Abstract
We retrospectively analyzed a rare Salmonella enterica serovar Manhattan outbreak that occurred in Italy in 2009 to evaluate the potential of new genomic tools based on differential single nucleotide polymorphism (SNP) analysis in comparison with the gold standard genotyping method, pulsed-field gel electrophoresis. A total of 39 isolates were analyzed from patients (n = 15) and food, feed, animal, and environmental sources (n = 24), resulting in five different pulsed-field gel electrophoresis (PFGE) profiles. Isolates epidemiologically related to the outbreak clustered within the same pulsotype, SXB_BS.0003, without any further differentiation. Thirty-three isolates were considered for genomic analysis based on different sets of SNPs, core, synonymous, nonsynonymous, as well as SNPs in different codon positions, by Bayesian and maximum likelihood algorithms. Trees generated from core and nonsynonymous SNPs, as well as SNPs at the second and first plus second codon positions detailed four distinct groups of isolates within the outbreak pulsotype, discriminating outbreak-related isolates of human and food origins. Conversely, the trees derived from synonymous and third-codon-position SNPs clustered food and human isolates together, indicating that all outbreak-related isolates constituted a single clone, which was in line with the epidemiological evidence. Further experiments are in place to extend this approach within our regional enteropathogen surveillance system.
INTRODUCTION
Salmonellosis is a major food-borne disease worldwide, with an estimated 93.8 million cases occurring each year, resulting in 155,000 deaths (1). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks (2) indicated that nontyphoid salmonellosis was the second most reported food-borne zoonosis in Europe in 2012, trailing only behind Campylobacter jejuni infection. The 2012 overall notification rate for human salmonellosis in the European Union (EU) was 22.2 episodes per 100,000 population, for a total of 91,034 confirmed cases, with hospitalization and mortality rates of 45.1% and 0.14%, respectively. The highest proportions of Salmonella-positive foodstuff samples were reported for fresh turkey, poultry, and pork at 4.4%, 4.1%, and 0.7%, respectively (2). In order to manage this food-borne infection and to limit its health and economic burdens, surveillance programs have developed and implemented DNA-based subtyping methods to identify outbreaks in a timely manner and to trace infections back to their food sources. Over the past decades, the two most intensively used protocols for Salmonella subtyping have been pulsed-field gel electrophoresis (PFGE) and multilocus variable-number tandem-repeat analysis (MLVA) (3). Unfortunately, these methods rely on just few features of the entire bacterial genome (rare restriction sites for PFGE or few polymorphic loci for MLVA) to assess the relatedness of different isolates. During epidemiological investigations of food-borne outbreaks, this limitation might lead to difficulties in distinguishing outbreak-related from outbreak-unrelated Salmonella enterica subsp. enterica isolates due to the high genetic homogeneity of this subspecies (4). Multilocus sequence typing (MLST) is another molecular tool for bacterial typing based on allelic differences in the loci of specified housekeeping genes (5). While proposed as an alternative to classical serotyping (6), MLST does not seem to be discriminatory enough when all isolates being tested belong to the same serotype (7). With the aim of improving resolution in molecular epidemiology, the technological advancements of whole-genome sequencing (WGS) may provide an unprecedented opportunity to access the entire genome information at a reasonable cost, as well as to set a new series of high-resolution standards in molecular epidemiology. As PFGE and MLVA are able to resolve more genotypes within a single serovar, WGS has already proved its resolution power to detect variations within otherwise undistinguishable bacterial clones (by PFGE or MLVA), as shown by recent examples in the literature (8, 9). Large studies based on WGS within S. enterica subspecies (10) and within serovars in S. enterica subsp. enterica (11, 12) contributed to the elucidation of Salmonella phylogenetic diversity and also accomplished important steps forward in the area of bacterial disease tracking. Moreover, serovar-specific studies on S. enterica subsp. enterica have highlighted microevolutionary differences among clinical, environmental, and food isolates in S. enterica serovars Montevideo (13, 14), Enteritidis (4), Newport (15), Typhimurium (16–18), and Heidelberg (12), which would have been missed by more traditional approaches.
While outbreaks of more common serovars, such as Salmonella Typhimurium and Salmonella Enteritidis, have been reported and investigated, only a few human outbreaks due to S. enterica serovar Manhattan have been reported (19, 20) worldwide in the past 60 years, and none have been characterized at the genomic level. Here, we present a WGS-based retrospective analysis of the only Salmonella Manhattan outbreak ever documented in Italy, which occurred from June to July 2009 in a relatively small geographic area in the province of Modena.
The outbreak investigation at the time of the event was carried out by international standard epidemiological techniques (21) and by PFGE on the isolates from patients and food, feed, animal, and environmental sources.
The aim of this study was 2-fold: (i) to evaluate the effectiveness of WGS to accurately identify the relationships among all the outbreak-related isolates with enough resolution to clarify the ambiguities that PFGE was not able to unravel, and (ii) to explore and test new genomic tools for bacterial molecular epidemiology based on synonymous and nonsynonymous single-nucleotide polymorphisms (SNPs) and SNPs in different codon positions.
We selected this specific Salmonella Manhattan outbreak to test our WGS pipeline because of three main features that made this outbreak a particularly suitable case study. First, Salmonella Manhattan is considered a rare serotype, as confirmed by the regional surveillance system for Salmonella of Emilia-Romagna, which over the past 3 years recorded a yearly average of only 5.6 sporadic cases over a total of 924 isolates per year, from a regional population of about 5,000,000 (M. Morganti, E. Scaltriti, L. Bolzoni, G. Casadei, and S. Pongolini; Enter-net Italia, unpublished data). This low prevalence of Salmonella Manhattan infection provides a reasonable confidence that virtually all isolates collected in the outbreak area at the time of the episode belonged to the outbreak, therefore preventing the noise effect due to unrelated isolates wrongly assigned to the epidemic. Second, the investigation conducted at the time of the outbreak was successful in tracing the infection back to a food point source using internationally coded epidemiological methods (21); bacterial isolates were also recovered not only from food (pork sausage) at the retail level but also along the food chain up to the raw meat used to prepare the implicated food (at the production establishment). Third, the regional surveillance system for Salmonella of Emilia-Romagna, hosted at the Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna (IZSLER), holds a full collection of Salmonella Manhattan strains covering the years 2001 to present. This set of isolates was pivotal in the conduct of a successful epidemiological investigation and for testing our WGS-based analyses of this rare serovar.
CASE REPORT
The diagnostic unit of Parma of IZSLER is the Regional Reference Center for Surveillance of Enteropathogens (Enter-net) of clinical, environmental, animal, and food origins. Within this activity, a cluster of 15 human infections caused by Salmonella Manhattan was detected in the province of Modena from June to July 2009. All 15 isolates showed the same PFGE profile, SXB_BS.0003, strengthening the hypothesis that the unusually high incidence of this rare serovar was due to an epidemic outbreak. Consequently, an epidemiological investigation was undertaken and, considering the rarity of the serovar involved, all 21 isolates of Salmonella Manhattan available from the surveillance collection of IZSLER were genotyped by PFGE to get possible clues about the source of the outbreak. Thirteen isolates from the collection had the same PFGE profile as that of the outbreak strain, but only three of them had been isolated just before the onset of the outbreak (May/June 2009). Two had been isolated from pork sausage at the establishment of an industrial producer that distributed in the outbreak area, while one had been recovered from swine intestine at an establishment near the outbreak area that processed guts for the salami industry. According to the epidemiological investigation, the gut processing establishment had no correlation with the outbreak. However, as its isolate presented the same PFGE pulsotype as that of the outbreak-related isolates, health authorities were left with a certain degree of uncertainty about its possible role. Following the results of the epidemiological and molecular analyses, food samples were collected at retail sources in the outbreak area and at the establishment producing the sausage in order to confirm the source and clonality of the outbreak strain. Two samples from retail-collected sausages, along with a sample from fresh pork supplies of the sausage producer, scored positive for the outbreak pulsotype. Based on these results, the sausage from the implicated producer was recalled, leading to the outbreak extinction.
