Abstract
Autophagy, the process for recycling cytoplasm in the lysosome, depends on membrane trafficking. We previously identified Drosophila Sbf as a Rab21 guanine nucleotide exchange factor (GEF) that acts with Rab21 in endosomal trafficking. Here, we show that Sbf/MTMR13 and Rab21 have conserved functions required for starvation-induced autophagy. Depletion of Sbf/MTMR13 or Rab21 blocked endolysosomal trafficking of VAMP8, a SNARE required for autophagosome–lysosome fusion. We show that starvation induces Sbf/MTMR13 GEF and RAB21 activity, as well as their induced binding to VAMP8 (or closest Drosophila homolog, Vamp7). MTMR13 is required for RAB21 activation, VAMP8 interaction and VAMP8 endolysosomal trafficking, defining a novel GEF-Rab-effector pathway. These results identify starvation-responsive endosomal regulators and trafficking that tunes membrane demands with changing autophagy status.
Keywords: autophagy, MTMR13, Rab21, Sbf, VAMP8
Introduction
Macroautophagy (hereafter autophagy), the process for degradation and recycling of cytoplasmic contents in the lysosome, is an important homeostatic response to cell stress 1. Autophagy relies on membrane trafficking, whereby cytoplasmic cargo encapsulated in a double-membrane-bound autophagosome is delivered to the lysosome by membrane fusion 2. Although basal levels of autophagy are ongoing in many cell types, various forms of cell stress such as nutrient starvation induce higher levels of autophagy to promote cell survival. Thus, in addition to the signaling components that sense and transduce a starvation-induced autophagy response, the underlying membrane trafficking also must be regulated to accommodate an increased autophagic membrane demand 3.
A conserved hierarchy of core autophagy functions directs autophagosome membrane formation, maturation and lysosomal delivery 1. In addition, there is a complexity of autophagy dependence on nearly all other major cellular membrane organelles and trafficking routes 2. A particularly important relationship exists with endosomes–lysosomes (endolysosomes) for numerous roles in both early and late stages of autophagy. Early endosomes serve as key cargo sorting stations, where endocytosed cargos are directed either through recycling endosomes for redelivery to the plasma membrane or to late endosomes for degradation upon lysosome fusion 4. Endocytic vesicles and recycling endosomes have been found to play roles in early steps of autophagy as additional membrane sources contributing to the pre-autophagosome 2, 5, 6, 7, while late endosomes and lysosomes play integral roles in the final steps of autophagosome degradation 8, 9. Selective membrane fusion has been described between autophagosomes and either multivesicular bodies or late endosomes to form amphisomes that subsequently fuse with lysosomes, or with lysosomes to form autolysosomes 10. Hereafter, we collectively refer to this delivery step as ‘autophagosome–lysosome fusion.’
Further evidence for the link between regulation of endolysosomal membrane identity and autophagy are the shared requirements for trafficking regulators 3. Functions that confer lysosomal membrane identity are similarly required for both endosome–lysosome fusion and autophagosome–lysosome fusion 10. Throughout the cell, dynamic membrane identity is defined by specific combination of Rab GTPases and phosphoinositide lipids under the tight control of regulatory enzymes 11. These in turn recruit specific effectors that mediate trafficking through membrane bending, tethering, fusion and fission. The lysosomal membrane identity is defined in part by Rab7 GTPase, Arl8B Arf-like GTPase, PtdIns(3)P and PtdIns(3,5)P2 12, 13, 14, 15. In both endolysosomal and autophagic trafficking, Rab7 GTPase plays a central role in recruiting effectors of the endosomal sorting complexes required for transport (ESCRT) and homotypic fusion and vacuole protein sorting (HOPS) complex that are involved in membrane tethering and fusion 9, 16, 17, 18, 19, 20. Although under constitutive demand for endosomal delivery to lysosomes, Rab7 GTPase is also the target of additional autophagy-specific regulation for induced levels of amphisome and autolysosome formation 21, 22, 23.
Another point for regulation of lysosomal membrane fusion is through the action of distinct soluble NSF [N-ethylmaleimide-sensitive factor] attachment receptors (SNAREs) 24. Specificity in membrane fusion is achieved by the combinatorial formation of trans-SNARE complexes, typically comprised of three Q-SNARE (a, b and c) and one R-SNARE motifs 25. For instance, the related VAMP7 and VAMP8 R-SNAREs found on endolysosomes are differentially involved in endosome–lysosome fusion or autophagosome–lysosome fusion, respectively 26, 27. In late steps of autophagy, the Syntaxin17 SNARE is recruited to mature autophagosomes where it forms a complex with endolysosomal VAMP8 and cytoplasmic SNAP-29 26, 28, 29, while VAMP8 within other SNARE complex combinations is involved in homotypic late endosome and exocytotic fusion events 28, 30, 31. SNARE trafficking must be actively controlled to ensure proper sites and timing of complex formation and activity 32, 33, 34, 35; however, it is not known how VAMP8 distribution may be regulated to mediate induced levels of autophagosome–lysosome fusion.
We previously characterized functions for Drosophila Sbf, a homolog of human MTMR13 pseudophosphatase, in a complex only found outside of yeast that has important roles to meet endosomal trafficking demands for cortical remodeling 36. Sbf/MTMR13 serves as both a Myotubularin (MTM) phosphoinositide phosphatase scaffold and a Rab GTPase guanine nucleotide exchange factor (GEF)36, 37, 38. We showed that Sbf regulates a PtdIns(3)P endosomal pool and acts as a Rab21 GEF to promote specific early endosomal trafficking 36. Here, we now show that starvation induces Sbf/MTMR13 and Rab21 endosomal activity, which promotes VAMP8 endolysosomal trafficking and autophagosome–lysosome fusion to accommodate induced levels of autophagy.
Results
Rab21 and Sbf are required for starvation-induced autophagy
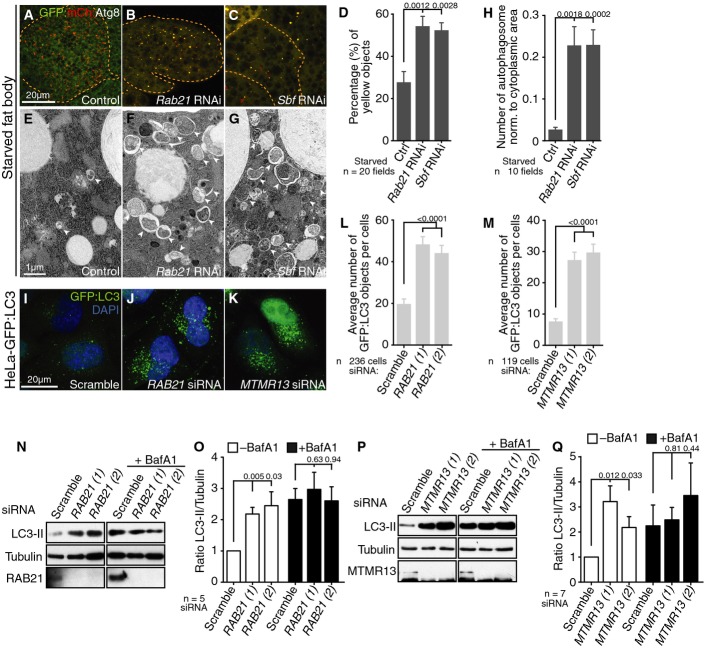
We used Drosophila third instar larval fat body as a starvation-inducible system to test the in vivo requirements for Rab21 and Sbf in autophagy. In this tissue, autolysosomes detected by the acidotropic probe, LysoTracker, are normally absent in larvae under well-fed conditions but are rapidly induced with autophagy in response to starvation 39, 40 (Fig1A, Supplementary Fig S1A and B). In contrast, RNAi-mediated depletion of Rab21 or Sbf with multiple independent hairpins each blocked the appearance of LysoTracker punctae in starved fat bodies (Fig1B–D; Supplementary Fig S1C–F). To confirm a role for Rab21 and Sbf in autophagy, we monitored protein levels of the autophagic cargo, p62 (known as Ref(2)P in flies), whose abundance correlates with autophagy status 41. Consistent with a block in autophagy, Ref(2)P accumulated with Rab21 or Sbf knockdown in fat bodies examined by immunofluorescence (Fig1E–H) or immunoblot (Fig1I–J). These results point to a requirement for Rab21 and Sbf functions in both basal and starvation-induced autophagy.
Figure 1.
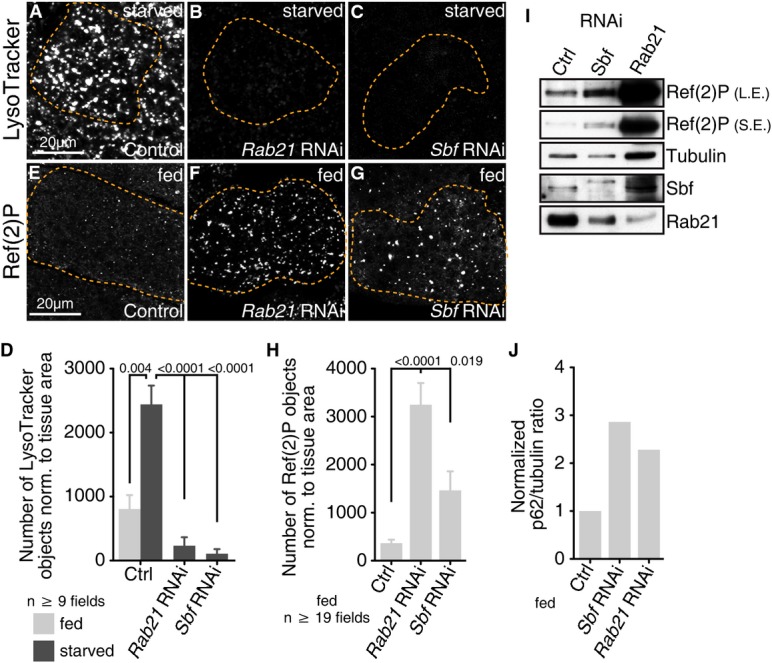
Rab21 and Sbf positively regulate autophagy
- A-D LysoTracker labels autolysosomes. (A) Control (LacZ) starved. (B, C) RNAi depletion of (B) Rab21 (VDRC 32941) or (C) Sbf (TRiP 32419) blocks starvation-induced autolysosomes. (D) Number of LysoTracker objects normalized to fat body area; SEM.
