Abstract
Background
We previously cloned a 1,3-specific lipase gene from the fungus Rhizomucor miehei and expressed it in methylotrophic yeast Pichia pastoris strain GS115. The enzyme produced (termed RML) was able to catalyze methanolysis of soybean oil and showed strong position specificity. However, the enzyme activity and amount of enzyme produced were not adequate for industrial application. Our goal in the present study was to improve the enzyme properties of RML in order to apply it for the conversion of microalgae oil to biofuel.
Results
Several new expression plasmids were constructed by adding the propeptide of the target gene, optimizing the signal peptide, and varying the number of target gene copies. Each plasmid was transformed separately into P. pastoris strain X-33. Screening by flask culture showed maximal (21.4-fold increased) enzyme activity for the recombinant strain with two copies of the target gene; the enzyme was termed Lipase GH2. The expressed protein with the propeptide (pRML) was a stable glycosylated protein, because of glycosylation sites in the propeptide. Quantitative real-time RT-PCR analysis revealed two major reasons for the increase in enzyme activity: (1) the modified recombinant expression system gave an increased transcription level of the target gene (rml), and (2) the enzyme was suitable for expression in host cells without causing endoplasmic reticulum (ER) stress. The modified enzyme had improved thermostability and methanol or ethanol tolerance, and was applicable directly as free lipase (fermentation supernatant) in the catalytic esterification and transesterification reaction. After reaction for 24 hours at 30°C, the conversion rate of microalgae oil to biofuel was above 90%.
Conclusions
Our experimental results show that signal peptide optimization in the expression plasmid, addition of the gene propeptide, and proper gene dosage significantly increased RML expression level and enhanced the enzymatic properties. The target enzyme was the major component of fermentation supernatant and was stable for over six months at 4°C. The modified free lipase is potentially applicable for industrial-scale conversion of microalgae oil to biodiesel.
Electronic supplementary material
The online version of this article (doi:10.1186/1754-6834-7-111) contains supplementary material, which is available to authorized users.
Keywords: Rhizomucor miehei, Lipase, Heterologous expression, Pichia pastoris, Microalgae oil, Biodiesel
Background
A fungal lipase (Rhizomucor miehei lipase; RML) from R. miehei is a highly versatile biocatalyst used in laboratory and commercial/ industrial applications [1]. RML is a single-chain α/β type protein, MW of approximately 31.6 kDa, comprised of 269 amino-acid residues [2]. Its high resolution three-dimensional structure has been determined, and the catalytic triad is Ser144, His257, Asp203 [3]. RML is synthesized as a precursor that includes a 70 amino-acid propeptide before the 269 amino-acid residues of the mature enzyme [4]. The crystal structure at 1.9 Å resolution and theoretical analysis of RML activation have been reported [3, 5].
Two forms of the enzyme (Palatase 2000 L in free form and Lipozyme RM IM in immobilized form) are currently commercially available from Novozymes (Novo Nordisk A/S Corp, Hillerod, Denmark) [6]. Because of its strong specificity, RML has been widely used in ester hydrolysis, ester synthesis, and transesterification reaction such as production of structured lipids [7], synthesis of butyl butyrate (pineapple flavor) [8], and synthesis of monoglyceryl esters from chiral and prochiral acid methyl esters [9]. Despite this wide applicability, cost remains a major obstacle to large-scale industrial use of RML [10]. Enhanced activity and reduced production cost of the enzyme are important goals.
Pichia pastoris is the most frequently used yeast system for heterologous protein production [11]. One advantage of this expression system is that its ability to perform eukaryotic post-translational modifications of heterologous proteins, such as glycosylation N-linked glycosylation is the most commonly when expressed heterologous proteins in Pichia pastoris, it was found that N-linked glycosylation also played an essential role in protein secretion [12, 13]. To date, over 350 recombinant proteins have been expressed in P. pastoris systems, with protein production of more than 10 g/L in some cases [14]. High-level expression in P. pastoris can be affected by factors such as nucleotide sequence properties, gene copy numbers, mRNA transcription, promoter choice, secretion signals, and protein folding in the endoplasmic reticulum (ER); these factors cannot be regulated solely through control of the fermentation process [15, 16]. Some studies have been successful in improving protein expression levels through regulation of the above factors [15, 17, 18].
There is increasing research and industrial interest in ‘biodiesel’ as a replacement for petroleum-derived diesel fuel [19]. Microalgae have many advantages as a source of biodiesel in comparison with food crops and non-food crops, and may be the only source that can be sustainably developed in the future [20, 21]. Previous studies have focused on the culture of microalgae and enhanced lipid content of microalgae cells [22–25], and conversion of microalgae oil to biodiesel [26–29]. The chemical conversion of microalgae oil to biodiesel using an acid or alkaline catalyst involves high energy consumption and pollution. Enzyme-catalyzed conversion reactions have obvious advantages and are a ‘hot’ research topic. Various studies have attempted to improve reaction efficiency through the discovery of new types of enzymes [30], molecular enzyme modification [31–33], and reaction process optimization [34, 35].
We previously cloned the gene of 1,3-specific RML from R. miehei and expressed it in P. pastoris strain GS115 (RML-GS115). The enzyme was used to catalyze methanolysis of soybean oil and displayed strong positional specificity. However, the enzyme activities obtained were only 50 U/mL in a flask with medium Buffered Glycerol-complex Medium (BMGY) and Buffered Methanol-complex Medium (BMMY) and 550 U/mL in a 7.5-L fermenter [34]. In order to increase RML activity and obtain recombinants with maximal expression capacity, we considered factors such as propeptide, codon usage of signal peptide, and gene dosage of target protein in the present study. We significantly enhanced enzyme activity by changing the expression system to P. pastoris strain X-33 and vector pPICZaA, adding one propeptide to RML, and selecting the optimal gene dosage.
Results
Addition of the propeptide of the target gene
We explored several strategies to modify RML and its heterologous expression system in order to improve enzyme activity and properties. Our first step was to examine the role of the target gene’s propeptide. We constructed two expression plasmids with carrier pPICZαA: one adding the target gene’s 70-amino-acid propeptide (prml) and one without propeptide (mrml). The recombinant plasmids were transformed separately into strain X-33. The positive strains, each containing one copy of the target gene, were termed zα-1mRML-X33 and zα-1pRML-X33. These two strains and a control strain (zα-X33, without target gene) were cultured in shaking flasks and sampled every day for cell growth and lipase activity. The screening results are shown in Figure 1. Cell growth of zα-1pRML-X33 and zα-X33 was higher than that of zα-1mRML-X33 (Figure 1A). The extracellular enzyme activity of zα-pRML-X33 (430 U/mL) was 7.7-fold higher than that of zα-1mRML-X33 (56 U/mL; Figure 1B). These findings indicate that in the absence of the propeptide, expression levels of the heterologous protein are consistent regardless of whether strain GS115 or X-33 is used as a host. Extracellular protein in fermentation supernatant was detected by Western blotting (Figure 1C). The content of secreted target protein for zα-1pRML-X33 (0.15 mg/mL) was approximately 7-fold higher than that for zα-1mRML-X33 (0.019 mg/mL).Quantitative real-time RT-PCR (qPCR) results confirmed that adding the propeptide resulted in upregulation of target gene expression (Figure 1D) at 48 and 96 hours. The propeptide evidently plays a promoting role in lipase activity and heterologous protein secretion.