MATERIALS AND METHODS
Bacterial isolates.
A total of 39 Salmonella Manhattan isolates were included in the study. Fifteen isolates were involved in the epidemic episode, another three isolates were collected within the epidemiological investigation, and 21 were collected between 2001 and 2009 during the surveillance activity of IZSLER (Table 1). The isolates were isolated and streak purified with standard microbiological techniques and stocked at −80°C. They were cultured on plates with Trypticase soy agar with 5% defibrinated sheep blood (TSA-SB) and incubated overnight at 37°C before being typed by pulsed-field gel electrophoresis, according to the PulseNet protocol (22). The isolates selected for WGS were inoculated into brain heart infusion broth and cultured overnight at 37°C with agitation (200 rpm).
TABLE 1.
Complete data set of Salmonella Manhattan isolates analyzed in this studya
| Lab no. | Isolate no. (this study) | Date of isolation (DD/MM/YYYY) | Isolation place (province) | Matrix | PFGE pulsotype |
|---|---|---|---|---|---|
| 160969_3 | SM1b | 06/30/2009 | Modena | Human | SXB_BS.0003 |
| 160969_5 | SM2b | 06/30/2009 | Modena | Human | SXB_BS.0003 |
| 160969_6 | SM3b | 06/30/2009 | Modena | Human | SXB_BS.0003 |
| 165051_2 | SM4b | 07/03/2009 | Modena | Human | SXB_BS.0003 |
| 165051_3 | SM5b | 07/03/2009 | Modena | Human | SXB_BS.0003 |
| 165051_5 | SM6b | 07/03/2009 | Modena | Human | SXB_BS.0003 |
| 165051_7 | SM7b | 07/30/2009 | Modena | Human | SXB_BS.0003 |
| 111113 | SM8b | 07/03/2009 | Modena | Human | SXB_BS.0003 |
| 165051_11 | SM9b | 07/03/2009 | Modena | Human | SXB_BS.0003 |
| 165051_12 | SM10b | 07/03/2009 | Modena | Human | SXB_BS.0003 |
| 180073_1 | SM11b | 07/22/2009 | Modena | Human | SXB_BS.0003 |
| 180073_2 | SM12b | 07/22/2009 | Modena | Human | SXB_BS.0003 |
| 180073_3 | SM13b | 07/22/2009 | Modena | Human | SXB_BS.0003 |
| 180073_4 | SM14b | 07/22/2009 | Modena | Human | SXB_BS.0003 |
| 180073_6 | SM15b | 07/22/2009 | Modena | Human | SXB_BS.0003 |
| 250920 | SM42b | 08/31/2009 | Milano | Pork | SXB_BS.0003 |
| 227021 | SM32b | 05/06/2009 | Milano | Pork sausage | SXB_BS.0003 |
| 188801 | SM52b | 05/06/2009 | Milano | Pork sausage | SXB_BS.0003 |
| 216630_1 | SM53b | 09/03/2009 | Modena | Pork sausage | SXB_BS.0003 |
| 216630_2 | SM54b | 09/03/2009 | Modena | Pork sausage | SXB_BS.0003 |
| 226957 | SM16 | 03/07/2006 | Mantova | Swine | SXB_PR.0753 |
| 226963 | SM17b | 03/20/2006 | Mantova | Swine | SXB_PR.0753 |
| 226972 | SM19b | 03/20/2006 | Sondrio | Pork salami | SXB_PR.0753 |
| 226979_1 | SM21b | 07/31/2006 | Cremona | Swine gut | SXB_BS.0003 |
| 226985 | SM23b | 08/03/2006 | Milano | Pork sausage | SXB_BS.0003 |
| 226987 | SM24b | 08/03/2006 | Milano | Pork sausage | SXB_BS.0003 |
| 226993 | SM26 | 01/22/2007 | Ravenna | Hamburger | SXB_BS.0003 |
| 226998 | SM27b | 06/29/2007 | Milano | Pork | SXB_BS.0003 |
| 227002 | SM28 | 09/18/2002 | Pavia | Surface water | SXB_BS.0003 |
| 227009 | SM29b | 09/02/2002 | Bologna | Bovine sausage | SXB_PR.0754 |
| 227015 | SM31 | 09/11/2001 | Pavia | Surface water | SXB_PR.0751 |
| 227033 | SM35b | 11/29/2008 | Ravenna | Swine stool | SXB_BS.0003 |
| 227039 | SM36b | 09/30/2008 | Brescia | Swine stool | SXB_PR.0752 |
| 227052 | SM38b | 09/24/2008 | Milano | Swine stool | SXB_BS.0003 |
| 188806 | SM48b | 06/03/2009 | Reggio Emilia | Swine intestine | SXB_BS.0003 |
| 188790 | SM47 | 10/01/2002 | Pavia | Surface water | SXB_BS.0003 |
| 188795 | SM49b | 03/09/2009 | Brescia | Chicken farm | SXB_PR.0753 |
| 188787 | SM51 | 09/17/2002 | Pavia | Surface water | SXB_BS.0003 |
| 188781 | SM50b | 07/31/2001 | Modena | Minced pork | SXB_PR.0751 |
The isolates above the line break are the outbreak-related isolates (15 human-origin and 5 food-origin isolates), and those below the line break are the 19 Salmonella Manhattan collection isolates. SM32 and SM52 were also collection isolates, but they were eventually attributed to the outbreak, following the results of this study.
These Salmonella Manhattan isolates were selected for whole-genome sequencing.
Pulsed-field gel electrophoresis.
All isolates were genotyped by PFGE, according to the PulseNet protocol (22). Genomic DNA underwent XbaI restriction before electrophoresis in a Chef Mapper XA system (Bio-Rad, CA, USA). The PFGE patterns were analyzed using the BioNumerics Software version 6.6 (Applied-Maths, Sint-Martens-Latem, Belgium) and associated with isolate information in our surveillance database. Clustering of the PFGE profiles was generated using the unweighted-pair group method using averages (UPGMA) based on the Dice similarity index (optimization, 1%; band matching tolerance, 1%). Following a comparison of the electrophoretic profiles, a PFGE pattern (pulsotype) was assigned to each isolate within the Regional Surveillance Database of Emilia-Romagna.
Whole-genome sequencing.
All outbreak-related isolates and a selection of the IZSLER Salmonella Manhattan collection, representative of the different pulsotypes detected, were subjected to WGS (Table 1), for a total of 33 isolates. Genomic DNA was extracted from overnight cultures using the Qiagen DNeasy blood and tissue kit (Qiagen) and quality controlled and quantified using a Synergy H1 hybrid multimode microplate reader (BioTek, Winooski, VT, USA). The sequencing libraries were prepared with the Nextera XT sample preparation kit (Illumina, San Diego, CA, USA), and sequencing was performed on the Illumina MiSeq platform, with a 2 × 250-bp paired-end run.