- E-H Ref(2)P (p62) protein detected by immunofluorescence in fed fat body. (E) Control (GFP). RNAi depletion of (F) Rab21 or (G) Sbf leads to Ref(2)P accumulation. (H) Number of Ref(2)P objects normalized to fat body area; SEM.
- I Ref(2)P protein levels in fat body lysates from control or RNAi conditions, as shown, with long (L.E.) and short exposures (S.E.) from same blot.
- J Ratio of Ref(2)P to tubulin integrated densities for the representative immunoblot in (I). See also Supplementary Fig S1.
Rab21 and Sbf share functions important for autophagosome–lysosome fusion
To identify at which step(s) in autophagy Rab21 and Sbf functions are needed, we assessed the GFP:mCherry:Atg8 flux reporter 42, 43. Atg8 is incorporated into the inner and outer membranes of expanding and mature autophagosomes, which ultimately fuse with endolysosomes for degradation in autolysosomes. In wild-type starved fat bodies, GFP:mCherry:Atg8 labels two stages of autophagic vesicles (AVs): a minor fraction is detected at autophagosomes that are double-positive for mCherry- and GFP-fluorescence, while the majority is detected at autolysosomes that, due to GFP pH-sensitivity, are marked solely by mCherry-fluorescence (Fig2A). In Rab21- or Sbf-depleted fat bodies, however, GFP:mCherry:Atg8 revealed an inverse accumulation of double-positive autophagosomes and reduction in mCherry-positive autolysosomes (Fig2B–D). With Rab21 or Sbf RNAi, the accumulated Ref(2)P autophagic cargo co-localized with Atg8 (Supplementary Fig S1G–J), consistent with a block in autophagic clearance at a step after autophagosome formation.
Figure 2.
Rab21 and Sbf/MTMR13 have a conserved function required for autophagosome–lysosome fusion
- A-D GFP:mCherry:Atg8 in Drosophila starved fat body. (A) Control (LacZ) shows predominantly autolysosomes (red punctae). (B, C) RNAi depletion of (B) Rab21 or (C) Sbf leads to an accumulation of autophagosomes (yellow punctae) and a depletion of autolysosomes (red punctae). (D) Percentage of autophagosomes (yellow) to total autophagic vacuoles (red + yellow) normalized to fat body area; SEM.
- E-H Transmission electron microscopy of starved larval fat body. (E) Control. (F) Rab21RNAi or (G) SbfRNAi leads to accumulation of autophagosomes (arrowheads). (H) Number of autophagosomes normalized to cytoplasmic area; SEM.
- I-M Stably expressed GFP:LC3 in HeLa cells grown in full media with siRNA. (I) Scramble (control). (J) RAB21 siRNA or (K) MTMR13 siRNA results in accumulation of GFP:LC3 autophagosomes. (L, M) Average number GFP:LC3 objects per cell from two independent siRNAs against (L) RAB21 or (M) MTMR13; SEM.
- N-Q RAB21 (N, O) and MTMR13 (P, Q) are required for LC3-II flux in HeLa cells. Anti-LC3 immunoblot of (N) RAB21 or (P) MTMR13 siRNA-depleted HeLa cells grown in full media with or without bafilomycin A1. (O, Q) Ratio of LC3-II to tubulin integrated densities from (O) five or (Q) seven independent experiments; SEM. See also Supplementary Figs S1 and S2.
The accumulation of autophagosomal markers in the knockdown conditions could result from a block in either autophagosome delivery to lysosomes or degradation within autolysosomes. To discriminate between these two possibilities, we examined starved fat bodies by transmission electron microscopy in order to distinguish autophagosomes from autolysosomes indicated by cytoplasmic contents within double- versus single-membrane compartments, respectively. In comparison to control fat bodies, Rab21 or Sbf knockdown exhibited an increased number and area of autophagosomes in the cytoplasm (Fig2E–H; Supplementary Fig S1K), as well as an increase in individual autophagosome size (Supplementary Fig S1L). These results implicate Rab21 and Sbf with shared roles needed for autophagosome delivery to and/or fusion with lysosomes.
Conserved roles for human RAB21 and Sbf/MTMR13 in autophagosome clearance
To generalize our findings to other cell types and across evolution, we tested autophagy requirements for the human orthologs, RAB21 and Sbf/MTMR13. As in flies, specific RAB21 or MTMR13 siRNA depletion in HeLa cell basal conditions (Supplementary Fig S2A and B) led to a similar abnormal increase in the number of autophagosomes indicated by GFP:LC3 (human Atg8; FigI–M). Consistent with this, knockdown of RAB21 or MTMR13 also exhibited increased levels of the endogenous lipidated LC3-II associated with autophagosomes (Fig2N–Q, white bars). To confirm that the observed increase in LC3-II was due to a clearance defect, we treated cells with bafilomycin A1, an inhibitor of lysosomal acidification 44. Bafilomycin treatment masked the effects of RAB21 or MTMR13 depletion such that now similar levels of LC3-II accumulated in the control and siRNA-treated cells (Fig2O and Q, black bars), suggesting a block in starvation-induced autophagy at the level of autophagosome degradation. In agreement with a role in clearance, there was no increase in the rate of autophagosome synthesis indicated by Atg5-marked isolation membranes in RAB21- or MTMR13-depleted cells (Supplementary Fig S2C–G)45. These results indicate conserved roles for RAB21 and MTMR13 in promoting autophagosome clearance at the level of autophagosome–lysosome fusion.
In mammals, the VARP GEF was found to activate RAB21 46, while the MTMR13 GEF was shown to be able to activate RAB28 38. Although there are no obvious fly homologs of VARP or RAB28, we explored whether their functions in human cells may contribute to RAB21 or MTMR13 roles in autophagy, respectively. In contrast to RAB21 or MTMR13 knockdown, VARP or RAB28 siRNA depletion had no effect on LC3-II processing or accumulation in bafilomycin A-treated cells (Supplementary Fig S2H–L). Moreover, both VARP and RAB28 knockdown led to fewer GFP:LC3 punctae (Supplementary Fig S2M–Q), suggesting a decrease in the number of autophagosomes. Together, our results support specific shared requirements for RAB21 and MTMR13 functions in autophagy at autophagosome–lysosome fusion.
Rab21 and Sbf/MTMR13 are not required for general lysosome biogenesis or function
Like other gene functions required for autophagy in the Drosophila larval fat body 39, 40, Rab21 or Sbf RNAi blocked the starvation-induced appearance of LysoTracker-marked autolysosomes with the block in autophagy flux (FigB–D). Lysosomal biogenesis, acidification and distribution also are required for normal autophagosome delivery to the lysosome 47, 48, 49, raising the question whether Rab21 and Sbf/MTMR13 promote autophagosome–lysosome fusion indirectly at the level of lysosomal maturation or function. However, normal number and distribution of LysoTracker-marked compartments were present with Rab21 or Sbf/MTMR13 knockdown, both in fed and starved conditions in fly macrophages (Fig3A–C) and in HeLa cells (Fig3D–F, Supplementary Fig S2R). Using the LysoSensor probe to quantify the extent of acidification, we found a similarly acidified average vesicular pH in control and Rab21 or MTMR13 siRNA cells (Fig3G–J). The presence of lysosomes in knockdown HeLa cells was confirmed by normal distribution and numbers of LAMP2 compartments (Fig3K–N). To address the potential for RAB21 and MTMR13 roles in general endolysosomal trafficking or function, we used an inducible assay for EGFR lysosomal delivery and degradation in human cells 20. Upon EGFR stimulation, there was no difference in the rate of EGFR degradation in either RAB21- or MTMR13-depleted cells (Fig3O–R). These results suggest that RAB21 and MTMR13 in mammals may have a direct role in the trafficking of a specific cargo required for autophagosome–lysosome fusion.
Figure 3.
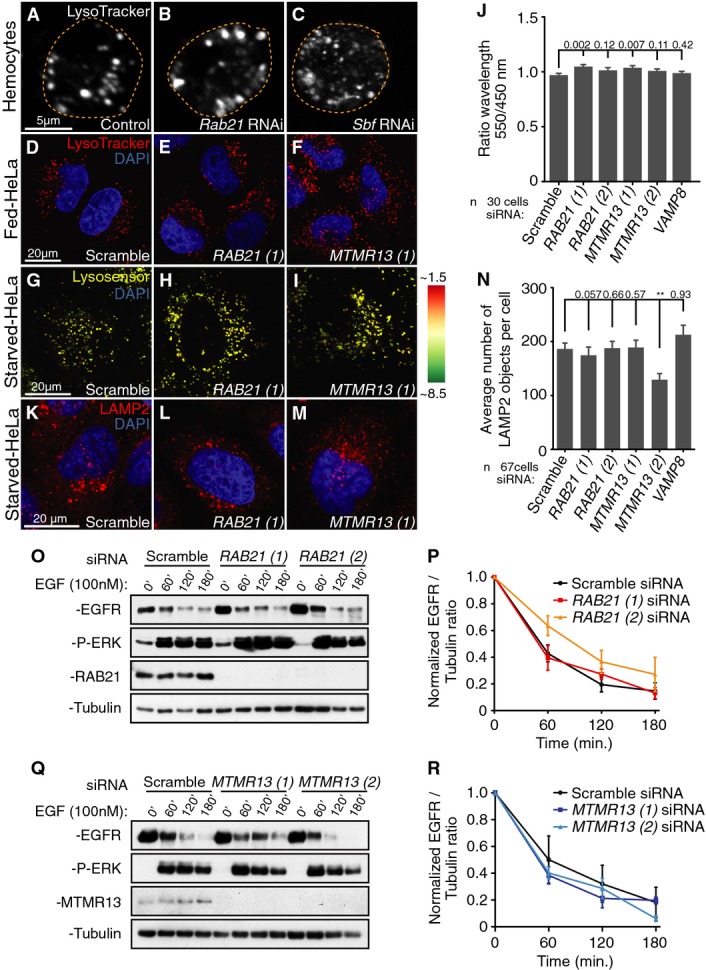
Rab21 and Sbf/MTMR13 are not required for general lysosomal function
- A-F Normal acidotropic LysoTracker staining in both fed (A–C) Drosophila hemocytes and (D–F) HeLa cells in (A, D) control, (B, E) Rab21 knockdown or (C, F) Sbf or MTMR13 knockdown conditions.
