and 100 μl of HRP-conjugated streptavidin (0.5 mg/ml; GenScript) was added and incubated at 37°C for 40 min. Wells were washed five times with PBS, and 100 μl of 3,3′,5,5′-tetramethylbenzidine liquid substrate solution was applied and incubated at room temperature with shaking for 10 min. The reaction was stopped by the addition of 50 μl of H2SO4 (2 M), and absorbance was read at 450 nm in an enzyme-linked immunosorbent assay (ELISA) plate reader. Wells incubated with supernatants from LAC* spa sbi, LAC* spa sbi(pRMC2), or Newman spa sbi were included to account for background, and absorbance readings for these wells were subtracted from the values obtained from the sample wells.
Whole-blood survival assay.
The ability of S. aureus to survive in whole human blood was studied as previously described (27). Bacteria were grown to an OD600 of 1.2 in BHI broth and washed twice in RPMI medium before being diluted to give 5 × 103 CFU/ml. Blood was obtained from healthy volunteers and treated with 50 μg/ml of the anticoagulant hirudin (Refludan; Pharmion). Twenty-five microliters of bacteria was added to 475 μl of blood. Immediately, 100 μl of each sample was added to 900 μl of ice-cold endotoxin-free water, and 100 μl was plated out on TSA in triplicate to calculate the number of input CFU. Tubes were incubated at 37°C with shaking (200 rpm) for 3 h. Following this, 100 μl of each sample was added to 900 μl of ice-cold endotoxin-free water, and 100 μl of each was plated out on TSA in triplicate to calculate the number of recovered CFU. The percentage increase in CFU counts was determined by dividing the mean CFU count after 3 h by the corresponding mean input CFU count. Three independent experiments were performed using blood from three different donors. Ethical approval for the use of human blood was obtained from the Trinity College Dublin (TCD) Faculty of Health Sciences ethics committee.
Statistical analysis.
Statistical analysis was performed using Prism GraphPad, version 5, software. P values were calculated using Student's t test.
RESULTS
Release of protein A by S. aureus strain LAC*.
Surface-displayed SpA becomes linked covalently to the S. aureus cell wall by sortase A (18, 28). Protein A is also released into culture supernatants (3–5). This study set out to investigate mechanisms of SpA release using an erythromycin-sensitive derivative of the USA300 strain LAC (LAC*) (29). Strain LAC expresses SpA at high levels (30, 31) and produces an additional Ig-binding protein (staphylococcal binder of immunoglobulin, Sbi) which associates with lipoteichoic acid in the cell envelope (32). Sbi is also found extracellularly (13).
In order to determine if LAC* releases SpA, proteins solubilized from the cell wall by lysostaphin treatment during protoplast formation (cell wall extract, CW) and culture supernatants (SN) were analyzed by Western immunoblotting probing with HRP-conjugated rabbit IgG (Fig. 2A). To distinguish between Sbi and SpA, isogenic spa and sbi mutants and a double mutant (LAC* spa sbi) were used. A single band corresponding to SpA was detected in cell wall extracts from LAC* and LAC* sbi and was absent in extracts from LAC* spa and the LAC* spa sbi mutant (Fig. 2A). Consistent with previous findings, Sbi was not detected in cell wall extracts (Fig. 2A) since it is not solubilized by lysostaphin during protoplast formation (13).
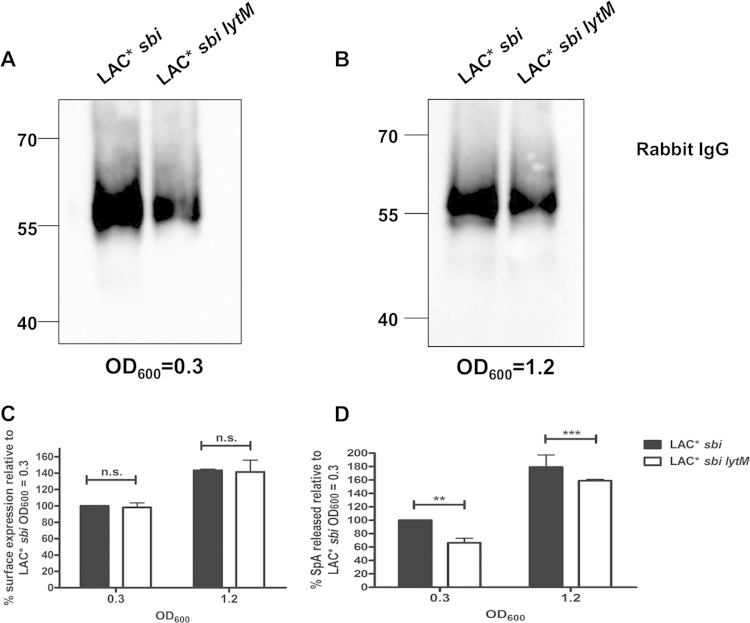
FIG 2.
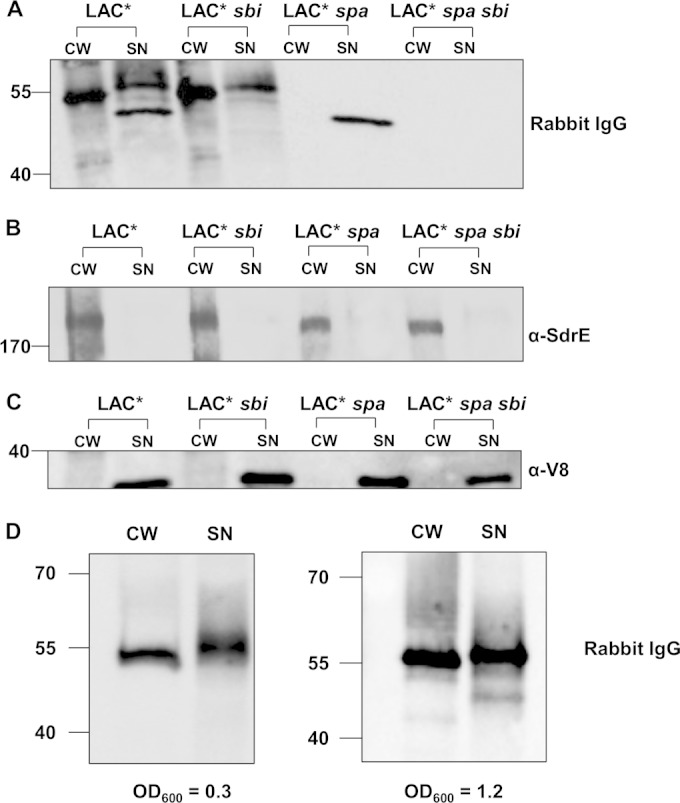
Staphylococcus aureus LAC* releases SpA. S. aureus was grown to an OD600 of 1.2. Cell wall extracts (CW) were diluted 1:5 prior to loading on the gel, and supernatant samples (SN) were not diluted. Protein A was detected using HRP-conjugated rabbit IgG (A), SdrE was detected using anti-SdrE IgG (B), and V8 was detected using anti-V8 serum (C). S. aureus LAC* sbi was grown to an OD600 of 0.3 or 1.2 as indicated (D). Cell wall extracts were diluted 1:20, and supernatant samples were not diluted. Size markers are indicated (kDa).