Read quality check and assembly.
All read sets were evaluated for sequence quality and read-pair length using the softwares FastQC and Flash (23). FastQC allowed us to assess the overall quality of the generated sequences, while Flash was used to measure the distance between the sequence read pairs. All the read sets that passed the quality check (visual check for FastQC and average read pair distance >100 nucleotides [nt] for Flash) were assembled with MIRA 4.0 (24) using accurate settings for de novo assembly mode.
In silico multilocus sequence typing.
In silico MLST was performed using the MLST scheme optimized by the University of Warwick (http://mlst.warwick.ac.uk/mlst/dbs/Senterica).
Comparative genomics by local variation calling.
In a previous work, we sequenced and published the first improved high-quality draft genome (25) of Salmonella Manhattan (strain 111113) (26). The 18 contigs of the Salmonella Manhattan 111113 genome, belonging to a human isolate of the outbreak presented here, were concatenated in a pseudochromosome and used as a reference for alignment of each of the other 32 genome assemblies included in this study, using progressiveMauve (27). A previously described bioinformatic pipeline (28) was then used to merge the results of all isolates for comparison and to extract the coordinates of all local variations spanning from SNPs to longer variations (mutations, insertions, and deletions), based on the annotation of the reference genome of strain 111113. Core SNPs were identified as single nondegenerate mutated bases flanked by identical bases and present in all 33 genomes (including that of strain 111113). Genes presenting at least one core SNP were selected and compared against the Virulence Factors Database (VFDB) (29–31), using a BLAST search with a 10−5 E value cutoff.
Analysis of variations.
Open reading frames (ORFs) were predicted and translated on all assembled genomes (including the previously published Salmonella Manhattan strain 111113 genome [26]) using Prodigal (32). Next, every genomic variation (SNPs, mutations, insertions, and deletions) was parsed in order to assign it to one of the following subsets of isolates: (i) all outbreak-related isolates, irrespective of the human, food, or raw meat origin; (ii) outbreak-related human-origin-only isolates; and (iii) outbreak-related food-origin-only isolates (including those from sausage and raw meat).
Phylogenetic analysis.
From the core SNP data set, different subsets were generated: (i) nonsynonymous SNPs, (ii) synonymous SNPs, and (iii) SNPs at the first, second, or third codon position. The core and subsets of SNPs were used as inputs for generating SNP-based phylogenies using the maximum likelihood (ML) or the Bayesian methods. Model choice was evaluated in JModelTest (33). Maximum likelihood analyses were run in PhyML (34), with a generalized time-reversible (GTR) substitution model and 100 bootstrap iterations, while Bayesian analyses were run in MrBayes (35, 36), using the same model for 2,000,000 generations, with chains sampled every 1,000 generations. The final parameter values and trees were summarized after discarding 25% of the posterior sample. The ML and Bayesian trees were displayed and edited for publication with FigTree version 1.4.0.
Nucleotide sequence accession numbers.
The genome sequences of Salmonella Manhattan (strain 111113; study identification [ID], SM8) contigs were previously deposited at EBI under the accession no. CBKW010000001 to CBKW010000021 (project PRJEB1854). The newly 32 sequenced genomes (contigs) of Salmonella Manhattan were deposited at EBI under the project number PRJEB5339 and are summarized here in the format isolate lab no./study identification no.: WGS accession number: 160969_3/SM1: CCBJ010000610 to CCBJ010000701, 160969_5/SM2: CCBJ010000175 to CCBJ010000212, 160969_6/SM3: CCBJ010000291 to CCBJ010000308, 165051_2/SM4: CCBJ010001977 to CCBJ010002069, 165051_3/SM5: CCBJ010002070 to CCBJ010000089, 165051_5/SM6: CCBJ010000001 to CCBJ010000100, 165051_7/SM7: CCBJ010004043 to CCBJ010004081, 165051_11/SM9: CCBJ010003194 to CCBJ010003512, 165051_12/SM10: CCBJ010000309 to CCBJ010000327, 180073_1/SM11: CCBJ010002338 to CCBJ010002378, 180073_2/SM12: CCBJ010003726 to CCBJ010003749, 180073_3/SM13: CCBJ010001070 to CCBJ010001515, 180073_4/SM14: CCBJ010001516 to CCBJ010001924, 180073_6/SM15: CCBJ010000702 to CCBJ010000770, 250920/SM42: CCBJ010000328 to CCBJ010000609, 227021/SM32: CCBJ010004870 to CCBJ010004957, 188801/SM52: CCBJ010002097 to CCBJ010002229, 216630_1/SM53: CCBJ010002817 to CCBJ010003193, 216630_2/SM54: CCBJ010000213 to CCBJ010000238, 226963/SM17: CCBJ010002257 to CCBJ010002337, 226972/SM19: CCBJ010002230 to CCBJ010002256, 226979_1/SM21: CCBJ010000101 to CCBJ010000174, 226985/SM23: CCBJ010003750 to CCBJ010004042, 226987/SM24: CCBJ010003702 to CCBJ010003725, 226998/SM27: CCBJ010000771 to CCBJ010001069, 227009/SM29: CCBJ010001925 to CCBJ010001976, 227033/SM35: CCBJ010000239 to CCBJ010000268, 227039/SM36: CCBJ010000269 to CCBJ010000290, 227052/SM38: CCBJ010002379 to CCBJ010002816, 188806/SM48: CCBJ010003540 to CCBJ010003701, 188795/SM49: CCBJ010004082 to CCBJ010004692, and 188781/SM50: CCBJ010004693 to CCBJ010004869.
RESULTS
We present here reanalysis by WGS of an outbreak caused by Salmonella Manhattan in the province of Modena (Italy) in 2009. The isolates from the human cases were SM1, -2, -3, -4, -5, -6, -7, -8, -9, -10, -11, -12, -13, -14, and -15. Out of the 21 collection isolates available, all were genotyped by PFGE to search for clues on the source of infection, and SM21, -23, -24, -26, -27, -28, -32, -35, -38, -47, -48, -51, and -52 showed the outbreak pulsotype; however, SM36, -16, -17, -19, -29, -31, -49, and -50 belonged to different pulsotypes, and a selection of them were included in this study as outgroup isolates. SM42, -53, and -54 were isolated during the microbiological follow-up of the episode and presented the outbreak pulsotype.
Pulsed-field gel electrophoresis.
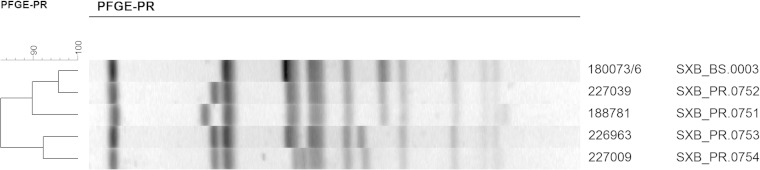
The 39 Salmonella Manhattan isolates of the study showed five different XbaI-PFGE profiles: SXB_BS.0003, SXB_PR.0753, SXB_PR.0754, SXB_PR.0751, and SXB_PR.0752 (Fig. 1). All the human isolates (SM1 to SM15) showed the same PFGE profile (SXB_BS.0003), supporting the hypothesis that the unusually high incidence of this rare serovar was due to a single epidemic clone.