- G-I Merged images of LysoSensor Blue acquired at two independent emissions spectra, with average pH indicated by colors depicted in the pseudocolored heatmap (right), in starved HeLa cells. Normal lysosomal pH in (G) control and (H) RAB21 or (I) MTMR13 knockdown conditions.
- J Per cell average ratio of LysoSensor detected at wavelength 550 over 450 nm; SEM. VAMP8 siRNA image data not shown.
- K-N Normal distribution of LAMP2 immunofluorescence in (K) control, (L) Rab21 siRNA or (M) MTMR13 siRNA-starved HeLa cells. (N) Per cell average number of LAMP2 objects in starved HeLa cells; SEM. VAMP8 siRNA image data not shown.
- O-R RAB21 or MTMR13 knockdown do not affect EGFR degradation following EGF stimulation in HeLa cells. (O, Q) EGFR and phospho-Erk levels in overnight serum-starved RAB21- or MTMR13-depleted HeLa cells, treated with 100 nM EGF. After EGF addition, cells were chased in cycloheximide and lysed at time points shown. Normalized EGFR to tubulin integrated densities ratio in (P) Rab21 or (R) MTMR13 siRNA-depleted HeLa cells from four independent experiments; SEM. See also Supplementary Fig S2.
Rab21 and Sbf affect Vamp7 and endolysosomal flux in flies
Others and we previously showed that Rab21 and Sbf/MTMR13 localize at endosomes and function in cargo trafficking 36, 37, 50, 51, 52, 53, 54, 55, 56. One possibility is that Rab21 and Sbf/MTMR13 endosomal functions are responsible for their roles in autophagy. The R-SNARE Vamp7, the single Drosophila homolog of mammalian VAMP7 and VAMP8 (76 and 63% similarity, respectively), represented a relevant endosomal cargo potentially affected by Rab21 and Sbf for several reasons. In mammals, VAMP7 forms a protein complex with RAB21 57, and VAMP8 is required on endolysosomes to mediate autophagosome–lysosome fusion 26, 28. In flies, Vamp7 was shown to fulfill the autophagy VAMP8 SNARE function required for autophagosome–lysosome fusion 29.
We validated that Drosophila Vamp7 is required for autophagy in starved fat body (Supplementary Fig S3A–C), then investigated whether Rab21 or Sbf function affects Vamp7 distribution. GFP:Vamp7 localizes on starvation-induced autolysosomes in control fat bodies (Fig4A). In contrast, Rab21- or Sbf-depleted fat body cells exhibited a striking increase in number and size of GFP:Vamp7 punctae with persistent lack in LysoTracker staining (Fig4B–D, Supplementary Fig S3D), suggesting a block in Vamp7 endosomal flux in the absence of autolysosome formation. Given distinct roles for mammalian VAMP7 and VAMP8 in mediating endosome–lysosome versus autophagosome–lysosome fusion, respectively 26, 30, we reasoned that disruption of fly Vamp7 distribution could impair both endolysosomal and autophagic trafficking. Consistent with a role in endolysosomal homeostasis, depletion of Rab21 or Sbf also led to an accumulation in the number of GFP:Rab7 and GFP:Lamp1 compartments (Supplementary Fig S3E–L). This was similar to that seen with depletion of Vamp7 or Rab7, and distinct from depletion of early endosomal Rab5 (Supplementary Fig S3M–P). Together, these results raise the possibility that Rab21 and Sbf functions in Vamp7 endolysosomal trafficking, either directly or indirectly, are required for SNARE-mediated autophagosome–lysosome fusion in flies.
Figure 4.
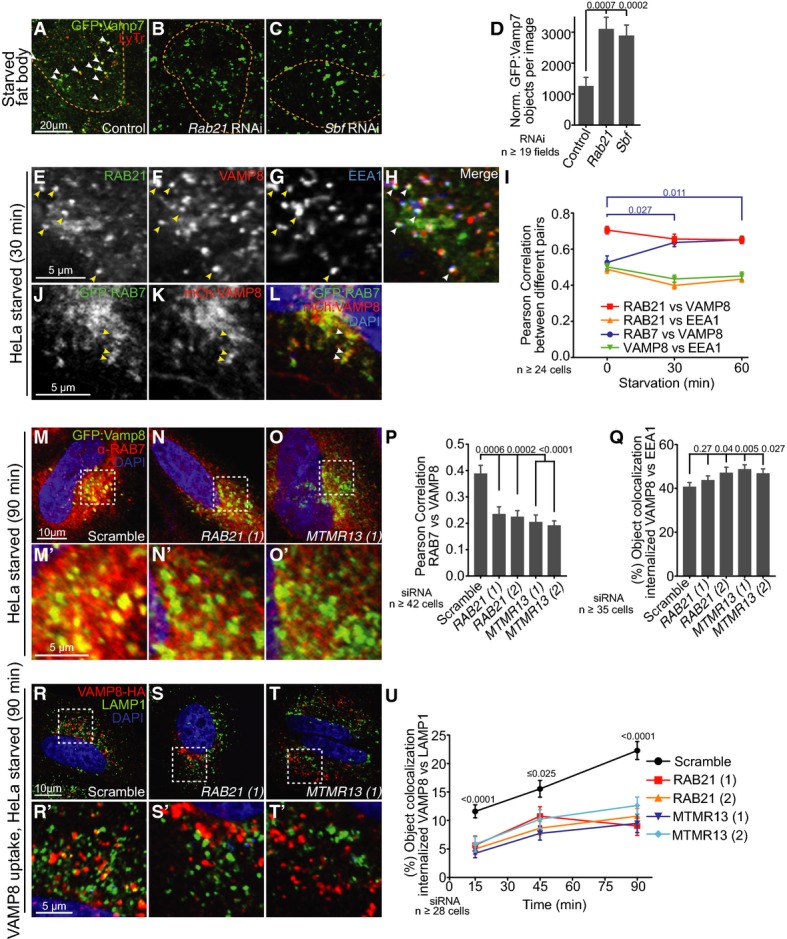
RAB21 and Sbf/MTMR13 regulate dVamp7/VAMP8 trafficking to lysosomes
- A-D GFP:Vamp7 in Drosophila starved larval fat body. (A) Control. Vamp7 trafficking is blocked with (B) Rab21RNAi or (C) SbfRNAi. (D) Number of GFP:Vamp7 punctae normalized to fat body area; SEM.
- E-H RAB21 and VAMP8 co-localize at EEA1 early endosomes. (E) GFP:RAB21, (F) VAMP8:3xHA and (G) EEA1 co-localize (arrowheads) in starved HeLa cells, merged in (H).
- I Per cell average Pearson Correlation of GFP:RAB21 with VAMP8:3xHA, GFP:RAB21 with EEA1, VAMP8:3xHA with EEA1 and GFP:RAB7 with mCherry:VAMP8 in response to starvation over time; SEM. Only VAMP8 co-localization with RAB7 was enhanced by starvation.
- J-L RAB7 and VAMP8 co-localize at late endosomes. (J) GFP:RAB7 and (K) mCherry:VAMP8 co-localize (arrowheads) in starved HeLa cells, merged in (L).
- M-P Starvation-enhanced co-localization between endogenous RAB7 and GFP:VAMP8 as in (M) scramble control requires (N) RAB21 and (O) MTMR13 function. (M', N' and O') Zooms of boxed regions shown above. (P) Per cell average Pearson Correlation between endogenous RAB7 and GFP:VAMP8 in siRNA-treated cells; SEM.
- Q VAMP8 exit from early endosomes is delayed in RAB21 or MTMR13 siRNA-depleted cells. Per cell average object co-localization between internalized VAMP8 and EEA1 in chloroquine-treated cells; SEM. See Supplementary Fig S4Q–S.
- R-U VAMP8 delivery to lysosomes is blocked in RAB21- or MTMR13-depleted cells. Antibody-uptake assay of VAMP8:3xHA (internalized anti-HA, red) with LAMP1 (green) and DAPI nuclei. (R) Scramble siRNA, (S) RAB21 siRNA and (T) MTMR13 siRNA-treated cells. (R', S' and T') Zooms of boxed regions shown above. (U) Per cell average object co-localization of internalized VAMP8:3xHA with LAMP1 over time; SEM. See also Supplementary Figs S3 and S4.