Figure 1.
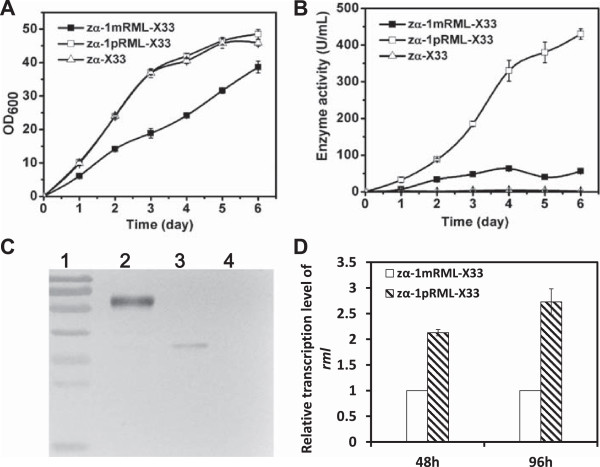
Effect of propeptide addition on cell growth and target enzyme activity. A: OD600 of recombinant strains in flask fermentation. Cell growth of control strain (zα-X33) and recombinant strain (zα-1pRML-X33) containing the propeptide sequence was much higher than that of the recombinant strain without the propeptide sequence (zα-1mRML-X33). B: Enzyme activity of recombinant strains in flask fermentation. Extracellular enzyme activity of zα-1pRML-X33 (430 U/mL) was 7.7-fold higher than that of zα-1mRML-X33 (56 U/mL). C: Extracellular protein production in fermentation supernatant detected by Western blotting. Lane 1: protein markers (top to bottom: 100, 70, 55, 40, 35, 25, 15 kDa). Lane 2: zα-1pRML-X33. Lane 3: zα-1mRML-X33. Lane 4: zα-X33. Each sample tested was 10 μL of 10 × diluted fermentation supernatant. The concentration of secreted extracellular target protein for zα-1pRML-X33 (0.15 mg/mL) was higher than for zα-1mRML-X33 (0.019 mg/mL). D: Comparison of the transcription level (by qPCR) of rml in zα-1pRML-X33 vs. zα-1mRML-X33. When the same signal peptide codons were present in the expression plasmid, adding the propeptide resulted in upregulation of target gene expression at both 48 and 96 hours.
Optimization of the signal peptide codons
The original expression plasmid (pPICZαA) and the modified signal peptide plasmid (pPICMαA) were kindly provided by Drs Yijun Huang and Jia Ban. We compared their effects on target enzyme activity and protein secretion. The nucleotide sequence of the modified signal peptide (mα) is shown in Additional file 1: Figure S1. We successfully constructed modified codons of the signal peptide recombinant strain (termed mα-1pRML-X33) and the original recombinant strain (termed zα-1pRML-X33). Each strain contained one copy of the target gene. Flask fermentation of the two strains is compared in Figure 2. Cell growth was essentially the same in mα-1pRML-X33, zα-1pRML-X33, and two control strains without the target gene (mα-X33, zα-X33); they all reached OD600 ≈ 50 (Figure 2A). However, the enzyme activity of mα-1pRML-X33 (600 U/mL) was higher than that of zα-1pRML-X33 (430 U/mL; Figure 2B). Optimization of the signal peptide codons resulted in a 1.4-fold increase in enzyme activity. The target protein was also detected by Western blotting. The amount of extracellular target protein (0.22 mg/mL) from mα-1pRML-X33 (Figure 2C, lane 3) was higher than that (0.16 mg/mL) from zα-1pRML-X33 (lane 2). Target gene transcription level was evaluated by qPCR. For a given dosage of the target gene in the cells, optimization of the signal peptide clearly increased enzyme activity and the amount of extracellular target protein. The transcription level of the target gene was 1.4-fold higher following optimization of the signal peptide codons (Figure 2D). These findings indicate that optimization of codons in the signal peptide to make it more suitable for transcription in host cells promoted expression and secretion of target heterologous protein.
Figure 2.

Cell growth and protein production of recombinant strain with modified α-factor signal peptide (mα-1pRML-X33), strain with original α-factor signal peptide (zα-1pRML-X33), and two control strains (mα-X33 and zα-X33). A: OD600 of the recombinants in flask fermentation. Cell growth did not differ notably among the four strains. B: Enzyme activity of the recombinants during fermentation. Activity of mα-1pRML-X33 (600 U/ml) was higher than that of zα-1pRML-X33 (430 U/ml). Activity increased 1.4-fold after optimization of signal peptide codons. C: Extracellular protein expression in fermentation supernatant detected by Western blotting. Lane 1: protein markers (same as in Figure 1). Lane 2: recombinant strain containing original α-factor signal peptide. Lane 3: strain containing modified α-factor signal peptide. The concentration of extracellular target protein for mα-1pRML-X33 (0.22 mg/mL) was higher than that for zα-1pRML-X33 (0.16 mg/mL). D: The transcription level of prml (detected by qPCR) in mα-1pRML-X33 vs. zα-1pRML-X33. Optimization of signal peptide (mα-1pRML-X33) increased target gene transcription at both 48 and 96 hours.
Optimization of the target gene dosage
We next investigated the effects of target gene copies on protein expression. We successfully constructed recombinants with 1, 2, 4, and 8 copies of the prml gene, termed respectively as mα-1pRML-X33, mα-2pRML-X33, mα-4pRML-X33 and mα-8pRML-X33. These recombinants were tested by qPCR. The standard curves of glyceraldehyde-3-phosphate dehydrogenase gene (gap; used as a reference gene) and rml detected by qPCR are shown in Additional file 2: Figure S2. C(t) values of the target gene (prml) and reference gene (gap) and copy numbers were calculated using the formula given in the Materials and Methods section ‘Screening for target transformants’, and results are shown in Additional file 3: Table S1.
The differential effects of the four recombinants on enzyme activity in flask fermentation are summarized in Figure 3A. When the target gene copy number was increased from 1 to 2, enzyme activity was doubled (from 600 to 1,200 U/mL) and extracellular protein content also increased approximately 2-fold (from 0.38 to 0.770 mg/mL). When copy number increased from 4 to 8, extracellular protein content was essentially unchanged (0.69 versus 0.73 mg/mL) and enzyme activity decreased from 713 to 503 U/mL. As copy number increased from 1 to 2, enzyme specific activity increased slightly from 1543 to 1684 U/mg, and then decreased for copy number 4 (1018 U/mg) and 8 (707 U/mg). The lower proportion of target protein in total protein may account for the reduced enzyme activity and specific activity for target gene copy numbers 4 and 8 in comparison with copy number 2. These findings indicate that 2 is the optimal copy number for prml expression in strain X-33. The 2-copy recombinant strain had the highest enzyme activity, extracellular protein content, and specific activity. Although qPCR analysis showed the highest target gene transcription level for the 4-copy strain (Figure 3B), our experiments showed that this strain did not have the maximal enzyme activity. Possible reasons for the advantage of the 2-copy strain over the 4-copy or 8-copy strain in terms of protein modification, transport pathways, and unfolded protein response (UPR)-related genes are considered in the ‘Discussion’ section. In conclusion, the lipase with maximal enzyme activity was secreted by the 2-copy strain (mα-2pRML-X33). This enzyme was termed Lipase GH2 and used in subsequent experiments.