Culture supernatants from LAC* and LAC* sbi contained a band corresponding to released SpA, and this was absent from LAC* spa and LAC* spa sbi (Fig. 2A). As a control, the same cell wall extracts and supernatants were probed for SdrE, a cell wall-associated protein (Fig. 2B), and V8, a secreted protease (Fig. 2C). The SdrE protein was detected only in the cell wall fraction (Fig. 2B), and V8 was detected only in supernatant samples, indicating purity of the samples and equal loading of protein (Fig. 2B and C).
A band corresponding to extracellular Sbi was detected in supernatants from LAC* and LAC* spa (Fig. 2A). Therefore, to study the release of SpA, it was important to use the LAC* sbi mutant to avoid interference from extracellular Sbi in culture supernatants. In order to determine the proportion of total SpA that is released during growth of LAC* sbi in brain heart infusion (BHI) broth, bands on a Western blot corresponding to SpA in cell wall extracts and supernatants were quantified using densitometry (Fig. 2D). Released SpA was expressed as a percentage of total SpA associated with the cell wall and supernatant. Released SpA represented 6.5% of total protein A from cultures grown to early exponential phase (OD600 of 0.3) and 7.3% of total protein A from cultures grown to an OD600 of 1.2 (Fig. 2D).
Previously Becker et al. (4) reported that the S. aureus endopeptidase LytM promotes the release of SpA by cleaving within the pentaglycine cross-bridge of peptidoglycan. The amount of SpA released into culture supernatants was reduced in an lytM-deficient mutant of strain Newman (4). In order to determine if release of SpA by LAC* is promoted by LytM, S. aureus culture supernatants from LAC* sbi and a LAC* sbi lytM mutant were examined by Western immunoblotting. Less released SpA was detected in the supernatant of the LAC* sbi lytM mutant (Fig. 3A and B). The amounts of SpA on the bacterial surface and in culture supernatants of an lytM-deficient mutant (LAC* sbi lytM) were quantified (Fig. 3C and D). Bacteria were incubated with FITC-labeled rabbit IgG to detect surface-located SpA by flow cytometry. There was no significant difference in the amounts of SpA displayed on the surface of LAC* sbi and LAC* sbi lytM strains grown to the same optical densities (OD600 of 0.3 or OD600 of 1.2) (Fig. 3C). However, there was a 34% reduction in the amount of SpA released by LAC* sbi lytM compared to that from LAC* sbi when supernatants were harvested from bacteria grown to an OD600 of 0.3 (Fig. 3D). LAC* sbi lytM grown to an OD600 of 1.2 released 11% less SpA than LAC* sbi grown to the same optical density (Fig. 3D). These results show that LytM contributes to the release of SpA by LAC*, similar to results with strain Newman (4) (see Fig. S1 in the supplemental material). Given that released SpA constitutes 7% of total SpA (Fig. 2), a 34% reduction in release would result in an undetectable change (∼2%) in the amount of surface-exposed SpA.
FIG 3.
LytM contributes to release of protein A by LAC*. (A and B) Culture supernatants from LAC* sbi and LAC* sbi lytM grown to the OD600 indicated were probed with HRP-labeled rabbit IgG in Western immunoblotting. Supernatants harvested at an OD600 of 0.3 were concentrated 8-fold before being loaded on a gel, and supernatants harvested at an OD600 of 1.2 were concentrated 2-fold. Size markers are indicated (kDa). (C) Protein A on the surface of LAC* sbi and LAC* sbi lytM was detected using FITC-labeled rabbit IgG, and the fluorescence intensity was measured by flow cytometry. Values are plotted as a percentage of the mean fluorescence intensity measured for LAC* sbi grown to an OD600 of 0.3. Bars represent the mean values, and error bars indicate the standard errors of the means of three independent experiments. (D) Protein was captured from culture supernatants using chicken anti-SpA polyclonal IgY and detected using biotin-conjugated mouse monoclonal anti-SpA IgG followed by streptavidin-HRP in an ELISA. Values are expressed as a percentage of total released SpA measured for LAC* sbi grown to an OD600 of 0.3. Bars represent the mean percent release from four independent experiments. Error bars represent the standard errors of the means. **, P = 0.006; ***, P = 0.0003; ns, not significant (P > 0.05).
Extracellular proteases are not required for the generation of released protein A.