FIG 1.
Similarity of Salmonella Manhattan isolates, examined in this study, inferred by pulsed-field gel electrophoresis profiles (PFGE-PR). The samples underwent XbaI restriction and pattern analysis according to the standard PulseNet protocol. The UPGMA dendrogram of all the profiles of the study is reported on the left; the ruler indicates the similarity values. The laboratory numbers of the isolates and their pulsotypes are reported on the right.
Another 13 isolates from the IZSLER surveillance collection belonged to genotype SXB_BS.0003. Among these, three (SM32, SM48, and SM52) dated back to just before the outbreak period (May/June 2009) and were pivotal in guiding the epidemiological investigation. SM48 originated from an establishment near the outbreak area that processed swine guts for the salami industry. Due to this microbiological and molecular finding, the establishment was suspected of having a role in the outbreak, although no evident correlation with the human infections was made. More significantly, SM32 and SM52 were isolated just before the onset of the episode from pork sausages produced at an industrial establishment that shipped to retail stores in the outbreak area. Consequently, sausages from this producer, which were on sale in the outbreak area, were sampled along with the pork purchased by the producer. Both the sausages and the pork were positive for Salmonella Manhattan with the outbreak pulsotype (SXB_BS.0003) (isolates SM53 and SM54 from the sausages and SM42 from pork). Interestingly, two Salmonella Manhattan isolates from our collection, isolated within the own-check hygiene procedures of the producer (SM23 and SM24) 3 years before the outbreak, presented the same genotype. Also, the surveillance collection isolates SM21, -26, -27, -28, -35, -38, -47, and -51 shared the outbreak pulsotype, but they did not seem to be correlated with the outbreak or source of infection.
Among the other nonoutbreak PFGE profiles detected, the pulsotype SXB_PR.0752 (isolate SM36) had 95% similarity with the outbreak pulsotype, while the genotypes SXB_PR.0751 (isolates SM31 and SM50), SXB_PR.0753 (isolates SM16, SM17, SM19, and SM49), and SXB_PR.0754 (isolate SM29) were less similar (90%, 84%, and 84%, respectively) (Fig. 1).
Whole-genome sequencing.
The genomes of the 33 Salmonella Manhattan isolates considered for genomic analysis, including the already deposited genome of strain 111113 (26), were sequenced, quality checked, and assembled to draft status, from an average of 2,593,738 MiSeq paired-end reads per genome. The average sequenced genome characteristics were 4,678,201 nt in length, 150 large (>1,000 nt) contigs, and an N50 of 212,360. The genome data for each isolate are listed in Table S1 in the supplemental material. The MLST profile was determined for all draft genomes, which were found to belong to the same sequence type (ST), ST18. All assembled genomes underwent comparative and phylogenetic analyses.
Analysis of variations.
A comparative genomic analysis was implemented to detect the differences between the Salmonella Manhattan genomes, in terms of nucleotide variations, exclusive to (i.e., present in all the isolates of a group and absent in all the others) the outbreak-related isolates, as divided into the following main groups: (i) all outbreak-related isolates, irrespective of the human, food, or raw meat origin; (ii) outbreak-related human-origin-only isolates; and (iii) outbreak-related food-origin-only isolates (including sausages and raw meat).
Of all the nondegenerate nucleotide variations (total 9,410) discovered by the progressiveMauve algorithm, 14 were outbreak specific, and all were core SNPs (two intergenic, two synonymous, and 10 nonsynonymous), divided as six variations exclusive to all outbreak-related isolates, three variations characteristic of the food-origin-only outbreak-related isolates, and five characteristic of the outbreak-related human-origin-only isolates (Table 2).
TABLE 2.
Characteristic SNPs of three groups of outbreak-related isolates
| Group of isolates | Amino acid change | Codon change | Position CDSa | Type of SNP | Gene | Locus →tag | Strand | Product name |
|---|---|---|---|---|---|---|---|---|
| All outbreak | C→R | TGT→CGT | 625 | Genic | cobT | SMA01→2283 | − | Nicotinate-nucleotide–dimethylbenzimidazole phosphoribosyltransferase |
| N→N | AAT→AAC | 156 | Genic | gntR | SMA01→3706 | − | Gluconate utilization system Gnt-transcriptional repressor | |
| A→T | GCC→ACC | 577 | Genic | ansB | SMA01→3765 | − | l-Asparaginase | |
| V→A | GTC→GCC | 988 | Genic | dcuC | SMA01→4465 | − | Putative cryptic C4-dicarboxylate transporter | |
| Intergenic | ||||||||
| K→E | AAA→GAA | 70 | Genic | betI | SMA01→1140 | + | Transcriptional regulator, TetR family | |
| Human origin | M→T | ATG→ACG | 584 | Genic | dsbI | SMA01→0572 | + | Thiol-disulfide oxidoreductase, DsbB-like |
| A→T | GCC→ACC | 310 | Genic | sthD | SMA01→3447 | − | β-fimbriae usher protein | |
| V→V | GTT→GTC | 465 | Genic | ispH | SMA01→3526 | + | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | |
| Q→STOP | CAA→TAA | 252 | Genic | rfbD | SMA01→4557 | + | UDP-galactopyranose mutase | |
| Intergenic | ||||||||
| Food origin | S→I | AGC→ATC | 872 | Genic | fliK | SMA01→2244 | + | Flagellar hook-length control protein FliK |
| P→L | CCT→CTT | 17 | Genic | SMA01→0101 | + | Hypothetical protein | ||
| A→V | GCC→GTC | 1544 | Genic | fdrA | SMA01→4374 | + | Protein FdrA: acyl-CoA synthetaseb |
CDS, coding sequence.
CoA, coenzyme.
Phylogenetic analysis.
Phylogeny was reconstructed using an SNP-based approach. SNPs were extracted from the assembled genomes using a bioinformatic pipeline (28) based on progressiveMauve (27). Of the 9,410 detected variations, 953 were core SNPs, with 224 being synonymous and 467 being nonsynonymous; the remaining 262 SNPs were marked as intergenic. Among the synonymous SNPs, 6% and 94% were located in the first and third codon positions, respectively, while among the nonsynonymous SNPs, 43% were in the first, 42% in the second (total, 85% for the two positions), and 15% in the third codon position. The number of synonymous and nonsynonymous core SNPs at the first, second, and third positions were 214, 194, and 283, respectively.