Figure 3.
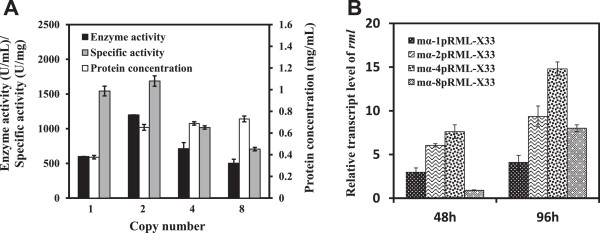
Comparison of recombinant strains containing 1, 2, 4, and 8 gene copies of prml. A: After 96 hours incubation at 30°C, mα-2pRML-X33 displayed higher enzyme activity (1200 U/mL), specific activity (1684 U/mg), and protein concentration (0.770 mg/mL) in the fermentation supernatant. B: rml transcription level in the 1-, 2-, 4, and 8-copy strains. At both 48 and 96 hours, transcription level was highest in the 4-copy strain and lower in the 8-copy strain.
Enzymatic characterization of Lipase GH2
Detection of glycosylation
Bioinformatics analysis indicated glycosylation sites in the propeptide region of the target gene. We therefore evaluated glycosylation level of the protein. zα-mRML-X33 (mRML) and mα-2pRML-X33 (pRML; Lipase GH2) were selected for the deglycosylation reaction. The extracellular target protein was analyzed by Western blotting. Because of the addition of its propeptide a broad molecular weight (MW) of 55 to 70 kDa was found for Lipase GH2 (Additional file 4: Figure S3, lane 2). Deglycosylation of Lipase GH2 by Endo Hf resulted in MW of approximately 43 kDa (Additional file 4: Figure S3, lane 3). The same MW is seen in lanes 4 and 5 of Additional file 4: Figure S3 because the enzyme (mRML) lack the propeptide; there is no effect on MW regardless of the deglycosylation treatment for the proteins.
Enzyme properties
Various properties (optimal temperature and pH, temperature and pH tolerance, methanol and ethanol tolerance) were compared among two of the newly constructed strains (mα-mRML-X33, mα-2pRML-X33) and a previously constructed strain (RML-GS115). The results are shown in Table 1. The most striking difference was the lower optimal temperature of Lipase GH2 (35°C) as compared with mRML (45°C). Lipase GH2, in comparison with mRML, also had approximately 2-fold higher values of temperature tolerance (0 to 70°C versus 0 to 40°C), methanol tolerance (0 to 40% versus 0 to 20%, v/v), and ethanol tolerance (0 to 20% versus 0 to 10%, v/v). These findings indicate adding the propeptide caused glycosylation that results in improved enzyme thermostability and ability to tolerate adverse environmental factors.
Table 1.
Enzymatic properties of R. miehei lipase expressed in three forms
| Optimal temperature (°C) | Temperature tolerance (°C) | Optimal pH | pH tolerance | Methanol tolerance (%) | Ethanol tolerance (%) | |
|---|---|---|---|---|---|---|
| Lipase GH2 (X-33) | 35 | 0-70 | 6.0 | 4-10 | 0-40 | 0-30 |
| mRML (X-33) | 45 | 0-40 | 8.6 | 4-10 | 0-20 | 0-10 |
| RML (GS115) | 50 | 0-50 | 8.0 | 4-9 | 0-30 | 0-20 |
Conversion of microalgae oil to biodiesel using Lipase GH2
Microalgae (Chlorella vulgaris) powder was kindly provided by Dr Peng Pu (State Key Laboratory of Catalytic Material and Reaction Engineering, Research Institute of Petroleum Processing, SINOPEC, Beijing, China). We extracted total lipids from the powder by the method of Tran et al. [36] and analyzed the fatty acid composition by gas chromatography [37]. Results are shown in Additional file 5: Table S2. The microalgae oil extracted from 1 g microalgae powder contained 508.26 mg fatty acids, of which C16 to C18 fatty acids comprised 487.52 mg (96% of the total). Therefore, microalgae oil is a potentially useful substrate for preparation of biodiesel.
We previously described a practical design for the conversion of soybean oil to biodiesel using a combination of two enzymes [34]. In the present study, we used two treatment strategies for the conversion of microalgae oil to biodiesel using Lipase GH2: (1) Lipase GH2 as a catalyst; (2) combined catalysis using two enzymes (Lipase GH2 and a non-specific mono/diacylglycerol lipase (MDL)) in a single reaction. The reaction conditions are described in the ‘Materials and Methods’ section. Gas chromatography (GC) and thin-layer chromatography (TLC) were used for product detection.
Substrate changes detected by GC at various times during methanolysis reaction of microalgae oil are shown in Figure 4A. For treatments 1 and 2 as above, substrate conversion rates after 24 hours reaction at 30°C were 91% and 89%, respectively. Most of the substrate (microalgae oil) was converted to biodiesel, and there was no significant difference between treatments 1 and 2.
Figure 4.
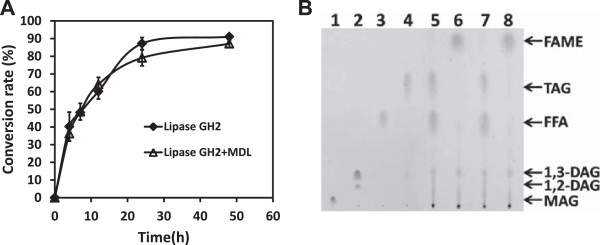
Detection of esterification reaction products by GC and TLC. A: GC detection. The substrate (microalgae oil) more than 90% conversion to biodiesel (FAME) after esterification reaction for 24 hours at 30°C. There was no significant difference between reactions catalyzed by a single lipase (Lipase GH2) vs. a combination of two lipases (Lipase GH2 and MDL). B: TLC detection. Lane: 1-monooleate (MAG). Lane 2: dioleoylglycerol (1,3- and 1,2-diacylglycerol). Lane 3: oleic acid (FFA). Lane 4: triacylglycerol (TAG). Lanes 5–6: single-lipase (Lipase GH2) treatment at 0 h and 24 h. Lanes 7–8: two-lipase (Lipase GH2 and MDL) treatment at 0 and 24 hours.