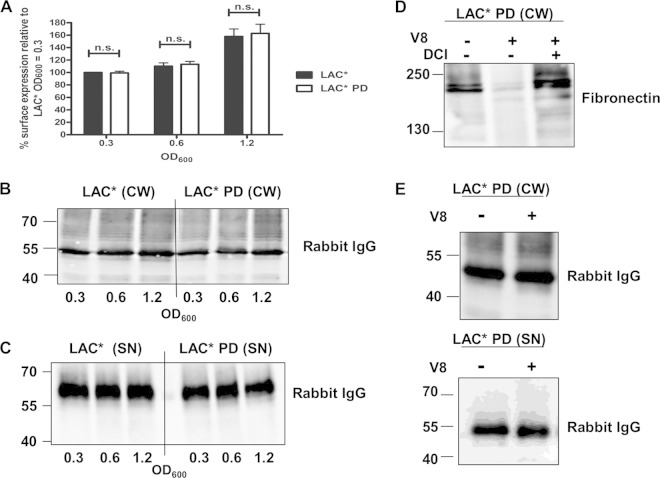
In agreement with studies performed by Becker et al. (4) using strain Newman, we found that release of SpA by the USA300 strain LAC* is partially dependent on the endopeptidase LytM (Fig. 3D). However, SpA release was not completely inhibited in an lytM-deficient mutant of LAC*, indicating that an alternative mechanism for the generation of released SpA exists. In order to investigate if the generation of extracellular protein A in LAC* requires the activity of proteases, the amounts of SpA on the surface of LAC* and an isogenic mutant (LAC* PD) lacking all extracellular proteases of S. aureus (V8, SplABCDEF, ScpA, SspB, and aureolysin) were compared. Surface-located SpA was detected with FITC-labeled IgG using flow cytometry. The amounts of SpA on the bacterial surface did not differ between LAC* and LAC* PD at any of the growth phases tested (Fig. 4A). Cell wall extracts were prepared from the same cultures and analyzed by Western immunoblotting. A band corresponding to SpA was detected in the cell wall of all cultures (Fig. 4B). In order to study the generation of released SpA by LAC* and LAC* PD, culture supernatants were examined by Western immunoblotting. Released SpA was detected in the culture supernatants of bacteria at all stages of growth (Fig. 4C). Less SpA was detected in the culture supernatant of an LytM-deficient mutant compared to the level in the wild-type LAC* under the same conditions (Fig. 3A). No major alterations in the level of released SpA were observed when supernatants from LAC* were compared to those of LAC* PD, indicating that extracellular proteases of S. aureus are not responsible for the generation of released SpA (Fig. 4C).
FIG 4.
Extracellular proteases are not required for the release of protein A. (A) Protein A on the surface of LAC* and LAC* PD was detected using FITC-labeled rabbit IgG, and the fluorescence intensity was measured by flow cytometry. Values are expressed as a percentage of the mean fluorescence intensity measured for LAC* harvested at an OD600 of 0.3. Bars represent the mean values, and error bars indicate the standard errors of the means of three independent experiments. ns, not significant (P > 0.05). (B and C) Cell wall extracts (CW) and culture supernatants (SN) from LAC* and LAC* PD grown to the OD600 indicated were probed with HRP-labeled rabbit IgG in a Western immunoblot. Supernatants harvested at an OD600 of 0.3 were concentrated 8-fold, those at an OD600 of 0.6 were concentrated 4-fold, and those at an OD600 of 1.2 were concentrated 2-fold before being loaded on a gel. (D) LAC* PD was grown to an OD600 of 0.8 in broth alone or in broth supplemented with V8 (1 U/ml) and DCI (200 μM), and cell wall extracts were probed with biotin-labeled fibronectin in a ligand affinity blot. Bound fibronectin was detected using streptavidin-HRP. (E) The same cell wall extracts and supernatants from the same cultures were probed with HRP-conjugated rabbit IgG to detect protein A. Size markers are indicated (kDa).
A study of Karlsson et al. (33) implicated the S. aureus serine protease V8 in the cleavage of SpA from the surface of the bacterium. However, McGavin et al. (34) could not replicate these findings. Since LAC* PD is deficient in the sspA gene encoding V8, it seemed unlikely that V8 protease was responsible for the generation of released SpA by LAC*. In order to determine if the addition of purified V8 protease could promote the release of SpA, the LAC* PD strain was grown in broth with or without added V8. The concentration of V8 used for this experiment was previously shown to be sufficient to remove fibronectin binding proteins A and B (FnBPs) from the surface of S. aureus (34). As a control for V8 activity, the same samples were probed with biotin-labeled fibronectin to detect FnBPs. Very faint bands corresponding to FnBPs were detected in cell wall extracts from bacteria grown in broth containing V8, confirming that the protease was active under the conditions used (Fig. 4D). The effect of V8 was inhibited by the serine protease inhibitor 3,4-dichloroisocoumarin (DCI), confirming that the serine protease activity of V8 is responsible for removing FnBPs from the surface of S. aureus. In order to determine if V8 promotes the release of SpA, the same cell wall extracts were probed with HRP-conjugated rabbit IgG in Western immunoblotting. The integrity or abundance of cell wall-associated and released SpA (Fig. 4E) was not affected by incubation with V8, indicating that V8 does not promote the removal of SpA from the surface of S. aureus.
Identification of a protein A precursor with an unprocessed sorting signal in S. aureus culture supernatants.
Extracellular proteases were not involved in the release of SpA by S. aureus LAC* (Fig. 4) while the glycyl-glycine endopeptidase LytM promoted some SpA release (Fig. 3). Since the release of SpA was not completely inhibited in an LytM-deficient mutant (Fig. 3D) (4) another mechanism of release must exist (4). In order to identify additional factors involved in SpA release, extracellular SpA was purified from the culture supernatant of strain LAC* sbi using affinity chromatography on IgG-Sepharose. The purified protein was analyzed using liquid chromatography tandem mass spectrometry (LC-MS/MS). The sequences of eight of the peptides identified mapped to the extreme C terminus of SpA (Table 2). One peptide terminated with four successive glycines following the sequence LPET. This was consistent with it originating from peptidoglycan-linked SpA and being released by LytM cleavage (4). Interestingly, seven unique peptides with an intact LPETG motif were identified (Table 2), indicating that the preprotein had not been processed by sortase A. This suggested that the protein did not derive from the cell wall and was not released by LytM-mediated cleavage of the pentaglycine cross-bridge of peptidoglycan and implied that SpA can be released by S. aureus prior to becoming covalently anchored to the peptidoglycan. The five N-terminal residues of purified extracellular SpA were identified as 37AQHDE41 by N-terminal sequencing, demonstrating that the signal peptide (residues 1 to 36) had been removed by signal peptidase. In agreement with this, none of the peptides identified by LC-MS/MS originated from the signal sequence (data not shown). Thus, extracellular SpA is processed by signal peptidase so that the N-terminal signal sequence is removed. A single band corresponding to total released SpA was detected by SDS-PAGE or Western blotting (see Fig. S1 in the supplemental material). Unprocessed SpA with an intact sorting signal has a predicted molecular weight of 51,928.9 while SpA released from the cell wall by LytM is linked to fragments of peptidoglycan of different lengths, with the most abundant forms having predicted molecular masses of between 52,555.4 and 54,151 (4). These different forms of extracellular SpA cannot be distinguished since they comigrate on an SDS-PAGE gel.
TABLE 2.