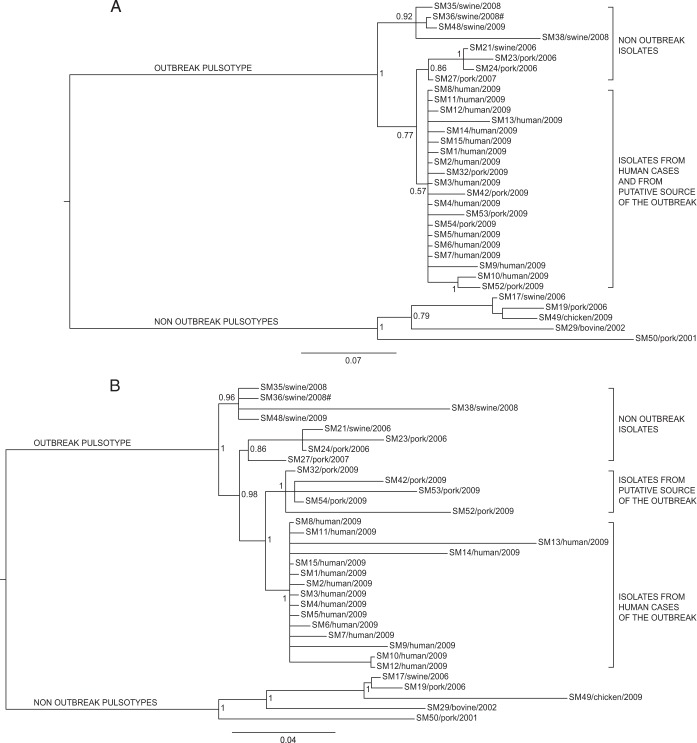
The phylogenetic analysis of the study isolates was performed separately based on the different subsets of SNPs considered, namely, core, synonymous, nonsynonymous, and different codon positions using both Bayesian (Fig. 2 to 4) and maximum likelihood algorithms (see Fig. S1 and S2 in the supplemental material). Both algorithms returned the same phylogenetic results on each subset.
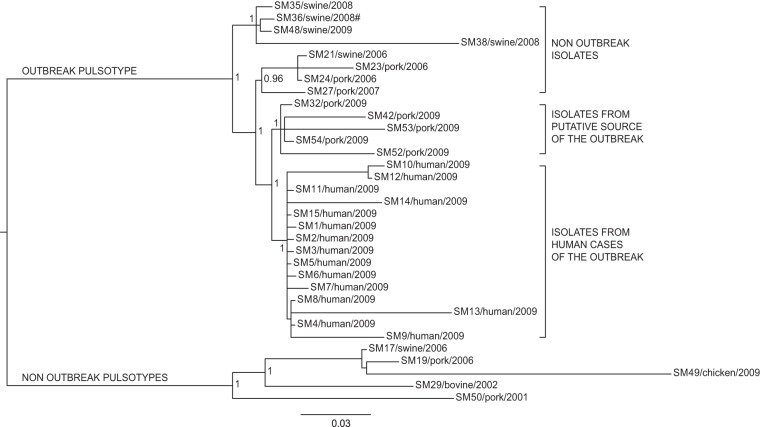
FIG 2.
Bayesian phylogeny of the 33 Salmonella Manhattan sequenced genomes based on core SNPs. The posterior probabilities are indicated in each principal node of the tree. The scale bar units are the nucleotide substitutions per site. #, WGS analyses clustered isolate SM36 (pulsotype SXB_PR.0752) together with the isolates of the outbreak pulsotype (SXB_BS0003).
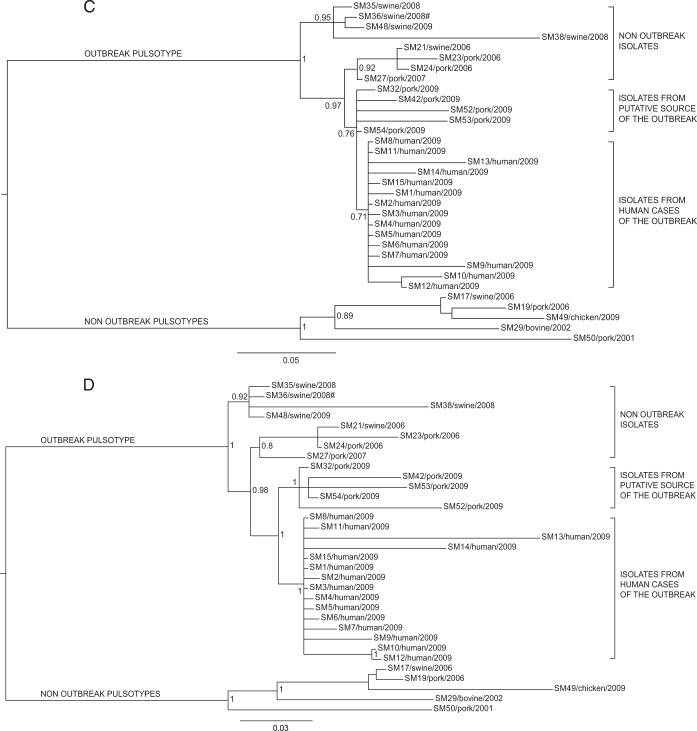
FIG 4.
Phylogenetic Bayesian analysis of the 33 Salmonella Manhattan sequenced genomes based on SNPs in first (A), second (B), third (C), and first plus second codon position (D) data sets. The posterior probabilities are indicated in each principal node of the tree. The scale bar units are the nucleotide substitutions per site. #, WGS analyses clustered isolate SM36 (pulsotype SXB_PR.0752) together with the isolates of the outbreak pulsotype (SXB_BS0003).
All data sets identified two major clades: one grouping all the isolates belonging to pulsotype SXB_BS.0003 and the highly related SXB_PR.0752 (95% similarity), and the other constituted by isolates with different pulsotypes (SXB_PR.0753, SXB_PR.0754, and SXB_PR.0751). Interestingly, WGS analyses clustered isolate SM36 (pulsotype SXB_PR.0752) together with the isolates of pulsotype SXB_BS0003, meaning they are highly related compared to isolates of the other pulsotypes of the study. Therefore, we considered SXB_PR.0752 together with SXB_BS.0003 for the subsequent analyses of phylogeny and presence of variants.
Phylogeny based on core SNPs revealed four main groups inside the outbreak pulsotype. Isolates that were not epidemiologically related to the outbreak formed two monophyletic clusters, with the outermost one grouping isolates from various locations and previous years but always from swine stool within the own-check procedures of pig farms (isolates SM35, SM36, and SM38) or at food processing plants (isolate SM48). The other group included isolates collected at the sausage-producing establishment within its hygiene monitoring system 3 years before the outbreak (SM23 and SM24), along with an isolate collected on a pig farm in the same period (SM21). Isolate SM27 originated from another food processing plant in the same area of the sausage producer, but that was never linked to the outbreak.