Similar results were obtained for each sample by TLC (Figure 4B). Standard samples (shown in lanes 1 to 4) were monoacylglycerol (MAG), diacylglycerol (DAG), free fatty acid (FFA) and triacylglycerol (TAG). Fatty acids are the major components of microalgae oil. Substrates (FFA and TAG) prior to esterification reaction are shown in lanes 5 and 7. After the final reaction (24 hours at 30°C), the substrates were almost completely converted to fatty acid methyl esters (FAME; lanes 6 and 8), and there was no significant difference between treatment 1 (lanes 5 and 6) and treatment 2 (lanes 7 and 8).
In conclusion, Lipase GH2 can be used as an effective catalyst for the conversion of microalgae oil to biodiesel with no requirement for a second enzyme as an auxiliary factor.
Discussion
Adding propeptide induced protein glycosylation and improved enzyme tolerance
Heterologous protein expression is affected by numerous factors, including the expression vector, host strain, gene structure, signal peptide, gene dosage, and culture conditions for recombinants [38, 39]. In this study, we successfully constructed a high-expressing recombinant strain through propeptide addition, optimized codon usage of the signal peptide, and optimized gene dosage. Glycosylation has been shown to promote enzyme stability [40]. In the case of subtilisins, the 77-residue propeptide acts as an intramolecular chaperone that organizes the correct folding of its own protease domain; the occurrence of a mutation (Ile (-48)-to-Val) in the propeptide results in two subtilisins differing in secondary structure, thermostability, and substrate specificity [41]. Based on these previous findings, we hypothesized that the 70-residue propeptide of R. miehei lipase (RML) may also have affected the enzyme folding or post translational modification. An advantage of P. pastoris as an expression system is its ability to undergo eukaryotic post-translational modifications, including glycosylation of heterologous proteins [12]. We found that the broad MW of Lipase GH2 observed on Western blotting is a result of glycosylation. Amino acid sequence analysis revealed two potential N-glycosylation sites (Asn-Xaa-Ser/Thr), both located in the propeptide sequence; they were identified by NetNGlyc 1.0 Server analysis (http://www.cbs.dtu.dk/services/NetNGlyc) at the 8th position with the NST(Asn-Ser-Thr) motif and the 58th position with the NAT(Asn-Ala-Thr) motif. Glycosylation enhanced the ability of Lipase GH2 to tolerate temperature, methanol, and ethanol, and to catalyze the enzymatic conversion of microalgae oil to biodiesel. Thus, in the case of Lipase GH2, addition of heterologous gene propeptide provides a dual benefit: it enhances enzyme tolerance and increases enzyme activity.
Why are two target gene copies better than four copies?
In secretory pathways, cells use specialized quality control systems to ensure that only properly folded and modified proteins are secreted into the extracellular space; unfolded or incorrectly folded (misfolded) proteins are reverse-transported to the cytoplasm by the ER degradation system or the lysosomal degradation system [42]. Correct folding of heterologous proteins is crucial in that it determines enzyme activity and functions.
How does integration of different numbers of target gene copies in recombinant strains affect protein transcription levels, folding, and secretion? We used qPCR detection to study expression of genes previously reported to be involved in protein folding, modification, and translocation pathways in yeast (see Additional file 6: Table S3). We focused on prml gene expression to examine the effect of target gene copy number.
We observed an association of four unfolded protein response (UPR)-related genes that have been described previously [43]. PDI, located in the ER membrane, encodes protein disulfide isomerase which is responsible for protein disulfide bond formation. ERO1, located in the ER membrane, is an ‘assistant’ of PDI, responsible for the glycoprotein required for oxidative protein folding in the ER. KAR2, located in the ER lumen, is a major Hsp70 chaperone. HAC1, located in the nucleus, is a transcription activator that regulates the UPR via UPRE (UPR element) binding and membrane biogenesis. Stress to the ER triggers increased expression of these genes and possible relief of the stress through the gene products.
Results for detection of these genes in strains zα-1mRML-X33 (not optimized) and mα-2pRML-X33 (optimized) are shown in Figure 5A. Transcriptional levels of all four genes were higher in zα-1mRML-X33 than that in mα-2pRML-X33. These findings suggest that some mRMLs may be expressed with abnormal structure or misfolded proteins, thereby inducing ER stress and initiating UPR.
Figure 5.

Transcription levels of four UPR-related genes in different recombinants. A: Transcription levels of HAC1, KAR2, PDI, and ERO1 were lower in mα-2pRML-X33 and higher in zα-1mRML-X33. B: Transcription levels of four UPR-related genes in the 2-copy vs. 4-copy strains. The transcriptional levels of four genes were all higher in 4-copy strain than that of 2-copy strain.
After finding that enzyme activity and target gene transcription level were lowest for the 1-copy and 8-copy recombinant strains (see Results - ‘Optimization of the target gene dosage’), we compared target gene expression in the 2-copy and 4-copy strains (Figure 5B). In the 4-copy strain, the transcriptional levels of four UPR-related genes were all higher than that of 2-copy strain, it seems that protein folding generated obstacles and was unable to improve enzyme activity or protein secretion. Thus, although the 4-copy strain (mα-4pRML-X33) had a higher transcription level of the target gene than did the 2-copy strain, the abnormal protein resulted in ER stress, abnormal protein assembly and modification, and triggering of UPR (upregulation of UPR-related genes). However, the UPR did not relieve the ER stress. These findings illustrate why 2 target gene copies in the host are better than 4 copies.
It is important to note that there was no positive correlation between gene dosage and the target protein yield in construction of a heterologous protein expression system. Rather, the bearing capacity of the host cell must be considered. Abnormal protein folding and a variety of stress factors may interfere with protein polymerization in the ER lumen, leading to accumulation of unfolded or misfolded proteins and consequent ER stress [44–46]. If the stress is too high, it cannot be relieved by upregulation of UPR-related genes.
Lipase GH2 is a useful catalyst for biodiesel synthesis
Lipase can be used in free or immobilized form in the biodiesel preparation process. The immobilized form increases the chances of re-usability of biocatalysts, preserving enzyme activities over several reaction cycles and can be removed easily from the mixture and can be reused in future. While the free enzyme (fermentation supernatant) in reaction process can be directly used which to avoid the cost of immobilized enzyme [47–49]. We previously described two successful methods for conversion of soybean oil to biodiesel catalyzed by free lipase: (i) a two-step reaction using Yarrowia lipolytica lipase which can be used by 25 cycles to produce fatty acid ethyl ester [35], and (ii) a one-step reaction using a combination of RML and MDL to produce FAME [34]. Soybean oil consists primarily of TAG. The RML lipase (RML-GS115) is 1,3-specific and cannot break a two-ester bond. MDL is able to break a two-ester bond, and the two enzymes are able to cooperate in the transesterification reaction. Use of soybean oil as substrate in a reaction system for biodiesel synthesis involves both hydrolysis reaction (hydrolysis of fat into fatty acids and glycerol) and esterification reaction (synthesis of fatty acids and methanol into FAME). Use of the new expression system (mα-2pRML-X33) constructed in this study resulted in a more than 20-fold increase in target protein expression. The enzymatic properties of the new system (improved temperature, methanol, and ethanol tolerance) are more suitable for application in biodiesel preparation. Microalgae oil consists of FFA, TAG, DAG, and MAG, with FFA being the major component. Biodiesel preparation requires methyl esterification by lipase, for which the newly constructed Lipase GH2 is suitable. The glycosylated protein strongly retains its enzyme activity; when we kept the fermentation supernatant at 4°C and sampled the lipase activity twice per month, we found no reduction in the activity after six months. For microalgae oil as substrate, the fermentation supernatant (free lipase) functions as a direct catalyst in the esterification and transesterification reaction, and achieves good results. This design has the advantages of simplifying the production process, saving time, and avoiding the expense of immobilized enzyme.