Peptides identified by liquid chromatography tandem mass spectrometry
| Peptide sequencec | No. of peptides identified | Peptide scorea | Mass (Da) | m/z | Position (aa)b |
|---|---|---|---|---|---|
| KAQALPETGEENPFI | 1 | 44.48 | 1,401.6411 | 701.8262 | 470–484 |
| KAQALPETGEENPFIGTT | 1 | 40.83 | 1,704.7842 | 853.3968 | 470–487 |
| KAQALPETGEENPFIGTTVFG | 1 | 39.57 | 2,019.9789 | 1,010.9971 | 470–490 |
| KAQALPETGEENPFIGTTVFGG | 1 | 40.09 | 2,077.0002 | 1,039.5046 | 470–491 |
| KAQALPETGEENPFIGTTVFGGL | 1 | 36.39 | 2,151.0483 | 1,076.5337 | 470–492 |
| KAQALPETGEENPFIGTTVFGGLSL | 2 | 47.19 | 2,351.1646 | 1,176.5889 | 470–494 |
| 47.76 | 2,372.0847 | 791.7029 | 470–494 | ||
| KAQALPETGGGG | 1 | 47.56 | 956.4563 | 479.2358 | 470–478 |
According to Peaks 7 user manual (Bioinformatic Solutions), calculated as −10 log10(P value).
aa, amino acids.
Sorting signal residues are underlined.
Replacing the SpA sorting signal with the sorting signal of SdrE reduces release of SpA.
The identification of SpA with an intact sorting signal in LAC* culture supernatants indicated that SpA can be released from the bacterium without being sorted to the cell wall. This strongly suggested a release mechanism independent of LytM activity. The sorting signal of cell wall-anchored surface proteins is essential for covalent linkage to cell wall peptidoglycan (17, 35). Since some released SpA harbors an unprocessed sorting signal (Table 2), we set out to determine if altering the sorting signal of SpA might influence release of the protein. The sorting signal from SdrE, another cell wall-anchored protein of S. aureus, was exchanged with the SpA sorting signal to generate a chimera, SpA-SdrESS. The SdrE protein is located exclusively in the cell wall fraction of S. aureus, and, in contrast to SpA, very little is detected in culture supernatants from LAC* (Fig. 2B).
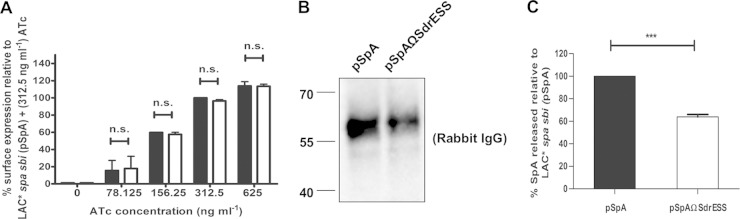
In order to facilitate the manipulation of spa, the full-length spa gene was cloned into the anhydrotetracycline (ATc)-inducible expression vector pRMC2 to generate plasmid pSpA. A variant was constructed where DNA encoding the SpA sorting signal was replaced with DNA encoding the sorting signal from SdrE (pSpAΩSdrESS). Both plasmids were introduced into the LAC* spa sbi mutant. The level of SpA displayed on the surface of the bacteria was detected using FITC-conjugated rabbit IgG, and fluorescence was measured using flow cytometry. Fluorescence was not detected in the absence of ATc, indicating that the promoter is tightly repressed when no inducer is present and SpA is not expressed (Fig. 5A). The amount of SpA displayed on the surface of S. aureus increased with increasing inducer concentration (Fig. 5A), and the level of SpA expressed by S. aureus carrying plasmid pSpA was identical to that of S. aureus carrying pSpAΩSdrESS at each ATc concentration tested (Fig. 5A). This indicated that replacing the SpA sorting signal with the sorting signal from SdrE did not alter the levels of SpA displayed on the surface of S. aureus. To study released SpA, culture supernatants from LAC* spa sbi(pSpA) and LAC* spa sbi(pSpAΩSdrESS) were examined by Western immunoblotting probing with HRP-conjugated rabbit IgG (Fig. 5B). Densitometric analysis of relative band intensities indicated that the LAC* spa sbi strain carrying plasmid pSpAΩSdrESS released less protein A, i.e., approximately 65% as much as LAC* spa sbi carrying plasmid pSpA (Fig. 5B). The relative amounts of released SpA in culture supernatants were then quantified by ELISA (Fig. 5C). The amount of SpA released by bacteria carrying the plasmid pSpAΩSdrESS was reduced to approximately 62% of the amount released by bacteria carrying pSpA (Fig. 5C). These data indicated that the release of SpA by S. aureus is reduced when the SpA sorting signal is replaced with the sorting signal of the wall-associated protein SdrE.
FIG 5.
Release of protein A into S. aureus culture supernatants can be inhibited by altering the sorting signal. (A) Protein A on the surface of LAC* spa sbi(pSpA) (black bars) and LAC* spa sbi(pSpAΩSdrESS) (white bars) was detected using FITC-labeled rabbit IgG, and the fluorescence intensity was measured using flow cytometry. Values are expressed as a percentage of the mean fluorescence intensity measured for LAC* spa sbi(pSpA) grown in broth supplemented with ATc (312.5 ng/ml). Bars represent the mean of three independent experiments. Error bars represent the standard errors of the means. ns, not significant (P > 0.05). (B) LAC* spa sbi(pSpA) and LAC* spa sbi(pSpAΩSdrESS) were grown in broth supplemented with ATc (312.5 ng/ml), and culture supernatants were probed with HRP-conjugated rabbit IgG. Size markers are indicated (kDa). (C) Quantification of SpA in culture supernatants of LAC* spa sbi(pSpA) and LAC* spa sbi(pSpAΩSdrESS) by ELISA. Bacteria were grown in broth supplemented with ATc (312.5 ng/ml), and SpA was captured from culture supernatants using chicken anti-SpA polyclonal IgY. Bound SpA was detected using biotin-labeled mouse monoclonal anti-SpA IgG and HRP-conjugated streptavidin in an ELISA. The absorbance at 450 nm was measured, and readings from wells incubated with culture supernatants from LAC* spa sbi(pRMC2) were subtracted from the mean readings for LAC* spa sbi(pSpA) and LAC* spa sbi(pSpAΩSdrESS) to account for background. Values for LAC* spa sbi(pSpA ΩSdrESS) are expressed as a percentage of the values measured for LAC* spa sbi(pSpA). Bars represent the mean of three independent experiments, and error bars represent the standard errors of the means. ***, P < 0.0001.
The C-terminal sorting signal of SpA allows release of cell wall-anchored proteins from the surface of S. aureus.