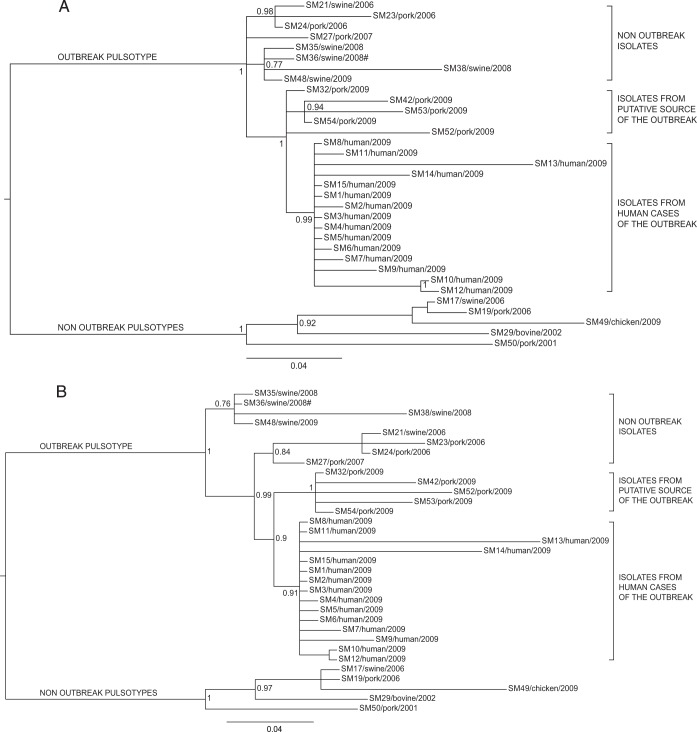
The two innermost clusters included all the outbreak-related isolates. Five strains isolated from sausages prepared by the implicated producer (SM32, SM42, SM52, SM53, and SM54), both at a retail locations in the outbreak area and at the establishment, which were distinct from the cluster of human isolates of the outbreak (from SM1 to SM15). All outbreak-related isolates are monophyletic, confirming their derivation from a common ancestor. In order to better investigate the relationships among those isolates, we performed additional analyses on specific subsets of the core SNPs to take into account the possible effects of selective evolutionary pressure. We separately considered nonsynonymous SNPs, synonymous SNPs, and SNPs at the first, second, and third codon positions as presumptively subjected to decreasing selective pressures (37). The trees corresponding to the different subsets of SNPs are shown in Fig. 3 and 4. The trees generated by nonsynonymous SNPs and SNPs at the first plus second and second codon positions showed the same topology described by the whole data set of core SNPs, with a clear distinction between outbreak-related isolates of human and food origins. The phylogenies generated by SNPs under minor selective pressure (i.e., third position) revealed different scenarios, with the loss of a node inside the outbreak cluster showing isolates of human origin as a subgroup within the food-origin outbreak isolates. Considering synonymous SNPs only, the outbreak isolates of human and food origins are grouped in one cluster, being indicative of a single circulating clone. The phylogenetic inferences made by Bayesian and maximum likelihood algorithms gave identical results (see Fig. S1 and S2 in the supplemental material).
FIG 3.
Phylogenetic Bayesian analysis of the 33 Salmonella Manhattan sequenced genomes based on synonymous (A) and nonsynonymous (B) SNP data sets. The posterior probabilities are indicated in each principal node of the tree. The scale bar units are the nucleotide substitutions per site. #, WGS analyses clustered isolate SM36 (pulsotype SXB_PR.0752) together with the isolates of the outbreak pulsotype (SXB_BS0003).
DISCUSSION
Microbiologists often need to determine the relatedness of bacterial isolates to define the network of relationships of an infectious outbreak and effectively assist epidemiological investigations. Standard protocols for typing Salmonella rely on internationally accepted methods, like PFGE and MLVA, which a few decades ago flanked the more limited serotyping. The possibility of accessing the vast amount of information provided by WGS of bacterial isolates promises to be the next frontier of subtyping methods, probably capable of surpassing PFGE and MLVA for molecular epidemiological purposes. In this study, we reanalyzed a well-defined Salmonella Manhattan outbreak detected in the summer of 2009 in the province of Modena (Italy) using WGS in order to test the power of this approach for resolving the ambiguities left by PFGE. The epidemic episode involved 15 human cases from June to July 2009, with all presenting the same PFGE profile (SXB_BS.0003). The molecular epidemiological investigation of the outbreak involved several isolates, some from the infectious episode and others from the historic collection of the regional surveillance system of the food chain. As expected, PFGE analysis attributed the same pulsotype (SXB_BS.0003) to all the outbreak-related isolates, but the same pulsotype was shared by many historic isolates as well. On the contrary, the WGS-based phylogeny inferred from the total core SNPs clearly showed the presence of four distinct groups of isolates (Fig. 2) within the outbreak pulsotype. The first branch of the tree, within the outbreak pulsotype, separates nonoutbreak historic isolates recovered from swine stool at different locations and times. Among these, we find isolate SM48, which was originally suspected of being implicated in the infectious episode, based on PFGE, and eventually cleared by WGS. Interestingly, isolate SM36, which does not belong to pulsotype SXB_BS.0003 but to the highly similar (95% similarity) pulsotype SXB_PR.0752, is included in this clade. This is a clear discrepancy between WGS and the more limited PFGE that relies on only few genomic loci (rare restriction sites) for its typing inferences. By placing SM36 together with pulsotype SXB_BS.0003 isolates, our WGS approach indicates that a limited genomic difference between isolates is able to jeopardize the typing outcome of PFGE. This observation confirms what Tenover et al. (38) already pointed out, the fact that as PFGE may be heavily influenced by a single mutational event (e.g., SNP occurring in a restriction site), isolates should be considered to be possibly related even if they differ by two or three bands. However, according to this conservative interpretation of PFGE results, the vast majority of the isolates of our study should be regarded as potentially belonging to the outbreak. This would have not been sufficiently discriminatory to help the epidemiological investigations. The interpretation criteria of Tenover et al. (38) are derived from logical considerations; as such, they are intrinsically valid, and our observations regarding isolate SM36 confirms their validity. At the same time, their use leaves molecular epidemiologists with considerable uncertainty about how to interpret PFGE results with regard to whether or not different pulsotypes are part of a single outbreak. In our case, WGS removed that uncertainty about SM36.
Moving deeper along the phylogenetic tree based on the total core SNPs, three other groups of isolates are evident. The outermost set of this node includes isolates (SM21, -23, -24, and -27) not related to the outbreak, as they were collected 3 years before (2006). It is interesting, however, to notice that WGS-based phylogeny indicates these strains to be closer to the outbreak node (inner branch) than was the previous set of swine-stool isolates. On a better look, we were struck by the fact that SM23 and SM24 were collected in 2006, within the own-check procedures of the sausage producer involved in the 2009 outbreak. Moreover, SM21, which is subbranched with SM23 and SM24, was routinely recovered from a local pig farm (from swine stool) at the same time as SM23 and SM24. While this specific molecular similarity was not inferred by PFGE, WGS highlighted a possible link between these two commercial entities. Moving one branch forward in the phylogenetic tree, WGS shows another bifurcation actually separating outbreak-related isolates of human origin from those of food origin. While still speculative, based on this WGS-based phylogeny, coupled with epidemiological data, we could argue that this outbreak was due to a persistent Salmonella Manhattan clone, which may have infected one or more pig farms and reached the food producer and the retail customers as animals arrived at the slaughterhouse in a nonclinical septic condition. This is a typical mode of transmission of Salmonella along the food chain, as it may asymptomatically persist (thus going unnoticed) within a herd of pigs for long periods of time (even years). Sporadically, animals carrying a high level of the pathogen arrive at the slaughterhouse and contaminate a defined set of food products, thus causing an infectious outbreak as the final consumers (39, 40) become exposed to it. In this scenario, WGS seems to depict a more detailed and articulated epidemiological story. In fact, the tree inferred from core SNPs (Fig. 2) leaves a certain level of uncertainty relative to the actual causative relationship between the isolates of food origin and of human origin within the outbreak, as they cluster in two distinct groups, although very closely to each other, as evidenced by the limited number of exclusive core SNPs accumulated by the two groups (3 for food and 5 for human isolates). In the absence of epidemiological insights, we argue that the two sets of isolates are very similar to each other but still are separate entities. This substantially