Human RAB21 and MTMR13 specifically control VAMP8 endolysosomal trafficking
We turned to human cells to better discern whether RAB21 and MTMR13 share requirements for a VAMP8-specific role in autophagosome–lysosome fusion 26, 28, 30. Both RAB21 and VAMP8 proteins shared a similar predominantly endosomal distribution profile (Supplementary Fig S3Q–Dd, and as previously reported in 36, 50, 51, 52, 53, 54, 55), with little obvious association with Golgi or (pre)-autophagosomal membranes in fed or starved conditions (Supplementary Fig S4A–I). RAB21 and VAMP8 exhibited extensive co-localization in both fed and starved conditions (up to 0.7 Pearson Correlation; FigE–I, red line), including at EEA1 early endosomes (Fig4I, green and orange lines) and shared distribution at RAB7 late endosomes (Fig4I–L, Supplementary Fig S3W and Dd). Only the RAB7 co-localization for both proteins was significantly modified in response to starvation, as seen by enhanced VAMP8 and decreased RAB21 association with RAB7, respectively (Fig4I, blue; Supplementary Fig S3W and Dd). Strikingly, the enhanced level of VAMP8 and RAB7 co-localization was dependent on both RAB21 and MTMR13 functions (Fig4M–P).
To directly test RAB21 and MTMR13 requirements in VAMP8 endosomal trafficking, we used a previously described antibody-uptake assay to monitor endocytic transit of cell surface-labeled VAMP8 34. We confirmed that this method specifically labels the VAMP8 pool at the plasma membrane that is then internalized with endocytosis over time (Supplementary Fig S4J–L). VAMP8 endocytic uptake and delivery to EEA1 early endosomes occurred with normal kinetics in RAB21 or MTMR13 knockdown cells (Supplementary Fig S4M–P). However, RAB21 or MTMR13 depletion led to an increased EEA1 co-localization when retrograde trafficking was also blocked with the addition of chloroquine (Fig4Q, Supplementary Fig S4Q–S), suggesting a block in normal VAMP8 endosomal sorting 58, 59. To test whether VAMP8 trafficking to endolysosomes where it is needed to mediate autophagosome fusion is affected, we repeated the uptake assay to follow VAMP8 delivery to LAMP1 lysosomes. At steady-state, VAMP8 showed a weak localization at lysosomes that decreased with starvation (Supplementary Fig S3Dd) 28, possibly due to dynamics with autophagic flux. Using the uptake assay in starved cells, however, we could detect internalized VAMP8 endosomal trafficking to lysosomes in a significant time-dependent manner (Fig4R–R' and U, black line). In contrast, both RAB21 and MTMR13 siRNA similarly blocked internalized VAMP8 delivery to LAMP1-positive endolysosomes in starved conditions (Fig4S–U). Together, these results indicate that RAB21 and MTMR13 function at endosomes to regulate VAMP8 endolysosomal transport required for SNARE-mediated autophagosome–lysosome fusion.
Active RAB21 and VAMP8 proteins interact in response to starvation
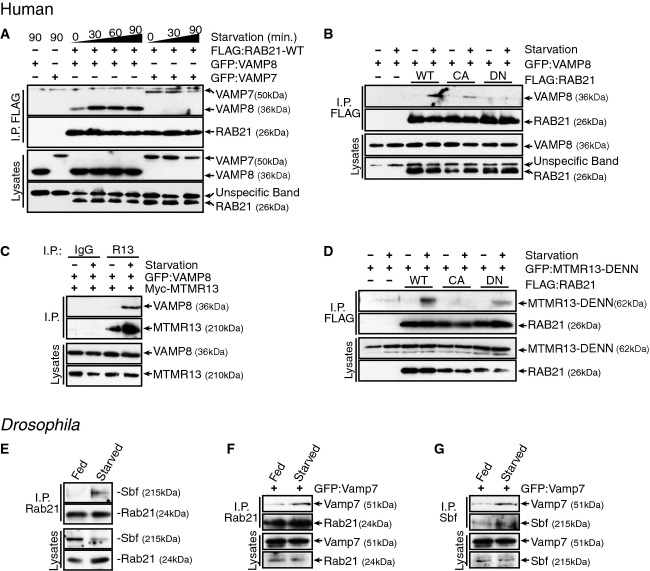
Membrane fusion events are commonly regulated by selective Rab GTPase activities that can tether membranes or target specific SNARE effectors 24, 60. We investigated whether a RAB21 protein interaction facilitates VAMP8 trafficking. We found that RAB21 co-immunoprecipitated VAMP8 (Fig5A), in addition to a previously reported interaction with VAMP7 57. However, in contrast to constitutive levels of interaction with VAMP7, the RAB21 interaction with VAMP8 was specifically and greatly enhanced in response to starvation (Fig5A). Enhancement of the RAB21 and VAMP8 interaction required full starvation, as neither serum-starvation nor glucose-starvation were sufficient to induce increased levels in protein binding (Supplementary Fig S5A). In line with a role in regulating VAMP8 trafficking, RAB21 interacted with only VAMP8 and not Syntaxin17 (Supplementary Fig S5B). The starvation-enhanced interaction was apparent between VAMP8 and RAB21 wild-type (WT) or constitutively activated (CA) forms, but not an inactivated (DN) form (Fig5B), suggesting a VAMP8 preference for GTP-bound RAB21. Likewise, greater co-localization was observed between VAMP8 and RAB21-WT or RAB21-CA than with RAB21-DN (Supplementary Fig S5C–F). These results suggest that starvation promotes RAB21 activity and enhanced protein association with VAMP8.
Figure 5.
Starvation promotes RAB21 and Sbf/MTMR13 interactions with dVamp7/VAMP8
- A RAB21 interacts with VAMP8 and VAMP7 in HeLa cells. RAB21 interaction with VAMP8 is increased by starvation. FLAG IP FLAG:Rab21-WT and immunoblot of co-expressed GFP-tagged VAMP8 or VAMP7.
- B VAMP8 interacts preferentially with active RAB21 forms in response to starvation (90 min). FLAG IP FLAG:RAB21-WT, RAB21-CA or RAB21-DN and immunoblot of co-expressed GFP-tagged VAMP8.
- C MTMR13 interacts with VAMP8 under starvation (60 min). MTMR13 IP of Myc:MTMR13 and immunoblot of co-expressed GFP:VAMP8.
- D MTMR13-DENN interacts preferentially with inactive RAB21 in response to starvation (90 min). FLAG IP FLAG:RAB21-WT, RAB21-CA or RAB21-DN and immunoblot of co-expressed GFP:MTMR13-DENN (GEF) domain.
- E-G Protein interactions in co-IP from fed or starved Drosophila third instar larvae. Starvation induces in vivo interaction between (E) endogenous Drosophila Rab21 (IP) and Sbf, (F) endogenous Rab21 (IP) and fat body-expressed GFP:Vamp7 (anti-GFP IB) and (G) endogenous Sbf (IP) and fat body-expressed GFP:VAMP7 (anti-GFP IB). See also Supplementary Fig S5.
Starvation induces MTMR13-dependent RAB21 GTPase activity and VAMP8 interaction
Previously, we showed in flies that Rab21 and Sbf directly interact in a protein complex and that the Sbf DENN domain acts as a Rab21 GEF 36. We found by co-immunoprecipitation that starvation induces MTMR13 as well as RAB21 association with VAMP8 (Fig5C). The complex of protein interactions raised the possibility that MTMR13 GEF activity promotes RAB21 activity and association with VAMP8. As in flies, we found that the human MTMR13 DENN GEF domain preferentially binds to GDP-bound RAB21 forms (WT and DN; Fig5D). Importantly, starvation enhanced the interaction between the MTMR13 DENN domain and RAB21 (Fig5D). Starvation of intact Drosophila larvae similarly enhanced the protein interaction between endogenous full-length Sbf and Rab21 (Fig5E) and between either Rab21 or Sbf and Vamp7 (Fig5F and G), revealing a conserved regulation of the Sbf/MTMR13, Rab21 and Vamp7/VAMP8 pathway.
Given that starvation promotes an interaction between Sbf/MTMR13 and RAB21, we hypothesized that starvation could lead to RAB21 GTPase activation. We found that starvation rapidly induced a transient RAB21 GTPase activity (Fig6A), and significantly, this activity depended on endogenous MTMR13 function (Fig6B and C). Consequently, MTMR13 depletion inhibited the starvation-induced response in RAB21 and VAMP8 interaction (Fig6D and E) and decreased RAB21 and VAMP8 co-localization (Supplementary Fig S5G–I). Together, our data support that starvation-regulated MTMR13 RAB21 GEF function promotes RAB21 GTPase activity and selective VAMP8 effector interaction, in order to increase VAMP8 endolysosomal flux critical to meet increased demands for SNARE-mediated autophagosome–lysosome fusion.
Figure 6.
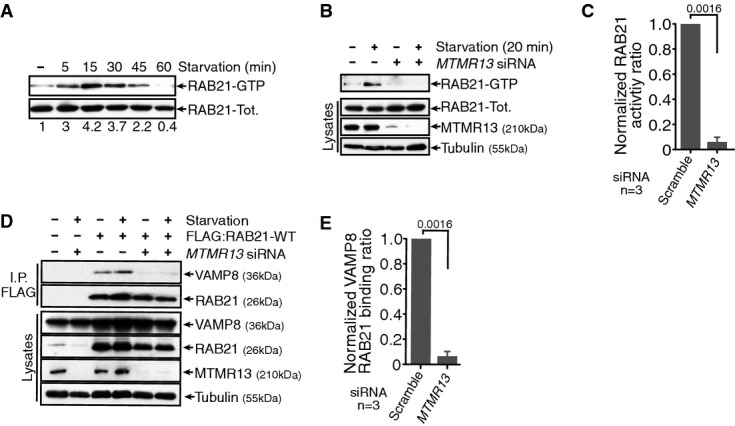
Starvation promotes MTMR13-dependent RAB21 activation and interaction with VAMP8
- A RAB21 GTPase activity is transiently activated following starvation. GST:APPL1 used as bait for RAB21-GTP pull-down. Immunoblot of GFP:RAB21 in pull-down (RAB21-GTP) and in cell lysates (RAB21-Tot.).