The use of Lipase GH2 as a catalyst in methanolysis of microalgae oil for biodiesel synthesis is economical for industrial applications and has great potential. Our ongoing studies are focused on further improvement and optimization of enzyme quality, reaction conditions, reaction rate, and industrial applicability.
Conclusions
Various strategies are available for improving the efficiency of heterologous protein production by P. pastoris. Integration of too many copies of the target gene in the host genome may lead to increased ER stress and reduced protein production. In this study, we significantly increased the RML expression level and modified enzymatic properties through optimal expression plasmid with signal peptide, addition of the target gene propeptide, and optimal exogenous gene dosage. Lipase GH2, the major component of the fermentation supernatant, is stable for over six months at 4°C and is an effective enzyme for catalytic conversion of microalgae oil to biodiesel. Further studies will lead to improved industrial productivity.
Materials and Methods
Gene cloning and construction of multimers in vitro
The prml and mrml genes were cloned from R. miehei lipase (GenBank accession number A02536.1) cDNA using two pairs of primers. R. miehei pro-mature lipase gene (prml) was amplified using the primer pairs pRML-f (5′-CCGGAATTCGTGCCAATCAAGAG-3′, EcoRI site) and RML-r (5′-CTAGTCTAGAGTACAGAGGCCTGTG-3′, Xba I site). Mature lipase gene (mrml) was amplified using the primer pairs mRML-f (5′-CCGGAATTCAGCATTGATGGTGG-3′, EcoR I site) and RML-r (5′-CTAGTCTAGAGTACAGAGGCCTGTG-3′, Xba I site). The italics indicate the enzymatic restriction sites. Two vectors were used: pPICZαA (Invitrogen; Carlsbad, California, United States), and pPICMαA (a pPICZαA-derived plasmid). The only difference between the two vectors is that the α-factor codons of pPICMαA are optimized, without change of the amino acid sequence. The sequences of the original and modified signal peptide were kindly provided by Dr Jia Ban; their differences are shown in Additional file 1: Figure S1. Single-copy recombinant plasmids pPICZa-1prml, pPICMa-1prml, and pPICZa-1mrml were constructed. pPICMaA plasmids containing various gene dosages (pPICMa-2prml, pPICMa-4prml, and pPICMa-8prml) were constructed as described by Menendez et al. [17]. The plasmids and strains used in this study are listed in Table 2.
Table 2.
Plasmids and strains used in this study
| Description | Source | |
|---|---|---|
| Plasmids | ||
| pPICZα A | Secretion expression vector with α-factor from S. cerevisiae | Donated by Dr Yijun Huang |
| pPICMα A | pPICZα A with α-factor optimized codons | Donated by Dr Jia Ban |
| Strains | ||
| X-33 | Host strain (WT Mut+) | Donated by Dr Yijun Huang |
| zα-1mRML-X33 | One copy mRML of the lipase without propeptide expressed in X-33 using pPICZα A | This study |
| zα-1pRML-X33 | One copy pPML of the lipase with propeptide expressed in X-33 using pPICZα A | This study |
| mα-1pRML-X33 | One copy pRML expressed in X-33 using pPICMα A | This study |
| mα-2pRML-X33 | Two copies pRML expressed in X-33 using pPICMα A | This study |
| mα-4pRML-X33 | Four copies pRML expressed in X-33 using pPICMα A | This study |
| mα-8pRML-X33 | Eight copies pRML expressed in X-33 using pPICMα A | This study |
Screening for target transformants
The recombinant plasmids were transformed into X-33 and screened with Yeast Extract Peptone Dextrose Medium with Sorbitol (YPDS) medium as described by Hu et al. [50], except that the multi-copy recombinant plasmids could not be linearized before transformation into X-33 by electroporation. For each transformation, 50 positive transformants were selected by tributyrin flat screening. Eight strains were selected for each transformant based on the size of the transparent circle in fermentation. Positive transformants were flask-cultured in BMGY/BMMY medium as described by Hu et al. [50]. Every day, 1% methanol (Sinopharm Chemical Reagent Beijing Co., Ltd, Beijing, China) was used to induce protein expression and activity and production were measured for each strain.
Absolute quantification of target gene copies was performed by qPCR using SYBR Green I Mix (Roche, Mannheim, Germany). qPCR was conducted in a LightCycler 480 RT-PCR System(Roche, Mannheim, Germany) using LightCycler 480 SYBR Green I Master Kit (Roche). Specific primers purified by HPLC (Invitrogen) were designed to yield 100 to 250 bp products. All primers (rml and gap) used for qPCR are listed in Additional file 6: Table S3. Genomic DNA of P. pastoris was used as template. The genomic DNA of the single-copy strain was used to establish the standard curve (Additional file 2: Figure S2). The C(t) values of the target gene (prml) and reference gene (gap) are shown in Additional file 3: Table S1. The numbers of target gene copies were calculated by absolute quantification as described by Abad et al. [51], using the following formula:
| 1 |
Transcription levels of genes related to various steps in protein expression, modification, and secretory pathways
There were a total of 14 genes related to the target lipase, UPR, protein transport, and protein secretion. RNA extraction and relative qPCR analysis were performed as described by Wang et al. [52]. The primers used for qPCR are listed in Additional file 6: Table S3.
Western blotting and glycosyl chain detection
The recombinant lipase was tested by Western blotting as described by Shen et al. [16]. sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 5% stacking gel and 12% resolving gel. Monoclonal mouse-anti-His-Tag antibody (Tiangen, Beijing, China) was used as the primary antibody and goat anti-mouse IgG antibody conjugated to alkaline phosphatase (Sigma-Aldrich, St. Louis, Missouri, United States) as the secondary antibody. The membrane was applied to a BCIP/NBT Chromogenic reagent kit (Tiangen) according to the manufacturer’s instructions.
Protein concentration was quantified by Bradford assay [53] using Bovine Serum Albumin (BSA) as a standard. Glycosyl chains in pRML were removed by Endo Hf (New England Biolabs; Beverly, Massachusetts, United States) according to the manufacturer’s instructions.