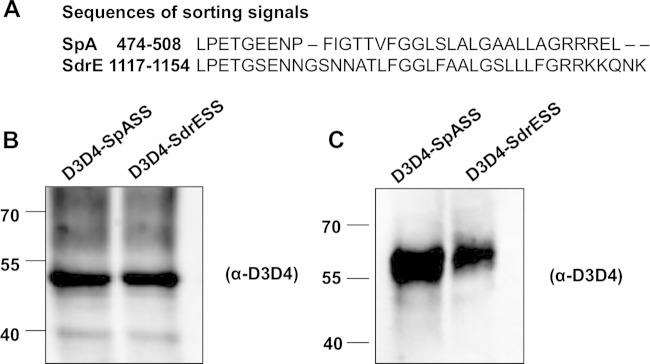
The C-terminal sorting signal comprises an LPXTG motif followed by a hydrophobic domain and a positively charged tail (Fig. 1) and is essential for efficient anchoring of proteins to peptidoglycan (35). Since the release of SpA was reduced when the SpA sorting signal was replaced with the sorting signal of the wall-associated protein SdrE, we hypothesized that the sequence of the sorting signal might influence the release of SpA. Rather than replacing the native SdrE sorting signal with the sorting signal from SpA, we instead generated chimeric proteins where the SpA or SdrE sorting signal was linked to a reporter protein. The advantage of this was that it allowed us to study the influence of the sorting signal alone on protein release. Chimeric proteins were generated where the D3D4 domains of Sbi were linked to the sorting signal of SpA or SdrE (Fig. 6A). Sbi is an envelope-associated protein which does not become anchored to cell wall peptidoglycan (13). Plasmids pD3D4-SpASS and pD3D4-SdrESS each carried DNA encoding the Sbi signal sequence and D3D4 domains and the sorting signal from either SpA and SdrE, respectively. Cell wall extracts and supernatants were prepared from cultures of LAC* spa sbi carrying plasmids pD3D4-SpASS and pD3D4-SdrESS and analyzed by Western immunoblotting using anti-D3D4 IgG. Both D3D4-SpASS and D3D4-SdrESS were detected in cell wall extracts (Fig. 6B), indicating that they had been sorted to the cell wall peptidoglycan. Less D3D4 protein was detected in culture supernatants from LAC* spa sbi(pD3D4-SdrESS) (43% reduction as estimated by densitometry) than from LAC* spa sbi(pD3D4-SpASS) (Fig. 6C). These data demonstrate that the release of a protein into S. aureus culture supernatants depends on the sequence of its sorting signal.
FIG 6.
Release of D3D4 reporter protein into S. aureus culture supernatants can be inhibited by altering the sorting signal. (A) Amino acid sequences of SpA and SdrE sorting signals. Amino acid coordinates are indicated. Cell wall extracts (B) and culture supernatants (C) from LAC* spa sbi(pD3D4-SpASS) and LAC* spa sbi(pD3D4-SdrESS) were probed with rabbit anti-D3D4 IgG. Bound antibody was detected using HRP-conjugated goat anti-rabbit IgG. Size markers are indicated (kDa).
The release of SpA by LAC* is mediated by both LytM and the native SpA sorting signal.
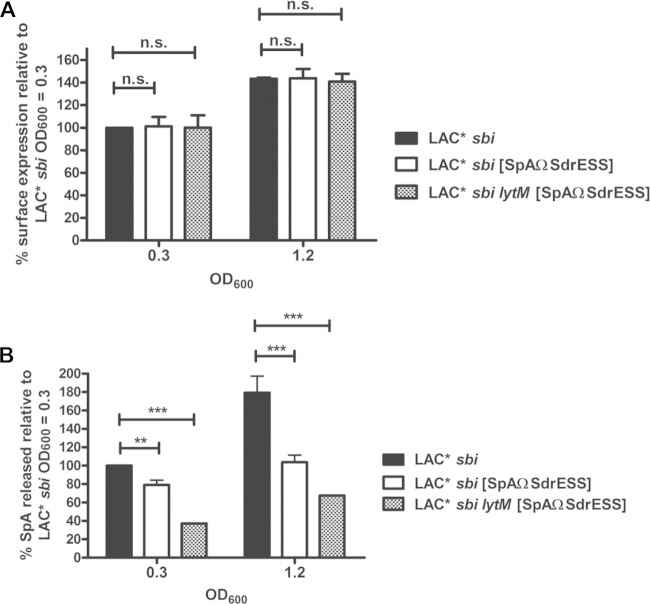
Taken together, our data suggested that both the SpA sorting signal and the activity of the glycyl-glycine endopeptidase LytM contribute to release of SpA by S. aureus. In order to study the relative contribution of these factors, the sorting signal from SdrE was exchanged with the SpA sorting signal on the chromosome of LAC* sbi and LAC* sbi lytM by allelic exchange to yield strains LAC* sbi [SpAΩSdrESS] and LAC* sbi lytM [SpAΩSdrESS], respectively. Protein A displayed on the surface of the bacteria was detected using FITC-conjugated rabbit IgG. The level of SpA expressed by LAC* sbi [SpAΩSdrESS] and LAC* sbi lytM [SpAΩSdrESS] was identical to the level expressed by LAC* sbi at each stage of growth tested (OD600s of 0.3 and 1.2) (Fig. 7A). The relative amount of released SpA in culture supernatants was quantified by ELISA. The amount of SpA released by S. aureus LAC* sbi [SpAΩSdrESS] grown to an OD600 of 0.3 was 21% less than the amount released by LAC* sbi grown to an equal optical density (Fig. 7B). At an OD600 of 1.2, LAC* sbi [SpAΩSdrESS] released 42% less SpA than LAC* sbi (Fig. 7B). When the sorting signal of SdrE replaced the SpA sorting signal in an LytM-deficient mutant (LAC* sbi lytM [SpAΩSdrESS]), the amount of SpA was reduced by 63% and 62% for bacteria grown to OD600s of 0.3 and 1.2, respectively, compared to the amount in LAC* sbi grown to the same optical density (Fig. 7B). These data demonstrate that the release of SpA by S. aureus is influenced by both the sorting signal and LytM activity and that together these factors account for up to 63% of released SpA in strain LAC*. Similar results were obtained using strain Newman sbi and Newman sbi [SpAΩSdrESS] (see Fig. S2 in the supplemental material).
FIG 7.