- B Starvation-induced RAB21 GTPase activity requires MTMR13, as tested in scramble (−) or MTMR13 siRNA (+)-treated HeLa cells. GST:APPL1 pull-down and immunoblot of activated and total RAB21.
- C Ratio of activated RAB21-GTP in pull-down versus lysate integrated densities from three independent experiments, normalized to scramble siRNA; SEM.
- D RAB21 interaction with VAMP8 requires MTMR13, as tested in scramble (−) or MTMR13 siRNA (+)-treated HeLa cells. FLAG IP FLAG:RAB21-WT and immunoblot of co-expressed GFP-tagged VAMP8.
- E Ratio of VAMP8 in IP versus lysate integrated densities from three independent experiments, normalized to scramble siRNA; SEM. See also Supplementary Fig S5.
Discussion
In both flies and human cells, we uncovered shared and conserved endosomal roles for Sbf/MTMR13 and Rab21 regulation in autophagy. We showed that starvation-inducible Sbf/MTMR13 and Rab21 GTPase activity specifically regulates endolysosomal trafficking of VAMP8 required for SNARE-mediated autophagosome–lysosome fusion. Our data suggest that a starvation-induced Sbf/MTMR13 GEF function promotes Rab21 activity, leading to enhanced protein interactions either directly or indirectly with VAMP8 on early endosomes. We envision that these interactions regulate VAMP8 endosomal sorting to promote an endolysosomal distribution. This is consistent with a starvation-induced increase in VAMP8 and RAB7 co-localization with upregulation of autophagy that is dependent on MTMR13 and RAB21 (Fig4I and P). Although Rab21 and Sbf/MTMR13 are required for VAMP8 endolysosomal trafficking and autophagy in both basal and starvation-induced conditions, the starvation conditions specifically enhanced regulatory protein interactions seen between Sbf/MTMR13, Rab21 and VAMP8. Altogether, our results indicate that Sbf/MTMR13 and Rab21 activities can tune VAMP8 endosomal distribution to accommodate changing membrane trafficking demands with autophagic status.
Rab21 and Sbf/MTMR13 are both only found outside yeast and shown to regulate early endosomal trafficking and recycling in cellular remodeling contexts with increased demands for membrane flux 36, 51. Previously, we showed that Drosophila Sbf acts as a Rab21 GEF for shared requirements in cortical remodeling. Here, we demonstrate that a regulatory relationship between Sbf/MTMR13 and RAB21 required for autophagy occurs in both fly and human cells. Similarly in mammals, the distinct VARP GEF (for which there is no obvious fly homolog) was shown to regulate RAB21 activity in a complex of protein interactions involved in VAMP7 exocytosis 46, 57. In contrast to VAMP8, however, we found that the RAB21 interaction with VAMP7 is not under starvation regulation and that RAB21 and VARP exhibit distinct autophagy defects. We propose that human RAB21 serves separable functions under the differential control of the MTMR13 GEF and VARP GEF, respectively.
We identified the R-SNARE, VAMP8, as an effector of Sbf/MTMR13 and Rab21 activity. Although VAMP8 is enriched on early endosomes, VAMP8 distribution is dynamically balanced between recycling to the plasma membrane, endocytosis and trafficking to late endosomes–lysosomes. VAMP8 trafficking to late endosomes is critical for its key functions within distinct SNARE complexes that with Stx7, Stx8 and Vti1b mediate late endosome homotypic fusion 61 or with Stx17 and SNAP29 mediate autophagosome–lysosome fusion 28, 29. It has not been determined, however, whether VAMP8 is required specifically on late endosomes and/or lysosomes for its role in autophagosome fusion.
How VAMP8 distribution is regulated is only partially understood in its early endocytic trafficking stages. The endocytic internalization of VAMP8 and other small R-SNAREs was shown to rely on the clathrin adaptor CALM, which binds to the VAMP8 SNARE motif to prevent precocious fusion 34. Sbf/MTMR13 and Rab21 are not required for Vamp7/VAMP8 internalization (Fig4U, Supplementary Fig S4P), indicating that they likely act downstream of CALM. One possibility is that VAMP8 on early endosomes is handed off from CALM to activated RAB21, which then promotes VAMP8 endolysosomal trafficking and/or prevents unwanted fusion events. Consistent with both of these scenarios, we observed enlarged Vamp7 and Rab7 endosomes in Rab21- or Sbf-depleted fly fat body cells (Fig4D, Supplementary Fig S3D and H), and increased VAMP8 accumulation at EEA1 endosomes in RAB21- or MTMR13-depleted human cells (Fig4Q).
Our results point to a role for RAB21 activity in VAMP8 early endosomal sorting. The presence of normal lysosomal identity, morphology, acidification, distribution and EGFR degradation indicated that RAB21 and MTMR13 are not required for general lysosomal function per se in human cells. SNARE sorting involves protein recruitment to nascent vesicles and/or tubules. At early and recycling endosomes, dynamic membrane tubules serve as sorting domains that enrich specific protein cargos with vesicle formation 62. RAB21 was shown to induce endosomal tubule formation upon depletion of phosphatidylinositol 3-phosphate (PtdIns(3)P) 52, and our work demonstrated that Sbf is required both for turnover of endosomal PtdIns(3)P and tubule formation 36. From this, we speculate that Sbf/MTMR13, through coordinated regulation of PtdIns(3)P turnover and Rab21 activation, promotes endosomal tubulation involved in VAMP8 sorting (in addition to possible PtdIns(3)P-mediated functions independent of Rab21). Thus, in the absence of Sbf/MTMR13 or Rab21 functions, VAMP8 would be retained at an endosomal compartment or misrouted to a retrograde or recycling pathway as suggested from our data (Fig4Q), with similar consequences to that seen with disruption of different mechanisms for VAMP7 endolysosomal sorting 59, 63.
How might VAMP8 endosomal retrieval be regulated? For VAMP7, a direct interaction between its longin domain and the AP-3 adaptor complex mediates its endosome to lysosome transport 32, 63, while an interaction with VARP and the retromer complex mediates its recycling to the plasma membrane 59. However, VAMP8 has neither a longin domain nor AP-3 consensus binding sequences 34. VAMP8 carrier vesicles could be specified by a sorting nexin (SNX, 64), even though a screen targeting mammalian SNXs did not identify any individually required for autophagosome–lysosome fusion 5. Alternatively, Rab21 activity could prevent VAMP8 sorting into endosomal tubules for retrograde and recycling trafficking, such that VAMP8 retention during endosomal maturation results in its accumulation on late endosomes. Since only low levels of VAMP8 unassociated with RAB21 were detected at the Golgi (Supplementary Fig S4C), it is unlikely that Sbf/MTMR13 and RAB21 regulate a VAMP8 biosynthetic route to endolysosomes. In contrast, VAMP8 shows starvation-induced endosomal enrichment and protein interaction with MTMR13 and RAB21, co-localizes with RAB21 at endosomes, and requires Sbf/MTMR13 and RAB21 for endolysosomal transit (Figs4 and 5). In addition to transport, Rabs also have roles in membrane tethering and SNARE complex formation 65. If Rab21 acts more directly in autophagosome–lysosome tethering or VAMP8 trans-SNARE complex formation 28, 29, then we might expect RAB21 to co-localize with pre-fusion autophagosomes or interact with the trans-SNARE complex. However, in neither case was this observed (Supplementary Figs S4F and S5B).
Our results show that MTMR13 enhanced binding to RAB21 and the VAMP8 effector is responsive to starvation. MTMR13 post-translational modifications are one possible mechanism for this specific regulated response. Phosphorylation of either Sbf/MTMR13 and/or an interacting protein, such as MTMR2 66, might regulate MTMR13 endosomal interactions, GEF activity or stability 56. In this regard, Drosophila Sbf and human MTMR13 both have conserved demonstrated phosphorylation sites 67, 68. The Rab21 response to full starvation (Supplementary Fig S5A) appears to be distinct from an early endosomal Rab5 response to growth factor deprivation leading to autophagy induction 69. Here, we demonstrate that Sbf/MTMR13 and Rab21 GTPase activity regulates VAMP8 endosomal sorting to lysosomes in a demand-dependent fashion, serving as a novel mechanism for autophagy regulation.
Materials and Methods
Drosophila strains
UAS-RNAi hairpins were targeted to fat body with Cg-GAL4. Autophagy flux was assessed in flies carrying UASp-GFP:mCherry:Atg8a2 (from I. Nezis and H. Stenmark). Genotypes used in this study include the following: (1) w;UAS-IR-Sbfv22317 (Sbf-1 RNAi), (2) y1 sc* v1; P{TRiP.HMS00414}attP2 (Bloomington 32419; Sbf-2 RNAi), (3) w; UAS-IR-Rab21v32941 (Rab21-1 RNAi), (4) w; UAS-IR-Rab21v109991 (Rab21-2 RNAi), (5) y1 v1; P{TRiP.JF03338}attP2 (Bloomington 29403; Rab21-3 RNAi), (6) w[1118]; P{w[+mC]+UAS-lacZ.B}Bg4-1-2, (7) w; UAS-2xEGFPAH2 (8) y[1] w[1118]; P{w[+mC]+UASp-GFP:mCherry:Atg8a}2 (from I. Nezis and H. Stenmark), (9) w;UAS-IR-Rab5v103945, (10) w;UAS-IR-Rab7v40338, (11) w;UAS-IR-CG1599v13317, (12) w; UAS-IR-CG1599R-1 (NIG-Fly), (13) w[1118]; P{w[+mC]+Cg-GAL4.A}2 and (14) w; Cg-GAL4, P{w[+mC]+UAS-2xEGFP}AH2.