Enzyme characterization
Lipase hydrolysis activity was measured as described in our previous study [35], except that we used the conditions 0.1 M sodium dihydrogen phosphate/citric acid, pH 6.0, 35°C for Lipase GH2 and 0.1 M CHES, pH 8.6, 45°C for mRML. Optimal pH values for Lipase GH2 and mRML were determined under standard conditions. The buffers used were 0.1 M sodium dihydrogen phosphate/citric acid (pH 3.12 to 8.17), 0.1 M Trihydroxymethyl aminomethane- hydrochloric acid (Tris-HCl) (pH 8.02-9.03), 0.1 M N-Cyclohexyl-2-aminoethanesulfonic acid (CHES) (pH 8.6-9.5), and 0.1 M N-Cyclohexyl-3-aminopropanesulfonic acid (CAPS) (pH 10.0-11.0). pH stability was determined at pH values ranging from 1.0 to 12.7 for 90 minutes, and residual activity was measured. Optimal temperatures were determined under standard conditions in the range 0 to 60°C. Thermostability was determined for 90 minutes at temperatures ranging from 0 to 80°C, and residual activity was measured.
Biodiesel preparation and detection
Lipids were extracted from microalgae powder as described by Tran et al. [36], except that chloroform/methanol (1:2 v/v) (Sinopharm Chemical Reagent Beijing Co., Ltd, Beijing, China) was used as solvent. Oil extracted from microalgae powder was used as a substrate to produce biodiesel using the organic solvent n-hexane (Sinopharm Chemical Reagent Beijing Co., Ltd, Beijing, China). Production conditions were: 0.1 g of microalgae oil/30 μL methanol, 1 mL n-hexane, 55 μL Lipase GH2 (2-copy recombinant strain) (1200 U/mL) and 5 μL MDL (1500 U/mL) (for Lipase GH2 alone, MDL was replaced by H2O), incubation at 30°C and shaking at 150 rpm for 24 hours.
FAME in the reaction mixture were analyzed using a GC system (model 7890A, Agilent Technologies (Santa Clara, California, United States) equipped with a capillary column (CP-FFAP CB; 25 m × 0.32 mm × 0.3 μm; Agilent Technology). Sample preparation and detection conditions were as described by Ren et al. [54]. TLC was performed as in our previous study [35].
Electronic supplementary material
Additional file 1: Figure S1: Sequence alignment of original signal peptide (zα) and modified signal peptide (mα) using DNAMAN program (Lynnon Corporation, United States). 46 codons in the modified signal peptide were optimized, resulting in changes of 50 nucleotides. G + C content increased from 41.1 to 47.0%. The modified signal peptide showed 82.5% identity with the original signal peptide (zα) after averaging the distribution of both the G + C content and the optimized codons. (PDF 186 KB)
Additional file 2: Figure S2: Standard curves of gap and rml detected by qPCR. (PDF 155 KB)
Additional file 3: Table S1: Target gene copy number in four recombinant strains. (PDF 144 KB)
Additional file 4: Figure S3: Detection of glycosylation of lipases. Deglycosylation treatment using Endo Hf showed that pRML (Lipase GH2) was a glycosylated protein, whereas mRML had no glycosylation. Lane 1: protein markers (top to bottom: 100, 70, 55, 40, 35, 25, 15 kDa). Lane 2: Lipase GH2 without Endo Hf (no deglycosylation). Lane 3: Lipase GH2 with Endo Hf (with deglycosylation). Lane 4: mRML with Endo Hf (with deglycosylation). Lane 5: mRML without Endo Hf (no deglycosylation). (PDF 112 KB)
Additional file 5: Table S2: Fatty acid analysis of microalgae oil by HPLC. (PDF 169 KB)
Additional file 6: Table S3: Genes and primers used for qPCR. (PDF 183 KB)
Acknowledgments
This study was supported by the Chinese High Technology Research and Development Program (Grant No. 2013AA065802), Chinese Scientific and Technical Supporting Programs (2013BAD10B01) and the Undergraduate Innovation Program of China Agricultural University (2013 to 2014). The authors are grateful to Drs Yijun Huang and Jia Ban for providing strain X-33 and plasmid pPICZαA, Professor Dehua Liu (Tsinghua University) for useful suggestions and help with product detection, and Dr S Anderson for English editing of the manuscript.
Abbreviations
- 1
2-DAG: 1,2-diacylglycerol
- 1
3-DAG: 1,3-diacylglycerol
- FAME
fatty acid methyl ester(s)
- FFA
free fatty acid
- MAG
monoacylglycerol
- TAG
triacylglycerold.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YL and JJH designed the study. JJH, JX, ZY, and FFG carried out molecular genetic studies. JJH, JX and DC performed the experiments of enzymatic properties. JJH and JX carried out the conversion of microalgae oil to biodiesel. GHG and WJ assisted with analysis and discussion of the results. JJH and YL wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jinjin Huang, Email: bingning521521@126.com.
Ji Xia, Email: sumdreamer1993@sina.com.
Zhen Yang, Email: 58yangzhen@163.com.
Feifei Guan, Email: guanfeifei@sina.com.
Di Cui, Email: 1002040312@cau.edu.cn.
Guohua Guan, Email: ghguan@cau.edu.cn.
Wei Jiang, Email: Jiangwei6712@sina.com.
Ying Li, Email: yingli528@vip.sina.com.