Replacing the SpA sorting signal with the sorting signal from SdrE reduces release of SpA by LAC*. (A) Protein A on the surface of LAC* sbi, LAC* sbi [SpAΩSdrESS], and LAC* sbi lytM [SpAΩSdrESS] was detected using FITC-labeled rabbit IgG, and the fluorescence intensity was measured using flow cytometry. Values are expressed as a percentage of the mean fluorescence intensity measured for LAC* sbi grown to an OD600 of 0.3. Bars represent the mean values, and error bars indicate the standard errors of the means of three independent experiments. (B) Protein A was captured from culture supernatants using chicken anti-SpA polyclonal IgY and detected using biotin-conjugated mouse monoclonal anti-SpA IgG followed by streptavidin-HRP in an ELISA. The absorbance at 450 nm was measured, and the mean reading from wells incubated with culture supernatants from LAC* spa sbi were subtracted from the readings for all other wells to account for background. Values are expressed as a percentage of total released SpA measured for LAC* sbi grown to an OD600 of 0.3. Bars represent the mean of four independent experiments. Error bars represent the standard errors of the means. **, P = 0.007; ***, P < 0.0001; ns, not significant (P > 0.05).
Both cell wall-anchored and released SpA contribute to the survival of LAC* in whole human blood.
Protein A contributes to the pathogenesis of invasive infection by protecting S. aureus from killing in blood (11, 13). Surface-located SpA inhibits bacterial uptake by neutrophils (13, 36, 37), but the ability of released SpA to promote S. aureus survival in blood has not been examined.
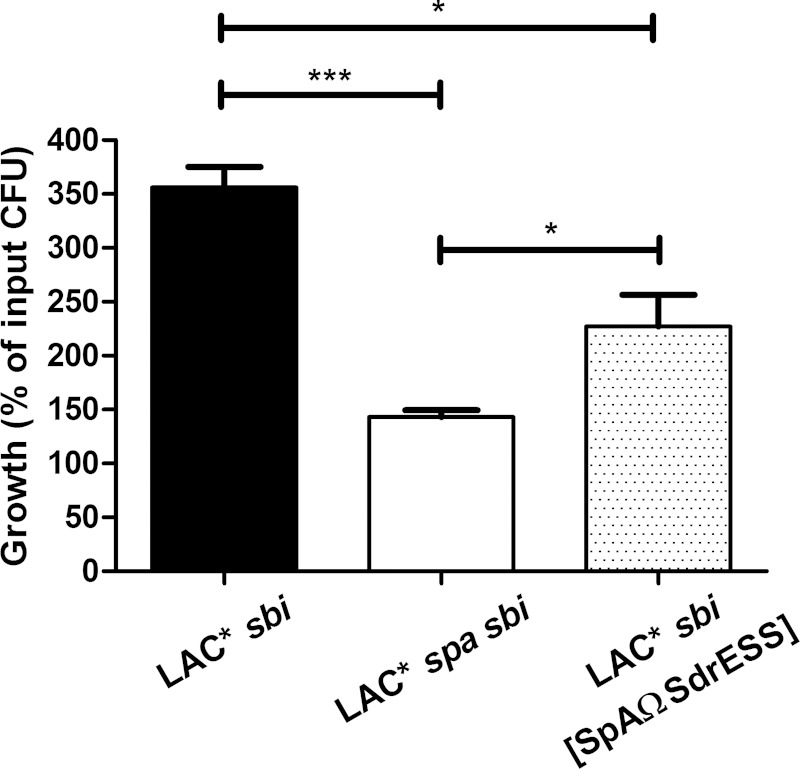
Previously, Malachowa et al. (38) demonstrated that transcription of the spa gene is highly upregulated when LAC is incubated in human blood. To investigate if SpA contributes to the ability of LAC* to resist killing in human blood, LAC* sbi and an LAC* spa sbi strain were incubated in blood, and viable counts were used to determine bacterial survival. The LAC* spa sbi mutant demonstrated a reduced ability to survive in whole blood (143% of input inoculum recovered) compared to that of LAC* sbi (356% of input inoculum recovered) (Fig. 8), showing that SpA protects LAC from phagocytic killing.
FIG 8.
Released SpA protects S. aureus from killing in human blood. Washed bacteria were incubated in blood for 3 h at 37°C, and the number of input and recovered bacteria was calculated by viable counting. The percentage increase in CFU counts (growth) of each strain was determined by dividing the mean CFU count after 3 h by the mean input CFU count. Bars represent the mean percentage increase in CFU counts from three independent experiments, and error bars indicate the standard errors of the means. ***, P < 0.001; *, P < 0.05.
LAC* sbi expresses both cell wall-associated and released SpA (Fig. 2). To investigate if the unprocessed form of released SpA increases the growth of S. aureus in blood, LAC* sbi [SpAΩSdrESS] was studied. This strain expresses similar levels of surface-associated SpA but less released SpA than LAC* sbi (Fig. 7). LAC* sbi [SpAΩSdrESS] grew significantly better in human blood than LAC* spa sbi (227% of input inoculum recovered), indicating that released SpA enhances the ability of S. aureus to survive and grow in blood (Fig. 8). The effect of LytM-mediated SpA release on blood survival was not examined here since LytM is proposed to contribute to the release of all proteins linked to the pentaglycine cross-bridge of peptidoglycan (4). Many of these proteins are important immune evasion factors and contribute to bacterial survival in blood (13, 39), and so the effects of LytM-released SpA could not be studied in isolation. In summary, these data indicate that released as well as cell wall-associated SpA protects S. aureus from killing in human blood.
DISCUSSION
Protein A is a virulence factor in murine models of S. aureus kidney abscess formation, skin infection, pneumonia, sepsis, and septic arthritis (15, 40–43). S. aureus produces both cell wall-associated and released forms of SpA. Protein A becomes covalently linked to peptidoglycan by the action of sortase A. Here, we demonstrate that SpA with an intact sorting signal is found in the culture supernatant, indicating that a portion of the protein can be released without being processed by sortase A.
Becker et al. (4) previously showed that the glycyl-glycine endopeptidase LytM cleaves within the pentaglycine cross-bridge of peptidoglycan to release SpA. An lytM mutant of strain Newman released less SpA than wild-type bacteria (4). In this study, we demonstrate that LytM also contributes to the release of SpA by strain LAC*. Culture supernatants from an lytM-deficient mutant contained 34% less SpA than supernatants from wild-type LAC* grown to an OD600 of 0.3 and 11% less SpA at an OD600 of 1.2 (Fig. 3D). Thus, it appears that LytM may have a greater influence on SpA release at early points in the growth phase.