New genotypes generated during this study include the following: (1) w; Cg-GAL4, P{w[+mC]+UASp-GFP:mCherry:Atg8a}2, (2) w; UAS-GFP:Vamp73/CyO, (3) w; Cg-GAL4, UAS-GFP:Vamp73/CyO, (4) w; Cg-GAL4, UAS-GFP:Lamp1 (UAS-GFP:Lamp1 from H. Krämer) and (5) w; Cg-GAL4, UAS-GFP:Rab72 (UAS-GFP:Rab7 from M. González-Gaitán).
Fly crosses and starvation protocol
Fly crosses were set up at room temperature. After 2 days, vials were shifted to 29°C and incubated for three more days. To avoid overcrowding, 20–30 early third instar larvae were transferred to a fresh vial for 16–24 h before dissection. Feeding third instar larvae were starved by transferring them onto a wet kimwipe paper for 3 h at 29°C.
Cell culture
HeLa-M cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Sigma) at 37°C and 5% CO2. Stable GFP:Atg5 and GFP:LC3 HeLa cells were provided by N. Fujita and T. Yoshimori. Plasmid transfection was carried out with either Fugene HD (Promega) or JetPRIME (Polyplus transfection) following manufacturer's instruction. Transfected cells were lysed or imaged 24–48 h post-transfection. siRNAs (7.5 nM) were transfected using Dharmafect 1 (Dharmacon) following manufacturer's instructions. siRNAs were purchased from Dharmacon: RAB21(1) (J-009450-05), RAB21(2) (J-009450-08), MTMR13(1) (J-014684-09), MTMR13(2) (J-014684-10), VAMP8 (J-013503-05), VARP (J-014788-09), RAB28 (J-008582-05) and non-targeting scrambled control siRNA (D-001810-01-05). Experiments were carried out 72 h following siRNA transfection.
Confocal fluorescence microscopy
For imaging in live fat bodies, fed or starved third instar larvae were dissected at room temperature in 1× PBS (pH 7.4) for detection of GFP:mCherry:Atg8, GFP:Vamp7, GFP:Lamp1, GFP:Rab7 or LysoTracker (1:5,000 for 5 min, Life Technologies), while hemocytes were stained with LysoTracker and imaged as described in 70. Fat body confocal images were acquired on a Zeiss LSM 700 microscope with a 40× oil Plan-NeoFluar/1.3 numerical aperture (NA) objective. Control fat bodies were always acquired first and used to set laser power and gain settings. These settings were kept and used for all subsequent experimental genotypes. For Ref(2)P and Atg8 immunofluorescence, fed third instar larvae were inverted (in order to have internal tissues facing outward) in 1× PBS and fixed in 8% paraformaldehyde 15 min. The paraformaldehyde was quenched in 50 mM NH4Cl at RT 10 min. Quenched larvae were permeabilized and blocked 30 min in PBSTB (1× PBS + 0.1% Triton X-100 + 0.03% BSA + 2% goat serum). Larvae were incubated with 1:600 rabbit anti-Ref(2)P (also called p62) and 1:300 rat anti-Atg8 (both kind gifts of Dr. G. Juhasz 29, 71) in PBSTB and incubated at 4°C overnight. Larvae were washed in PBSTB 5× 5 min and incubated at RT 2 h in secondary antibody (1:500 with anti-Rat Alexa 488 and anti-Rabbit Alexa 546). Larvae were washed 5×, fat bodies were dissected and mounted in mounting medium (50% glycerol, 50% 1× PBS) and imaged as described above.
HeLa cells were grown on #1.5 cover glass. Cells were starved in Earle's Balanced Salt Solution (EBSS) at 37°C for 2 h. For GFP:LC3, GFP:Atg5, GFP:RAB21 and mCherry:VAMP8 imaging, cells were fixed in 3.7% formaldehyde in 1× PBS at RT for 10 min. Cells were then washed 3× with 1× PBS at RT for 5 min each. Nuclei were stained with DAPI (1 μg/ml in 1× PBS) at RT 10 min. Finally, cells were washed 3× with 1× PBS then mounted in Slow-fade reagent (Life Technologies). Images were acquired on a FV 1000 confocal microscope with a 60x/1.42 NA Plan Apo N objective at 1.5× zoom setting, yielding a 0.138 μm pixel size (in X and Y). For immunofluorescence analysis, Cell Signaling immunostaining protocol was followed. Antibodies used were rabbit anti-RAB5 (1:200, Cell Signaling #3547), rabbit anti-RAB7 (1:100, Cell Signaling #9367), mouse anti-RAB11 (1:250, BD Biosciences #610656), rabbit anti-LAMP1 (1:100, Cell Signaling #9091), rabbit anti-Clathrin heavy chain (1:50, Cell Signaling #4796), rabbit anti-Syntaxin 6 (1:100, Cell Signaling #2869), mouse anti-LAMP2 (1:100, Developmental Studies Hybridoma Bank #H4B4) and mouse anti-FLAG (1:250, Sigma #F1804). For Golgi co-localization, GFP:GOLPH3 and GFP:SialT (kindly provided by Dr. S. Field) were cotransfected with FLAG:RAB21-WT and VAMP8-3xHA. Cells were stained with DAPI and mounted as for GFP or mCherry fusion proteins. Images were acquired on a FV 1000 confocal microscope as described above.
In vivo pH measurements were performed as in 72. Briefly, HeLa cells were washed twice with EBSS and incubated with 5 μM LysoSensor Yellow/Blue DND-160 (Life Technologies) diluted in EBSS for 45 min. Following this incubation, cells were washed 3× with EBSS and imaged immediately on a FV1000 spectral scanning confocal microscope in an environmental chamber with CO2 at 37°C. Images were acquired sequentially using the 405 laser line for excitation, and emissions were gated between 420–500 nm (channel 1) and 500–600 nm (channel 2). Probe calibration was performed using a free 25 μM LysoSensor solution at different pH. Laser power and gain settings were kept constant for both calibration and experimental images. To calculate average vesicular pH values, Cell Profiler was first used to identify and measure object intensities. For each object, its integrated intensity from channel 2 (550 nm) was divided by the integrated intensity of channel 1 (450 nm). LysoTracker images were acquired on live HeLa cells stained with 1/20,000 LysoTracker for 5 min prior to immediate imaging on a FV1000 confocal microscope. LysoTracker-positive punctae were identified and counted using Cell Profiler.
VAMP8 trafficking
VAMP8-3xHA uptake assays were performed using a slightly modified protocol from 34. Briefly, siRNA-treated cells expressing VAMP8-3xHA grown on #1.5 cover glass were washed 2× with ice-cold DMEM and incubated on ice for 60 min in DMEM containing rabbit anti-HA antibody (1:200, Abcam #9110) in order to selectively label VAMP8-3xHA at the cell surface. Cells were washed 3× with ice-cold media and washed 2× with 37°C EBSS and incubated for different times at 37°C to permit VAMP8-3xHA internalization and trafficking. Cells were then washed 2× in ice-cold 1× PBS and fixed in 4% paraformaldehyde at RT for 15 min. For inhibition of endosome to trans-Golgi traffic, cells were incubated with chloroquine (100 μM) during the 4°C antibody incubation step and for the whole 60-min uptake experiment for a total of 2 h chloroquine treatment. Controls confirmed that anti-HA staining selectively labeled VAMP8-3xHA at the cell surface at time 0 (after 60 min on ice before 37°C shift) and that labeled VAMP8-3xHA was internalized over time (Supplementary Fig S4J–L). Following fixation, co-immunofluorescence analysis was performed as described above using mouse anti-LAMP1 (1:100, Developmental Studies Hybridoma Bank #H4A3) or mouse anti-EEA1 (1:1,000, BD biosciences #610457) antibodies. Images were acquired on a FV 1000 confocal microscope with a 60× objective as described above, and the percentage of object co-localization was measured as detailed bellow.
Transmission electron microscopy
Fat bodies from starved third instar larvae were dissected directly in modified 2× Karnovsky's fixative (5% glutaraldehyde and 4% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4) and fixed for at least 24 h at 4°C. Fat bodies were post-fixed in 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 h and stained en bloc in 4% uranyl acetate for 1 h. Fat bodies were then dehydrated in ethanol, embedded in epoxy resin, sectioned at 60 nm and picked up on Formvar- and carbon-coated copper grids. Grids were stained with uranyl acetate and Sato's lead stain. Images were acquired on a transmission electron microscope (FEI Tecnai Spirit G2 BioTWIN) and photographed by a bottom mount Eagle 4K digital camera.
Immunoprecipitations and immunoblots
A total of 8 × 104 HeLa cells were plated per well of a six-well plate, transfection was carried out the following day, and cell lysis was performed 24 h following transfection. When siRNA and plasmid were transfected in the same experiment, siRNAs were transfected first following the usual protocol and plasmids were transfected 48 h later. Cell lysis in CoIP buffer (25 mM Tris–HCl pH 7.4, 1 mM EDTA, 0.1 mM EGTA, 15 mM MgCl2, 150 mM NaCl, 2 mM Na3VO4, 10% glycerol, 1% NP-40) was performed 24 h following plasmid transfection. FLAG-M2 affinity beads (Sigma) or Ab116 (Anti-MTMR13, kindly provided by Fred Robinson) linked to protein A were used for immunoprecipitations. Antibodies used for immunoblots were mouse anti-GFP (1:1,000, Santa Cruz Biotechnology), rabbit anti-phospho-Akt S473 (1:500, Cell Signaling #9018), rabbit anti-phospho-S6K T389 (1:500, Cell Signaling #9234), mouse anti-tubulin (1:2,500, Sigma DM1A), rabbit anti-MTMR13 (1:250 Ab116), anti-RAB21 (1:1,000 Sigma #R4405), rabbit anti-phospho-Erk Thr 202/204 (1:1,000, Cell Signaling #4370), rabbit anti-EGFR (1:1,000, Cell Signaling #4267), rabbit anti-LC3 (1:1,000 Cell Signaling #12741), rabbit anti-Sbf (1:4,000, 36) and rabbit anti-Rab21 (1:1,000, 36). Anti-RAB5, anti-RAB7, anti-RAB11, anti-Ref(2)P and anti-FLAG were the same as described for immunofluorescence. For the assessment of Ref(2)P levels in the fat body by immunoblot, we dissected fat bodies from four individual larvae. Fat bodies were homogenized in 100 μl of lysis buffer (25 mM Tris–HCl pH 7.4, 1 mM EDTA, 0.1 mM EGTA, 5 mM MgCl2, 150 mM NaCl, 2 mM Na3VO4, 10% glycerol, 1% NP-40 and 0.1% sodium dodecyl sulfate with 2× protease and phosphatase inhibitors) on ice for 20 min. Lysates were centrifuged at 16,100 g at 4°C for 10 min. Supernatants were collected, and equal amounts of protein were loaded on SDS–PAGE gels. For the representative gel shown, a minimum of three independent experiments was performed. For LC3-II processing, siRNA-transfected HeLa cells were kept in full medium with or without bafilomycin A1 (10 μg/ml) at 37°C. After 2 h, cells were lysed and subjected to SDS–PAGE electrophoresis and Western blotting. A minimum of four independent repeats were performed and quantified.