contradicts the epidemiological evidence that the two sets of isolates belong to the same outbreak clone. Therefore, we further investigated this apparent inconsistency of the WGS-based results by comparing new alternative phylogenies based on two different subsets of polymorphisms, synonymous and nonsynonymous, instead of the total core SNPs. The trees generated from these two subsets of SNPs were different (Fig. 3A and B). Phylogenetic analysis based on nonsynonymous SNPs (Fig. 3B) still divided the outbreak isolates of food and human origins, as in the approach based on total core SNPs. On the contrary, the tree obtained from synonymous SNPs (Fig. 3A) clustered the human isolates together with the food isolates, indicating that all outbreak-related Salmonella Manhattan strains constituted a single clone, in line with epidemiological evidence. While intriguing, this new outcome may have been the misleading effect of the smaller amount of data present in these new subsets than that with the total set of core SNPs, of which there were 953, whereas the number of synonymous and nonsynonymous SNPs were 224 and 467, respectively. Therefore, to confirm these results, we took a step forward in this approach by considering not just synonymous versus nonsynonymous SNPs but also taking into account the different codon position of each SNP in the core genome. Salmonella Manhattan synonymous SNPs were at the 3rd codon position 94% of the time, while nonsynonymous SNPs were at the 2nd 42% and at the first position 43% of the time (total, 85%). In this study, 1st, 2nd, and 3rd position SNPs accounted for 214, 194, and 283 nucleotide substitutions, respectively. The comparison of subsets of SNPs based on their codon site would then not be impaired by too-large differences in the amount of data processed by the phylogenetic algorithms. The tree obtained from second codon position (Fig. 4B) was comparable to that of the nonsynonymous SNPs, as expected, whereas the tree obtained from third codon position showed human isolates as a subgroup of the food isolates (Fig. 4C), essentially confirming the tree based on synonymous SNPs. These results show that at least limited to our outbreak, synonymous and third-position SNPs were the only ones able to describe the causal relationship between food (source of the outbreak) and clinical isolates in a way that was consistent with the epidemiological evidence. At the same time, our results indicate that nonsynonymous and total core SNPs may have led to misleading conclusions about the relationships between the human and food isolates of the outbreak. One last aspect that caught our attention by deciphering topologies of this WGS-based retrospective analysis was that SNP-based clustering of isolates separated human from food outbreak-related isolates when considering total core SNPs (Fig. 2). As we just discussed, this topology was mainly influenced by nonsynonymous mutations, which means it is possible to find distinctive nonsynonymous SNPs for each group of isolates (human versus food). Using progressiveMauve, we identified a set of 953 core SNPs, among which we selected those that were exclusive to specific clusters of interest: six SNPs exclusive to all outbreak isolates (human and food origin), three exclusive to all food origin outbreak isolates, and only five exclusive to all human origin outbreak isolates (Table 2). The extremely limited number of exclusive SNPs in food and human isolates within the outbreak is an additional compelling element indicative of the fact that these two groups of isolates did not have enough evolutionary time to significantly differentiate, indicating they belong to the same clone. A BLAST analysis of these SNPs against the Virulence Factors Database revealed three genes of particular interest: (i) fliK, coding for a flagellar hook-length control protein (41), (ii) sthD, a gene coding for a fimbrial outer membrane usher protein (42), and (iii) rfbD, coding for a UDP-galactopyranose mutase precursor involved in the synthesis of the O antigen of the lipopolysaccharide (LPS). All three proteins are virulence determinants in Salmonella (43–46). WGS has already proved its usefulness for elucidating the evolutionary diversity of large populations of bacterial isolates (11, 47, 48). In the specific case of Salmonella, WGS was successfully applied to illuminate the diversity of the pathogen within a vast epidemic episode, allowing highly efficient traceback of clinical and food isolates (4, 13). The results obtained in this study underscore the power of WGS-based methods, when applied together with the most appropriate phylogenetic tools, to resolve small outbreaks characterized by few and highly clonal bacterial isolates. Our comparative genomics approach was able to correctly cluster the clinical isolates within the composite scenario of outbreak-related and collection isolates. Accurate backtracking to the source of infection at the retail and industrial levels was made possible while flagging an originally overlooked suspicious correlation with a farm supplier and clearing an originally suspect food operator. Moreover, by selectively choosing the different types of detected nucleotide variations, we were able to read the message hidden within neutral mutations as opposed to the general use of total core SNPs. Further use of the differential analysis of synonymous and nonsynonymous mutations will test the validity of this approach in deciphering the details of infection transmission in the context of other outbreaks caused by Salmonella and, potentially, other pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Elena Carra for providing some of the isolates included in the study. We thank Roberto Alfieri for technical assistance in remote analysis at the University of Parma Department of Physics and Earth Sciences.
This study was supported by Regione Lombardia grant delibera regionale 001051/22122000 and by the Italian Ministry of Health grant IZSLER-PRC2012/006.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02930-14.
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies . 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.European Food Safety Authority (EFSA), European Centre for Disease Prevention and Control (ECDC). 2013. Scientific report of EFSA and ECDC: the European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J 11:3129–3378. doi: 10.2903/j.efsa.2013.3129. [DOI] [Google Scholar]
- 3.Wattiau P, Boland C, Bertrand S. 2011. Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives. Appl Environ Microbiol 77:7877–7885. doi: 10.1128/AEM.05527-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allard MW, Luo Y, Strain E, Pettengill J, Timme R, Wang C, Li C, Keys CE, Zheng J, Stones R, Wilson MR, Musser SM, Brown EW. 2013. On the evolutionary history, population genetics and diversity among isolates of Salmonella Enteritidis PFGE pattern JEGX01.0004. PLoS One 8:e55254. doi: 10.1371/journal.pone.0055254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urwin R, Maiden MCJ. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol 11:479–487. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S, S. enterica MLST Study Group . 2012. Multilocus sequencing typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fakhr MK, Nolan LK, Logue CM. 2005. Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar Typhimurium. J Clin Microbiol 43:2215–2219. doi: 10.1128/JCM.43.5.2215-2219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardy JL, Johnston JC, Sui SJH, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJM, Brinkman FSL, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 10.Desai PT, Porwollik S, Long F, Cheng P, Wollam A, Clifton SW, Weinstock GM, McClelland M. 2013. Evolutionary genomics of Salmonella enterica subspecies. mBio 4:e00579-12. doi: 10.1128/mBio.00579-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timme RE, Pettengill JB, Allard MW, Strain E, Barrangou R, Wehnes C, Van Kessel JS, Karns JS, Musser SM, Brown EW. 2013. Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol 5:2109–2123. doi: 10.1093/gbe/evt159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M, Zhao S, Pettengill J, Luo Y, Monday SR, Abbott J, Ayers SL, Cinar HN, Muruvanda T, Li C, Allard MW, Whichard J, Meng J, Brown EW, McDermott PF. 2014. Comparative genomic analysis and virulence differences in closely related Salmonella enterica serotype Heidelberg isolates from humans, retail meats, and animals. Genome Biol Evol 6:1046–1068. doi: 10.1093/gbe/evu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allard MW, Luo Y, Strain E, Li C, Keys CE, Son I, Stones R, Musser SM, Brown EW. 2012. High resolution clustering of Salmonella enterica serovar Montevideo strains using a next-generation sequencing approach. BMC Genomics 13:32. doi: 10.1186/1471-2164-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lienau EK, Strain E, Wang C, Zheng J, Ottesen AR, Keys CE, Hammack TS, Musser SM, Brown EW, Allard MW, Cao G, Meng J, Stones R. 2011. Identification of a salmonellosis outbreak by means of molecular sequencing. N Engl J Med 364:981–982. doi: 10.1056/NEJMc1100443. [DOI] [PubMed] [Google Scholar]