We demonstrate that extracellular proteases of S. aureus do not mediate the release of SpA (Fig. 4). Despite conflicting evidence in the literature (34), it has been assumed that the S. aureus serine protease V8 cleaves SpA from the surface of S. aureus (44). However, this assumption was based on a single study with an uncharacterized strain (33). Our data demonstrate that V8 does not mediate SpA release. However, a previous study showed that there was a 2.1-fold increase in the amount of SpA in the cell wall of LAC* PD compared to that in wild-type LAC* (44) when the surface proteome of stationary-phase cultures (15 h of growth) was examined. Thus, it is likely that V8 can affect the stability of SpA on the S. aureus surface to some degree.
The factors promoting SpA release reported in this study and by Becker et al. (4) do not account for all released SpA. Replacing the SpA sorting signal with that of SdrE in an LytM-deficient mutant of LAC* reduced the amount of released SpA by up to 63% (Fig. 7B). As proposed by Becker et al. (4), it is possible that an unidentified autolysin or another factor might be involved. A portion of SpA is released into the culture supernatant without being processed by sortase. Replacing the native sorting signal from SpA with the sorting signal of SdrE reduced the release of SpA. Why is a protein with the SpA sorting signal released abundantly into the S. aureus culture supernatant while a protein with the SdrE sorting signal is released to a lesser extent? Schneewind et al. (35) demonstrated that removing the charged tail of the sorting signal of SpA resulted in the release of the protein into the culture medium instead of it becoming linked to the cell wall. Substitution of two arginine residues with serine in the charged tail region resulted in a dramatic reduction in the sorting of SpA to the cell wall (17). The spacing of the LPETG motif and positively charged tail was also important since reducing the number of residues in the hydrophobic domain of the sorting signal from 25 to 23 resulted in less protein being released into culture supernatants (17). The SdrE sorting signal contains more positively charged residues at the C terminus (five rather than three) and has a slightly longer hydrophobic domain (one residue longer) than the sorting signal from SpA (Fig. 5A). Thus, it seems reasonable to hypothesize that this is the reason why less protein is released when the native SpA sorting signal is replaced with the sorting signal from SdrE. Released SpA can protect S. aureus from killing in human blood (Fig. 8). Therefore, the release of SpA is likely to contribute to the ability of S. aureus LAC* to survive in the human bloodstream. The mechanism by which released SpA protects bacteria from killing in blood warrants further investigation. Surface-located SpA has long been assumed to protect bacteria from opsonophagocytosis through its ability to bind to the Fc region of IgG. However, recent work by Nordenfelt et al. (45) suggested that the majority of IgG is likely to be bound to SpA via the Fab region when bacteria are in the bloodstream. Falugi et al. (11) showed that a variant of SpA lacking the ability to recognize IgG Fc survived poorly in mouse blood. Therefore, the ability of SpA to bind to IgG or to another ligand which shares the same binding site on SpA can protect bacteria from killing in blood. Further study will allow the mechanisms involved to be elucidated fully.
The release of SpA by S. aureus is likely to contribute to the ability of S. aureus to interfere with adaptive immunity through its binding to the Fab region of human IgM on B cells. Becker et al. (4) proposed that the removal of the amino sugars from cell wall-derived SpA is necessary so that released SpA will not be recognized by NOD2. If this is the case, then the release of SpA with an unmodified C terminus (intact sorting signal) represents a second strategy for the production of a soluble form of SpA that will not trigger host innate immune responses.
In summary, we report that a portion of released SpA does not originate from the cell wall and is released following processing by signal peptidase but prior to cleavage by the sortase A enzyme. Released SpA protects S. aureus from killing in human blood, and the bacterium employs at least two independent strategies to ensure that SpA will be released into the culture supernatant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported financially by Trinity College Dublin.
We thank Tim Foster for helpful discussions and for offering comments on the manuscript. We are grateful to Martin McGavin for providing V8 antiserum and Alex Horswill for providing the LAC* and LAC* PD strains.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03122-14.
REFERENCES
- 1.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core surveillance (ABCs) MRSA investigators . 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Lachica RV, Genigeorgis CA, Hoeprich PD. 1979. Occurrence of protein A in Staphylococcus aureus and closely related Staphylococcus species. J Clin Microbiol 10:752–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Movitz J. 1976. Formation of extracellular protein A by Staphylococcus aureus. Eur J Biochem 68:291–299. doi: 10.1111/j.1432-1033.1976.tb10788.x. [DOI] [PubMed] [Google Scholar]
- 4.Becker S, Frankel MB, Schneewind O, Missiakas D. 2014. Release of protein A from the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A 111:1574–1579. doi: 10.1073/pnas.1317181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsgren A, Nordstrom K. 1974. Protein A from Staphylococcus aureus: the biological significance of its reaction with IgG. Ann N Y Acad Sci 236:252–266. doi: 10.1111/j.1749-6632.1974.tb41496.x. [DOI] [PubMed] [Google Scholar]
- 6.Lindmark R, Movitz J, Sjoquist J. 1977. Extracellular protein A from a methicillin-resistant strain of Staphylococcus aureus. Eur J Biochem 74:623–628. doi: 10.1111/j.1432-1033.1977.tb11431.x. [DOI] [PubMed] [Google Scholar]
- 7.Moks T, Abrahmsen L, Nilsson B, Hellman U, Sjoquist J, Uhlen M. 1986. Staphylococcal protein A consists of five IgG-binding domains. Eur J Biochem 156:637–643. doi: 10.1111/j.1432-1033.1986.tb09625.x. [DOI] [PubMed] [Google Scholar]
- 8.Sjodahl J. 1977. Repetitive sequences in protein A from Staphylococcus aureus. Arrangement of five regions within the protein, four being highly homologous and Fc-binding. Eur J Biochem 73:343–351. [DOI] [PubMed] [Google Scholar]
- 9.Deisenhofer J. 1981. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-Å resolution. Biochemistry 20:2361–2370. doi: 10.1021/bi00512a001. [DOI] [PubMed] [Google Scholar]