For endogenous co-immunoprecipitations, 20 third instar larvae were either kept on food (fed) or starved in PBS for 3 h. For GFP:Vamp7 co-immunoprecipitations, larvae were raised at room temperature to achieve low GFP:Vamp7 expression levels. Larvae were homogenized in 800 μl of CoIP buffer. The homogenate was incubated on ice for 20 min and spun down at 16,100 g at 4°C for 10 min. Supernatants were collected, and care was taken to avoid lipid contamination. For each condition, 200 μl of supernatant was incubated for 2 h at 4°C on a rotating wheel with anti-Rab21 or anti-Sbf antibodies (2 μg) 36 pre-bound to Optima F beads (Santa Cruz Biotechnology). After the incubation, beads were washed four times with CoIP buffer. Co-immunoprecipitations were blotted for GFP, Sbf and Rab21 using Optima F secondary antibodies (Santa Cruz Biotechnology) following manufacturer's instruction.
EGFR degradation assay
siRNA-transfected cells were serum-starved overnight. Cells were incubated in serum-free DMEM containing 100 ng/ml EGF and 25 μg/ml cycloheximide for the indicated times 20, 73. Cells were lysed and subjected to SDS–PAGE electrophoresis and immunoblotting. Four independent experiments were performed and quantified.
RAB21 activity assay
5 × 105 HeLa cells were plated in 100-mm dishes (1 for each time point) and transfected the following day with GFP:RAB21-WT cDNA using JetPRIME. On the next day, cells were starved in EBSS for the indicated amount of time and lysed in 750 μl of modified MLB buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 1% NP-40, 10% glycerol, 20 mM MgCl2, 1 mM EDTA, 1 mM sodium orthovanadate, 25 μM GTP and protease inhibitor cocktail) 74. Lysates were incubated on ice for 20 min and spun down at 16,100 g for 10 min at 4°C. For each time point, the supernatant was collected and 700 μl was added to 4 μg of purified GST:APPL1 (5-419) beads and incubated on a rotating wheel for 1 h at 4°C. GST:APPL1 beads were washed four times with MLB buffer and bound RAB21 was eluted with 2× SDS–PAGE loading buffer. Samples were heated at 95°C for 5 min and subjected to SDS–PAGE electrophoresis and Western blot.
For RAB21 activity assays in MTMR13-depleted cells, 1.75 × 105 GFP:RAB21-WT stable cells were plated in 100-mm dishes. On the following day, non-targeting or MTMR13(2) siRNAs were transfected, and the activity assay was performed as described above 72 h later.
Protein purification
GST:APPL1 (5-419), a well-described RAB21-GTP interacting protein region 75, was expressed in Escherichia coli BL21 using the pGEX system. Following protein expression, bacteria were lysed by sonication and cell debris was pelleted by a 10-min centrifugation at 9,300 g at 4°C. GST:APPL1 was purified on glutathione–Sepharose 4B beads (GE healthcare), and protein concentration was established by comparing GST:APPL1 band intensities (stained by Coomassie) to known BSA quantities on an SDS–PAGE gels. Beads were stored at 4°C in 1× PBS.
Generation of DNA constructs and stable cells
Full-length Drosophila Vamp7 (CG1599) was PCR-amplified from cDNA isolated from Kc167 cells, cloned into pENTR/D-TOPO (Life Technologies) and subcloned by an LR recombination into the Gateway destination vector pTGW-1075 to generate pUASt-EGFP:Vamp7. Full-length human RAB21, SYNTAXIN17, SNAP29, VAMP7 and VAMP8 were PCR-amplified from HeLa cDNAs and each cloned into pEGFP-C1, pmCherry-C1 or pCDNA3.1-FLAG using the In Fusion HD kit (Clontech). Human RAB21-Q78L (constitutively active, referred to as RAB21-CA) and RAB21-T33N (dominant negative, referred to as RAB21-DN) were both generated from the parental pCDNA3.1-FLAG:RAB21-WT and pmCherry-C1 RAB21-WT plasmids using the Quickchange mutagenesis kit (Agilent Technologies). VAMP8-3xHA was generated using the In Fusion HD kit. VAMP8 was PCR-amplified from pEGFPC1-VAMP8 while 3xHA was PCR-amplified from pTHW. Both fragments were recombined into the BamH1 site of pCDNA3.1. GST:APPL1 (5-419) was generated using the In Fusion HD system. The APPL1 coding sequence corresponding to amino acids 5-419 was PCR-amplified from HEK293T cDNA and subcloned into pGEX5-X3.
Stably expressing GFP:RAB21-WT cells were generated by transfecting the pEGFP-hRAB21WT vector into parental HeLa-M cells. Twenty-four hours post-transfection, transfected cells were selected by the addition of 400 μM Geneticin (Invitrogen). GFP-positive clones were picked and expanded 7 days following selection.
Quantification and statistical methods
CellProfiler software (www.cellprofiler.org) was used to quantify the number or co-localization of objects in fat body or HeLa cells. For object number quantification in fat body, individual pipelines were created with optimized object segmentation and identification for LysoTracker, Ref(2)P, anti-Atg8a, GFP:mCherry:Atg8, GFP:Lamp1 and GFP:Rab7. The fly data were normalized to the fraction of the image covered by fat body tissue. For all the quantifications, three to four independent experiments were performed. Quantification of GFP:LC3, GFP:ATG5, LAMP2 and LysoTracker object number in HeLa cells was performed with CellProfiler. Pipelines were created in which cells were first identified by their nuclei (DAPI staining) and their total area determined by fluorescence background. Objects (dots) were then segmented, identified and related to their parental cell, yielding number of objects per cell. The percentage of object co-localization was calculated following the recommendations found at (http://www.cellprofiler.org/linked_files/ExampleColocalization_Tutorial.pdf). Briefly, internalized VAMP8, LAMP1 and EEA1 objects were segmented and identified using specific primary objects identification modules. All these objects were related and averaged per cell as explained above. To calculate co-localization, identified objects were shrunken to a single centrally located pixel for each object. These single pixels were then uniformly expanded by 2 pixels to generate objects of 5 pixels diameter. Objects in both channels were filtered, and only objects touching another object in the other channel were counted. The percentage of internalized VAMP8 objects touching either LAMP1 or EEA1 objects was calculated in Excel by dividing filtered VAMP8 objects with total number of VAMP8 objects per cell. Pearson's correlation measurements for co-localization quantification were performed on Volocity 6.1 (Perkin Elmer) as shown in Fig4I and P and in Supplementary Figs S3W, S3Dd, S4C, S4F, S4I, S5F and S5I. Individual HeLa cells were manually selected, and the Pearson correlation coefficient was measured per cell. For the TEM quantification, Image-J software was used. Autophagosomes were manually selected as region of interest (ROI) and their surface area measured. The number or the area occupied by autophagosomes in each image was normalized to the cytoplasmic area. The graphs presented are from eight independent fat bodies, each imaged at two different positions. Immunoblots were analyzed by measuring the mean intensities of unsaturated bands in inverted images. Band intensities were normalized by subtracting mean intensities of adjacent areas in the same lanes of each band. In all cases, Prism software (GraphPad Software) was used to calculate the mean, the standard error and Student's t- or Mann–Whitney tests.
Acknowledgments
We thank S. Field, M. González-Gaitán, G. Juhasz, H. Krämer, I. Nezis, F. Robinson, H. Stenmark, T. Yoshimori, P. Luzio, Bloomington Drosophila Stock Center, DGRC, and VDRC for reagents. Imaging was conducted in part at the UCSD Neuroscience Microscopy Facility (NIH P30 NS047101). We thank T. Meerloo and M. Farquhar for use of the CMM Electron Microscope Facility. This research was supported by FRSQ, AHA and CRS postdoctoral fellowships to SJ, and NIH RO1 GM078176 and support from the SDCSB NIH P50 GM085764 to AAK.