- 15.Cao G, Meng J, Strain E, Stones R, Pettengill J, Zhao S, McDermott P, Brown E, Allard M. 2013. Phylogenetics and differentiation of Salmonella Newport lineages by whole genome sequencing. PLoS One 8:e55687. doi: 10.1371/journal.pone.0055687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mather AE, Reid SWJ, Maskell DJ, Parkhill J, Fookes MC, Harris SR, Brown DJ, Coia JE, Mulvey MR, Gilmour MW, Petrovska L, De Pinna E, Kuroda M, Akiba M, Izumiya H, Connor TR, Suchard MA, Lemey P, Mellor DJ, Haydon DT, Thomson NR. 2013. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 341:1514–1517. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang S, Octavia S, Feng L, Liu B, Reeves PR, Lan R, Wang L. 2013. Genomic diversity and adaptation of Salmonella enterica serovar Typhimurium from analysis of six genomes of different phage types. BMC Genomics 14:718. doi: 10.1186/1471-2164-14-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leekitcharoenphon P, Friis C, Zankari E, Svendsen CA, Price LB, Rahmani M, Herrero-Fresno A, Fashae K, Vandenberg O, Aarestrup FM, Hendriksen RS. 2013. Genomics of an emerging clone of Salmonella serovar Typhimurium ST313 from Nigeria and the Democratic Republic of Congo. J Infect Dev Ctries 7:696–706. doi: 10.3855/jidc.3328. [DOI] [PubMed] [Google Scholar]
- 19.Noël H, Dominguez M, Weill FX, Brisabois A, Duchazeaubeneix C, Kerouanton A, Delmas G, Pihier N, Couturier E. 2006. Outbreak of Salmonella enterica serotype Manhattan infection associated with meat products, France, 2005. Euro Surveill 11:270–273. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=660. [PubMed] [Google Scholar]
- 20.Fisher I, Crowcroft N. 1998. Enter-net/EPIET investigation into the multinational cluster of Salmonella Livingstone. Euro Surveill 2:pii=1271 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=1271. [Google Scholar]
- 21.European Food Safety Authority. 2012. Technical report: manual for reporting of food-borne outbreaks in accordance with Directive/99/EC from the year 2011. Supporting publication 2012:EN-265. European Safety Food Authority, Parma, Italy: http://www.efsa.europa.eu/en/supporting/doc/265e.pdf. [Google Scholar]
- 22.PulseNet. 2010. One-day (24–28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei, by pulsed field gel electrophoresis (PFGE). Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/pulsenet/protocols/ecoli_salmonella_shigella_protocols.pdf. [Google Scholar]
- 23.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–56. In Computer science and biology. Proceedings of the German Conference on Bioinformatics, GCB '99 GCB, Hannover, Germany. [Google Scholar]
- 25.Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, Cole JR, Ding Y, Dugan S, Field D, Garrity GM, Gibbs R, Graves T, Han CS, Harrison SH, Highlander S, Hugenholtz P, Khouri HM, Kodira CD, Kolker E, Kyrpides NC, Lang D, Lapidus A, Malfatti SA, Markowitz V, Metha T, Nelson KE, Parkhill J, Pitluck S, Qin X, Read TD, Schmutz J, Sozhamannan S, Sterk P, Strausberg RL, Sutton G, Thomson NR, Tiedje JM, Weinstock G, Wollam A, Genomic Standards Consortium Human Microbiome Project Jumpstart Consortium, Detter JC . 2009. Genome Project standards in a new era of sequencing. Science 326:236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sassera D, Gaiarsa S, Scaltriti E, Morganti M, Bandi C, Casadei G, Pongolini S. 2013. Draft genome sequence of Salmonella enterica subsp. enterica serovar Manhattan strain 111113, from an outbreak of human infections in northern Italy. Genome Announc 1:e00632-13. doi: 10.1128/genomeA.00632-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaiarsa S, Comandatore F, Gaibani P, Corbella M, Dalla Valle C, Epis S, Scaltriti E, Carretto E, Farina C, Labonia M, Landini MP, Pongolini S, Sambri V, Bandi C, Marone P, Sassera D. 2014. Genomic epidemiology of Klebsiella pneumoniae in Italy and novel insights into the origin and global evolution of its resistance to carbapenem antibiotics. Antimicrob Agents Chemother 59:389–396. doi: 10.1128/AAC.04224-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Chen L, Sun L, Yu J, Jin Q. 2007. VFDB 2008 release: an enhanced Web-based resource for comparative pathogenomics. Nucleic Acids Res 36:D539–D542. doi: 10.1093/nar/gkm951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Xiong Z, Sun L, Yang J, Jin Q. 2011. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res 40:D641–D645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 35.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 36.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 37.Bofkin L, Goldman N. 2006. Variation in evolutionary processes at different codon positions. Mol Biol Evol 24:513–521. doi: 10.1093/molbev/msl178. [DOI] [PubMed] [Google Scholar]
- 38.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rostagno MH. 2009. Can stress in farm animals increase food safety risk? Foodborne Pathog Dis 6:767–776. doi: 10.1089/fpd.2009.0315. [DOI] [PubMed] [Google Scholar]
- 40.Rostagno MH, Callaway TR. 2012. Pre-harvest risk factors for Salmonella enterica in pork production. Food Res Int 45:634–640. doi: 10.1016/j.foodres.2011.04.041. [DOI] [Google Scholar]
- 41.Uchida K, Aizawa SI. 2014. The flagellar soluble protein FliK determines the minimal length of the hook in Salmonella enterica serovar Typhimurium. J Bacteriol 196:1753–1758. doi: 10.1128/JB.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waters RC, O'Toole PW, Ryan KA. 2007. The FliK protein and flagellar hook-length control. Protein Sci 16:769–780. doi: 10.1110/ps.072785407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suez J, Porwollik S, Dagan A, Marzel A, Schorr YI, Desai PT, Agmon V, McClelland M, Rahav G, Gal-Mor O. 2013. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PLoS One 8:e58449. doi: 10.1371/journal.pone.0058449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Baumler AJ. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun 73:3358–3366. doi: 10.1128/IAI.73.6.3358-3366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komoriya K, Shibano N, Higano T, Azuma N, Yamaguchi S, Aizawa S-I. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella Typhimurium. Mol Microbiol 34:767–779. doi: 10.1046/j.1365-2958.1999.01639.x. [DOI] [PubMed] [Google Scholar]
- 46.Köplin R, Brisson J-R, Whitfield C. 1997. UDP-galactofuranose precursor required for formation of the lipopolysaccharide O antigen of Klebsiella pneumoniae serotype O1 is synthesized by the product of the rfbDKPO1 gene. J Biol Chem 272:4121–4128. doi: 10.1074/jbc.272.7.4121. [DOI] [PubMed] [Google Scholar]
- 47.Leekitcharoenphon P, Lukjancenko O, Friis C, Aarestrup F, Ussery D. 2012. Genomic variation in Salmonella enterica core genes for epidemiological typing. BMC Genomics 13:88. doi: 10.1186/1471-2164-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lienau EK, Blazar JM, Wang C, Brown EW, Stones R, Musser S, Allard MW. 2013. Phylogenomic analysis identifies gene gains that define Salmonella enterica subspecies I. PLoS One 8:e76821. doi: 10.1371/journal.pone.0076821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.