References
- 1.Rodrigues RC, Fernandez-Lafuente R. Lipase from Rhizomucor miehei as a biocatalyst in fats and oils modification. J Mol Catal B: Enzym. 2010;66:15–32. doi: 10.1016/j.molcatb.2010.03.008. [DOI] [Google Scholar]
- 2.Stobiecka A. Acrylamide-quenching of Rhizomucor miehei lipase. J Photochem Photobiol B. 2005;80:9–18. doi: 10.1016/j.jphotobiol.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Brady L, Brzozowski AM, Derewenda ZS, Dodson E, Dodson G, Tolley S, Turkenburg JP, Christiansen L, Hugejensen B, Norskov L, Thim L, Menge U. A serine protease triad forms the catalytic center of a triacylglycerol lipase. Nature. 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 4.Boel E, Huge-Jensen B, Christensen M, Thim L, Fiil NP. Rhizomucor miehei triglyceride lipase is synthesized as a precursor. Lipids. 1988;23:701–706. doi: 10.1007/BF02535672. [DOI] [PubMed] [Google Scholar]
- 5.Norin M, Olsen O, Svendsen A, Edholm O, Hult K. Theoretical-studies of Rhizomucor-miehei lipase activation. Protein Eng. 1993;6:855–863. doi: 10.1093/protein/6.8.855. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues RC, Fernandez-Lafuente R. Lipase from Rhizomucor miehei as an industrial biocatalyst in chemical process. J Mol Catal B: Enzym. 2010;64:1–22. doi: 10.1016/j.molcatb.2010.02.003. [DOI] [Google Scholar]
- 7.Palla CA, Pacheco C, Carrin ME. Production of structured lipids by acidolysis with immobilized Rhizomucor miehei lipases: selection of suitable reaction conditions. J Mol Catal B: Enzym. 2012;76:106–115. doi: 10.1016/j.molcatb.2011.11.022. [DOI] [Google Scholar]
- 8.De Diego T, Lozano P, Abad MA, Steffensky K, Vaultier M, Iborra JL. On the nature of ionic liquids and their effects on lipases that catalyze ester synthesis. J Biotechnol. 2009;140:234–241. doi: 10.1016/j.jbiotec.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Acosta A, Filice M, Fernandez-Lorente G, Palomo JM, Guisan JM. Kinetically controlled synthesis of monoglyceryl esters from chiral and prochiral acids methyl esters catalyzed by immobilized Rhizomucor miehei lipase. Bioresour Technol. 2011;102:507–512. doi: 10.1016/j.biortech.2010.08.095. [DOI] [PubMed] [Google Scholar]
- 10.Huang DF, Han SY, Han ZL, Lin Y. Biodiesel production catalyzed by Rhizomucor miehei lipase-displaying Pichia pastoris whole cells in an isooctane system. Biochem Eng J. 2012;63:10–14. doi: 10.1016/j.bej.2010.08.009. [DOI] [Google Scholar]
- 11.Gasser B, Prielhofer R, Marx H, Maurer M, Nocon J, Steiger M, Puxbaum V, Sauer M, Mattanovich D. Pichia pastoris: protein production host and model organism for biomedical research. Future Microbiol. 2013;8:191–208. doi: 10.2217/fmb.12.133. [DOI] [PubMed] [Google Scholar]
- 12.De Pourcq K, De Schutter K, Callewaert N. Engineering of glycosylation in yeast and other fungi: current state and perspectives. Appl Microbiol Biotechnol. 2010;87:1617–1631. doi: 10.1007/s00253-010-2721-1. [DOI] [PubMed] [Google Scholar]
- 13.Tian B, Chen Y, Ding SJ. A combined approach for improving alkaline acetyl xylan esterase production in Pichia pastoris, and effects of glycosylation on enzyme secretion, activity and stability. Protein Expr Purif. 2012;85:44–50. doi: 10.1016/j.pep.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Yu MR, Wen S, Tan TW. Enhancing production of Yarrowia lipolytica lipase Lip2 in Pichia pastoris. Eng Life Sci. 2010;10:458–464. doi: 10.1002/elsc.200900102. [DOI] [Google Scholar]
- 15.Sha C, Yu XW, Li F, Xu Y. Impact of gene dosage on the production of Lipase from Rhizopus chinensis CCTCC M201021 in Pichia pastoris. Appl Biochem Biotechnol. 2013;169:1160–1172. doi: 10.1007/s12010-012-0050-9. [DOI] [PubMed] [Google Scholar]
- 16.Shen Q, Wu M, Wang HB, Naranmandura H, Chen SQ. The effect of gene copy number and co-expression of chaperone on production of albumin fusion proteins in Pichia pastoris. Appl Microbiol Biotechnol. 2012;96:763–772. doi: 10.1007/s00253-012-4337-0. [DOI] [PubMed] [Google Scholar]
- 17.Menendez C, Martinez D, Trujillo LE, Mazola Y, Gonzalez E, Perez ER, Hernandez L. Constitutive high-level expression of a codon-optimized beta-fructosidase gene from the hyperthermophile Thermotoga maritima in Pichia pastoris. Appl Microbiol Biotechnol. 2013;97:1201–1212. doi: 10.1007/s00253-012-4270-2. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Wang D, Wang B, Mei ZH, Liu J, Yu HW. Enhanced activity of Rhizomucor miehei lipase by directed evolution with simultaneous evolution of the propeptide. Appl Microbiol Biotechnol. 2012;96:443–450. doi: 10.1007/s00253-012-4049-5. [DOI] [PubMed] [Google Scholar]
- 19.Taher H, Al-Zuhair S, Al-Marzouqi AH, Haik Y, Farid MM. A review of enzymatic transesterification of microalgal oil-based biodiesel using supercritical technology. Enzyme Research. 2011;2011:1–25. doi: 10.4061/2011/468292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad AL, Yasin NHM, Derek CJC, Lim JK. Microalgae as a sustainable energy source for biodiesel production: a review. Renew Sust Energ Rev. 2011;15:584–593. doi: 10.1016/j.rser.2010.09.018. [DOI] [Google Scholar]
- 21.Demirbas MF. Biofuels from algae for sustainable development. Appl Energy. 2011;88:3473–3480. doi: 10.1016/j.apenergy.2011.01.059. [DOI] [Google Scholar]
- 22.Hong-Yu R, Bing-Feng L, Chao M, Lei Z, Nan-Qi R. A new lipid-rich microalga Scenedesmus sp. strain R-16 isolated using Nile red staining: effects of carbon and nitrogen sources and initial pH on the biomass and lipid production. Biotechnol Biofuels. 2013;6:143. doi: 10.1186/1754-6834-6-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaimes GG, Vikas K. Microalgal biomass production pathways: evaluation of life cycle environmental impacts. Biotechnol Biofuels. 2013;6:88. doi: 10.1186/1754-6834-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caroline C, Boris M, Hoa Mai N, Audrey B-A, Séverine B, Stéphan C, Fred B, Gilles P, Yonghua L-B. Development of a forward genetic screen to isolate oil mutants in the green microalga Chlamydomonas reinhardtii. Biotechnol Biofuels. 2013;6:178. doi: 10.1186/1754-6834-6-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonardo Brantes Bacellar M, Alane Beatriz V. Allelopathy as a potential strategy to improve microalgae cultivation. Biotechnol Biofuels. 2013;6:152. doi: 10.1186/1754-6834-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguirre AM, Bassi A. Investigation of biomass concentration, lipid production, and cellulose content in Chlorella vulgaris cultures using response surface methodology. Biotechnol Bioeng. 2013;110:2114–2122. doi: 10.1002/bit.24871. [DOI] [PubMed] [Google Scholar]
- 27.Dillschneider R, Steinweg C, Rosello-Sastre R, Posten C. Biofuels from microalgae: photoconversion efficiency during lipid accumulation. Bioresour Technol. 2013;142:647–654. doi: 10.1016/j.biortech.2013.05.088. [DOI] [PubMed] [Google Scholar]
- 28.Lai JQ, Hu ZL, Wang PW, Yang Z. Enzymatic production of microalgal biodiesel in ionic liquid [BMIm] [PF6] Fuel. 2012;95:329–333. doi: 10.1016/j.fuel.2011.11.001. [DOI] [Google Scholar]