- 10.Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Charbonnier JB, Silverman GJ. 2000. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U S A 97:5399–5404. doi: 10.1073/pnas.97.10.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falugi F, Kim HK, Missiakas DM, Schneewind O. 2013. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio 4(5):e00575-00513. doi: 10.1128/mBio.00575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodyear CS, Silverman GJ. 2003. Death by a B cell superantigen: in vivo VH-targeted apoptotic supraclonal B cell deletion by a staphylococcal toxin. J Exp Med 197:1125–1139. doi: 10.1084/jem.20020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith EJ, Visai L, Kerrigan SW, Speziale P, Foster TJ. 2011. The Sbi protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infect Immun 79:3801–3809. doi: 10.1128/IAI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez MI, O'Seaghdha M, Magargee M, Foster TJ, Prince AS. 2006. Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J Biol Chem 281:20190–20196. doi: 10.1074/jbc.M601956200. [DOI] [PubMed] [Google Scholar]
- 15.Martin FJ, Gomez MI, Wetzel DM, Memmi G, O'Seaghdha M, Soong G, Schindler C, Prince A. 2009. Staphylococcus aureus activates type I IFN signaling in mice and humans through the Xr repeated sequences of protein A. J Clin Invest 119:1931–1939. doi: 10.1172/JCI35879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sibbald MJ, Ziebandt AK, Engelmann S, Hecker M, de Jong A, Harmsen HJ, Raangs GC, Stokroos I, Arends JP, Dubois JY, van Dijl JM. 2006. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol Mol Biol Rev 70:755–788. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneewind O, Mihaylova-Petkov D, Model P. 1993. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J 12:4803–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazmanian SK, Liu G, Ton-That H, Schneewind O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 19.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem 277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- 20.Mazmanian SK, Ton-That H, Schneewind O. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol 40:1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- 21.Yung SC, Parenti D, Murphy PM. 2011. Host chemokines bind to Staphylococcus aureus and stimulate protein A release. J Biol Chem 286:5069–5077. doi: 10.1074/jbc.M110.195180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li MZ, Elledge SJ. 2012. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol Biol 852:51–59. doi: 10.1007/978-1-61779-564-0_5. [DOI] [PubMed] [Google Scholar]
- 23.Corrigan RM, Foster TJ. 2009. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61:126–129. doi: 10.1016/j.plasmid.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3(2):e00277–11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lofblom J, Kronqvist N, Uhlen M, Stahl S, Wernerus H. 2007. Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J Appl Microbiol 102:736–747. doi: 10.1111/j.1365-2672.2006.03127.x. [DOI] [PubMed] [Google Scholar]
- 26.O'Brien L, Kerrigan SW, Kaw G, Hogan M, Penades J, Litt D, Fitzgerald DJ, Foster TJ, Cox D. 2002. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol Microbiol 44:1033–1044. doi: 10.1046/j.1365-2958.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- 27.Visai L, Yanagisawa N, Josefsson E, Tarkowski A, Pezzali I, Rooijakkers SH, Foster TJ, Speziale P. 2009. Immune evasion by Staphylococcus aureus conferred by iron-regulated surface determinant protein IsdH. Microbiology 155:667–679. doi: 10.1099/mic.0.025684-0. [DOI] [PubMed] [Google Scholar]
- 28.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci U S A 96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ventura CL, Malachowa N, Hammer CH, Nardone GA, Robinson MA, Kobayashi SD, DeLeo FR. 2010. Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS One 5:e11634. doi: 10.1371/journal.pone.0011634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, Albrecht D, Novick R, Gotz F. 2010. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun 78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith EJ, Corrigan RM, van der Sluis T, Grundling A, Speziale P, Geoghegan JA, Foster TJ. 2012. The immune evasion protein Sbi of Staphylococcus aureus occurs both extracellularly and anchored to the cell envelope by binding lipoteichoic acid. Mol Microbiol 83:789–804. doi: 10.1111/j.1365-2958.2011.07966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson A, Saravia-Otten P, Tegmark K, Morfeldt E, Arvidson S. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect Immun 69:4742–4748. doi: 10.1128/IAI.69.8.4742-4748.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGavin MJ, Zahradka C, Rice K, Scott JE. 1997. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun 65:2621–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneewind O, Model P, Fischetti VA. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267–281. doi: 10.1016/0092-8674(92)90101-H. [DOI] [PubMed] [Google Scholar]
- 36.Kim HK, Emolo C, DeDent AC, Falugi F, Missiakas DM, Schneewind O. 2012. Protein A-specific monoclonal antibodies and prevention of Staphylococcus aureus disease in mice. Infect Immun 80:3460–3470. doi: 10.1128/IAI.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson PK, Verhoef J, Sabath LD, Quie PG. 1977. Effect of protein A on staphylococcal opsonization. Infect Immun 15:760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malachowa N, Whitney AR, Kobayashi SD, Sturdevant DE, Kennedy AD, Braughton KR, Shabb DW, Diep BA, Chambers HF, Otto M, DeLeo FR. 2011. Global changes in Staphylococcus aureus gene expression in human blood. PLoS One 6:e18617. doi: 10.1371/journal.pone.0018617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. 2009. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J Exp Med 206:2417–2427. doi: 10.1084/jem.20090097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel AH, Nowlan P, Weavers ED, Foster T. 1987. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect Immun 55:3103–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmqvist N, Foster T, Tarkowski A, Josefsson E. 2002. Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb Pathog 33:239–249. doi: 10.1006/mpat.2002.0533. [DOI] [PubMed] [Google Scholar]
- 42.Gomez MI, Lee A, Reddy B, Muir A, Soong G, Pitt A, Cheung A, Prince A. 2004. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat Med 10:842–848. doi: 10.1038/nm1079. [DOI] [PubMed] [Google Scholar]
- 43.Kim HK, Cheng AG, Kim HY, Missiakas DM, Schneewind O. 2010. Nontoxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J Exp Med 207:1863–1870. doi: 10.1084/jem.20092514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolar SL, Ibarra JA, Rivera FE, Mootz JM, Davenport JE, Stevens SM, Horswill AR, Shaw LN. 2013. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2:18–34. doi: 10.1002/mbo3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nordenfelt P, Waldemarson S, Linder A, Morgelin M, Karlsson C, Malmstrom J, Bjorck L. 2012. Antibody orientation at bacterial surfaces is related to invasive infection. J Exp Med 209:2367–2381. doi: 10.1084/jem.20120325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wormann ME, Reichmann NT, Malone CL, Horswill AR, Grundling A. 2011. Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J Bacteriol 193:5279–5291. doi: 10.1128/JB.00369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.