Author contributions
SJ designed, performed and analyzed all experiments; SC performed subset of HeLa cell experiments; SN generated and tested human RAB21 constructs; AAK designed and analyzed experiments; SJ and AAK co-wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Legends
Review Process File
References
- Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- Bento CF, Puri C, Moreau K, Rubinsztein DC. The role of membrane-trafficking small GTPases in the regulation of autophagy. J Cell Sci. 2013;126:1059–1069. doi: 10.1242/jcs.123075. [DOI] [PubMed] [Google Scholar]
- Scott CC, Vacca F, Gruenberg J. Endosome maturation, transport and functions. Semin Cell Dev Biol. 2014;31:2–10. doi: 10.1016/j.semcdb.2014.03.034. [DOI] [PubMed] [Google Scholar]
- Knævelsrud H, Søreng K, Raiborg C, Håberg K, Rasmuson F, Brech A, Liestøl K, Rusten TE, Stenmark H, Neufeld TP, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol. 2013;202:331–349. doi: 10.1083/jcb.201205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader CM, Sanchez D, Furlan M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttinen JMT, Niittykoski M, Salminen A, Kaarniranta K. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta. 2013;1833:503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- Jean S, Kiger AA. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat Rev Mol Cell Biol. 2012;13:463–470. doi: 10.1038/nrm3379. [DOI] [PubMed] [Google Scholar]
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, et al. Mutation of FIG 4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann I, Munro S. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J Cell Sci. 2006;119:1494–1503. doi: 10.1242/jcs.02958. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci U S A. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Numrich J, Ungermann C. Endocytic Rabs in membrane trafficking and signaling. Biol Chem. 2014;395:327–333. doi: 10.1515/hsz-2013-0258. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K, Matsunaga K, Sakane A, Sasaki T, Noda T, Yoshimori T. Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol Biol Cell. 2010;21:4162–4172. doi: 10.1091/mbc.E10-06-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll B, Mohd-Naim N, Maximiano F, Frasa MA, McCormack J, Finelli M, Thoresen SB, Perdios L, Daigaku R, Francis RE, et al. The TBC/RabGAP armus coordinates Rac1 and Rab7 functions during autophagy. Dev Cell. 2013;25:15–28. doi: 10.1016/j.devcel.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18:3463–3471. doi: 10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–1010. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Takats S, Nagy P, Varga A, Pircs K, Karpati M, Varga K, Kovacs AL, Hegedus K, Juhasz G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201:531–539. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor PR, Mullock BM, Bright NA, Lindsay MR, Gray SR, Richardson SC, Stewart A, James DE, Piper RC, Luzio JP. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5:590–595. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CC, Shi H, Guo K, Ng CP, Li J, Gan BQ, Chien Liew H, Leinonen J, Rajaniemi H, Zhou ZH, et al. VAMP8/endobrevin as a general vesicular SNARE for regulated exocytosis of the exocrine system. Mol Biol Cell. 2007;18:1056–1063. doi: 10.1091/mbc.E06-10-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arca S, Rudge R, Vacca M, Raposo G, Camonis J, Proux-Gillardeaux V, Daviet L, Formstecher E, Hamburger A, Filippini F, et al. A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc Natl Acad Sci U S A. 2003;100:9011–9016. doi: 10.1073/pnas.1431910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Collins BM, McCoy AJ, Robinson MS, Owen DJ. A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature. 2007;450:570–574. doi: 10.1038/nature06353. [DOI] [PubMed] [Google Scholar]
- Miller SE, Sahlender DA, Graham SC, Honing S, Robinson MS, Peden AA, Owen DJ. The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell. 2011;147:1118–1131. doi: 10.1016/j.cell.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor PR, Jackson L, Gray SR, Edeling MA, Thompson A, Sanderson CM, Evans PR, Owen DJ, Luzio JP. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell. 2008;134:817–827. doi: 10.1016/j.cell.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean S, Cox S, Schmidt EJ, Robinson FL, Kiger A. Sbf/MTMR13 coordinates PI(3)P and Rab21 regulation in endocytic control of cellular remodeling. Mol Biol Cell. 2012;23:2723–2740. doi: 10.1091/mbc.E12-05-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson FL, Dixon JE. The phosphoinositide-3-phosphatase MTMR2 associates with MTMR13, a membrane-associated pseudophosphatase also mutated in type 4B Charcot-Marie-Tooth disease. J Biol Chem. 2005;280:31699–31707. doi: 10.1074/jbc.M505159200. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjørkøy G, Johansen T, Rusten TE, Brech A, Baehrecke EH, Stenmark H. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010;190:523–531. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, He X, Fu XY, Chang Z. Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J Cell Sci. 2006;119:1053–1062. doi: 10.1242/jcs.02810. [DOI] [PubMed] [Google Scholar]
- Kawai A, Uchiyama H, Takano S, Nakamura N, Ohkuma S. Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy. 2007;3:154–157. doi: 10.4161/auto.3634. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Simpson JC, Griffiths G, Wessling-Resnick M, Fransen JA, Bennett H, Jones AT. A role for the small GTPase Rab21 in the early endocytic pathway. J Cell Sci. 2004;117:6297–6311. doi: 10.1242/jcs.01560. [DOI] [PubMed] [Google Scholar]
- Pellinen T, Arjonen A, Vuoriluoto K, Kallio K, Fransen JA, Ivaska J. Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of beta1-integrins. J Cell Biol. 2006;173:767–780. doi: 10.1083/jcb.200509019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egami Y, Araki N. Characterization of Rab21-positive tubular endosomes induced by PI3K inhibitors. Exp Cell Res. 2008;314:729–737. doi: 10.1016/j.yexcr.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Tower-Gilchrist C, Lee E, Sztul E. Endosomal trafficking of the G protein-coupled receptor somatostatin receptor 3. Biochem Biophys Res Commun. 2011;413:555–560. doi: 10.1016/j.bbrc.2011.08.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Holroyd C, Tikkanen R, Honing S, Jahn R. The R-SNARE endobrevin/VAMP-8 mediates homotypic fusion of early endosomes and late endosomes. Mol Biol Cell. 2000;11:3289–3298. doi: 10.1091/mbc.11.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Zhang T, Xu Y, Subramaniam VN, Griffiths G, Hong W. Endobrevin, a novel synaptobrevin/VAMP-like protein preferentially associated with the early endosome. Mol Biol Cell. 1998;9:1549–1563. doi: 10.1091/mbc.9.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AA, Logan AM, Schmidt EJ, Robinson FL. The CMT4B disease-causing phosphatases Mtmr2 and Mtmr13 localize to the Schwann cell cytoplasm and endomembrane compartments, where they depend upon each other to achieve wild-type levels of protein expression. Hum Mol Genet. 2013;22:1493–1506. doi: 10.1093/hmg/dds562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgo A, Proux-Gillardeaux V, Sotirakis E, Bun P, Casano A, Verraes A, Liem RK, Formstecher E, Coppey-Moisan M, Galli T. A molecular network for the transport of the TI-VAMP/VAMP7 vesicles from cell center to periphery. Dev Cell. 2012;23:166–180. doi: 10.1016/j.devcel.2012.04.019. [DOI] [PubMed] [Google Scholar]
- Chapman RE, Munro S. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO J. 1994;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh GG, Pérez-Dorado I, Jackson LP, Wartosch L, Schäfer IB, Gray SR, McCoy AJ, Zeldin OB, Garman EF, Harbour ME, et al. VARP is recruited on to endosomes by direct interaction with retromer, where together they function in export to the cell surface. Dev Cell. 2014;29:591–606. doi: 10.1016/j.devcel.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Holroyd C, Fasshauer D, Pabst S, Von Mollard GF, Jahn R. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ. Endosomal sorting and signalling: an emerging role for sorting nexins. Nat Rev Mol Cell Biol. 2008;9:574–582. doi: 10.1038/nrm2427. [DOI] [PubMed] [Google Scholar]
- Kent HM, Evans PR, Schäfer IB, Gray SR, Sanderson CM, Luzio JP, Peden AA, Owen DJ. Structural basis of the intracellular sorting of the SNARE VAMP7 by the AP3 adaptor complex. Dev Cell. 2012;22:979–988. doi: 10.1016/j.devcel.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Lewis MJ, Black MW, Pelham HR. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–557. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya T, Miaczynska M, Coskun U, Lommer B, Runge A, Drechsel D, Kalaidzidis Y, Zerial M. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–1097. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- Franklin NE, Bonham CA, Xhabija B, Vacratsis PO. Differential phosphorylation of the phosphoinositide 3-phosphatase MTMR2 regulates its association with early endosomal subtypes. J Cell Sci. 2013;126:1333–1344. doi: 10.1242/jcs.113928. [DOI] [PubMed] [Google Scholar]
- Oppermann FS, Grundner-Culemann K, Kumar C, Gruss OJ, Jallepalli PV, Daub H. Combination of chemical genetics and phosphoproteomics for kinase signaling analysis enables confident identification of cellular downstream targets. Mol Cell Proteomics. 2012;11:0111.012351. doi: 10.1074/mcp.O111.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Malmstrom J, Gerrits B, Campbell D, Lam H, Schmidt A, Rinner O, Mueller LN, Shannon PT, Pedrioli PG, et al. PhosphoPep–a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Pan JA, Dbouk HA, Ballou LM, DeLeon JL, Fan Y, Chen JS, Liang Z, Li G, Backer JM, et al. Class IA PI3K p110beta subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell. 2013;50:29–42. doi: 10.1016/j.molcel.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichkova M, Juan J, Kadandale P, Jean S, Ribeiro I, Raman V, Stefan C, Kiger AA. Drosophila Mtm and class II PI3K coregulate a PI(3)P pool with cortical and endolysosomal functions. J Cell Biol. 2010;190:407–425. doi: 10.1083/jcb.200911020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS ONE. 2012;7:e44214. doi: 10.1371/journal.pone.0044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak AL, Griffin S, Rowlands D, Harris M, Yi M, Lemon SM, Weinman SA. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 2010;6:e1001087. doi: 10.1371/journal.ppat.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco I, Gulluni F, Campa CC, Costa C, Margaria JP, Ciraolo E, Martini M, Monteyne D, De Luca E, Germena G, et al. PI3K class II alpha controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev Cell. 2014;28:647–658. doi: 10.1016/j.devcel.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Chen J, Liu J, Brunzelle JS, Huang B, Wakeham N, Terzyan S, Li X, Rao Z, Li G, et al. Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5. EMBO J. 2007;26:3484–3493. doi: 10.1038/sj.emboj.7601771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Legends
Review Process File