- 29.Prathima DM, Swamy YV, Venkata MS. Nutritional mode influences lipid accumulation in microalgae with the function of carbon sequestration and nutrient supplementation. Bioresour Technol. 2013;142:278–286. doi: 10.1016/j.biortech.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Qing P, Xue Z, Meng S, Xu W, Guili W, Bingxue L, Guohua G, Ying L, Youshao W. A novel esterase gene cloned from a metagenomic library from neritic sediments of the South China Sea. Microb Cell Fact. 2011;10:95. doi: 10.1186/1475-2859-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao-Wei Y, Chong S, Yong-Liang G, Rong X, Yan X. High-level expression and characterization of a chimeric lipase from Rhizopus oryzae for biodiesel production. Biotechnol Biofuels. 2013;6:29. doi: 10.1186/1754-6834-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helge Jans J, Alexander S. Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol Biofuels. 2014;7:7. doi: 10.1186/1754-6834-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korman TP, Bobby S, Charbonneau DM, Huang GL, Marc B, Bowie JU. Dieselzymes: development of a stable and methanol tolerant lipase for biodiesel production by directed evolution. Biotechnol Biofuels. 2013;6:70. doi: 10.1186/1754-6834-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan FF, Peng P, Wang GL, Yin T, Peng Q, Huang JJ, Guan GH, Li Y. Combination of two lipases more efficiently catalyzes methanolysis of soybean oil for biodiesel production in aqueous medium. Process Biochem. 2010;45:1677–1682. doi: 10.1016/j.procbio.2010.06.021. [DOI] [Google Scholar]
- 35.Yonghong M, Guili W, Na Y, Zhiqi Z, Yuejuan L, Xiaomei L, Jinnan C, Ying L, Jilun L. Two-step synthesis of fatty acid ethyl ester from soybean oil catalyzed by Yarrowia lipolytica lipase. Biotechnol Biofuels. 2011;4:6. doi: 10.1186/1754-6834-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran DT, Yeh KL, Chen CL, Chang JS. Enzymatic transesterification of microalgal oil from Chlorella vulgaris ESP-31 for biodiesel synthesis using immobilized Burkholderia lipase. Bioresour Technol. 2012;108:119–127. doi: 10.1016/j.biortech.2011.12.145. [DOI] [PubMed] [Google Scholar]
- 37.Li ZS, Yuan HL, Yang JS, Li BZ. Optimization of the biomass production of oil algae Chlorella minutissima UTEX2341. Bioresour Technol. 2011;102:9128–9134. doi: 10.1016/j.biortech.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Gasser B, Sauer M, Maurer M, Stadlmayr G, Mattanovich D. Transcriptomics-based identification of novel factors enhancing heterologous protein secretion in Yeasts. Appl Environ Microbiol. 2007;73:6499–6507. doi: 10.1128/AEM.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Idiris A, Tohda H, Kumagai H, Takegawa K. Engineering of protein secretion in yeast: strategies and impact on protein production. Appl Microbiol Biotechnol. 2010;86:403–417. doi: 10.1007/s00253-010-2447-0. [DOI] [PubMed] [Google Scholar]
- 40.Potvin G, Ahmad A, Zhang Z. Bioprocess engineering aspects of heterologous protein production in Pichia pastoris: a review. Biochem Eng J. 2012;64:91–105. doi: 10.1016/j.bej.2010.07.017. [DOI] [Google Scholar]
- 41.Shinde UP, Liu JJ, Inouye M. Protein memory through altered folding mediated by intramolecular chaperones. Nature. 1997;389:520–522. doi: 10.1038/39097. [DOI] [PubMed] [Google Scholar]
- 42.Trombetta ES, Parodi AJ. Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- 43.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelham HRB. Control of protein exit from the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:1–23. doi: 10.1146/annurev.cb.05.110189.000245. [DOI] [PubMed] [Google Scholar]
- 45.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 46.Ma Y, Hendershot LM. The unfolding tale of the unfolded protein response. Cell. 2001;107:827–830. doi: 10.1016/S0092-8674(01)00623-7. [DOI] [PubMed] [Google Scholar]
- 47.Xu WD, Jing Z, Dehua Liu Y. Conversion of soybean oil to biodiesel fuel using lipozyme TL IM in a solvent-free medium. Biocatal Biotransfor. 2004;22:45–48. doi: 10.1080/10242420410001661222. [DOI] [Google Scholar]
- 48.Poppe JK, Garcia-Galan C, Matte CR, Fernandez-Lafuente R, Rodrigues RC, Ayub MAZ. Optimization of synthesis of fatty acid methyl esters catalyzed by lipase B from Candida antarctica immobilized on hydrophobic supports. J Mol Catal B-Enzym. 2013;94:51–56. doi: 10.1016/j.molcatb.2013.05.010. [DOI] [Google Scholar]
- 49.Zhang B, Weng Y, Xu H, Mao Z. Enzyme immobilization for biodiesel production. Appl Microbiol Biotechnol. 2012;93:61–70. doi: 10.1007/s00253-011-3672-x. [DOI] [PubMed] [Google Scholar]
- 50.Hu H, Gao J, He J, Yu B, Zheng P, Huang Z, Mao X, Yu J, Han G, Chen D. Codon optimization significantly improves the expression level of a keratinase gene in Pichia pastoris. PLoS One. 2013;8:3. doi: 10.1371/annotation/03b700e2-348b-4b1c-b068-4760c32de19e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abad S, Kitz K, Hormann A, Schreiner U, Hartner FS, Glieder A. Real-time PCR-based determination of gene copy numbers in Pichia pastoris. Biotechnol J. 2010;5:413–420. doi: 10.1002/biot.200900233. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q, Liu JX, Zhang WJ, Zhang TW, Yang J, Li Y. Expression patterns of key iron and oxygen metabolism genes during magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. FEMS Microbiol Lett. 2013;347:163–172. doi: 10.1111/1574-6968.12234. [DOI] [PubMed] [Google Scholar]
- 53.Bradford MM. Rapid and sensitive method for the quantitation ofmicrogram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 54.Ren HJ, Du W, Lv LL, Liu DH. Study on free lipase-catalyzed ethanolysis for biodiesel preparation in an oil/water biphasic system. J American Oil Chem Soc. 2011;88:1551–1555. doi: 10.1007/s11746-011-1813-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1: Sequence alignment of original signal peptide (zα) and modified signal peptide (mα) using DNAMAN program (Lynnon Corporation, United States). 46 codons in the modified signal peptide were optimized, resulting in changes of 50 nucleotides. G + C content increased from 41.1 to 47.0%. The modified signal peptide showed 82.5% identity with the original signal peptide (zα) after averaging the distribution of both the G + C content and the optimized codons. (PDF 186 KB)
Additional file 2: Figure S2: Standard curves of gap and rml detected by qPCR. (PDF 155 KB)
Additional file 3: Table S1: Target gene copy number in four recombinant strains. (PDF 144 KB)
Additional file 4: Figure S3: Detection of glycosylation of lipases. Deglycosylation treatment using Endo Hf showed that pRML (Lipase GH2) was a glycosylated protein, whereas mRML had no glycosylation. Lane 1: protein markers (top to bottom: 100, 70, 55, 40, 35, 25, 15 kDa). Lane 2: Lipase GH2 without Endo Hf (no deglycosylation). Lane 3: Lipase GH2 with Endo Hf (with deglycosylation). Lane 4: mRML with Endo Hf (with deglycosylation). Lane 5: mRML without Endo Hf (no deglycosylation). (PDF 112 KB)
Additional file 5: Table S2: Fatty acid analysis of microalgae oil by HPLC. (PDF 169 KB)
Additional file 6: Table S3: Genes and primers used for qPCR. (PDF 183 KB)


